Abstract
Members of the P53 transcription factor family, P53, P63, and P73, play important roles in normal development and in regulating the expression of genes that control apoptosis and cell cycle progression in response to genotoxic stress. P53 is involved in the DNA damage response pathway that is activated by hydroxyurea in organogenesis-stage murine embryos. The extent to which P63 and P73 contribute to this stress response is not known. To address this question, we examined the roles of P53, P63, and P73 in mediating the response of Trp53-positive and Trp53-deficient murine embryos to a single dose of hydroxyurea (400 mg/kg) on gestational day 9. Hydroxyurea treatment downregulated the expression of Trp63 and upregulated Trp73 in the absence of effects on the levels of Trp53 transcripts; Trp73 upregulation was P53-dependent. At the protein level, hydroxyurea treatment increased the levels and phosphorylation of P53 in the absence of effects on P63 and P73. Upregulation of the expression of genes that regulate cell cycle arrest and apoptosis, Cdkn1a, Rb1, Fas, Trp53inp1, and Pmaip1, was P53-dependent in hydroxyurea-treated embryos. The increase in cleaved caspase-3 and cleaved mammalian sterile-20-like-1 kinase levels induced by hydroxyurea was also P53-dependent; in contrast, the increase in phosphorylated H2AX, a marker of DNA double-strand breaks, in response to hydroxyurea treatment was only partially P53-dependent. Together, our data show that P53 is the principal P53 family member that is activated in the embryonic DNA damage response.
Keywords: P53 family proteins, hydroxyurea, teratogen, embryonic stress, developmental toxicity, birth defects
Hydroxyurea (HU), a drug used to treat sickle cell anemia and cancer, inhibits ribonucleotide reductase, causing DNA-strand breaks as a result of replication fork stalling, leading to cell cycle arrest and apoptosis (Kovacic, 2011). In mice, exposure to HU during organogenesis causes DNA damage (Banh and Hales, 2013) and leads to gross and skeletal malformations in the fore- and hind-limbs, tail and craniofacial regions (Yan and Hales, 2005). Doses of HU that are teratogenic in organogenesis-stage murine embryos increase P53 phosphorylation and activate the P53 signaling pathway (El Husseini et al., 2016); using microarray and pathway analysis, we demonstrated that genes that control DNA damage repair, cell cycle arrest and apoptosis are upregulated. Fitting to its proposed role as a “Guardian of the Genome,” previous studies have reported that P53 activation protects the developing conceptus against congenital malformations after exposure to DNA damaging agents such as benzo[a]pyrene, cyclophosphamide, or radiation (Mikheeva et al., 2004; Nicol et al., 1995; Norimura et al., 1996). Nevertheless, the ocular teratogenic effects of 2-chloro-2′-deoxyadenosine were reduced in P53 null transgenic mice (Wubah et al., 1996). It is clear that the role of P53 in mediating how embryos respond to a stressor is complex.
P53 is a member of a family of proteins that includes the homologs P63 and P73 (Levrero et al., 2000). Each of the P53 family proteins has unique functions during embryonic development. Under normal conditions, P53 expression is widespread until gestation day (GD) 11 in organogenesis-stage mouse embryos but it is largely transcriptionally inactive; at later stages of development, P53 expression is restricted to differentiating tissues, such as specific regions of the brain (Choi and Donehower, 1999; Molchadsky et al., 2010). P63 is essential for proper limb and epithelial development, while P73 is involved in the development of the nervous and immune systems (Yang et al., 1999, 2000). Both P63 and P73 have numerous isoforms which have been reported to have divergent and even antagonistic functions (Levrero et al., 2000). P63 and P73 each have 2 transcription start sites which produce protein isoforms with transactivation domains (TAs), or with truncated N-termini (ΔN) that lack the TA domain (Murray-Zmijewski et al., 2006). Each of these isoforms also has several splice variants (P63: α-γ, P73: α-ζ) (Levrero et al., 2000); the functions of these isoforms during development are not well-understood. The TA isoforms of P63 and P73 exhibit significant functional homology to P53 in response to stress stimuli, including DNA damage (Levrero et al., 1999; Yang et al., 2002).
Because the DNA binding domains of P63 and P73 share considerable homology to that of P53 they bind to many P53 DNA binding sites and upregulate a number of the same downstream target genes (Riley et al., 2008). All 3 P53 family proteins are activated in response to various stress stimuli and act as transcription factors, translocating from the cytoplasm to the nucleus and upregulating the expression of transcripts that are involved in regulating cell cycle arrest (eg, Cdkn1a, Rb1) and apoptosis (eg, Fas, Pmaip1, Trp53inp1) (Costanzo et al., 2002; Kruse and Gu, 2009; Petitjean et al., 2008). Furthermore, under certain stress conditions, the ability of P53 to promote apoptosis appears to rely on the activation of P63 and P73 (Fatt et al., 2014; Flores et al., 2002). However, there are some gene targets that are unique to each family member (Fontemaggi et al., 2002; Wu et al., 2003; Yang et al., 2010). Trp53inp1, a direct downstream transcriptional target of P53, is involved in mediating apoptosis and the response to oxidative stress (Seillier et al., 2012). Although they are not direct targets, the expression levels of Pax9 and Tbx5, which are involved in limb and skeletal development, have been negatively correlated with the upregulation of P63 and P73, respectively (Holembowski et al., 2014; Wang et al., 2011). In addition, Jag2 expression is upregulated directly by P63 and P73 but is either mildly upregulated or not upregulated at all by P53 (Sasaki et al., 2002; Wu et al., 2003).
One of the main mechanisms by which P53 family proteins induce apoptosis is by activation of the caspase cascade. All 3 transcription factors are capable of inducing proapoptotic factors (eg, Pmaip1, Puma, Bax, Bak) which interact with the mitochondrial membrane to release cytochrome c into the cytoplasm and activate the caspase cascade (Borrelli et al., 2009; Chipuk et al., 2004; Gong et al., 1999; Gressner et al., 2005; Jones et al., 2007; Schuler et al., 2000). This involves the promotion of procaspase-3 cleavage into its proteolytic form, cleaved caspase-3, the effector caspase that drives the process of apoptotic cell death by the specific cleavage of key cellular proteins (Porter and Janicke, 1999). One of the targets of caspase-3 is the mammalian sterile-20-like-1 (MST-1) kinase (Lee et al., 2001). Mammalian sterile-20-like-1 is a serine/threonine kinase that is involved in promoting DNA damage repair and apoptosis (Pefani and O’Neill, 2016); after cleavage by caspase-3, MST-1 translocates to the nucleus where it phosphorylates the tail of the H2AX histone variant on Ser139, contributing to the formation of γH2AX foci (Teraishi et al., 2006). These foci are important for the modification of chromatin structure and the recruitment of the DNA damage repair machinery (eg, the MRE11/RAD50/NBS1 complex) required to repair DNA double-strand breaks (Furuta et al., 2003; Rogakou et al., 2000). Furthermore, γH2AX foci are essential for nuclear condensation and DNA fragmentation that are hallmarks of the commitment of a cell to apoptosis (Wen et al., 2010). Previously, it has been shown that HU exposure increases the levels of γH2AX foci in the caudal regions of the organogenesis-stage embryo (Banh and Hales, 2013). The contributions of P53, P63, and P73 to the activation of caspase-3 and MST-1 and the increase in γH2AX foci that is triggered in the organogenesis-stage embryo by DNA damage remain to be determined.
Although genotoxic agents and oxidative stress have been reported to increase the protein levels of both P63 and P73 in in vivo and in vitro models (Flores et al., 2002; Gonfloni et al., 2009; Klein et al., 2011; Petitjean et al., 2008), the contributions of these proteins in the response of embryos to DNA damage is not known. Here, we elucidated the impact of exposure to HU on the activation of P63 and P73 in early embryos in the presence and absence of P53.
MATERIALS AND METHODS
Experimental animals
Trp53 transgenic (B6.129S2-Trp53tm1Tyj/J) mice were purchased from The Jackson Laboratory (Bar Harbor, Maine) and housed in the McIntyre Animal Resource Centre (Montreal, QC, Canada). Animal treatments were conducted in accordance with the guidelines outlined in the Canadian Guide to the Care and Use of Experimental Animals. To produce timed pregnant mice heterozygote females were mated with heterozygote males overnight; the mice were separated at 10 am the following day (designated as GD 0). Between 8 and 10 am on GD 9 pregnant dams were treated with either saline (control) or 400 mg/kg HU (Aldrich Chemical Co, Madison, Wisconsin) by intraperitoneal injection.
