Abstract
Targeting tumor-initiating, drug-resistant populations of cancer stem cells (CSC) with phytochemicals is a novel paradigm for cancer prevention and treatment. We herein employed a phenotypic drug discovery approach coupled to mechanism-of-action profiling and target deconvolution to identify phenolic components of extra virgin olive oil (EVOO) capable of suppressing the functional traits of CSC in breast cancer (BC). In vitro screening revealed that the secoiridoid decarboxymethyl oleuropein aglycone (DOA) could selectively target subpopulations of epithelial-like, aldehyde dehydrogenase (ALDH)-positive and mesenchymal-like, CD44+CD24−/low CSC. DOA could potently block the formation of multicellular tumorspheres generated from single-founder stem-like cells in a panel of genetically diverse BC models. Pretreatment of BC populations with noncytotoxic doses of DOA dramatically reduced subsequent tumor-forming capacity in vivo. Mice orthotopically injected with CSC-enriched BC-cell populations pretreated with DOA remained tumor-free for several months. Phenotype microarray-based screening pointed to a synergistic interaction of DOA with the mTOR inhibitor rapamycin and the DNA methyltransferase (DNMT) inhibitor 5-azacytidine. In silico computational studies indicated that DOA binds and inhibits the ATP-binding kinase domain site of mTOR and the S-adenosyl-l-methionine (SAM) cofactor-binding pocket of DNMTs. FRET-based Z-LYTE™ and AlphaScreen-based in vitro assays confirmed the ability of DOA to function as an ATP-competitive mTOR inhibitor and to block the SAM-dependent methylation activity of DNMTs. Our systematic in vitro, in vivo and in silico approaches establish the phenol-conjugated oleoside DOA as a dual mTOR/DNMT inhibitor naturally occurring in EVOO that functionally suppresses CSC-like states responsible for maintaining tumor-initiating cell properties within BC populations.
EVOO, a central part of the health-promoting Mediterranean diet, contains a biophenol that functionally depletes tumor-initiating CSC-like states within heterogeneous cancer cell populations by operating as a dual inhibitor of mTOR—a major sensor of cellular metabolites—and DNMTs—a major category of epigenetic writers.
Introduction
Cancer relapse and metastatic dissemination can occur after the primary tumor has been eradicated by surgical removal, chemotherapy, radiation or targeted therapy. Such life-threatening phenomena can be largely attributed to the incomplete elimination of so-called cancer stem cells (CSC), a particularly aggressive type of malignant cell defined in terms of functional traits of self-renewal, differentiation, therapy resistance and tumor/metastasis-initiating capacity (1–5). Accordingly, the relative abundance of CSC populations correlates with unfavorable outcomes and is an independent risk factor for tumor recurrence and post therapy progression.
The CSC model has created new opportunities for cancer therapy. In the last decade, more than 150 therapeutic approaches have been envisioned to deplete the CSC pool via targeting of CSC surface antigens, CSC-associated oncoproteins, stemness regulation pathways or inhibiting CSC-related drug resistance pathways (6,7). Unfortunately, progress in the medical development of CSC-direct approaches has been disappointing, and no bona fide anti-CSC drugs have entered the clinical use. One reason for such failure might relate to the widely accepted belief that genetically predefined populations of treatment-refractory CSC should be viewed as the sole source of minimal residual disease, tumor recurrence and metastasis. While the actual contribution of phenomena such as epithelial-to-mesenchymal transition and dedifferentiation/reprogramming plasticity to the de novo generation of CSC during carcinogenesis remains a matter of debate (8–13), it is well accepted that conventional therapies would enrich cancer tissues with stem cell-like cancer cell populations that remain largely refractory to existing therapeutics. Accordingly, the sole credible target that could be exploited to prevent the manifestation of CSC would be the biological machinery in charge of the epigenetic proclivity of cancer cell populations to generate, maintain and perpetuate the so-called CSC-like states.
