Abstract
Primary human hepatocytes (PHHs) are commonly used for in vitro studies of drug-induced liver injury. However, when cultured as 2D monolayers, PHH lose crucial hepatic functions within hours. This dedifferentiation can be ameliorated when PHHs are cultured in sandwich configuration (2Dsw), particularly when cultures are regularly re-overlaid with extracellular matrix, or as 3D spheroids. In this study, the 6 participating laboratories evaluated the robustness of these 2 model systems made from cryopreserved PHH from the same donors considering both inter-donor and inter-laboratory variability and compared their suitability for use in repeated-dose toxicity studies using 5 different hepatotoxins with different toxicity mechanisms. We found that expression levels of proteins involved in drug absorption, distribution, metabolism, and excretion, as well as catalytic activities of 5 different CYPs, were significantly higher in 3D spheroid cultures, potentially affecting the exposure of the cells to drugs and their metabolites. Furthermore, global proteomic analyses revealed that PHH in 3D spheroid configuration were temporally stable whereas proteomes from the same donors in 2Dsw cultures showed substantial alterations in protein expression patterns over the 14 days in culture. Overall, spheroid cultures were more sensitive to the hepatotoxic compounds investigated, particularly upon long-term exposures, across testing sites with little inter-laboratory or inter-donor variability. The data presented here suggest that repeated-dosing regimens improve the predictivity of in vitro toxicity assays, and that PHH spheroids provide a sensitive and robust system for long-term mechanistic studies of drug-induced hepatotoxicity, whereas the 2Dsw system has a more dedifferentiated phenotype and lower sensitivity to detect hepatotoxicity.
Keywords: cytotoxicity, hepatocytes, predictive toxicology, cell culture
Before a new medicine can reach the market, significant safety considerations must be met. However, despite rigorous testing regimens, some severe adverse drug reactions (ADRs) are still only identified in the late stages of drug development and even post marketing, resulting in increased patient morbidity and costly drug withdrawals. Hepatotoxicity remains one of the most frequent causes of compound attrition and late stage failures (Cook et al., 2014). Current models to predict hepatotoxicity before a compound enters clinical stages include animal models and in vitro assays. The predictive power of animal models, however, is limited due to substantial inter-species differences in expression and activities of enzymes and transporters involved in drug absorption, distribution, metabolism, and excretion (ADME), resulting in discrepant pharmacokinetic parameters and metabolic fates of the tested compounds (Martignoni et al., 2006). Consequently, a large-scale investigation of human and animal toxicity events associated with 150 compounds found that rodent (primarily rat) and non-rodent (primarily dog) animal models predicted only around 50% of the human hepatotoxicity events attributed to these drugs (Olson et al., 2000).
In contrast to animal studies, in vitro assays, which utilize human cells, represent models that offer the potential to investigate human-specific toxicity profiles (Ewart et al., 2018; Soldatow et al., 2013). For the evaluation of drug-induced liver injury (DILI), primary human hepatocytes (PHHs) have been considered the gold standard model (Gómez-Lechón et al., 2014; Sison-Young et al., 2017). In conventional 2D monolayer cultures, however, PHH rapidly dedifferentiate within hours after plating and lose hepatic functions, such as albumin production and drug metabolizing enzyme activity (den Braver-Sewradj et al., 2016), which severely limits their predictive capacity, particularly where chemically reactive metabolites are implicated in the toxicity mechanisms (Heslop et al., 2016; Lauschke et al., 2016b).
To prevent or ameliorate the dedifferentiation of PHH in vitro, a variety of methods have been presented, including sandwich cultures, spheroid models, and microfluidic systems (Lauschke et al., 2016a). Overlaying 2D cultures of PHH with a thin layer of extracellular matrix (ECM) proteins after attachment mimics the physiological microenvironment of hepatocytes in vivo, in which cells interact with ECM components in the space of Disse, and improves polarization into distinct apical and basolateral domains (Treyer and Müsch, 2013). Although PHH in such a sandwich configuration exhibit improved stability of cellular phenotypes and form functional bile canalicular networks, dedifferentiation, however, remains, as evidenced by indications of epithelial-to-mesenchymal transition and decreases in albumin secretion and urea production after 2 weeks in culture (Rowe et al., 2013). Thus, such sandwich cultures are primarily used for short-term studies of hepatic uptake and transporter function and have been successfully applied to delineate mechanisms underlying cholestatic toxicity (Burbank et al., 2016; Chatterjee et al., 2014; De Bruyn et al., 2013; Oorts et al., 2016). However, sandwich cultures in which the ECM overlay is renewed every 3–4 days, hereafter referred to as sandwich cultures (2Dsw), reduce dedifferentiation and extend culture (and exposure) times up to 14 days, thus providing a promising tool to investigate long-term hepatotoxicity (Bellwon et al., 2015; Parmentier et al., 2013).
Culturing PHH as spheroidal structures generated by gravitational aggregation in hanging-drops or on ultralow attachment surfaces represents another emerging high-throughput compatible method for assessing compound liabilities. Importantly, whole proteome analyses revealed that the molecular phenotypes of in vivo liver tissues are conserved in spheroid cultures, thus rendering this model system suitable to study liver functions and DILI in a microphysiological setting (Bell et al., 2016). Hepatic spheroids are phenotypically and functionally stable over at least 5 weeks in culture and maintain endogenous hepatic functions, such as albumin and urea production as well as glycogen storage (Bell et al., 2016; Messner et al., 2017). Furthermore, PHH spheroids retain drug metabolizing capacities and metabolic profiles of freshly isolated or cryopreserved hepatocytes, enabling the emulation of pharmacokinetic differences between donors, rendering them suitable for in vitro studies of long-term drug metabolism (Vorrink et al., 2017).
Despite available characterization data of 2Dsw and 3D spheroid cultures, no comparison of the 2 systems has been reported in long-term cultures. In this study, we, therefore, comprehensively compared the changes in cellular phenotypes during long-term cultures using global proteomic analyses and benchmarked cryopreserved PHH in 3D spheroid and 2Dsw cultures in repeated dose toxicity assays. In addition, we assessed the robustness of the 2 model systems considering both inter-donor and inter-laboratory variability and compared their suitability for use in repeated-dose toxicity studies in a multi-center study across both academic and industrial laboratories within the IMI—MIP-DILI project.
MATERIALS AND METHODS
A harmonized protocol was generated and circulated to all 6 participating laboratories. Three cryopreserved PHH donors were used in the study, and the experimental setup was designed with the aim of repeating the 3D spheroid incubations at least 3 times across different laboratories for each of these donors. In addition, 2 laboratories performed 3D spheroid cytotoxicity experiments for all 3 donors at the same site (Table 1).
Table 1.
Distribution of Experimental Work Among the Participating Laboratories
| Proteomics |
CYP Activity |
Cytotoxicity |
||||
|---|---|---|---|---|---|---|
| Laboratory | 2Dsw | 3D | 2Dsw | 3D | 2Dsw | 3D |
| AstraZeneca | 1, 2, 2, 3 | |||||
| Janssen | 1 | 1 | 1 | 1 | ||
| Orion | 1 | |||||
| GSK | 1, 3 | 1, 3 | ||||
| KI | 1, 2, 3 | 1, 2, 3 | ||||
| KaLy Cell | 1, 2, 3 | 2 | ||||
1= Donor 1; 2 = Donor 2; 3= Donor 3.
Chemicals and Reagents
Matrigel was obtained from Corning (Avon, France), bosentan was purchased from Sequoia (Pangbourne, UK) and fialuridine from Carbosynth (Berkshire, UK). Pioglitazone and troglitazone were obtained from AstraZeneca Compound Management, AstraZeneca R&D (Macclesfield, UK). Hyclone fetal bovine serum (FBS) was obtained from Thermofisher (Sweden). All other reagents were purchased from Sigma-Aldrich (St-Quentin-Fallavier, France) unless otherwise stated. Cell culture maintenance media comprised Williams’ E medium supplemented with 10 µg/ml insulin, 5.5 µg/ml transferrin, 6.7 ng/ml sodium selenite, 100 U/ml penicillin, 100 µg/ml streptomycin, and 100 nM dexamethasone. Wherever possible, reagents were obtained from the same batch/lot.
Cell Culture
Cryopreserved PHHs from the 3 donors used in the present study were provided by KaLy Cell (Plobsheim, France) (Table 2). They were pre-characterized for long-term culture in a re-overlayed sandwich (2Dsw) configuration by KaLy Cell and for 3D-spheroid aggregation and culture by Karolinska Institutet. The human biological samples were acquired in agreement with ethical regulations and their research use was in accord with the terms of the informed consents. The experimental outline for the comparison of 2Dsw and 3D spheroid cultures is depicted in Figure 1.