All dams were euthanized by CO2 asphyxiation and cervical dislocation 3 h after HU treatment. Tail tips were taken for DNA extraction and genotyping. The uteri were removed and embryos were explanted in Hanks’ balanced salt solution (Invitrogen Canada, Inc, ON, Canada). At the time of collection, single whole embryos representing each genotype from every litter were separated for future sample processing for genotyping, Western blot analysis, and real-time Quantitative Reverse Transcriptase (qRT)-PCR experiments.
DNA, RNA, and protein extraction from single embryos
Embryos were placed in RNAlater Stabilization Reagent (Qiagen, Mississauga, ON, Canada) at the time of collection and stored at −80°C. RNA, DNA, and protein were extracted from single embryos using the RNeasy Plus Mini Kit and the DNeasy Blood and Tissue Kit with a modified protocol (Qiagen). RNA and DNA were extracted using the manufacturer’s protocol and the concentration and purity of each sample were assessed by spectrophotometry using a NanoDrop1000 spectrophotometer (Fisher Scientific, Wilmington, Delaware). Proteins were precipitated from the remaining embryonic lysate using ice-cold 100% acetone and dissolved in protein lysis buffer (8 M urea, 2 M thiourea, 0.5% [3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate]). Total protein content from each sample was quantified using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Ltd, Mississauga, ON, Canada).
Genotyping by high resolution melt-curve analysis
DNA from individual embryos was extracted as described earlier for genotyping. Tail tip samples obtained from adult male and female mice were used to extract DNA using a modified version of the ethanol precipitation method (Zeugin, 1985). Briefly, tissue samples were kept overnight at 55°C in 250 μl of DNA extraction buffer (50 mM Tris-HCL, 10 mM EDTA, 100 mM NaCl, 1% SDS) and 0.4 mg/ml proteinase K (Sigma-Aldrich, St Louis, Missouri). Samples were then centrifuged at 20 000 × g for 10 min to remove fur and debris, followed by the addition of 250 μl 100% ethanol to the supernatant. Lastly, the samples were centrifuged to isolate the precipitated DNA, the ethanol was discarded and the pellet left to air dry. The DNA was then dissolved in distilled water and the concentration was determined using a NanoDrop1000 spectrophotometer (Fisher Scientific).
The Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems, Foster City, California) and the StepOnePlus Real-Time PCR System (Applied Biosystems) were used to do high resolution melt-curve analysis to determine the zygosity of adult and embryonic DNA samples (Pryor and Wittwer, 2006). Each reaction was composed of 5 μl of 25 ng/μl DNA mixed with 10 μl SYBR Green Master Mix, 0.3 μl of each of the 10 μM forward and reverse primers, and completed to a total reaction volume of 20 μl with DNase and RNase free water. Samples were run in single-plex for each allele. Temperature cycling was determined per the supplier’s recommended protocol: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Subsequently, the melt-curve stage was as follows: 95°C for 15 s, 60°C for 1 min, followed by incremental increases of 0.3°C up to 95°C. The peak melting temperatures for the mutant and wild-type amplicons matched those of the supplier (approximately 78°C and 84°C, respectively). The sequences of the forward and reverse primers for the wild type and mutant alleles of Trp53 were provided by the supplier (wild type forward: AGGCTTAGA GGTGCAAGCTG; mutant forward: CAGCCTCTGTTCCACATACACT; common reverse: TGGATGGTGGTATACTCAGAGC) and were synthesized by Alpha DNA (Montreal, QC, Canada).
Western blotting using SDS-PAGE and Phos-tag gels
Single embryos from across all treatment groups and genotypes were used for the detection and quantification of proteins by Western blot analysis. The phosphorylated forms of proteins were detected using Phos-tag acrylamide gel electrophoresis (Waco Chemicals USA Inc, Richmond, Virginia). Proteins (10 μg) were loaded and separated on 10% SDS-PAGE gels. Phos-tag gels were prepared in the same manner as for regular acrylamide gels but with the addition of 20 μM Phos-tag and 20 μM MgCl2. A negative control for the Phos-tag gel was done by incubating 20 μM Phos-tag and 20 μM MgCl2 with 10 mM EDTA before preparing the acrylamide gel. After gel electrophoresis, Phos-tag gels were washed twice in 10 mM EDTA for 5 min, and twice in transfer buffer. Proteins from regular and Phos-tag gels were then transferred to PVDF membranes at 14 volts overnight at 4°C (Bio-Rad Laboratories, Ltd). Afterwards, membranes were blocked with 5% milk in 1X tris-buffered saline (TBS) containing 0.1% Tween-20 (TBS-T) for 1 h at room temperature. Membranes were incubated overnight, at 4°C, with primary antibodies against P53 (1:1000, cs2524, Cell Signaling Technology, Inc, Danvers, Massachusetts), P63 (1:1000, PA121739, Thermo Fisher Scientific, Waltham, Massachusetts), P73 (1:1000, ab189896, Abcam), cleaved caspase-3 (1:1000, cs9661, Cell Signaling Technology, Inc), γH2AX (1:1000, cs9718, Cell Signaling Technology, Inc), MST-1 (1:1000, cs3682, Cell Signaling Technology, Inc), or actin (1:1000, sc-1616, Santa Cruz Biotechnology, Texas). Membranes were washed 3 times for 5 min intervals with 1X TBS-T and then incubated for 1 h at room temperature with horseradish peroxidase conjugated anti-rabbit (1:5000, cs7074, Cell Signaling Technology, Inc), anti-mouse (1:5000, cs7076, Cell Signaling Technology, Inc), or anti-goat (1:5000, sc-2020, Santa Cruz Biotechnology) secondary antibodies.
Membranes were washed and proteins were detected using the enhanced chemiluminescence technique (ECL Prime, RPN2236, GE Healthcare). Protein bands were visualized using the Amersham Imager 600 (General Electric Healthcare, Mississauga, ON, Canada) and quantified using ImageJ software (National Institutes of Health, Maryland) where the area under the curve represents band intensity. All intensities were normalized to that of the actin loading control. An internal control was run on each blot to account for interblot variation.
Real-time qRT-PCR
RNA extracted from individual embryos was diluted to a working solution of 4 ng RNA/µl and transcripts were quantified using the Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems) and the StepOnePlus Real-Time PCR System (Applied Biosystems). Each reaction was composed of 10 μl SYBR Green Master Mix, 1–2 μl forward/reverse primer, 0.16 μl Reverse Transcriptase mix, 5 µl sample and completed to 20 µl with RNase-DNase-free water. The PCR reactions were conducted under the following conditions: 48°C for 30 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. The following primer sets were purchased from Qiagen: transformation related protein 53 (Trp53, QT00101906); transformation related protein 63 (Trp63, QT00197904); transformation related protein 73 (Trp73, QT00123487); cyclin-dependent kinase inhibitor 1A (Cdkn1a, QT00137053); Fas cell surface death receptor (Fas, QT00095333); phorbol-12-myristate-13-acetate-induced protein 1 (Pmaip1, QT00163506); retinoblastoma 1 (Rb1, QT00164255); transformation related protein 53 inducible nuclear protein 1 (Trp53inp1, QT00112910); jagged 2 (Jag2, QT01043819); paired box 9 (Pax9, QT00110040); T-box transcription factor 5 (Tbx5, QT00124362); and hypoxanthine phosphoribosyltransferase 1 (Hprt1, QT00166768). Serial dilutions of embryonic RNA, pooled from all treatment groups and genotypes, were used as an internal reference and to create a standard curve for optimizing primer efficiency and concentration. Each reaction was done in triplicate, and was averaged and normalized to the amounts of Hprt1 RNA transcripts. The levels of Hprt1 were found to be stable in all treatment groups. The relative quantity of each transcript was determined using the StepOnePlus Software (version 2.3).
Statistical analyses
All data were analyzed using GraphPad Prism Software (version 5, Graph Pad Software Inc, La Jolla, California). Data were tested by 2-way ANOVA, followed by a Bonferroni post hoc multiple comparison correction, to detect statistical differences between the control and HU-treated groups and the different genotypes. Three embryos were evaluated per genotype treatment group in all experiments. The litter was used to define the statistical unit (n). The level of significance for all statistical tests was set to P < .05.