Plant-derived polyphenols whose consumption has been epidemiologically, clinically and experimentally implicated in the dietary protection against aging-related chronic diseases, including cancer, are potentially useful leads to develop new families of anti-CSC drugs (14–18). For instance, curcumin, the main polyphenol in turmeric, has been shown to target functional properties of chemotherapy-resistant colon and breast CSC subpopulations (19). Sulforaphanes, a family of isothiocyanates enriched in cruciferous vegetables such as broccoli, cauliflower, kale and cabbage, can inhibit the self-renewal and tumor-initiating capacity of CSC (20). Likewise, resveratrol, a natural stilbene from a wide variety of plant species including grapes, mulberries and peanuts, can inhibit CSC traits (21). Another example is the polyphenol genistein, the predominant isoflavone in soybean-enriched foods, which has been found to reduce the tumor-initiating capacity of CSC (22). Epigallocatechin gallate, the most abundant catechin in tea, has been also found to reduce CSC-related attributes in various cancers (23).
The ability of the so-called Mediterranean diet to significantly decrease the risk of several chronic diseases, including breast cancer (BC), has been largely attributed to the unique characteristics of extra virgin olive oil (EVOO), the juice from the fruits of olive trees obtained solely by mechanical means and consumed without further refinement (24–26). In addition to a favorable fat composition due to its high content (60–80%) of the monounsaturated fatty acid, oleic acid, the other fundamental health-related characteristic of EVOO is the presence of a large number of phenolic-like compounds (27–29). Of note, unlike phenolic acids, phenolic alcohols, tocopherols, flavonoids and lignans, which can also be found in many fruits and vegetables belonging to different botanical families, the group of complex phenol-conjugated compounds named oleosidic secoiridoids or oleosides is present only in plants of the Oleacea family including the European olive tree (Olea europaea L.). However, despite the considerable effort expended on identifying new oleosides in O.europaea L. and determining their chemistry during processing and storage of EVOO (30), no studies have explored whether such secoiridoids contained in the phenolic fraction of cold-pressed EVOO juice have anti-CSC properties.
We now report a phenotypic drug discovery approach coupled to mechanism-of-action profiling and computational-experimental target deconvolution aimed at identifying and characterizing phenol-conjugated oleosidic secoiridoids as a new family of EVOO bioactive phytochemicals capable of specifically and potently suppressing the functional traits of CSC.
Materials and methods
Cell lines
HMLER cells expressing a control shRNA (shCntrl) or an shRNA targeting E-cadherin (shEcad) were generated as described (10,31) and maintained in a 1:1 mixture of DMEM + 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), insulin, hydrocortisone and Clonetics™ MEGM™ (Mammary Epithelial Cell Growth Medium). MCF10DCIS.com and SUM-159 cells were purchased from Asterand (Detroit, MI), authenticated using short tandem repeat DNA fingerprint and passaged in our laboratory by starting a low-passage cell stock every month up to 6 months after resuscitation. DCIS.com cells were cultured in DMEM/F12 with l-glutamine supplemented with 5% horse serum and penicillin/streptomycin. SUM-159 cells were cultured in Ham’s F12 with 5% (vol/vol) FBS, 5 μg/mL insulin and 1 μg/mL hydrocortisone. HER2-overexpressing MCF-7/HER2 (clone 18) cells and their matched isogenic control (empty vector transfected) MCF-7/neo cells were kindly provided by Mien-Chie Hung (The University of Texas MD Anderson Cancer Center, Houston, TX) and routinely grown in improved minimum essential medium (IMEM) containing 5% (vol/vol) FBS and 2 mmol/L l-glutamine. T47D and ZR-75-1 were purchased from American Type Culture Collection (Manassas, VA), authenticated according to the ATCC guidelines before used in this study and grown in IMEM containing 5% (vol/vol) FBS and 2 mmol/L l-glutamine. Cell lines were regularly screened for Mycoplasma contamination using a MycoAlert Mycoplasma Detection Kit (Lonza, Verviers, Belgium).
Metabolic status assessment
Cell viability was determined using standard colorimetric MTT-based reduction assays.
Mammosphere-forming efficiency
Mammosphere formation assays were conducted as described previously (32). Mammosphere-forming efficiency (MSFE) of cells growing in suspension cultures was calculated as the number of mammospheres-like structures formed within 7 days divided by the original number of cells seeded and expressed as percentages. Refeeding of mammospheres cultures with compounds and/or sphere medium was performed on days 3 and 5.