Table 2.
Clinical and Demographic Information for the 3 Hepatocyte Donors Used in This Study
| Donor | Sex | Age | Ethnicity | Pathology | Medications |
|---|---|---|---|---|---|
| Donor 1 | Male | 45 | Caucasian | Liver metastasis from neuroendocrine tumor | No medications |
| Donor 2 | Male | 62 | Caucasian | Liver metastasis from sigmoid adenocarcinoma | Lemaline, Lovazepam, Ramipril |
| Donor 3 | Male | 69 | Caucasian | n/a | n/a |
Abbreviation: n/a, not available.
Figure 1.
Scheme of experimental procedure. Primary human hepatocytes (PHHs) were seeded in either 2Dsw or 3D spheroid configuration. Due to the time taken for spheroids to form (7–10 days) day 0 for 3D experiments is 7–10 days after seeding. Medium changes (± test compound) were performed every 2–3 days (grey boxes) and for 2Dsw cultures matrigel was re-overlaid as shown (black boxes). Sampling points for proteomics (triangles) and ATP measurements (circles) are indicated. Toxicity assays were done with cells from 3 different donors. The morphologies of cells from donor 1 in each of the systems at days 3, 7, and 14 are illustrated. These images were generated at Janssen.
2Dsw culture
Cryopreserved PHHs were thawed at 37°C (1–2 min in water bath), transferred to 50 ml of Universal Cryopreservation Recovery Media (IVAL, Colombia) and centrifuged at 100 × g for 10 min. Viable cells were counted by Trypan Blue exclusion and diluted to the appropriate concentration in PHH maintenance medium supplemented with 10% FBS. A minimum viability of 80% was required to proceed with the experiment. Cells were seeded at a density of 70 000 viable cells/well on Collagen I-Biocoat 96-well plates for cytotoxicity and CYP activity assessment and 400 000 viable cells/well on Collagen I- Biocoat 24-well plates for proteomic assessment. After 4 to 6 hours attachment at 37°C with 5% CO2, PHH culture medium was renewed and cells were incubated overnight. The following day, cells were overlaid with ice-cold matrigel (0.25 mg/ml) diluted in PHH maintenance medium. From the first day of compound incubation, FBS-free PHH culture medium was renewed every 2/3 days and cells were overlaid with matrigel every 3/4 days as previously described (Parmentier et al., 2017) (Figure 1).
3D spheroid culture
Cryopreserved PHHs were thawed as described above and seeded in PHH maintenance medium supplemented with 10% FBS as previously reported (Bell et al., 2016). Briefly, 1500 viable cells/well were seeded in 96-well ultra-low attachment plates (Corning) and centrifuged for 2 min at 125 × g. After 4–5 days, a 50% medium change was performed with FBS-free medium. Medium was then exchanged daily until spheroids were fully formed (days 7–10) so that by the time compound exposures began negligible levels of FBS remained in the well. Dosing medium was renewed every 2/3 days, according to the experimental design provided in Figure 1. Prior to performing the main ring-trial a “pilot” ring trial was performed in which each of the laboratories generated and maintained spheroids for 14 days, measuring ATP periodically. They also treated their spheroids with troglitazone and pioglitazone. The experimental setup was outlined in a detailed protocol provided by Karolinska to all participating partners. This had to be completed successfully with regard to spheroid aggregation, morphology and viability before continuing to the full ring trial.
Finally for data to be included in the overall analysis, ATP levels in control spheroids/sandwich cultures needed to remain within an acceptable range that is >50% of the levels at day 0.
Proteomics
The phenotype of cryopreserved PHH cultured in the 2 cell models was determined by semi-quantitative proteomics. Samples from 3 donors were prepared at day 0 and day 14 for each of the respective in vitro culture formats to have comparable data relating to the cytotoxicity experiments. Furthermore, freshly thawed samples were snap-frozen for comparative proteomic assessment. For 2Dsw samples, cells from 3 wells of a 24-well plate were washed twice in PBS and harvested by gentle scraping. Cells were then centrifuged at 900 × g for 7 min (4°C) and snap-frozen. All 2Dsw samples were generated at KaLy Cell. For 3D samples, spheroids from 7 plates were pooled, washed in PBS, pelleted and snap-frozen. All samples were stored at –70°C prior to analysis. All 3D samples were generated at KI. For iTRAQ analysis, each cell pellet was thawed and lysed immediately by sonication in an equal volume of 0.5 M triethylammonium bicarbonate/0.1% sodium dodecyl sulfate (SDS). The cell lysate was centrifuged at 14 000 × g for 15 min at 4°C and the supernatant collected. Protein concentration was determined by the Bradford assay. Sample labeling was performed according to the manufacturer’s instructions (AB Sciex). Briefly, 100 μg of protein in 20 μl was denatured, reduced and capped with MMTS according to the protocol. Samples were subsequently digested with trypsin overnight, and labeled with iTRAQ isobaric tags 113–121. Samples were subjected to cation exchange chromatography, to remove unbound trypsin and reagent. The digests were diluted to 4 ml with 10 mM potassium dihydrogen phosphate/25% ACN (w/v). The pH of the samples was adjusted to <3 using phosphoric acid prior to fractionation on a Polysulfoethyl A strong cation-exchange column (200 × 4.6 mm, 5 μm, 300 Å; Poly LC, Columbia, MD). Fractions of 2 ml were collected and dried by centrifugation under vacuum (SpeedVac, Eppendorf). Fractions were reconstituted in 1 ml of 0.1% TFA and were subsequently desalted using a mRP Hi Recovery protein column 4.6 × 50 mm (Agilent) on an Infinity 1260 HPLC system (Agilent) prior to mass spectrometric analysis. Desalted fractions were reconstituted in 40 μl 0.1% formic acid and 5 μl aliquots were delivered into a Triple TOF 6600 (Sciex) via an Eksigent NanoLC 400 System (Sciex) mounted with a NanoAcquity 5 µm, 180 µm × 20 mm C18 trap and 1.7 µm, 75 µm × 250 mm analytical column (Waters). A NanoSpray III source was fitted with a 10 μm inner diameter PicoTip emitter (New Objective). A gradient of 2–50% ACN/0.1% formic acid (v/v) over 90 min was applied to the columns at a flow rate of 300 nl/min. The mass spectrometer was operated in positive ion mode (Analyst TF1.7) with survey scans of 250 ms, MS/MS accumulation time of 100 ms and with monitoring of the 25 most intense ions (total cycle time 2.5 s). Data were searched using ProteinPilot 5 software (Sciex) against the latest UniProt database with iTRAQ as a variable modification and MMTS as the cysteine alkylating reagent. The reversed database was used as a decoy to determine the false discovery rate (FDR) for protein identification, and only those proteins identified at >1% FDR were evaluated further. Principle component analyses were performed using Qlucore Omics Explorer 3.1 (Qlucore, Lund, Sweden). Differentially affected pathways in 2Dsw and 3D spheroid culture were identified using overrepresentation enrichment analyses using WebGestalt (Wang et al., 2013). For whole proteome analyses, Benjamini-Hochberg multiple testing correction was applied with an FDR of 5%. For comparisons between groups, p-values refer to two-tailed heteroscedastic t-tests unless specified otherwise. Results were considered significant when p < .05.