RESULTS
Hydroxyurea Exposure Affects the Transcript Levels of Trp63 and Trp73 but not Trp53
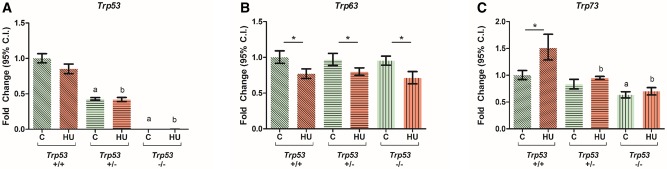
Hydroxyurea treatment did not affect the transcript levels of Trp53 in Trp53+/+ embryos (Figure 1A). As expected, the levels of Trp53 were 50% lower in the heterozygote embryos (Trp53+/−) compared with Trp53+/+ and Trp53 was not detected in the homozygote knockout (Trp53−/−) embryos in either the control or HU-treated groups. Transcript levels of Trp63 were significantly reduced in all drug-treated embryos (Figure 1B). This effect was independent of the Trp53 gene as the levels of Trp63 were not statistically different between HU-treated embryos that expressed or lacked Trp53. The transcript levels of Trp73 were significantly increased by HU exposure in Trp53+/+ embryos (Figure 1C). Interestingly, HU-increased Trp73 expression was dependent on the gene dosage of Trp53; the induction of Trp73 expression by HU was ablated in both Trp53+/− and Trp53−/− embryos. In addition, the levels of Trp73 in saline-treated Trp53−/− embryos were significantly less than in their Trp53+/+ littermates. Thus, the regulation of Trp73 transcription in the organogenesis-stage embryo relies on P53.
Figure 1.
Effects of HU on the relative expression of Trp53, Trp63, and Trp73 in embryos. A–C, Relative expression levels of Trp53, Trp63, and Trp73. Each bar represents the fold change of the mean quantity of the transcript relative to Hprt. C, control; HU, hydroxyurea. P < .05, 2-way ANOVA followed by a Bonferroni post hoc test, n = 3. (*) denotes significant change compared with control group with the same genotype, (a) denotes significant change compared with the Trp53+/+ control group, and (b) denotes significant change compared with the Trp53+/+ HU-treated group.
Hydroxyurea Exposure Increases the Protein Levels and Phosphorylation of P53 but not Those of P63 and P73
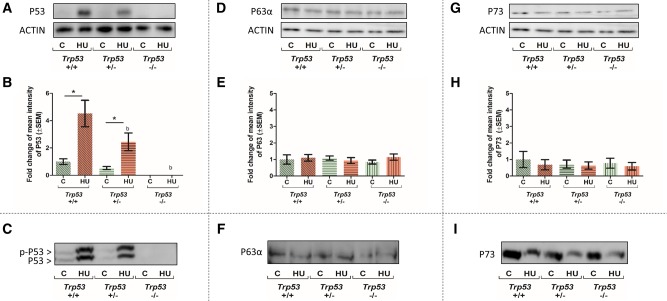
Low or no P53 immunoreactivity was detected in control GD 9 embryos from all genotypes (Figure 2A). Hydroxyurea treatment of Trp53+/+ embryos induced a significant increase in P53 protein levels (Figs. 2A and 2B). The amounts of P53 detected in HU-exposed embryos reflected the Trp53 gene dosage; P53 decreased by 50% in Trp53+/− embryos and was not detected in Trp53−/− embryos. Because an essential component of P53 stabilization is its posttranslational phosphorylation, Phos-tag gels were used to determine if P53 was phosphorylated. Phos-tag causes phosphorylated proteins to migrate at a slower rate during electrophoresis, due to their relatively higher molecular weights and altered electric charge (Kosako et al., 2009). In HU-treated Trp53+/+ and Trp53+/− embryos a higher molecular weight band (approximately 55 kDa), representing phosphorylated P53, appeared above the P53 band (approximately 53 kDa) (Figure 2C). As expected, no detectable levels of phosphorylated P53 were found in Trp53−/− embryos.
Figure 2.
Effects of HU treatment on P53, P63, and P73 protein expression and phosphorylation status in embryos. Top: Representative Western blots and quantification of P53 (A and B), P63 (D and E), and P73 (G and H) immunoreactivity, normalized to the loading control, actin, across all treatment groups, and Trp53 genotypes. Each bar represents the fold change of the mean quantity of the protein relative to actin. Bottom: Representative Phos-tag blots of P53 (C), P63 (F), P73 (I). Only P53 showed a higher molecular weight band, representing phosphorylated P53, p-P53. C, control; HU, hydroxyurea. P < .05, 2-way ANOVA followed by a Bonferroni post hoc test, n = 3. (*) denotes significant change compared with control group with the same genotype and (b) denotes significant change compared with the Trp53+/+ HU-treated group.
TAP63α, the P63 full-length isoform, is shown in Figure 2D. In contrast to P53, P63 was readily detected in control embryos from all genotypes. Furthermore, HU treatment did not significantly alter the expression of TAP63α (Figure 2E). Moreover, no higher molecular weight TAP63α band was observed in Phos-tag gels, suggesting that HU treatment did not induce P63 phosphorylation (Figure 2F).
P73 protein immunoreactivity was detected in control embryos (Figure 2G); HU treatment did not alter P73 protein concentrations in embryos with or without Trp53 (Figure 2H). In addition, no higher molecular weight form of P73 was observed in Phos-tag gels (Figure 2I). Thus, both P63 and P73 proteins are present during organogenesis; however, they are not responsive to HU treatment and their expression is not affected by the absence of P53.
P53 Family Proteins and Regulation of the Expression of Genes Involved in Skeletal and Limb Development or Cell Cycle Arrest and Apoptosis in Response to HU-Induced Stress in the Organogenesis-Stage Embryo
In our previous study, we identified a number of transcripts with altered expression in HU-treated embryos that are either directly or indirectly associated with the transcriptional activity of one or more of the P53 family proteins (El Husseini et al., 2016). These transcripts are involved in cell cycle regulation (Cdkn1a, Rb1), apoptosis (Fas, Pmaip1, Tp53inp1), or skeletal and limb development (Pax9, Tbx5, Jag2) (Table 1).
Table 1.
Prediction Analysis of Unique and Common Markers of P53, P63, and P73 Transcriptional Activity
| Protein | Gene | Prediction | References |
|---|---|---|---|
| P53 | Trp53inp1 | ↑ | Kenzelmann Broz et al. (2013); Okamura et al. (2001) |
| Cdkn1a | ↑ | Kenzelmann Broz et al. (2013); Nayak and Das (2002) | |
| Fas | ↑ | Lima et al. (2011); Munsch et al. (2000); Thiery et al. (2005) | |
| Pmaip1 | ↑ | Hamard et al. (2013); Zhu et al. (2010) | |
| Rb1 | ↑ | Hammond et al. (2006); Kenzelmann Broz et al. (2013); Porrello et al. (2000) | |
| P63 | Pax9 | ↓ | Wang et al. (2011) |
| Cdkn1a | ↑ | Helton et al. (2008); Petitjean et al. (2008); Sen et al. (2011) | |
| Fas | ↑ | Gressner et al. (2005); Petitjean et al. (2008) | |
| Pmaip1 | ↑ | Kerr et al. (2012) | |
| Jag2 | ↑ | Candi et al. (2007); Sasaki et al. (2002); Wu et al. (2003) | |
| P73 | Tbx5 | ↓ | Holembowski et al. (2014) |
| Cdkn1a | ↑ | Jung et al. (2001); Nozell and Chen (2002); Ozaki et al. (1999) | |
| Fas | ↑ | Dicker et al. (2006); Terrasson et al. (2005); Wei et al. (2008) | |
| Rb1 | ↑ | De Laurenzi et al. (2000) | |
| Jag2 | ↑ | Fontemaggi et al. (2002); Sasaki et al. (2002) |
Genes associated with the transcriptional activity of each of the P53 family proteins. Arrows indicate the predicted changes in expression in response to HU if each of the P53 proteins is activated; upregulation (upward arrow) and downregulation (downward arrow). Prediction analysis is based on Ingenuity Pathway Analysis (IPA) software using microarray data from El Husseini et al. (2016).