Tumor xenograft studies
Approximately 2 × 106 SUM-159 cells were subcutaneously injected into the dorsal flanks of female athymic nude mice (4- to 5-week-old mice, 23–25 g; Harlan Laboratories). The cells were incubated with or without DOA or vehicle for 3 days with daily DOA replenishment before injection. In both pretreatment regimens, the body weight and diet consumption were determined weekly after dosing; tumor size was measured daily with electronic calipers, and tumor volume was calculated using the following formula: volume (mm3) = length × width2 × 0.5. The experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the Institut d’Investigació Biomèdica de Bellvitge (IDIBELL; Animal Use Protocol #6302 authorized by the Animal Experimental Commission of the Catalan Government, Barcelona, Spain). Mice were euthanized by cervical dislocation.
Tumor orthotopic studies
SUM-159 cells cultured in nonadherent and nondifferentiating conditions for 48 h were collected by centrifugation, and cells that were visibly anoikis-resistant with intact plasma membranes (assessed by trypan blue exclusion) were treated with or without graded concentrations of DOA for 2 h before being orthotopically implanted (5 × 103 cells) into the second right mammary fat pad of female SCID/Beige mice. The mice were palpated twice per week, and the tumor volumes were calculated as described above.
ALDEFLUOR® activity assay
The ALDEFLUOR® assay was performed as per the manufacturer’s instructions (StemCell Technologies, Vancouver, BC, Canada), with or without the addition of EVOO-PE or purified OA/DOA.
Docking calculations
All docking calculations were performed using Itzamna and Kin (www.mindthebyte.com), classical docking and blind-docking software tools. Protein structures from RSCB PDB were directly employed for docking calculations using the cavities defined by available crystallographic ligands where available. Two runs were carried out for each calculation to avoid false positives.
Molecular dynamics calculations
Short (1 ns) molecular dynamics (MD) simulations were performed using NAMD version 2.10 over the best-docked complexes, which were selected based on the interaction energy. The Ambers99SB-ILDN and the GAFF forcefield set of parameters were employed for receptors and ligands, respectively. The ligand GAFF parameters were obtained using Acpype software, whereas the receptor structures were modeled using the leap module of Amber Tools. Simulations were carried out in explicit solvent using the TIP3P water model with the imposition of periodic boundary conditions via a cubic box. Electrostatic interactions were calculated by the particle-mesh Ewald method using constant pressure and temperature conditions. Each complex was solvated with a minimum distance of 10 Å from the surface of the complex to the edge of the simulation box. Na+ or Cl− ions were also added to the simulation to neutralize the overall charge of the systems. The temperature was maintained at 300 K using a Langevin thermostat, and the pressure was maintained at 1 atm using a Langevin Piston barostat. The time step employed was 2 fs. Bond lengths to hydrogens were constrained with the SHAKE algorithm. Before production runs, the structure was energy minimized followed by a slow heating-up phase using harmonic position restraints on the heavy atoms of the protein. Subsequently, the system was energy minimized until volume equilibration, followed by the production run without any position restraints.
Binding free energy analysis
Molecular mechanics/generalized Borne surface area (MM/GBSA) calculations were performed to calculate the binding free energy (∆Gbind) of DOAgly and related inhibitors against selected molecular targets. MM/GBSA scoring was performed using the MMPBSA.py algorithm within AmberTools. The snapshots generated in the 1 ns MD simulation were imputed into the post-simulation MM/GBSA calculations of binding free energy. Graphical representations were prepared using Visual Molecular Dynamics version 1.9.1, LigPlot+ and PLIP version 1.3.0.
mTOR activity/inhibition assays
To characterize the mTOR kinase potency of DOA, PP242 and Torin 2, IC50 determinations for FRAP1 (mTOR) were outsourced to Invitrogen (Life Technologies) using the FRET-based Z-LYTE™ SelectScreen Kinase Profiling Service. To evaluate the phosphorylation status of P70S6K1 at T389, SUM-159 cells grown to 80–90% confluence were exposed to 20 μmol/L DOAgly for 0, 30, 180 and 360 min; alternatively, SUM-159 cells grown to 80–90% confluence were exposed to graded concentrations of DOA (5, 10 and 20 μmol/L) for 1 h. Cells were then collected and subjected to western blot analysis with antibodies against phospho-p70S6K1 (Thr389) and total p70S6K1 (Cell Signaling Technology, Danvers, MA). F062-actin (Sigma–Aldrich) was used as loading control.