Cytochrome P450 Activity Quantifications
A comparison of CYP450 activity in the 2 culture formats was performed at Janssen. PHH cultures from donor 1 were treated with a cocktail consisting of the probe substrates phenacetin (100 µM), amodiaquine (10 µM), tolbutamide (100 µM), dextromethorphan (15 µM), and midazolam (10 µM), and to determine the activities of CYP1A2, CYP2C8, CYP2C9, CYP2D6, and CYP3A4, respectively. For spheroid cultures, probes were incubated for 2h and for sandwich cultures, probes were incubated for 30 minutes (based on the sensitivity of the analysis and linearity of the enzymatic reaction from a pilot experiment; not shown). Supernatants were stored at -20°C until further analysis. Metabolite formation (acetaminophen, desethyl-amodiaquine, 4-OH-tolbutamide, dextrorphan, and 1-OH-midazolam) was quantified on a Shimadzu Nexera SIL-30AC system (Shimadzu Corp., Kyoto, Japan) connected to a SCIEX QTRAP 6500 mass spectrometer (SCIEX, Ontario, Canada) (for determining 4-OH-tolbutamide and phenacetin) or a SCIEX API-4000 (SCIEX) (for determining 1-OH-midazolam, desethyl-amodiaquine, and dextrorphan) equipped with an ionization source operated in negative (4-OH-tolbutamide) or positive (desethyl-amodiaquine, 1-OH-midazolam, dextrorphan, acetaminophen) mode. Separation was carried out at 50°C using either a Waters Acquity C18 BEH 1.7 µm-50 × 2.1 mm ID (Waters Corp., MA—for midazolam and dextromethorphan) or a Waters Acquity HSS T3 1.8 µm-2.1 × 100 mm (Waters Corp., MA—for phenacetin, amodiaquine or tolbutamide). All data collection, processing, and analysis were performed with Analyst software (v1.6.2; SCIEX). The lower limit of quantification was set at 0.5, 2.0, 0.1, 0.1, and 0.5 ng/ml for acetaminophen, desethyl-amodiaquine, 4-OH-tolbutamide, dextrorphan, and 1-OH-midazolam, respectively. All results are expressed as pmol metabolite formed/h/million seeded cells from single spheroids or 2Dsw culture wells.
Incubations With Test Compounds
Six test compounds were selected for hepatotoxicity investigation from the MIP-DILI training set (Dragovic et al., 2016) (acetaminophen [APAP], bosentan, diclofenac, fialuridine, pioglitazone, and troglitazone) and cells from 3 different donors were dosed in 2Dsw and 3D spheroid culture as indicated (Figure 1 and Table 1). Compound concentrations tested are provided in Table 3. These represented hepatotoxic compounds with diverse toxicity mechanisms. Pioglitazone was included as a structural analog of troglitazone with reduced hepatic liabilities. Stock solutions were made in DMSO and diluted in FBS-free PHH culture medium to yield a maximum final concentration of DMSO of 0.5%. APAP and diclofenac were dissolved directly in PHH culture medium. On each treatment day, 100 µl of medium from cells in 2Dsw culture was removed and replaced with 100 µl of the drug solution. For 3D spheroids, 70 µl of medium was removed and replaced with 70 µl of the drug solution. The spheroids were allowed to settle, prior to performing a second 70 µl medium change with the drug solution.
Table 3.
Test Compound Concentrations Utilized in the Cytotoxicity Studies
| Test Compound | Final Concentration (µM) |
|---|---|
| Acetaminophen | 10 000, 5000, 2000, 1000, 500, 200, 100 |
| Bosentan | 400, 200, 100, 40, 20, 10, 4 |
| Diclofenac | 500, 250, 100, 50, 25, 10, 5 |
| Fialuridine | 300, 100, 30, 10, 3, 1, 0.3 |
| Pioglitazone | 40, 20, 10, 4, 2, 1, 0.4 |
| Troglitazone | 40, 20, 10, 4, 2, 1, 0.4 |
Viability Measurements
At 72 h (single treatment), day 7 (3 treatments), and day 14 (6 treatments), viability was determined by measuring cellular ATP using the CellTiter-Glo kit (Promega, Sweden or France). Blank values were subtracted from all wells. Curve-fitting was performed at a single site using the sigmoidal dose response function in GraphPad Prism, constrained at both 0 and 100. EC50 values corresponding to the concentration of the compound causing a 50% reduction in viability were computed for each compound and time point. p-Values refer to heteroscedastic two-tailed t-tests unless otherwise stated. Values of p < .05 were considered significant. For data to be included in the analysis, ATP values in control wells should not decrease by >50% over the 14 day culture period.
RESULTS
Primary Human Hepatocytes Cultured in 3D Spheroids and 2Dsw Cultures Exhibit Distinct Molecular Phenotypes
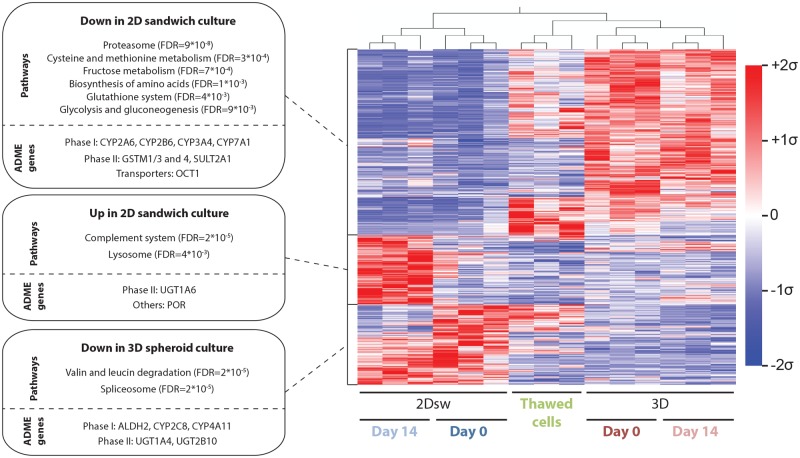
For our comparative study, we used the treatment protocol for re-overlaid 2D sandwich (2Dsw) and 3D spheroid cultures as illustrated in Figure 1 and monitored phenotypic changes over the course of 2 weeks using global proteomic analyses. The morphology of 2Dsw and 3D spheroids generated from 1 donor in 5 different laboratories is shown in Supplementary Figure 1. Compared with freshly thawed cells, PHH in sandwich configuration rapidly lose proteins involved in various metabolic pathways, including amino acid metabolism and biosynthesis, glycolysis and gluconeogenesis as well as homeostasis of the glutathione system, whereas these proteins are generally upregulated after aggregation in 3D spheroids (Figure 2). Furthermore, compared with freshly thawed cells and 3D spheroids, PHH in 2Dsw culture exhibit significantly lower levels of various ADME proteins, such as CYP2A6, CYP2B6, and CYP3A4, various glutathione S-transferases (GSTs) and the thiamine transporter OCT1 (Figure 2) (Chen et al., 2014; den Braver-Sewradj et al., 2016). Conversely, 2Dsw cultures demonstrated a significant increase in complement system components (FDR = 2 × 10−5) and lysosomal proteins (FDR = 4 × 10−3) particularly after later culture stages (day 14), indicative of an increase in degradative processes and cellular demise. Levels of spliceosome components (FDR = 2 × 10−5) and proteins involved in valine and leucine degradation (FDR = 2 × 10−5) were significantly reduced in 3D spheroid cultures. Furthermore, 3D spheroids showed reduced levels of ALDH2, CYP2C8, CYP4A11, and the UGTs UGT1A4 and UGT2B10 as compared with freshly thawed cells.
Figure 2.
Proteomic signatures of primary human hepatocytes are strongly influenced by the choice of in vitro culture paradigm. Heatmap visualization of mean-centered, sigma-normalized hierarchically clustered whole proteome data from freshly thawed PHH as well as from cultures from the same 3 donors in 2Dsw and 3D spheroid culture is shown (FDR = 5%). Enrichment analyses of the identified gene clusters revealed significantly affected pathways. In addition, ADME genes within each cluster are shown separately.