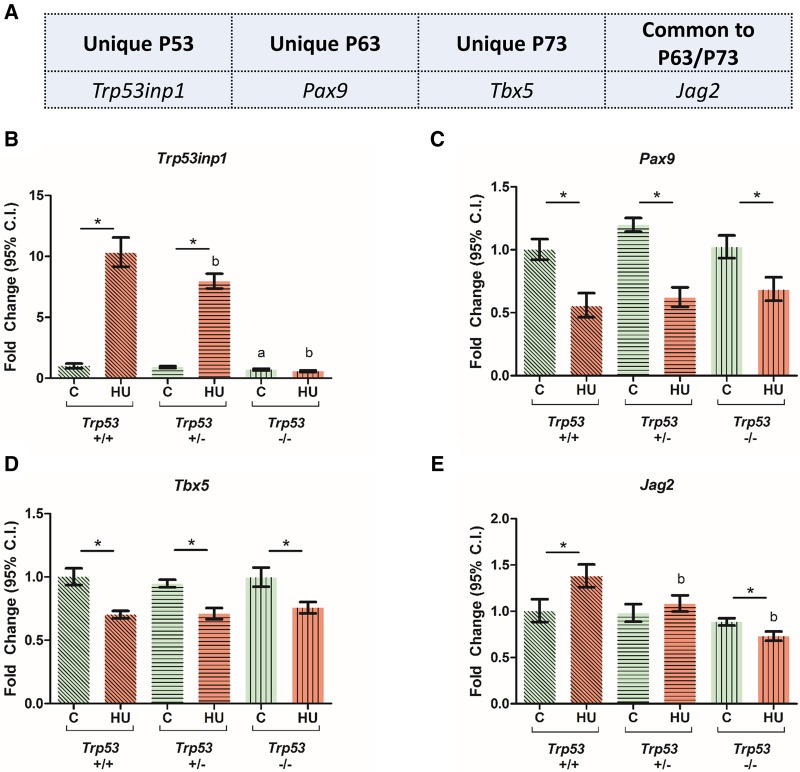
Using qRT-PCR, we assessed the levels of transcripts that are specifically associated with the transcriptional activity of P53 (Trp53inp1), P63 (Pax9), P73 (Tbx5), or both P63 and P73 (Jag2) (Figure 3A), under control and HU-treated conditions, in the presence and absence of Trp53. Trp53inp1 transcript levels were significantly upregulated by HU in Trp53+/+ embryos; this effect was abolished in Trp53 null embryos (Figure 3B). In contrast, the transcript levels of Pax9 and Tbx5 decreased in response to HU treatment and were not affected by the presence or absence of Trp53 (Figs. 3C and 3D). Lastly, HU treatment-induced Jag2 transcript levels in a Trp53-dependent manner (Figure 3E).
Figure 3.
Effects of HU exposure in embryos on the relative expression of genes that are downstream targets of unique P53 family proteins. A, Selected candidates of unique downstream targets of P53, P63, and P73. B–E, Relative expression levels of Trp53inp1, Pax9, Tbx5, and Jag2. Each bar represents the fold change of the mean quantity of the transcript relative to Hprt. C, control; HU, hydroxyurea. P < .05, 2-way ANOVA followed by a Bonferroni post hoc test, n = 3. (*) denotes significant change compared with control group with the same genotype and (b) denotes significant change compared with the Trp53+/+ HU-treated group.
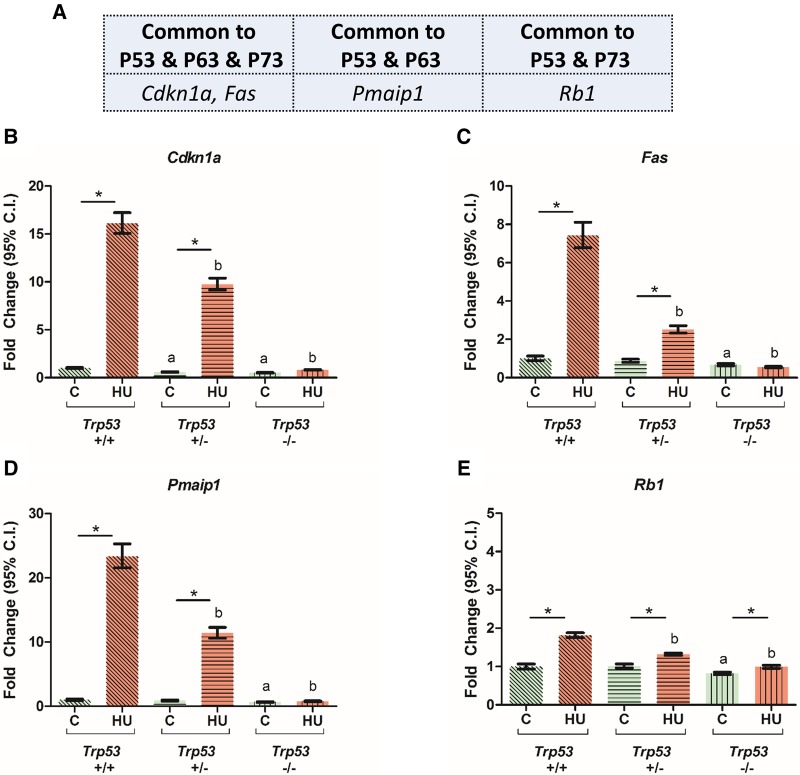
We then assessed the level of transcripts that are commonly regulated by all 3 P53 family members (Cdkn1a, Fas), by P53 and P63 (Pmaip1) or by P53 and P73 (Rb1) (Figure 4A). For each of these downstream target genes, transcript levels were significantly increased in embryos exposed to HU; this induction was ablated by the deletion of Trp53 (Figs. 4B–E). In addition, the levels of each of these transcripts were significantly lower in the saline-treated Trp53−/− embryos than in their Trp53+/+ littermates. Thus, the induction of these downstream markers showed a strong dependence on P53 transcriptional activity, rather than on P63 or P73.
Figure 4.
Effects of HU exposure in embryos on the relative expression of transcripts that control cell cycle progression and apoptosis and are regulated by P53 family proteins. A, Selected candidates of common downstream targets of P53, P63, and P73. B–E, Relative expression levels of Cdkn1a, Fas, Pmaip1, and Rb1. Each bar represents the fold change of the mean quantity of the transcript relative to Hprt. C, control; HU, hydroxyurea. P < .05, 2-way ANOVA followed by a Bonferroni post hoc test, n = 3. (*) denotes significant change compared with control group with the same genotype, (a) denotes significant change compared with the Trp53+/+ control group, and (b) denotes significant change compared with the Trp53+/+ HU-treated group.
Together, P53 transcriptional activity has a dominant impact on gene expression in embryos in response to HU treatment. Conversely, it appears that neither P63 nor P73 have active roles as transcription factors in regulating the expression of P53 family signaling pathway genes in the organogenesis-stage embryo exposed to a DNA damaging agent.
Hydroxyurea Treatment Induces P53-Dependent Caspase-3 and MST-1 Cleavage and H2AX Phosphorylation in Organogenesis-Stage Embryos
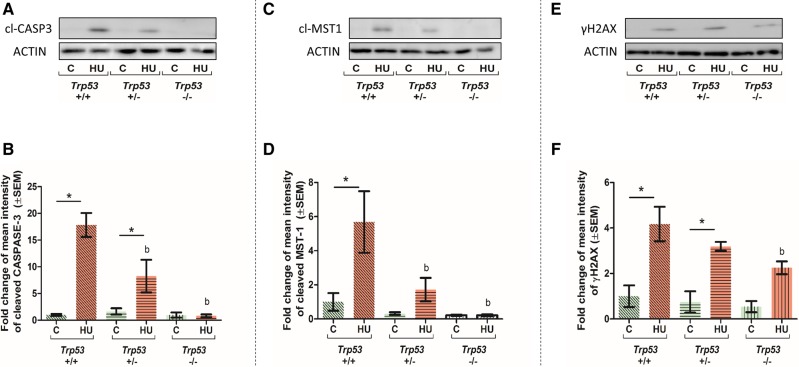
The cleavage of caspase-3 was investigated in control and HU-treated embryos as a marker of apoptosis. Cleaved caspase-3 was not detected in control embryos from any genotype (Figure 5A). A significant increase in cleaved caspase-3 was observed in HU-treated Trp53+/+ embryos and Trp53+/− embryos (Figure 5B). However, no appreciable cleaved caspase-3 was observed in HU-treated Trp53−/− embryos (Figure 5A), indicating that HU-induced caspase-3 cleavage is dependent on the presence of P53.
Figure 5.
Effects of HU exposure on the cleavage of caspase-3 and mammalian sterile-20-like-1 (MST-1) and H2AX phosphorylation in embryos. Representative Western blots and quantification of cleaved caspase-3 (A and B), cleaved MST-1 (C and D) and γH2AX (E and F) protein expression levels, normalized to the loading control, actin. Each bar represents the fold change of the mean quantity of the protein relative to actin. C, control, HU, hydroxyurea. P < .05, 2-way ANOVA followed by a Bonferroni post hoc test, n = 3. (*) denotes significant change compared with control group with the same genotype and (b) denotes significant change compared with the Trp53+/+ HU-treated group.