DNMT activity/inhibition assays
DNMT activity was evaluated in nuclear extracts using the DNMT activity/inhibition assay (Active Motif, Carlsbad, CA). Briefly, 10 μg of nuclear extracts, obtained with the Nuclear Extract Kit (Active Motif, Carlsbad, CA), were incubated with 20 μmol/L DOA for 2 h. Optical density (OD) was measured on a microplate reader at 450 nm, and DNMT activity (OD/h/mg) was calculated according to the following formula: DNMT activity = 1000 × (average sample OD −average blank OD)/[protein amount (μg) × incubation time (h)]. All samples were assayed in duplicate. As positive control, nuclear extracts were incubated with 5 μmol/L 5-azacytidine. The effect of DOA on the enzymatic activities of recombinant human DNMTs using in vitro enzymatic assays was outsourced to BPS Bioscience and carried out by following the protocol described in BPS Bioscience DNMT Universal Assay Kit (BPS#52035) using a DNMT substrate precoated plate.
Statistical analysis
All statistical analyses were performed using XLSTAT 2010 (Addinsoft™). For all experiments, at least two independent biological replicates were performed with n ≥ 3 technical replicates per experiment. No statistical method was used to predetermine sample size. Investigators were not blinded to data allocation. Experiments were not randomized. Data are presented as mean ± SD. Two-group comparisons were performed using Student’s t-test for paired and unpaired values. Comparisons of means of ≥3 groups were performed by ANOVA, and the existence of individual differences, in case of significant F values at ANOVA, was tested by Scheffé’s multiple contrasts. P values < 0.05 were considered to be statistically significant (denoted as *). All statistical tests were two sided.
Further details
See Supplementary Material, available at Carcinogenesis Online file for full description of details concerning isolation/purification/analytical characterization of EVOO-derived oleosides, synergism studies, mTOR/DNMT activity/inhibition assays, RNA isolation, reverse transcription and stem cell transcription factors assays.
Results
A crude phenolic extract derived from EVOO suppresses epithelial- and mesenchymal-like tumorsphere-initiating CSC states
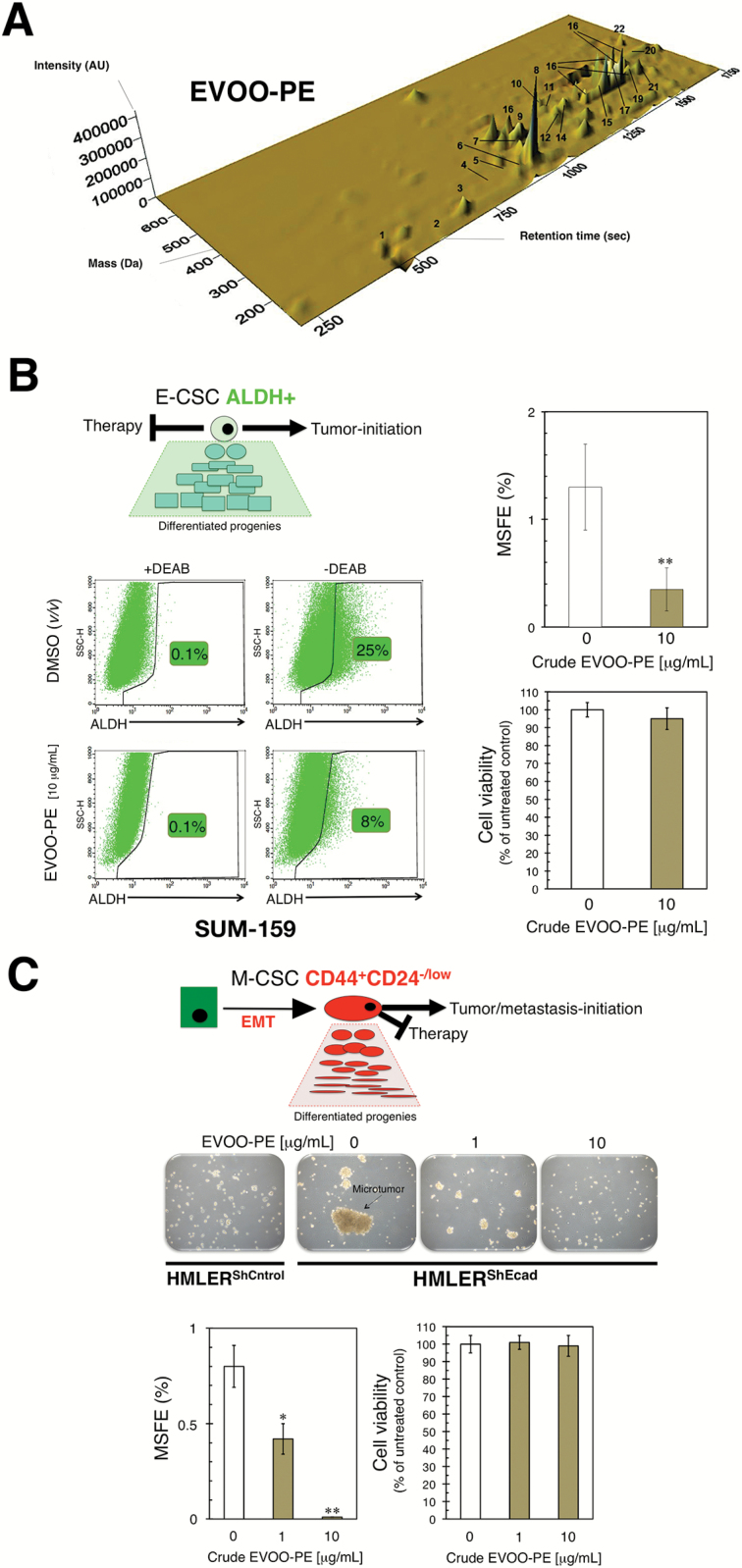