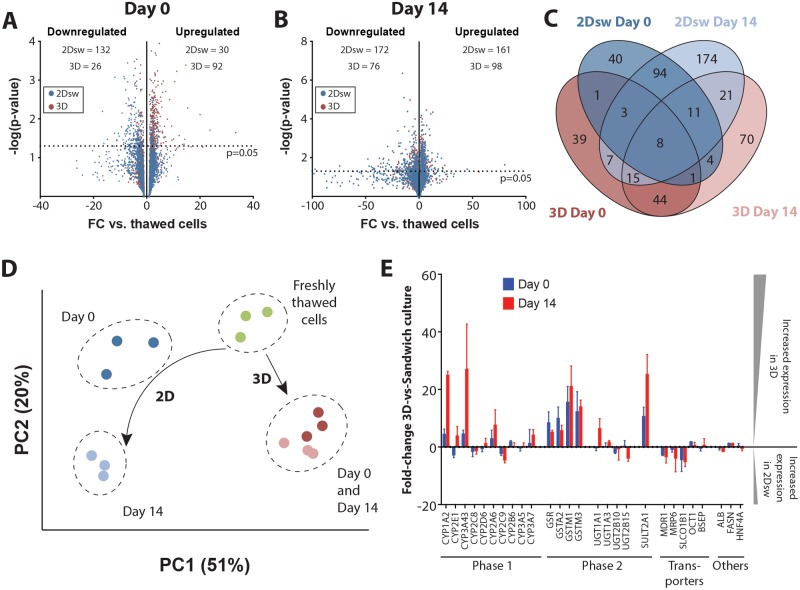
Overall, we found that 4.6% (n = 132) and 6% (n = 172) of the 2847 proteins, we identified were significantly downregulated with a fold-change >3 compared with freshly thawed cells in 2Dsw culture at day 0 and day 14, respectively (Figs. 3A and 3B). In contrast, levels of only 0.9% (n = 26) and 2.7% (n = 76) proteins were significantly reduced in 3D spheroids at day 0 and day 14, respectively. Downregulated ADME proteins included CYP3A4 (28-fold downregulation) and CYP2E1 (17-fold downregulation) in 2Dsw culture as well as CYP2A6 (42- and 6.8-fold downregulation in 2Dsw and 3D spheroids, respectively) and CYP2D6 (6.3- and 4.6-fold downregulation in 2Dsw and 3D spheroids, respectively) in both culture paradigms (Table 4). Importantly, the affected gene sets in 2Dsw and 3D spheroid culture overlapped only marginally with 8 proteins being dysregulated in all conditions (Figure 3C). Most prominent among these was the downregulation of the alcohol dehydrogenase ADH1A (8- and 74-fold in 2Dsw and 3- and 13-fold in 3D spheroids at day 0 and day 14, respectively) and of the serine dehydratase/threonine deaminase SDS (7- and 25-fold in 2Dsw and 9- and 22-fold in 3D spheroids at day 0 and day 14, respectively) as well as the upregulation of the fatty acid desaturase FADS2 (3- and 17-fold in 2Dsw and 11- and 8-fold in 3D spheroids at day 0 and day 14, respectively). Furthermore, levels of apolipoproteins, such as APOB, APOC1, APOC2, and APOE, were strongly increased mostly in 3D spheroid culture, suggesting that the molecular constituents necessary for the formation of chylomicrons remain present in vitro. The translational elongation factor EEF1A1, which delivers tRNAs to the ribosome, was strongly downregulated in 2Dsw culture (17- and 9-fold at day 0 and day 14, respectively), suggesting deficits in protein biosynthesis, whereas its levels were unaffected in 3D spheroids. Similar trends were observed for the peroxiredoxins PRDX1 (15- and 33-fold downregulation in 2Dsw at day 0 and day 14, respectively), PRDX2 (16- and 42-fold downregulation), and PRDX6 (12- and 53-fold downregulation) in 2Dsw configuration but not in 3D spheroids (all changes <2-fold at all time points). Proteins that were significantly affected in 3D spheroid but not in 2Dsw culture include upregulation of the aldoketoreductases AKR1B10 (33- and 56-fold upregulation in 3D spheroids at day 0 and day 14, respectively), AKR1C1 (17- and 12-fold upregulation) and AKR1C2 (21- and 24-fold upregulation) as well as the thioredoxin reductase TXNRD1 (10- and 11-fold upregulation).
Figure 3.
PHH undergo distinct molecular changes in 2Dsw and 3D spheroid culture that substantially influence their phenotype and functionality. Semi-log volcano plots showing the distribution of up- and downregulated proteins in 2D sandwich or 3D spheroid culture at day 0 (A) and after 14 days in culture (B). The dashed lines indicate the significance threshold of p = .05. C, Venn diagram visualizing the overlap of dysregulated proteins at day 0 and day 14 in 2Dsw and 3D spheroid cultures compared with freshly thawed cells. Note that the vast majority of proteins are either affected in 2Dsw or in 3D culture but only few in both. 2Dsw = 2D sandwich culture. D, Principal component analysis of differentially expressed genes reveals that samples cluster by in vitro culture paradigm. Furthermore, the plot indicates that the molecular phenotypes of PHH in 3D spheroid cultures remain stable over time, whereas substantial changes occur in 2Dsw configuration. Thawed cells are also depicted. E, Targeted analysis of ADME proteins and other factors with importance for hepatic functionality indicates that their levels are overall increased in 3D spheroids compared with 2Dsw cultures from the same donor (ie, positive fold-change) throughout the culture time. Values on the ordinate indicate the fold change (FC), defined as protein abundance in 3D spheroids divided by protein abundance in 2Dsw cultures if FC > 1 and the negative inverse if FC < 1. All samples for proteomic analysis were generated at KaLyCell (2Dsw) and Karolinska Institutet (3D) and processed and analyzed at the University of Liverpool.
Table 4.
Significantly Downregulated ADME Proteins in 2Dsw and 3D Spheroid Cultures as Compared With Freshly Thawed Cells
| 2Dsw |
3D Spheroids |
|||
|---|---|---|---|---|
| ADME Protein | Day 0 | Day 14 | Day 0 | Day 14 |
| ADH1A | −74-fold | −3-fold | −13-fold | |
| ADH3 | −8.5-fold | −24-fold | ||
| CYP2A6 | −42-fold | −6.8-fold | ||
| CYP2D6 | −6.3-fold | −4.6-fold | ||
| CYP2E1 | −17-fold | |||
| CYP4A11 | −5.2-fold | −5.8-fold | −38-fold | |
| GSS | −6.1-fold | −15-fold | ||
| GSTA2 | −97-fold | −18-fold | ||
| GSTM1 | −47-fold | |||
| SLC27A5 | −14-fold | −3.9-fold | −19-fold | |
| UGT1A4 | −10-fold | |||
| UGT2B10 | −3.3-fold | −3.7-fold | ||
Only ADME proteins that are downregulated >3-fold in at least one time point and culture paradigm are shown. Values in the table indicate average fold changes compared with freshly thawed cells from the same donors (n = 3 donors).
Importantly, although spheroids remained overall phenotypically stable after aggregation, the proteomes of 2Dsw cultures changed considerably during 2 weeks in culture, an effect that was observed for all 3 donors tested (Figure 3D). To estimate the effects of culture format on drug ADME, we focused on 25 proteins with importance for hepatic functionality (Figure 3E). Overall, the expression of pivotal phase I and phase II enzymes was elevated in 3D spheroids compared with 2Dsw cultures at both time points. The largest discrepancies between 2Dsw and 3D spheroids were seen in the expression of CYP3A4, whose expression was elevated by 5- and 27-fold in 3D spheroids at day 0 and day 14, respectively. Similarly, the expression of CYP1A2 was 4-fold (day 0) and 25-fold (day 14) higher in 3D spheroids. In contrast, levels of CYP2C8 and CYP2C9 as well as of the drug transporters MDR1, MRP6, and OATP1B1 were increased in sandwich configuration. The expression of the canalicular efflux transporter bile salt export pump (BSEP), inhibition of which must be investigated prior to a drug’s regulatory approval, was however indistinguishable between the 2 models.
Phase I Metabolic Activity is Overall Higher and More Stable in Long-term 3D Spheroids Compared With 2D Sandwich Cultures
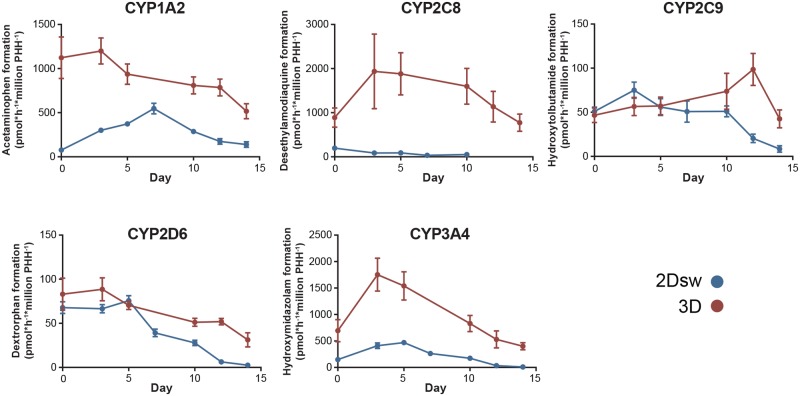
Hepatic pharmacokinetics is a key factor determining drug clearance. Moreover, chemically reactive metabolites are frequently formed and cited as a major cause of ADRs and hepatotoxicity (Park et al., 2011; Thompson et al., 2016). We therefore analyzed the enzymatic activity of CYP1A2, CYP2C8, CYP2C9, CYP2D6, and CYP3A4 that jointly account for >70% of all clinically relevant phase I reactions (Evans and Relling, 1999) by monitoring the formation of metabolites via LC-MS/MS throughout 14 days after spheroid or sandwich formation (Figure 4).
Figure 4.