To determine if activation of caspase-3 by P53 leads to the cleavage of MST-1 and the subsequent phosphorylation of H2AX, cleaved MST-1 and γH2AX were examined using Western blots. A significant increase in cleaved MST-1 occurred in HU-treated Trp53+/+ embryos; the levels of cleaved MST-1 decreased in a dose-dependent manner relative to Trp53 concentration (Figs. 5C and 5D), reflecting the trend observed with cleaved caspase-3. In contrast, all HU-treated embryos, including the Trp53+/− and Trp53−/− embryos, exhibited strong upregulation of γH2AX levels compared with controls (Figs. 5E and 5F). However, after HU treatment, γH2AX levels were significantly lower in Trp53−/− embryos compared with their Trp53+/+ littermates (Figure 5F).
DISCUSSION
The exposure of organogenesis-stage embryos to a DNA damaging agent, HU, induces a stress response that is complex. Hydroxyurea treatment does not affect steady state concentrations of the Trp53 transcript; in contrast, Trp63 and Trp73 expression profiles are differentially affected, suggesting that their transcriptional regulation differs in response to genotoxic stress during organogenesis. Although Trp63 levels decrease with HU treatment, Trp73 levels are significantly increased. In addition, the levels of Trp73, but not Trp63, are significantly affected by the absence of Trp53, suggesting a dependence of Trp73 transcriptional regulation on P53. Indeed, it has been shown that P53 binds to a promoter site that is upstream of the Trp73 gene and can upregulate Trp73 in response to stress (Daily et al., 2011; Wang et al., 2007).
At the protein level, P53 is not detected in control embryos; in contrast, both P63 and P73 proteins are easily detected. P53 is minimally expressed under normal conditions due to its constant ubiquitination by MDM2, an E3 ubiquitin ligase which targets P53 for proteasomal degradation (Kruse and Gu, 2009). The mechanisms by which P63 and P73 are regulated are less well-understood, although they do seem to share some mechanisms of regulation with P53. Under normal conditions, both have been reported to be targeted for proteasomal degradation in certain cells (Rossi et al., 2006; Satija and Das, 2016). However, unlike P53, appreciable amounts of P63 and P73 are found in organogenesis-stage embryos. It is not surprising that the regulation of these 2 proteins during organogenesis differs from that of P53 because P63 and P73 are essential for the maintenance of normal embryo development (Yang et al., 1999, 2000).
Hydroxyurea treatment increases the steady state concentrations and phosphorylation of P53 in the embryo, in the absence of effects on the levels or phosphorylation of P63 or P73. Under stress conditions, P53, P63, and P73 are all phosphorylated by a variety of upstream kinases; phosphorylation promotes their stabilization and transactivation in certain cell types, leading to the activation of cell cycle arrest and cell death pathways (Agami et al., 1999; Gonfloni et al., 2009; Jones et al., 2007; Petitjean et al., 2008; Sanchez-Prieto et al., 2002; Satija and Das, 2016; Shi and Gu, 2012). How P53 family proteins respond to a DNA damaging agent may be determined by the stressor (Levrero et al., 1999, 2000); indeed, various genotoxicants have been reported to have different effects on the protein concentrations and activation of individual P53 family members, resulting in altered cellular stress responses (Kaghad et al., 1997; Liu et al., 1996; Mirkes et al., 2000; Yang et al., 2002). In the embryo, P53 may be more sensitive or more responsive to a wider range of stress stimuli than P63 or P73 (Appella and Anderson, 2001; Gottlieb et al., 1997; Kruse and Gu, 2009; Moallem and Hales, 1998).
Several studies have shown that all P53 family proteins can translocate to the nucleus under stress conditions and act as transcription factors, upregulating the expression of downstream transcripts (Kruse and Gu, 2009; Petitjean et al., 2008; Satija and Das, 2016). We found that HU altered the expression of several genes involved in limb and skeletal development that are predicted to be regulated by one or more of the P53 family members. The dysregulation of the expression of Pax9, Tbx5, and Jag2 suggests that they may be partially involved in mediating the embryotoxicity of HU; Pax9 is involved in skeletal development (McGlinn et al., 2005; Peters et al., 1998), Tbx5 is essential for limb bud initiation (Agarwal et al., 2003), while Jag2 codes for an important factor of the Notch signaling pathway and is required for proper limb bud development and digit formation (Jiang et al., 1998; Xu et al., 2010). It is clear that Pax9 and Tbx5 are not affected by the absence of P53, whereas Jag2 is. The lack of P63 and P73 upregulation in HU-exposed embryos and the concomitant decrease in Pax9 and Tbx5 transcript levels suggest that other upstream regulators (ie, not P63 or P73) may be involved in the transcriptional suppression of these genes. Indeed, Pax9 transcription may be regulated by the sonic hedgehog signaling pathway (LeClair et al., 1999), while that of Tbx5 may be regulated by Hox proteins, HOXB1, HOXC4, and HOXB9, all of which play pivotal roles in embryonic development (Minguillon et al., 2012).
The expression of genes that regulate cell cycle arrest and apoptosis was also affected in embryos exposed to HU. Hydroxyurea strongly induced the expression of a unique P53 marker, Trp53inp1, as well as cell cycle arrest (Cdkn1a, Rb1) and apoptosis (Fas, Pmaip1) genes that may be activated by all 3 P53 family members. This induction was ablated in the absence of Trp53, indicating that P53 is the main transcriptional activator in their response to HU during organogenesis; P63 and P73 are incapable of compensating for the lack of P53 in regulating these genes. This finding is consistent with the observation that in wild-type mouse embryonic fibroblasts P63 binds to the Cdkn1a promoter only when P53 is present; when Trp53 is knocked out, neither P63 nor P73 can induce Cdkn1a transcription, suggesting that P53 is required to drive Cdkn1a transcriptional activity (Flores et al., 2002). Taken together, we conclude that P53 is the main transcriptional activator of genes involved in cell cycle arrest and apoptosis in response to HU in the organogenesis-stage embryo. Indeed, P53 responds to HU-induced stress by mediating cell cycle arrest to allow for DNA damage repair or, failing this, to instigate apoptosis (Gatz and Wiesmuller, 2006; Vousden and Prives, 2009).
The accumulation of cells with irreparable DNA breaks instigates cell death in the form of caspase-3 mediated apoptosis. Here, we report that cleaved caspase-3 is almost undetectable in the Trp53−/− embryos. The relationship between P53 accumulation and caspase-mediated apoptosis due to genotoxic stress is well-established (Pekar et al., 2007; Schuler et al., 2000). There are conflicting reports regarding the ability of P63 and P73 to induce apoptosis in the absence of P53. In tissues lacking Trp53, but containing Trp63 and Trp73, apoptosis has been reported to be either decreased or increased in response to γ-irradiation-induced DNA damage (Flores et al., 2002; Senoo et al., 2004). Our data suggest that P53 is the main driver of apoptosis in response to DNA damage during organogenesis. Trp53−/− embryos exposed to HU may undergo a form of “less regulated” cell death if caspase-3 dependent apoptosis is not activated; indeed Moallem and Hales (1998) reported that Trp53−/− embryonic limbs underwent necrosis, rather than apoptosis, after exposure to a preactivated metabolite of cyclophosphamide, a DNA damaging anticancer drug.
Hydroxyurea induces replication fork stalling, causing DNA-strand breaks that result in the accumulation of phosphorylated H2AX at the sites of damage (Zeman and Cimprich, 2014). An increase in γH2AX was observed in all HU-exposed embryos; however, γH2AX accumulation was significantly lower in Trp53−/− embryos treated with HU than in their Trp53+/+ littermates. This decrease in γH2AX in the absence of P53 may be due to the absence of appreciable cleaved caspase-3, resulting in low levels of cleaved MST-1. Caspase cleavage activates MST-1 (Lee et al., 2001; Oh et al., 2009) and decreased MST-1 activation leads to less H2AX phosphorylation (Wen et al., 2010). However, other kinases, including the DNA damage response proteins, Ataxia Telangiectasia Mutated and Ataxia Telangiectasia And Rad3-Related Protein, may phosphorylate H2AX in response to genotoxic stress and, at least partially, compensate for the decrease in MST-1 activation (Burma et al., 2001; Ward and Chen, 2001).
Although P63 and P73 are essential for crucial developmental processes under normal conditions (Yang et al., 1999, 2000), our study shows that P53 is the main member of the P53 family that drives the early embryonic stress response to DNA damage.