Breast CSC appear to exist in two different, but reversible and therefore interchangeable, epithelial (E)- and mesenchymal (M)-like states. The former is characterized by the expression of aldehyde dehydrogenase (ALDH+), and the latter is characterized by a CD44+CD24−/low immunophenotype (13,33). Based on our previous studies demonstrating that an EVOO-derived crude phenolic extract (EVOO-PE) containing at least twenty different phenolic compounds could regulate some stemness traits (29,34,35), we speculated that such EVOO-PE (Figure 1A) might target CSC subpopulations irrespective of their E- or M-like traits.
Figure 1.
A phenolic-enriched crude olive oil extract inhibits the mammosphere-initiating capacity of BC CSC-like states. (A) Three-dimensional map of phenolic compound separation obtained by HPLC-ESI-TOF in a crude EVOO-PE obtained via isolation protocols described in Supplementary material, Materials and Methods, available at Carcinogenesis Online. (B) Representative ALDEFLUOR® assay to identify SUM-159 cells with high ALDH activity (ALDH+) in the absence or presence of 10 μg/mL of the crude EVOO-PE for 3 days. The ALDH inhibitor diethylaminobenzaldehyde (DEAB) was used as negative control. Monolayer cultures were fed with the crude EVOO-PE every other day. Results are representative of two technical replicates per n; n = 3 biological replicates. MSFE is expressed as percentages means (columns) ± SD (bars); three technical replicates per n; n = 3 biological replicates. MTT uptake-based measurement of cell viability is expressed as percentages uptake (OD570) relative to untreated control cells (=100% cell viability). The results are expressed as percentages means (columns) ± SD (bars); three technical replicates per n; i = 3 biological replicates. (C) Figure shows representative light microscope representations (×20 magnification) of mammospheres formed by HMLERShControl and HMLERShEcad cells growing in sphere medium for 7 days in the absence or presence of graded concentrations of EVOO-PE. MSFE and MTT calculations were performed as described for SUM-159 cells in the left panels; three technical replicates per n; n = 3 biological replicates. (*P < 0.01 and **P < 0.001, statistically significant differences from the untreated (control) group).