CYP catalyzed drug metabolism in 3D spheroids and 2Dsw cultures from the same donor. PHHs from donor 1 were cultured as either 3D spheroids or 2Dsw cultures and the activities of CYP1A2, CYP2C8, CYP2C9, CYP2D6, and CYP3A4 were quantified by LC-MS/MS using phenacetin, amodiaquine, tolbutamide, dextromethorphan, and midazolam as probe substrates, respectively. As cells were incubated with probe substrates for different times (2h for spheroids and 30min for 2Dsw), activity data are shown as normalized per time and number of cells. Data represent the average of three 2Dsw incubations or 6 spheroids. Error bars indicate SD.
CYP1A2, CYP3A4, and CYP2C8 activities, when expressed per million seeded cells, were substantially higher in 3D spheroid cultures throughout the entire culture time. Particularly desethylamodiaquine formation was increased more than 100-fold in 3D spheroids compared with 2Dsw culture. In contrast, activities of CYP2D6 and CYP2C9 were similar between the 2 culture methods during the first 10 days of culture and PHH in spheroid cultures exhibited higher activities only after longer culture periods (12 days and more). Overall, metabolic activities in 3D spheroids remained high and stable during 2 weeks and none of the tested enzymes decreased >2.5-fold in activity. In contrast, activities of CYP2C8, CYP2C9, CYP2D6, and CYP3A4 decreased 4- to 30-fold over the course of 2 weeks when cryopreserved PHH from the same donor were cultured in 2Dsw configuration. Combined, these data indicate that the choice of culture method substantially affects the metabolic capacity of PHH, which might translate into differences in drug metabolism and susceptibility to DILI.
Differential Sensitivity of Cryopreserved PHH Cultured in 2Dsw and 3D Spheroids to Chronic Exposure With Model DILI Compounds
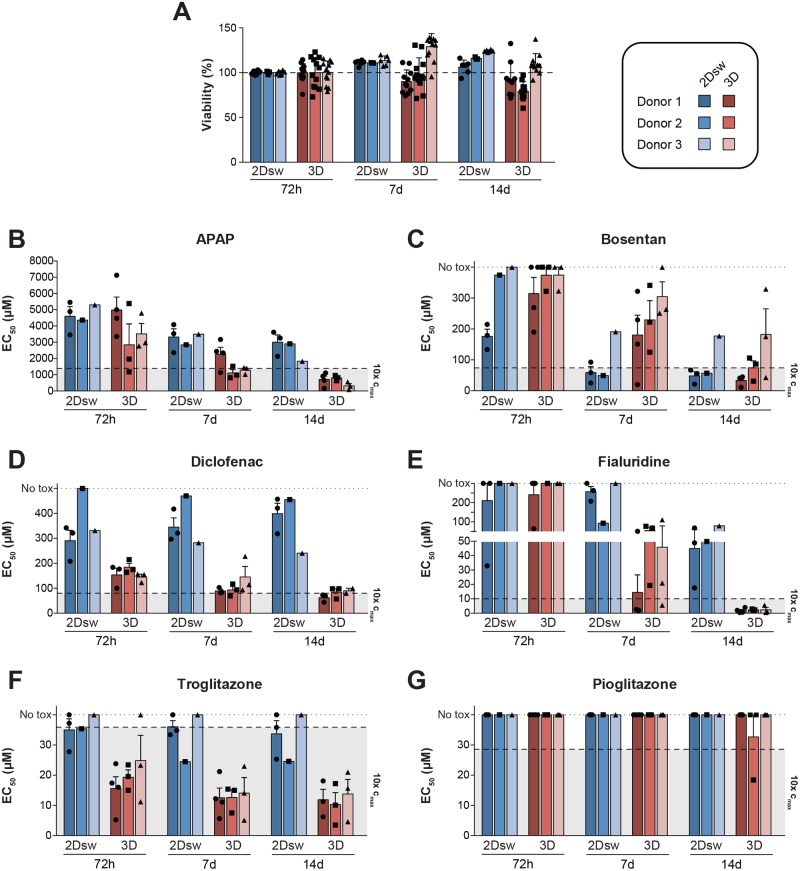
We next evaluated the effects of the choice of culture format on the sensitivity of cells to model hepatotoxic compounds in a multi-center study. To allow for an accurate comparison, we first established that cryopreserved PHH in 2Dsw and 3D spheroid culture formats remained viable throughout the entire 2-week period (Figure 5A). Cryopreserved PHHs from each donor, cultured in either 2Dsw or 3D spheroid configuration, were then treated with hepatotoxins that exert their toxicity through different mechanisms for 72 hours (single treatment), 7 days (3 treatments), and 14 days (6 treatments). Specifically, we chose (1) acetaminophen (APAP), as a model compound for toxicity via reactive metabolites (Figure 5B), (2) the BSEP inhibitor bosentan (Figure 5C), (3) diclofenac, as an inhibitor of mitochondrial ATP synthesis (Figure 5D), (4) fialuridine that causes depletion of mitochondrial DNA (Figure 5E), and (5) troglitazone that inhibits β-oxidation by blocking long-chain acyl-CoA synthase (Figure 5F) (Lauschke and Ingelman-Sundberg, 2016). PHHs in 3D spheroid cultures were overall more sensitive and detected toxicity of all 5 hepatotoxins at clinically relevant concentrations (<10× therapeutic exposure levels) after repeated exposures. In contrast, cryopreserved PHH in 2Dsw configuration exhibited substantially lower sensitivities to APAP, diclofenac, fialuridine, and troglitazone after long-term exposure when using the chosen treatment regimen. Thus, EC50 values could not be attained in any of the cryopreserved PHH donors in 2Dsw culture with diclofenac and fialuridine. The sensitivity of PHH in 2Dsw culture toward the cholestatic agent bosentan was, however, higher as compared with 3D spheroids after 7 days, whereas at 14 days an equal sensitivity was registered for the 2 systems.
Figure 5.
The choice of culture format influences the sensitivity to selected hepatotoxins in a compound-specific manner. Hepatocytes from 3 different donors were cultured in 2Dsw and 3D spheroid configuration at 6 different laboratories. A, Hepatocyte viability remains stable in both culture methods for up to 2 weeks in culture as quantified by ATP measurements in untreated cells. Viability is represented as normalized to ATP levels from the same donor at day 3. For 2Dsw incubations, n = 6, for 3D spheroids n = 10–12. PHH from 3 donors cultured in either 3D spheroid or 2Dsw configuration were exposed to the hepatotoxic compounds acetaminophen (APAP, B), bosentan (C), diclofenac (D), fialuridine (E), or troglitazone (F) for up to 14 days. Furthermore, we determined the toxicity of troglitazone’s less-toxic structural analog pioglitazone (G). EC50 values were determined by quantification of intracellular ATP levels. Dashed lines indicate 10× therapeutic Cmax concentrations for the respective compound (human Cmax values: APAP = 139 µM [Sevilla-Tirado et al., 2003], bosentan = 7.4 µM [Gutierrez et al., 2013], diclofenac = 8 µM [G.D. Searle LLC Division of Pfizer Inc., FDA professional drug information—Arthrotec (2015) available at: http://www.drugs.com/pro/arthrotec.html (accessed: 4 September 2017)], fialuridine = 1 µM [Bowsher et al., 1994], pioglitazone = 2.9 µM [Wong et al., 2004], troglitazone = 3.6 µM corresponding to 400 mg recommended daily dose [Loi et al., 1999]). Dotted line indicates EC50 values above the highest tested concentration. All 3D spheroid experiments were performed at a minimum of 3 different laboratories (as indicated in Table 1), whereas for 2Dsw experiments, donor 1 was compared across 3 sites, while incubations for donors 2 and 3 were performed only at a single site. Error bars indicate SEM.
Cryopreserved PHH Spheroids Better Discriminate Between Toxicity of Troglitazone and Its Less Toxic Analog Pioglitazone
We evaluated the specificity of the model systems by comparing the toxicity of the hepatotoxic thiazolidinedione troglitazone to its less toxic structural analog pioglitazone, whose exposure levels in vivo are very similar (troglitazone = 3.6 µM at recommended daily dose of 400 mg; pioglitazone = 2.9 µM at recommended daily dose of 30 mg [Loi et al., 1999; Wong et al., 2004]). The toxicity of troglitazone was successfully identified in 3D spheroids at relevant concentrations already after 72 hours by all 3 donors tested (Figure 5F), whereas in the 2Dsw culture only 1 of the 3 donors indicated troglitazone toxicity. Pioglitazone was non-toxic in both 3D spheroid and 2Dsw cultures (Figure 5G). Furthermore, the ratio between concentrations at which toxicity of pioglitazone and troglitazone is observed, is significantly higher in 3D culture compared with 2Dsw (p < .001), indicative of elevated specificity of PHH in spheroid configuration (Supplementary Figure 2).