ACKNOWLEDGMENTS
We thank Shafqat Rasool (McGill University, Montreal) for his advice on the preparation of the Phos-tag SDS-PAGE gels.
FUNDING
This work was supported by funding from the Canadian Institutes of Health Research (grant number: MOP-57867). N.EL.H. is the recipient of a student stipend award from the CIHR Training Program in Reproduction, Early Development, and the Impact on Health (CIHR-REDIH), and the Graduate Excellence Fellowship from McGill University. B.F.H. is a James McGill Professor.
REFERENCES
- Agami R., Blandino G., Oren M., Shaul Y. (1999). Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature 399, 809–813.http://dx.doi.org/10.1038/21697 [DOI] [PubMed] [Google Scholar]
- Agarwal P., Wylie J. N., Galceran J., Arkhitko O., Li C., Deng C., Grosschedl R., Bruneau B. G. (2003). Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 130, 623–633. [DOI] [PubMed] [Google Scholar]
- Appella E., Anderson C. W. (2001). Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268, 2764–2772.http://dx.doi.org/10.1046/j.1432-1327.2001.02225.x [DOI] [PubMed] [Google Scholar]
- Banh S., Hales B. F. (2013). Hydroxyurea exposure triggers tissue-specific activation of p38 mitogen-activated protein kinase signaling and the DNA damage response in organogenesis-stage mouse embryos. Toxicol. Sci. 133, 298–308.http://dx.doi.org/10.1093/toxsci/kft069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli S., Candi E., Alotto D., Castagnoli C., Melino G., Vigano M. A., Mantovani R. (2009). p63 regulates the caspase-8-FLIP apoptotic pathway in epidermis. Cell Death Differ. 16, 253–263.http://dx.doi.org/10.1038/cdd.2008.147 [DOI] [PubMed] [Google Scholar]
- Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. (2001). ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276, 42462–42467. [DOI] [PubMed] [Google Scholar]
- Candi E., Rufini A., Terrinoni A., Giamboi-Miraglia A., Lena A. M., Mantovani R., Knight R., Melino G. (2007). DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc. Natl. Acad. Sci. U.S.A. 104, 11999–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk J. E., Kuwana T., Bouchier-Hayes L., Droin N. M., Newmeyer D. D., Schuler M., Green D. R. (2004). Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303, 1010–1014. [DOI] [PubMed] [Google Scholar]
- Choi J., Donehower L. A. (1999). p53 in embryonic development: Maintaining a fine balance. Cell. Mol. Life Sci. 55, 38–47.http://dx.doi.org/10.1007/s000180050268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo A., Merlo P., Pediconi N., Fulco M., Sartorelli V., Cole P. A., Fontemaggi G., Fanciulli M., Schiltz L., Blandino G. et al. , (2002). DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell 9, 175–186. [DOI] [PubMed] [Google Scholar]
- Daily K., Patel V. R., Rigor P., Xie X., Baldi P. (2011). MotifMap: Integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinformatics 12, 495.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurenzi V., Raschella G., Barcaroli D., Annicchiarico-Petruzzelli M., Ranalli M., Catani M. V., Tanno B., Costanzo A., Levrero M., Melino G. (2000). Induction of neuronal differentiation by p73 in a neuroblastoma cell line. J. Biol. Chem. 275, 15226–15231. [DOI] [PubMed] [Google Scholar]
- Dicker F., Kater A. P., Prada C. E., Fukuda T., Castro J. E., Sun G., Wang J. Y., Kipps T. J. (2006). CD154 induces p73 to overcome the resistance to apoptosis of chronic lymphocytic leukemia cells lacking functional p53. Blood 108, 3450–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Husseini N., Schlisser A. E., Hales B. F. (2016). Editor’s highlight: Hydroxyurea exposure activates the P53 signaling pathway in murine organogenesis-stage embryos. Toxicol. Sci. 152, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatt M. P., Cancino G. I., Miller F. D., Kaplan D. R. (2014). p63 and p73 coordinate p53 function to determine the balance between survival, cell death, and senescence in adult neural precursor cells. Cell Death Differ. 21, 1546–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores E. R., Tsai K. Y., Crowley D., Sengupta S., Yang A., McKeon F., Jacks T. (2002). p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416, 560–564. [DOI] [PubMed] [Google Scholar]
- Fontemaggi G., Kela I., Amariglio N., Rechavi G., Krishnamurthy J., Strano S., Sacchi A., Givol D., Blandino G. (2002). Identification of direct p73 target genes combining DNA microarray and chromatin immunoprecipitation analyses. J. Biol. Chem. 277, 43359–43368. [DOI] [PubMed] [Google Scholar]
- Furuta T., Takemura H., Liao Z. Y., Aune G. J., Redon C., Sedelnikova O. A., Pilch D. R., Rogakou E. P., Celeste A., Chen H. T. et al. , (2003). Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 278, 20303–20312. [DOI] [PubMed] [Google Scholar]
- Gatz S. A., Wiesmuller L. (2006). p53 in recombination and repair. Cell Death Differ. 13, 1003–1016.http://dx.doi.org/10.1038/sj.cdd.4401903 [DOI] [PubMed] [Google Scholar]
- Gonfloni S., Di Tella L., Caldarola S., Cannata S. M., Klinger F. G., Di Bartolomeo C., Mattei M., Candi E., De Felici M., Melino G. et al. , (2009). Inhibition of the c-Abl-TAp63 pathway protects mouse oocytes from chemotherapy-induced death. Nat. Med. 15, 1179–1185. [DOI] [PubMed] [Google Scholar]
- Gong J. G., Costanzo A., Yang H. Q., Melino G., Kaelin W. G. Jr, Levrero M., Wang J. Y. (1999). The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399, 806–809. [DOI] [PubMed] [Google Scholar]
- Gottlieb E., Haffner R., King A., Asher G., Gruss P., Lonai P., Oren M. (1997). Transgenic mouse model for studying the transcriptional activity of the p53 protein: Age- and tissue-dependent changes in radiation-induced activation during embryogenesis. EMBO J. 16, 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressner O., Schilling T., Lorenz K., Schulze Schleithoff E., Koch A., Schulze-Bergkamen H., Lena A. M., Candi E., Terrinoni A., Catani M. V. et al. , (2005). TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 24, 2458–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamard P. J., Barthelery N., Hogstad B., Mungamuri S. K., Tonnessen C. A., Carvajal L. A., Senturk E., Gillespie V., Aaronson S. A., Merad M. et al. , (2013). The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev. 27, 1868–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond E. M., Mandell D. J., Salim A., Krieg A. J., Johnson T. M., Shirazi H. A., Attardi L. D., Giaccia A. J. (2006). Genome-wide analysis of p53 under hypoxic conditions. Mol. Cell. Biol. 26, 3492–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helton E. S., Zhang J., Chen X. (2008). The proline-rich domain in p63 is necessary for the transcriptional and apoptosis-inducing activities of TAp63. Oncogene 27, 2843–2850.http://dx.doi.org/10.1038/sj.onc.1210948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holembowski L., Kramer D., Riedel D., Sordella R., Nemajerova A., Dobbelstein M., Moll U. M. (2014). TAp73 is essential for germ cell adhesion and maturation in testis. J. Cell Biol. 204, 1173–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R., Lan Y., Chapman H. D., Shawber C., Norton C. R., Serreze D. V., Weinmaster G., Gridley T. (1998). Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 12, 1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. V., Dickman M. J., Whitmarsh A. J. (2007). Regulation of p73-mediated apoptosis by c-Jun N-terminal kinase. Biochem. J. 405, 617–623.http://dx.doi.org/10.1042/BJ20061778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M. S., Yun J., Chae H. D., Kim J. M., Kim S. C., Choi T. S., Shin D. Y. (2001). p53 and its homologues, p63 and p73, induce a replicative senescence through inactivation of NF-Y transcription factor. Oncogene 20, 5818–5825. [DOI] [PubMed] [Google Scholar]