The highly aggressive basal/mesenchymal SUM-159 BC cell line is commonly employed to test CSC activity of oncology drugs because it naturally harbors a significant proportion of ALDH+ E-CSC-like cells (36). We found that in the presence of a bioactive but noncytotoxic concentration of crude EVOO-PE, the proportion of ALDH+ cells in the SUM-159 cell population (approx. 25% in nontreated cells) was selectively and significantly reduced to levels as low as 8% (Figure 1B). The ALDH+ population of SUM-159 cells is known to be solely responsible for initiating and maintaining tumorspheres when grown under nonadherent/nondifferentiating conditions (37). Thus, we tested the relationship between the ability of crude EVOO-PE to reduce the number of ALDH+ cells and the intrinsic capacity of SUM-159 cells to form mammospheres (scored as MSFE) in suspension culture. As expected, the MSFE of SUM-159 cells was significantly reduced by more than 80% by treatment with EVOO-PE (Figure 1B).
We then exploited the landmark observation by Robert Weinberg that BC epithelial cell populations experimentally induced into epithelial–mesenchymal transition dramatically and stably increase the proportion of M-like CD44+CD24−/low CSCs (10), which constitutes a valuable screening method to identify agents capable of targeting M-like CSC (31). The MSFE of CD44+CD24−/low-enriched HMLERshECad cells was considerably greater than that of HMLERshCntrl cells, which mostly failed to form bona fide mammospheres (Figure 1C). This acquired ability of HMLERshECad to form CD44+CD24−/low-initiated mammospheres was completely prevented by EVOO-PE (Figure 1C).
The ability of the various phenolic compounds contained in the crude EVOO-PE to suppress BC cell survival and proliferation as mammospheres was not due to nonspecific cytotoxic effects because cell viability of monolayer cultures of SUM-159 and HMLERshECad cells remained as high as 95% in its presence (Figure 1B and C).
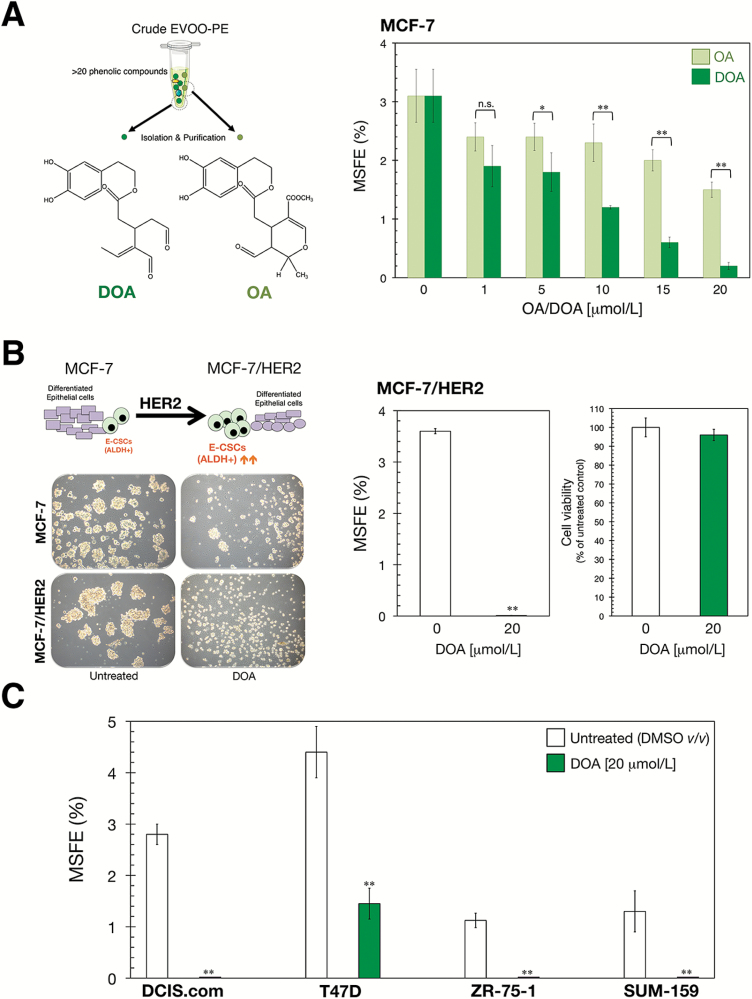
A purified form of the complex polyphenol DOA strongly suppresses mammosphere formation in a panel of established BC cell lines
We have previously performed a qualitative and quantitative characterization of the EVOO-PE employed in this study and identified that the secoiridoids derivatives of hydroxytyrosol linked to elenolic acid and its decarboxymethylated form, namely oleuropein aglycone (OA) and decarboxymethyl oleuropein aglycone (DOA), were the most abundant compounds as they constituted up to 23 and 46% of total phenolics, respectively (29,35). Based on these findings, we hypothesized that the purified forms of OA and DOA may recapitulate the anti-CSC activity of crude EVOO-PE. Procedures for the isolation and purification of OA and DOA from EVOO can be found in Supplementary Material, Figure S1, available at Carcinogenesis Online.