Robustness of 2Dsw and 3D Spheroid Culture Assays
Inter-individual differences can impact upon both the efficacy and safety of new medicines and are therefore important to consider already during the early stages of drug development. In particular, genetic polymorphisms in drug metabolizing enzymes can influence both the bioactivation of compounds to chemically reactive metabolites as well as detoxification and clearance of these products. To assess the effects of inter-individual variability, we evaluated hepatocytes from 3 different donors in 3D spheroids and 2Dsw cultures (Figs. 5B–G). We observed that the variability in sensitivity between the donors evaluated in this study was overall minor and outweighed by differences due to the choice of culture method. Notably, 1 donor (donor 3) exhibited a higher sensitivity to APAP but a lower sensitivity to bosentan compared with the other donors, suggesting that variability between donors might be compound specific (Figs. 5B and 5C).
We moreover evaluated the robustness of both culture methods across participating laboratories by comparing EC50 values generated at multiple sites for donor 1. Most importantly, data that were generated across multiple sites were highly similar for both culture systems, with an inter-laboratory coefficient of variation of 31 ± 18% SD and 53 ± 36% SD for 2Dsw cultures and 3D spheroids, respectively.
DISCUSSION
DILI continues to present a significant challenge for the pharmaceutical industry, with late-stage and post-marketing attritions constituting serious consequences of failure to detect hepatotoxicity pre-clinically. Approximately 18% of compounds withdrawn from the market between 1953 and 2013 were due to hepatotoxicity (Onakpoya et al., 2016), making the liver the most frequent site of ADRs leading to compound failure. In vitro systems that accurately predict the hepatotoxicity of new compounds are important tools for improving pre-clinical safety. In particular, systems in which repeated dose regimes can be accommodated and in which toxicity can be observed at concentrations approximating in vivo exposure levels would provide an important step toward better identification of compounds that pose a risk to patient safety.
To constitute a suitable system for prediction of human drug metabolism and drug toxicity, it is of fundamental importance that the culture method supports the long-term maintenance of relevant cellular phenotypes. We here compared the effects of 3D spheroid and 2Dsw culture on the proteome signatures of PHH from the same donors. Importantly, we found that the choice of culture method has pronounced influence on protein levels and functionality. Cells in 2Dsw culture showed perturbed glycolysis and gluconeogenesis, reminiscent of PHH in 2D monolayer culture (Bell et al., 2016), indicating that certain aspects of hepatocyte dedifferentiation are not prevented in sandwich configuration in agreement with previous studies (Rowe et al., 2010). Furthermore, we found that cryopreserved PHH in sandwich configuration displayed significant perturbations of the redox system, indicated by a downregulation of various GSTs as well as rapid decline of multiple peroxiredoxins, suggesting a lower capacity to detoxify reactive metabolic intermediates. Besides causing differences in molecular phenotypes, the choice of culture method also influenced the stability thereof, as cryopreserved PHH cultured in 2Dsw but not in 3D spheroid configuration underwent significant alterations of protein signatures in long-term culture, which further translated into less stable metabolic activities. These findings are in agreement with previous studies that described sudden decreases in phase I and phase II activities in PHH upon plating (Heslop et al., 2016). Metabolic activity determinations have, however, shown a relative stability in 2Dsw culture when regularly re-overlayed up to 14 days of culture (Bellwon et al., 2015), when activities were normalized to protein amount instead of seeded cell numbers. The data from the present study might suggest that proteomic signature is a more sensitive marker of the differentiated status of PHH cultures.
In general, different cells from different donors can differ in quality and to be able to draw correct conclusions from long-term experiments, it is important to continuously monitor cellular viability, for example by measuring ATP and ensuring that it does not drop below 50% of initial values at the first day of spheroid culture.
Importantly, the illustrated variables likely contribute to the observed differences in sensitivity toward the hepatotoxic model compounds included in our long-term cytotoxicity studies. PHH in 3D spheroid configuration showed enhanced sensitivity to compounds that exert toxicity via mitochondrial perturbation involving uncoupling of oxidative phosphorylation, blocking of β-oxidation or depletion of mitochondrial DNA, at clinically relevant concentrations, whereas cells from the same donors in 2D cultures were much less sensitive. Notably, there is considerable evidence that phase I bioactivation is a requisite step in the sequence of events leading to liver damage following exposure to many drugs with DILI liabilities. We detected robustly higher levels of CYP1A2 and CYP3A4 activity (Figure 2) in 3D versus 2D culture and both of these P450s have been implicated in the bioactivation of acetaminophen to its reactive and toxic intermediate, NAPQI (Laine et al., 2009). This is consistent with our finding of increased APAP sensitivity in 3D spheroid versus 2Dsw culture after long-term exposure. Moreover, the 3D configuration was also more sensitive to diclofenac exposure, again in agreement with higher activities of CYP3A4 and CYP2C8 in 3D cultures, which are both implicated in diclofenac toxicity (Bort et al., 1999). Similarly, formation of reactive metabolites from troglitazone can occur through the activity of CYP3A4 and CYP2C8, which may explain increased sensitivity in 3D versus 2D in this study (Yamamoto et al., 2002; Yamazaki et al., 1999). Nevertheless, all of these associations will need to be verified through pharmacological intervention, for example through 1-aminobenzotriazole-mediated pan-CYP inhibition.
In the case of fialuridine, the proteomic dataset does not support an obvious correlation of the expression levels of various proteins known to play a role in the uptake of fialuridine into the mitochondria and its metabolism to the active triphosphate moiety, such as ENT1, TK2, and TMPK, with differences in sensitivity. Nevertheless, the differences in activity of these proteins, or indeed in the overall physiology of the mitochondria between 3D and 2Dsw, are possible and need to be tested to gain further understanding of how these models can be used for particular DILI liabilities. In contrast, 2Dsw cultures accurately detected toxicity of the cholestatic agent bosentan, which is supported by the presence of functional bile canalicular networks and maintained physiologically relevant expression levels of bile transporters, including BSEP and members of the MRP and OATP families of transporters. These findings are in agreement with previous studies that demonstrated the 2Dsw cultures are reliable tools for the prediction of cholestatic liabilities (Chatterjee et al., 2014; Oorts et al., 2016). However, for all of the test compounds, clearance data will eventually be required in follow-up studies to bridge sensitivity to chemical exposure.
A crucial prerequisite for more wide-spread dissemination of any platform for toxicity testing are large-scale validation studies, ideally performed in a multi-center setting. In the present study, we assessed the robustness of the platforms toward inter-individual variability between donors and variability between participating laboratories. A variety of patient-specific factors, such as physiological parameters, environmental exposures, or genetic predisposition, have been described that influence risk and severity of ADRs. Previous studies have reported only minor inter-individual differences in PHH 2D monolayer cultures when using EC50 values as readout (Sison-Young et al., 2017) and, in agreement with these findings, we did not detect significant differences between donors for any of the compounds tested. When applying a daily treatment protocol and using the highest non-cytotoxic concentration as readout, inter-donor variability was previously seen with APAP and diclofenac after short-term (72 h) treatment of PHH in 2Dsw (Richert et al., 2016). This was not seen in the present study after long-term treatment, most probably due the gradual loss of CYP activities that play a role in the cytotoxicity of both compounds (Figure 4). As CYP genes are highly variable between populations (Zhou et al., 2017) and previous studies already showed that genotypic differences can alter the pharmacokinetics of the respective substrates (Vorrink et al., 2017), it is likely that larger scale investigations with donors from different ethnic backgrounds and with selected genotypes of interest will reveal more noticeable differences in drug sensitivities between donors also in vitro. Importantly, using the harmonized protocol that was adhered to by all partners in this study, inter-laboratory variability was overall quite similar between 2Dsw and 3D spheroid cultures when considering the EC50 values obtained for the hepatoxic compounds (CV between laboratories [donor 1]: 31 ± 18% SD in 2Dsw and 53 ± 36% SD in 3D spheroids; p = .054).