- Kaghad M., Bonnet H., Yang A., Creancier L., Biscan J. C., Valent A., Minty A., Chalon P., Lelias J. M., Dumont X. et al. , (1997). Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90, 809–819. [DOI] [PubMed] [Google Scholar]
- Kenzelmann Broz D., Spano Mello S., Bieging K. T., Jiang D., Dusek R. L., Brady C. A., Sidow A., Attardi L. D. (2013). Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 27, 1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J. B., Hutt K. J., Michalak E. M., Cook M., Vandenberg C. J., Liew S. H., Bouillet P., Mills A., Scott C. L., Findlay J. K. et al. , (2012). DNA damage-induced primordial follicle oocyte apoptosis and loss of fertility require TAp63-mediated induction of Puma and Noxa. Mol. Cell 48, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A., Maldonado C., Vargas L. M., Gonzalez M., Robledo F., Perez de Arce K., Munoz F. J., Hetz C., Alvarez A. R., Zanlungo S. (2011). Oxidative stress activates the c-Abl/p73 proapoptotic pathway in Niemann-Pick type C neurons. Neurobiol. Dis. 41, 209–218. [DOI] [PubMed] [Google Scholar]
- Kosako H., Yamaguchi N., Aranami C., Ushiyama M., Kose S., Imamoto N., Taniguchi H., Nishida E., Hattori S. (2009). Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat. Struct. Mol. Biol. 16, 1026. [DOI] [PubMed] [Google Scholar]
- Kovacic P. (2011). Hydroxyurea (therapeutics and mechanism): Metabolism, carbamoyl nitroso, nitroxyl, radicals, cell signaling and clinical applications. Med. Hypotheses 76, 24–31.http://dx.doi.org/10.1016/j.mehy.2010.08.023 [DOI] [PubMed] [Google Scholar]
- Kruse J. P., Gu W. (2009). Modes of p53 regulation. Cell 137, 609–622.http://dx.doi.org/10.1016/j.cell.2009.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClair E. E., Bonfiglio L., Tuan R. S. (1999). Expression of the paired-box genes Pax-1 and Pax-9 in limb skeleton development. Dev. Dyn. 214, 101–115.http://dx.doi.org/10.1002/(SICI)1097-0177(199902)214:2<101::AID-AJA1>3.0.CO;2-4 [DOI] [PubMed] [Google Scholar]
- Lee K. K., Ohyama T., Yajima N., Tsubuki S., Yonehara S. (2001). MST, a physiological caspase substrate, highly sensitizes apoptosis both upstream and downstream of caspase activation. J. Biol. Chem. 276, 19276–19285. [DOI] [PubMed] [Google Scholar]
- Levrero M., De Laurenzi V., Costanzo A., Gong J., Melino G., Wang J. Y. (1999). Structure, function and regulation of p63 and p73. Cell Death Differ. 6, 1146–1153. [DOI] [PubMed] [Google Scholar]
- Levrero M., De Laurenzi V., Costanzo A., Gong J., Wang J. Y., Melino G. (2000). The p53/p63/p73 family of transcription factors: Overlapping and distinct functions. J. Cell Sci. 113(Pt 10), 1661–1670. [DOI] [PubMed] [Google Scholar]
- Lima R. T., Busacca S., Almeida G. M., Gaudino G., Fennell D. A., Vasconcelos M. H. (2011). MicroRNA regulation of core apoptosis pathways in cancer. Eur. J. Cancer 47, 163–174. [DOI] [PubMed] [Google Scholar]
- Liu Z. G., Baskaran R., Lea-Chou E. T., Wood L. D., Chen Y., Karin M., Wang J. Y. (1996). Three distinct signalling responses by murine fibroblasts to genotoxic stress. Nature 384, 273–276. [DOI] [PubMed] [Google Scholar]
- McGlinn E., van Bueren K. L., Fiorenza S., Mo R., Poh A. M., Forrest A., Soares M. B., Bonaldo M. D. F., Grimmond S., Hui C.-C. et al. , (2005). Pax9 and Jagged1 act downstream of Gli3 in vertebrate limb development. Mech. Dev. 122, 1218–1233. [DOI] [PubMed] [Google Scholar]
- Mikheeva S., Barrier M., Little S. A., Beyer R., Mikheev A. M., Kerr M. K., Mirkes P. E. (2004). Alterations in gene expression induced in day-9 mouse embryos exposed to hyperthermia (HS) or 4-hydroperoxycyclophosphamide (4CP): Analysis using cDNA microarrays. Toxicol. Sci. 79, 345–359. [DOI] [PubMed] [Google Scholar]
- Minguillon C., Nishimoto S., Wood S., Vendrell E., Gibson-Brown J. J., Logan M. P. O. (2012). Hox genes regulate the onset of <em>Tbx5</em> expression in the forelimb. Development 139, 3180–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkes P. E., Wilson K. L., Cornel L. M. (2000). Teratogen-induced activation of ERK, JNK, and p38 MAP kinases in early postimplantation murine embryos. Teratology 62, 14–25.http://dx.doi.org/10.1002/1096-9926(200007)62:1<14::AID-TERA6>3.0.CO;2-9 [DOI] [PubMed] [Google Scholar]
- Moallem S. A., Hales B. F. (1998). The role of p53 and cell death by apoptosis and necrosis in 4-hydroperoxycyclophosphamide-induced limb malformations. Development 125, 3225–3234. [DOI] [PubMed] [Google Scholar]
- Molchadsky A., Rivlin N., Brosh R., Rotter V., Sarig R. (2010). p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis 31, 1501–1508. [DOI] [PubMed] [Google Scholar]
- Munsch D., Watanabe-Fukunaga R., Bourdon J. C., Nagata S., May E., Yonish-Rouach E., Reisdorf P. (2000). Human and mouse Fas (APO-1/CD95) death receptor genes each contain a p53-responsive element that is activated by p53 mutants unable to induce apoptosis. J. Biol. Chem. 275, 3867–3872. [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F., Lane D. P., Bourdon J. C. (2006). p53/p63/p73 isoforms: An orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 13, 962–972. [DOI] [PubMed] [Google Scholar]
- Nayak B. K., Das G. M. (2002). Stabilization of p53 and transactivation of its target genes in response to replication blockade. Oncogene 21, 7226–7229.http://dx.doi.org/10.1038/sj.onc.1205889 [DOI] [PubMed] [Google Scholar]
- Nicol C. J., Harrison M. L., Laposa R. R., Gimelshtein I. L., Wells P. G. (1995). A teratologic suppressor role for p53 in benzo[a]pyrene-treated transgenic p53-deficient mice. Nat. Genet. 10, 181–187. [DOI] [PubMed] [Google Scholar]
- Norimura T., Nomoto S., Katsuki M., Gondo Y., Kondo S. (1996). p53-dependent apoptosis suppresses radiation-induced teratogenesis. Nat. Med. 2, 577–580. [DOI] [PubMed] [Google Scholar]
- Nozell S., Chen X. (2002). p21B, a variant of p21(Waf1/Cip1), is induced by the p53 family. Oncogene 21, 1285–1294.http://dx.doi.org/10.1038/sj.onc.1205191 [DOI] [PubMed] [Google Scholar]
- Oh S., Lee D., Kim T., Kim T. S., Oh H. J., Hwang C. Y., Kong Y. Y., Kwon K. S., Lim D. S. (2009). Crucial role for Mst1 and Mst2 kinases in early embryonic development of the mouse. Mol. Cell. Biol. 29, 6309–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura S., Arakawa H., Tanaka T., Nakanishi H., Ng C. C., Taya Y., Monden M., Nakamura Y. (2001). p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell 8, 85–94.http://dx.doi.org/10.1016/S1097-2765(01)00284-2 [DOI] [PubMed] [Google Scholar]
- Ozaki T., Naka M., Takada N., Tada M., Sakiyama S., Nakagawara A. (1999). Deletion of the COOH-terminal region of p73alpha enhances both its transactivation function and DNA-binding activity but inhibits induction of apoptosis in mammalian cells. Cancer Res. 59, 5902–5907. [PubMed] [Google Scholar]
- Pefani D. E., O’Neill E. (2016). Hippo pathway and protection of genome stability in response to DNA damage. FEBS J. 283, 1392–1403. [DOI] [PubMed] [Google Scholar]
- Pekar O., Molotski N., Savion S., Fein A., Toder V., Torchinsky A. (2007). p53 regulates cyclophosphamide teratogenesis by controlling caspases 3, 8, 9 activation and NF-kappaB DNA binding. Reproduction 134, 379–388. [DOI] [PubMed] [Google Scholar]
- Peters H., Neubuser A., Kratochwil K., Balling R. (1998). Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 12, 2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A., Ruptier C., Tribollet V., Hautefeuille A., Chardon F., Cavard C., Puisieux A., Hainaut P., Caron de Fromentel C. (2008). Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with DeltaNp73. Carcinogenesis 29, 273–281.http://dx.doi.org/10.1093/carcin/bgm258 [DOI] [PubMed] [Google Scholar]