The luminal BC cell line MCF-7, which has a high MSFE level (32), was used to compare the ability of OA and DOA to inhibit tumorsphere formation. Micromolar concentrations of OA and DOA inhibited MCF-7 mammosphere formation in a dose-dependent manner, with the highest dose of OA and DOA (20 μmol/L) eliciting greater inhibitory effects than the lowest dose (1 μmol/L; Figure 2A). Moreover, whereas OA reduced MSFE by up to 50%, DOA dramatically suppressed such efficiency >90% at the highest concentration tested (Figure 2A). We therefore focused our attention on the ability of DOA to target CSC in a wider range of BC models. We first examined whether exposure to DOA was sufficient to target and suppress the well-recognized ability of the HER2 oncogene to augment the tumor-initiating capacity of BC cell populations via expansion of the E-like ALDH+ CSC population (13,33,37–39). The greater MSFE acquired upon HER2 overexpression was completely prevented when cultures of MCF-7/HER2 cells were supplemented with DOA (Figure 2B). We then explored the effects of DOA for mammosphere inhibition in four additional BC cell lines: DCIS.com (a basal-like model for ductal carcinoma in situ), T47D and ZR-75-1 (two luminal-like BC models) and SUM-159. In all cases, DOA significantly reduced MSFE relative to untreated controls (Figure 2C).
Figure 2.
Purified forms of phenolic oleosides inhibit the mammosphere-initiating capacity of BC CSC-like states. (A) MSFE is expressed as percentages means (columns) ± SD (bars); three technical replicated per n; n = 2 biological replicates. (*P < 0.01 and **P < 0.001, statistically significant differences between groups; n.s. not statistically significant). (B) Figure shows representative light microscope images (×20 magnification) of mammospheres formed by MCF-7 and MCF-7/HER2 cells growing in sphere medium for 7 days in the absence or presence of 20 μmol/L DOA. MSFE and MTT calculations of suspension/monolayer cultures of MCF-7/HER2 cells growing in the absence or presence of 20 μmol/L DOA were performed as described for MCF-7 cells in A; three technical replicates per n; n = 2 biological replicates. (C) MSFE calculations were performed as described for MCF-7 cells in A; three technical replicates per n; n = 3 biological replicates. (*P < 0.01 and **P < 0.001, statistically significant differences from the untreated (control) group; n.s. not statistically significant).