In this study, we evaluated the sensitivity of 2Dsw and 3D spheroid models using cytotoxicity as an endpoint. However, more complex endpoints, which can resolve more subtle alterations in cellular health or molecular phenotypes, might better separate the cell models. Indeed, gene expression alterations specific for cholestatis, steatosis, or genotoxicity have been identified in cryopreserved PHH spheroids that precede cytotoxicity and inform about underlying toxicity mechanisms (Bell et al., 2017). Furthermore, image-based screening approaches that automatically integrate various endpoints, such as nuclear and cell shape and size, mitochondrial membrane potential, phospholipid accumulation, apoptosis or cytoskeleton integrity have been shown to improve hepatotoxicity predictions (Sirenko et al., 2014).
In conclusion, we benchmarked PHH cultured in 3D spheroids against 2Dsw cultures established from the same donors and found that 3D spheroids were more functionally stable and exhibited increased sensitivity after long-term repeated exposures for the detection of hepatotoxicity presumably due to reactive metabolites or mitochondrial perturbations, including inhibition of β-oxidation or mitochondrial respiration, uncoupling of oxidative phosphorylation or mitochondrial DNA depletion, such as that induced by APAP, diclofenac, fialuridine, and troglitazone, whereas both paradigms performed equally well for the detection of cholestatic liabilities, as exemplified by bosentan. Furthermore, our proteomic and functional analyses revealed important culture method-induced differences in phase I and phase II enzymes as well as drug transporters, which are underlying the differential sensitivity to hepatotoxins. Lastly, this study was successful in setting up a robust culture protocol at multiple sites, allowing to exploit the phenotype of hepatocytes cultured in 3D spheroid configuration for future screening applications as well as mechanistic studies.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
We express our gratitude to Prof. P. Bachellier (Hôpital de Hautepierre, Avenue Molière 67100 Strasbourg, France) and Prof. B. Heyd (Hôpital Jean Minjoz, 3 Boulevard Alexandre Fleming 25000 Besançon, France) for providing liver samples. V.M.L. and M.I.-S. are co-founders and owners of HepaPredict, A.B., C.C.B., D.W., S.R., and A.J.F. are employed at AstraZeneca, L.R. is a co-founder of KaLy Cell, L.R. and A.B. are employed at KaLy Cell, S.J. and T.L. are employees of Orion OY, T.W. and C.S. are employed at GSK, and A.W.M.v.d.V., F.J., J.v.H., and J.S. are employed at Janssen Pharmaceutical Companies of Johnson & Johnson.
FUNDING
Innovative Medicine Initiative project MIP-DILI [115336] and Swedish Research Council [2015-02760, 2016-06222, 2016-01153, and 2016-01154].
AUTHORS’ CONTRIBUTIONS
Project lead and planning: C.C.B., L.R., J.S., M.I-S. Experimental work: A.B., A.C.A.D., A.W.M.v.d.V., F.J., J.v.H., T.W., S.R., C.S., A.J.F., R.S-Y., C.C.B. Data analyses: V.M.L., C.C.B., T.L., S.J., R.J., A.J.F., D.W., C.R., A.B., L.R., M.I-S. Manuscript writing and/or editing: V.M.L., C.C.B., K.P., A.J.F., D.W., A.B., L.R., M.I-S.
REFERENCES
- Bell C. C., Hendriks D. F. G., Moro S. M. L., Ellis E., Walsh J., Renblom A., Fredriksson Puigvert L., Dankers A. C. A., Jacobs F., Snoeys J. et al. , (2016). Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Scientific Reports 6, 25187.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C. C., Lauschke V. M., Vorrink S. U., Palmgren H., Duffin R., Andersson T. B., Ingelman-Sundberg M. (2017). Transcriptional, functional, and mechanistic comparisons of stem cell-derived hepatocytes, HepaRG cells, and three-dimensional human hepatocyte spheroids as predictive in vitro systems for drug-induced liver injury. Drug Metabolism and Disposition 45, 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellwon P., Truisi G. L., Bois F. Y., Wilmes A., Schmidt T., Savary C. C., Parmentier C., Hewitt P. G., Schmal O., Josse R. et al. , (2015). Kinetics and dynamics of cyclosporine A in three hepatic cell culture systems. Toxicology in Vitro 30, 62–78. [DOI] [PubMed] [Google Scholar]
- Bort R., Macé K., Boobis A., Gómez-Lechón M. J., Pfeifer A., Castell J. (1999). Hepatic metabolism of diclofenac: Role of human CYP in the minor oxidative pathways. Biochemical Pharmacology 58, 787–796. [DOI] [PubMed] [Google Scholar]
- Bowsher R. R., Compton J. A., Kirkwood J. A., Place G. D., Jones C. D., Mabry T. E., Hyslop D. L., Hatcher B. L., DeSante K. A. (1994). Sensitive and specific radioimmunoassay for fialuridine: Initial assessment of pharmacokinetics after single oral doses to healthy volunteers. Antimicrobial Agents and Chemotherapy 38, 2134–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbank M. G., Burban A., Sharanek A., Weaver R. J., Guguen-Guillouzo C., Guillouzo A. (2016). Early alterations of bile canaliculi dynamics and the rho kinase/myosin light chain kinase pathway are characteristics of drug-induced intrahepatic cholestasis. Drug Metabolism and Disposition 44, 1780–1793. [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Richert L., Augustijns P., Annaert P. (2014). Hepatocyte-based in vitro model for assessment of drug-induced cholestasis. Toxicology and Applied Pharmacology 274, 124–136. [DOI] [PubMed] [Google Scholar]
- Chen L., Shu Y., Liang X., Chen E. C., Yee S. W., Zur A. A., Li S., Xu L., Keshari K. R., Lin M. J. et al. , (2014). OCT1 is a high-capacity thiamine transporter that regulates hepatic steatosis and is a target of metformin. Proceedings of the National Academy of Sciences of the United States of America 111, 9983–9988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D., Brown D., Alexander R., March R., Morgan P., Satterthwaite G., Pangalos M. N. (2014). Lessons learned from the fate of AstraZeneca’s drug pipeline: A five-dimensional framework. Nature Reviews Drug Discovery 13, 419–431. [DOI] [PubMed] [Google Scholar]
- De Bruyn T., Chatterjee S., Fattah S., Keemink J., Nicolaï J., Augustijns P., Annaert P. (2013). Sandwich-cultured hepatocytes: Utility for in vitro exploration of hepatobiliary drug disposition and drug-induced hepatotoxicity. Expert Opinion on Drug Metabolism & Toxicology 9, 589–616. [DOI] [PubMed] [Google Scholar]
- den Braver-Sewradj S. P., den Braver M. W., Vermeulen N. P. E., Commandeur J. N. M., Richert L., Vos J. C. (2016). Inter-donor variability of phase I/phase II metabolism of three reference drugs in cryopreserved primary human hepatocytes in suspension and monolayer. Toxicology in Vitro 33, 71–79. [DOI] [PubMed] [Google Scholar]
- Dragovic S., Vermeulen N. P. E., Gerets H. H., Hewitt P. G., Ingelman-Sundberg M., Park B. K., Juhila S., Snoeys J., Weaver R. J. (2016). Evidence-based selection of training compounds for use in the mechanism-based integrated prediction of drug-induced liver injury in man. Archives of Toxicology doi: 10.1007/s00204-016-18451,1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans W. E., Relling M. V. (1999). Pharmacogenomics: Translating functional genomics into rational therapeutics. Science 286, 487–491.http://dx.doi.org/10.1126/science.286.5439.487 [DOI] [PubMed] [Google Scholar]
- Ewart L., Dehne E.-M., Fabre K., Gibbs S., Hickman J., Hornberg E., Ingelman-Sundberg M., Jang K.-J., Jones D. R., Lauschke V. M. et al. , (2018). Application of microphysiological systems to enhance safety assessment in drug discovery. Annual Review of Pharmacology and Toxicology 58. [DOI] [PubMed] [Google Scholar]
- Gómez-Lechón M. J., Tolosa L., Conde I., Donato M. T. (2014). Competency of different cell models to predict human hepatotoxic drugs. Expert Opinion on Drug Metabolism & Toxicology 10, 1553–1568. [DOI] [PubMed] [Google Scholar]
- Gutierrez M. M., Nicolas L. B., Donazzolo Y., Dingemanse J. (2013). Relative bioavailability of a newly developed pediatric formulation of bosentan vs. the adult formulation. International Journal of Clinical Pharmacology and Therapeutics 51, 529–536. [DOI] [PubMed] [Google Scholar]