- Porrello A., Cerone M. A., Coen S., Gurtner A., Fontemaggi G., Cimino L., Piaggio G., Sacchi A., Soddu S. (2000). p53 regulates myogenesis by triggering the differentiation activity of pRb. J. Cell Biol. 151, 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter A. G., Janicke R. U. (1999). Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 6, 99–104.http://dx.doi.org/10.1038/sj.cdd.4400476 [DOI] [PubMed] [Google Scholar]
- Pryor R. J., Wittwer C. T. (2006). Real-time polymerase chain reaction and melting curve analysis. Methods Mol. Biol. 336, 19–32. [DOI] [PubMed] [Google Scholar]
- Riley T., Sontag E., Chen P., Levine A. (2008). Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412.http://dx.doi.org/10.1038/nrm2395 [DOI] [PubMed] [Google Scholar]
- Rogakou E. P., Nieves-Neira W., Boon C., Pommier Y., Bonner W. M. (2000). Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J. Biol. Chem. 275, 9390–9395. [DOI] [PubMed] [Google Scholar]
- Rossi M., De Simone M., Pollice A., Santoro R., La Mantia G., Guerrini L., Calabro V. (2006). Itch/AIP4 associates with and promotes p63 protein degradation. Cell Cycle 5, 1816–1822. [DOI] [PubMed] [Google Scholar]
- Sanchez-Prieto R., Sanchez-Arevalo V. J., Servitja J. M., Gutkind J. S. (2002). Regulation of p73 by c-Abl through the p38 MAP kinase pathway. Oncogene 21, 974–979. [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Ishida S., Morimoto I., Yamashita T., Kojima T., Kihara C., Tanaka T., Imai K., Nakamura Y., Tokino T. (2002). The p53 family member genes are involved in the Notch signal pathway. J. Biol. Chem. 277, 719–724. [DOI] [PubMed] [Google Scholar]
- Satija Y. K., Das S. (2016). Tyr99 phosphorylation determines the regulatory milieu of tumor suppressor p73. Oncogene 35, 513–527.http://dx.doi.org/10.1038/onc.2015.111 [DOI] [PubMed] [Google Scholar]
- Schuler M., Bossy-Wetzel E., Goldstein J. C., Fitzgerald P., Green D. R. (2000). p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J. Biol. Chem. 275, 7337–7342. [DOI] [PubMed] [Google Scholar]
- Seillier M., Peuget S., Dusetti N.J., Carrier A. (2012). Antioxidant Role of p53 and of Its Target TP53INP1 In Antioxidant Enzyme (El-Missiry M. A., Ed.), pp. 117–138, Ch. 05. InTech, Rijeka. Available at: https://www.intechopen.com/books/antioxidant-enzyme/antioxidant-role-of-p53-and-of-its-target-tp53inp1. [Google Scholar]
- Sen T., Sen N., Huang Y., Sinha D., Luo Z. G., Ratovitski E. A., Sidransky D. (2011). Tumor protein p63/nuclear factor kappaB feedback loop in regulation of cell death. J. Biol. Chem. 286, 43204–43213.http://dx.doi.org/10.1074/jbc.M111.257105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M., Manis J. P., Alt F. W., McKeon F. (2004). p63 and p73 are not required for the development and p53-dependent apoptosis of T cells. Cancer Cell 6, 85–89.http://dx.doi.org/10.1016/j.ccr.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Shi D., Gu W. (2012). Dual roles of MDM2 in the regulation of p53: Ubiquitination dependent and ubiquitination independent mechanisms of MDM2 repression of p53 activity. Genes Cancer 3, 240–248.http://dx.doi.org/10.1177/1947601912455199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teraishi F., Guo W., Zhang L., Dong F., Davis J. J., Sasazuki T., Shirasawa S., Liu J., Fang B. (2006). Activation of sterile20-like kinase 1 in proteasome inhibitor bortezomib-induced apoptosis in oncogenic K-ras-transformed cells. Cancer Res. 66, 6072–6079.http://dx.doi.org/10.1158/0008-5472.CAN-06-0125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrasson J., Allart S., Martin H., Lule J., Haddada H., Caput D., Davrinche C. (2005). p73-dependent apoptosis through death receptor: Impairment by human cytomegalovirus infection. Cancer Res. 65, 2787–2794. [DOI] [PubMed] [Google Scholar]
- Thiery J., Abouzahr S., Dorothee G., Jalil A., Richon C., Vergnon I., Mami-Chouaib F., Chouaib S. (2005). p53 potentiation of tumor cell susceptibility to CTL involves Fas and mitochondrial pathways. J. Immunol. 174, 871–878. [DOI] [PubMed] [Google Scholar]
- Vousden K. H., Prives C. (2009). Blinded by the light: The growing complexity of p53. Cell 137, 413–431.http://dx.doi.org/10.1016/j.cell.2009.04.037 [DOI] [PubMed] [Google Scholar]
- Wang J., Liu Y. X., Hande M. P., Wong A. C., Jin Y. J., Yin Y. (2007). TAp73 is a downstream target of p53 in controlling the cellular defense against stress. J. Biol. Chem. 282, 29152–29162.http://dx.doi.org/10.1074/jbc.M703408200 [DOI] [PubMed] [Google Scholar]
- Wang X., Ouyang H., Yamamoto Y., Kumar P. A., Wei T. S., Dagher R., Vincent M., Lu X., Bellizzi A. M., Ho K. Y. et al. , (2011). Residual embryonic cells as precursors of a Barrett’s-like metaplasia. Cell 145, 1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I. M., Chen J. (2001). Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276, 47759–47762.http://dx.doi.org/10.1074/jbc.C100569200 [DOI] [PubMed] [Google Scholar]
- Wei J., O’Brien D., Vilgelm A., Piazuelo M. B., Correa P., Washington M. K., El-Rifai W., Peek R. M., Zaika A. (2008). Interaction of Helicobacter pylori with gastric epithelial cells is mediated by the p53 protein family. Gastroenterology 134, 1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Zhu F., Zhang J., Keum Y. S., Zykova T., Yao K., Peng C., Zheng D., Cho Y. Y., Ma W. Y. et al. , (2010). MST1 promotes apoptosis through phosphorylation of histone H2AX. J. Biol. Chem. 285, 39108–39116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Nomoto S., Hoque M. O., Dracheva T., Osada M., Lee C. C., Dong S. M., Guo Z., Benoit N., Cohen Y. et al. , (2003). DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 63, 2351–2357. [PubMed] [Google Scholar]
- Wubah J. A., Ibrahim M. M., Gao X., Nguyen D., Pisano M. M., Knudsen T. B. (1996). Teratogen-induced eye defects mediated by p53-dependent apoptosis. Curr. Biol. 6, 60–69. [DOI] [PubMed] [Google Scholar]
- Xu J., Krebs L. T., Gridley T. (2010). Generation of mice with a conditional null allele of the Jagged2 gene. Genesis 48, 390–393.http://dx.doi.org/10.1002/dvg.20626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Hales B. F. (2005). Activator protein-1 (AP-1) DNA binding activity is induced by hydroxyurea in organogenesis stage mouse embryos. Toxicol. Sci. 85, 1013–1023.http://dx.doi.org/10.1093/toxsci/kfi148 [DOI] [PubMed] [Google Scholar]
- Yang A., Kaghad M., Caput D., McKeon F. (2002). On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 18, 90–95.http://dx.doi.org/10.1016/S0168-9525(02)02595-7 [DOI] [PubMed] [Google Scholar]
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C. et al. , (1999). p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718.http://dx.doi.org/10.1038/19539 [DOI] [PubMed] [Google Scholar]
- Yang A., Walker N., Bronson R., Kaghad M., Oosterwegel M., Bonnin J., Vagner C., Bonnet H., Dikkes P., Sharpe A. et al. , (2000). p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404, 99–103. [DOI] [PubMed] [Google Scholar]
- Yang A., Zhu Z., Kettenbach A., Kapranov P., McKeon F., Gingeras T. R., Struhl K., Tora L. (2010). Genome-wide mapping indicates that p73 and p63 co-occupy target sites and have similar DNA-binding profiles in vivo. PLoS One 5, e11572.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman M. K., Cimprich K. A. (2014). Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeugin J. A. J. J. A. (1985). Ethanol precipitation of DNA. Focus 7, 1–2. [Google Scholar]
- Zhu F., Dolle M. E., Berton T. R., Kuiper R. V., Capps C., Espejo A., McArthur M. J., Bedford M. T., van Steeg H., de Vries A. et al. , (2010). Mouse models for the p53 R72P polymorphism mimic human phenotypes. Cancer Res. 70, 5851–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]