- Heslop J. A., Rowe C., Walsh J., Sison-Young R., Jenkins R., Kamalian L., Kia R., Hay D., Jones R. P., Malik H. Z. et al. , (2016). Mechanistic evaluation of primary human hepatocyte culture using global proteomic analysis reveals a selective dedifferentiation profile. Archives of Toxicology doi:10.1007/s00204-016-1694-y,1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine J. E., Auriola S., Pasanen M., Juvonen R. O. (2009). Acetaminophen bioactivation by human cytochrome P450 enzymes and animal microsomes. Xenobiotica 39, 11–21. [DOI] [PubMed] [Google Scholar]
- Lauschke V. M., Hendriks D. F. G., Bell C. C., Andersson T. B., Ingelman-Sundberg M. (2016a). Novel 3D culture systems for studies of human liver function and assessments of the hepatotoxicity of drugs and drug candidates. Chemical Research in Toxicology 29, 1936–1955. [DOI] [PubMed] [Google Scholar]
- Lauschke V. M., Ingelman-Sundberg M. (2016). The importance of patient-specific factors for hepatic drug response and toxicity. International Journal of Molecular Sciences 17, 1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauschke V. M., Vorrink S. U., Moro S. M. L., Rezayee F., Nordling Å., Hendriks D. F. G., Bell C. C., Sison-Young R., Park B. K., Goldring C. E. et al. , (2016b). Massive rearrangements of cellular MicroRNA signatures are key drivers of hepatocyte dedifferentiation. Hepatology 64, 1743–1756. [DOI] [PubMed] [Google Scholar]
- Loi C. M., Alvey C. W., Vassos A. B., Randinitis E. J., Sedman A. J., Koup J. R. (1999). Steady-state pharmacokinetics and dose proportionality of troglitazone and its metabolites. Journal of Clinical Pharmacology 39, 920–926. [DOI] [PubMed] [Google Scholar]
- Martignoni M., Groothuis G. M. M., de Kanter R. (2006). Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opinion on Drug Metabolism & Toxicology 2, 875–894. [DOI] [PubMed] [Google Scholar]
- Messner S., Fredriksson L., Lauschke V. M., Roessger K., Escher C., Bober M., Kelm J. M., Ingelman-Sundberg M., Moritz W. (2017). Transcriptomic, proteomic, and functional long-term characterization of multicellular three-dimensional human liver microtissues. Applied in Vitro Toxicology aivt.2017.0022–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson H., Betton G., Robinson D., Thomas K., Monro A., Kolaja G., Lilly P., Sanders J., Sipes G., Bracken W. et al. , (2000). Concordance of the toxicity of pharmaceuticals in humans and in animals. Regulatory Toxicology and Pharmacology 32, 56–67. [DOI] [PubMed] [Google Scholar]
- Onakpoya I. J., Heneghan C. J., Aronson J. K. (2016). Post-marketing withdrawal of 462 medicinal products because of adverse drug reactions: A systematic review of the world literature. BMC Medicine doi: 10.1186/s12916-016-0553-2,1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oorts M., Baze A., Bachellier P., Heyd B., Zacharias T., Annaert P., Richert L. (2016). Drug-induced cholestasis risk assessment in sandwich-cultured human hepatocytes. Toxicology in Vitro 34, 179–186. [DOI] [PubMed] [Google Scholar]
- Park B. K., Boobis A., Clarke S., Goldring C. E. P., Jones D., Kenna J. G., Lambert C., Laverty H. G., Naisbitt D. J., Nelson S. et al. , (2011). Managing the challenge of chemically reactive metabolites in drug development. Nature Reviews Drug Discovery 10, 292–306. [DOI] [PubMed] [Google Scholar]
- Parmentier C., Couttet P., Wolf A., Zaccharias T., Heyd B., Bachellier P., Uteng M., Richert L. (2017). Evaluation of transcriptomic signature as a valuable tool to study drug-induced cholestasis in primary human hepatocytes. Archives of Toxicology 91, 2879–2893. [DOI] [PubMed] [Google Scholar]
- Parmentier C., Truisi G. L., Moenks K., Stanzel S., Lukas A., Kopp-Schneider A., Alexandre E., Hewitt P. G., Mueller S. O., Richert L. (2013). Transcriptomic hepatotoxicity signature of chlorpromazine after short- and long-term exposure in primary human sandwich cultures. Drug Metabolism and Disposition 41, 1835–1842. [DOI] [PubMed] [Google Scholar]
- Richert L., Baze A., Parmentier C., Gerets H. H. J., Sison-Young R., Dorau M., Lovatt C., Czich A., Goldring C., Park B. K. et al. , (2016). Cytotoxicity evaluation using cryopreserved primary human hepatocytes in various culture formats. Toxicology Letters 258, 207–215. [DOI] [PubMed] [Google Scholar]
- Rowe C., Gerrard D. T., Jenkins R., Berry A., Durkin K., Sundstrom L., Goldring C. E., Park B. K., Kitteringham N. R., Hanley K. P. et al. , (2013). Proteome-wide analyses of human hepatocytes during differentiation and dedifferentiation. Hepatology 58, 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe C., Goldring C. E. P., Kitteringham N. R., Jenkins R. E., Lane B. S., Sanderson C., Elliott V., Platt V., Metcalfe P., Park B. K. (2010). Network analysis of primary hepatocyte dedifferentiation using a shotgun proteomics approach. Journal of Proteome Research 9, 2658–2668. [DOI] [PubMed] [Google Scholar]
- Sevilla-Tirado F. J., González-Vallejo E. B., Leary A. C., Breedt H. J., Hyde V. J., Fernández-Hernando N. (2003). Bioavailability of two new formulations of paracetamol, compared with three marketed formulations, in healthy volunteers. Methods and Findings in Experimental and Clinical Pharmacology 25, 531–535. [DOI] [PubMed] [Google Scholar]
- Sirenko O., Hesley J., Rusyn I., Cromwell E. F. (2014). High-content assays for hepatotoxicity using induced pluripotent stem cell-derived cells. Assay and Drug Development Technologies 12, 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sison-Young R. L., Lauschke V. M., Johann E., Alexandre E., Anthérieu S., Aerts H., Gerets H. H. J., Labbe G., Hoët D., Dorau M. et al. , (2017). A multicenter assessment of single-cell models aligned to standard measures of cell health for prediction of acute hepatotoxicity. Archives of Toxicology 91, 1385–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatow V. Y., LeCluyse E. L., Griffith L. G., Rusyn I. (2013). In vitro models for liver toxicity testing. Toxicology Research 2, 23–39.http://dx.doi.org/10.1039/C2TX20051A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. A., Isin E. M., Ogese M. O., Mettetal J. T., Williams D. P. (2016). Reactive metabolites: Current and emerging risk and hazard assessments. Chemical Research in Toxicology doi: 10.1021/acs.chemrestox.5b00410, acs.chemrestox.5b00410-29.http://dx.doi.org/10.1021/acs.chemrestox.5b00410 [DOI] [PubMed] [Google Scholar]
- Treyer A., Müsch A. (2013). Hepatocyte polarity. Comprehensive Physiology 3, 243–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorrink S. U., Ullah S., Schmidt S., Nandania J., Velagapudi V., Beck O., Ingelman-Sundberg M., Lauschke V. M. (2017). Endogenous and xenobiotic metabolic stability of primary human hepatocytes in long-term 3D spheroid cultures revealed by a combination of targeted and untargeted metabolomics. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology doi: 10.1096/fj.201601375R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Duncan D., Shi Z., Zhang B. (2013). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Research 41, W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H., Ozalp Y., Lainesse A., Alpan R. S. (2004). In vivo bioequivalence of oral antidiabetic agents: Pioglitazone tablets. Arzneimittel-Forschung 54, 618–624. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Yamazaki H., Ikeda T., Watanabe T., Iwabuchi H., Nakajima M., Yokoi T. (2002). Formation of a novel quinone epoxide metabolite of troglitazone with cytotoxic to HepG2 cells. Drug Metabolism and Disposition: The Biological Fate of Chemicals 30, 155–160. [DOI] [PubMed] [Google Scholar]
- Yamazaki H., Shibata A., Suzuki M., Nakajima M., Shimada N., Guengerich F. P., Yokoi T. (1999). Oxidation of troglitazone to a quinone-type metabolite catalyzed by cytochrome P-4502C8 and P-450 3A4 in human liver microsomes. Drug Metabolism and Disposition: The Biological Fate of Chemicals 27, 1260–1266. [PubMed] [Google Scholar]
- Zhou Y., Ingelman-Sundberg M., Lauschke V. M. (2017). Worldwide distribution of cytochrome P450 alleles: A meta-analysis of population-scale sequencing projects. Clinical Pharmacology & Therapeutics 102, 688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.