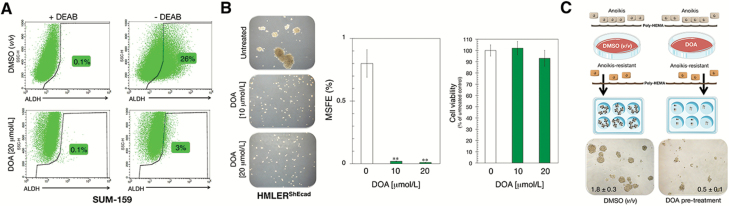
To corroborate the ability of DOA to target CSC subpopulations regardless of their E- or M-like trait, we first confirmed that the ALDH+ cell content of E-like CSC in SUM-159 cell populations was reduced by approximately 97% by DOA (Figure 3A). The ability of CD44+CD24−/low (M-CSC)-enriched HMLERshECad cells to form mammospheres was also completely prevented by DOA (Figure 3B). To substantiate the bona fide anti-CSC effects of DOA, we induced anoikis in SUM-159 cells by plating them in culture dishes coated with poly-2-hydroxyethyl methacrylate in the absence or presence of DOA for 48 h prior to replating a viable fraction of the cell suspension in serum- and DOA-free mammosphere medium (Figure 3A). The ability of DOA-treated anoikis-resistant cultures to form tight spheroids was reduced >70% after removal of DOA, and these anoikis-resistant cells were characterized by a high prevalence of irregular cell aggregates with lower sphericity than that found in untreated cultures (Figure 3C). All the MSFE-reducing effects were not due to nonspecific, cytotoxic effects of DOA, as it had no impact on cell viability in any tested BC cell line growing in adherent culture (Figures 2B and 3B, and data not shown).
Figure 3.
DOA specifically and potently decreases the mammosphere-initiating capacity of BC CSC-like states. (A) Representative ALDEFLUOR® assay to identify SUM-159 cells with high ALDH activity (ALDH+) in the absence or presence of 20 μmol/L DOA for 3 days. The ALDH inhibitor DEAB was used as negative control. Monolayer cultures were fed with the DOA every other day. Results are representative of two technical replicates per n; n = 3 biological replicates. (B) Figure shows representative light microscope representations (×20 magnifications) of mammospheres formed by HMLERShEcad cells growing in sphere medium for 7 days in the absence or presence of graded concentrations of DOA. MSFE and MTT calculations were performed as described in Figure 2; three technical replicates per n; n = 3 biological replicates. (*P < 0.01 and **P < 0.001, statistically significant differences from the untreated (control) group; n.s. not statistically significant). (C) Anoikis-resistant cells obtained as described in Supplementary material, Materials and Methods, available at Carcinogenesis Online were cultured in DOA-free mammosphere medium for 7 days, and MSFE were calculated following the same procedure as that described in Figures 1 and 2; three technical replicates per n; n = 2 biological replicates.
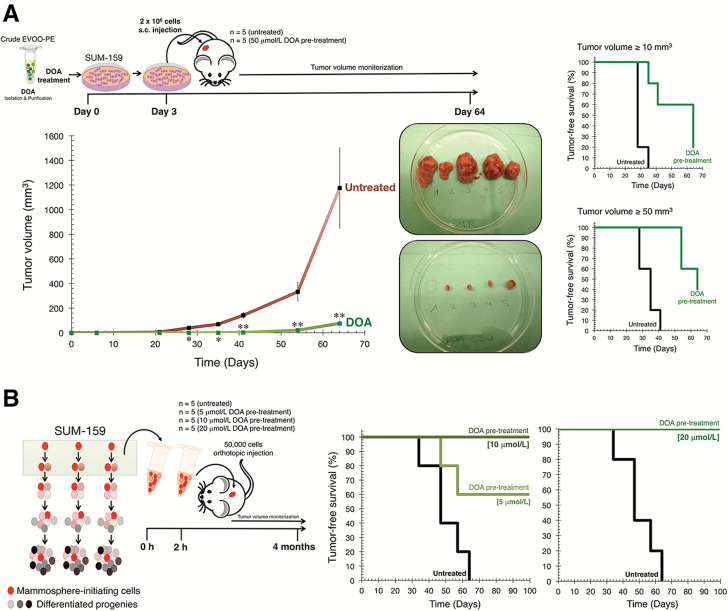
DOA reduces the in vivo tumorigenicity of BC cell populations
Because CSC are believed to be responsible for tumor initiation, we assessed the impact of DOA for the tumor-seeding ability of heterogeneous BC populations in vivo. To do this, we pretreated monolayer cultures of SUM-159 cells with either vehicle or 20 μmol/L DOA for 3 days with daily refeeding of the compound (Figure 4A). Subsequently, cells that were fully viable as determined by trypan-blue exclusion were subcutaneously injected into the left rear flanks of female nude mice at a saturating concentration (2 × 106), aimed to ensure the development of tumor masses within a few weeks. Examination of palpable tumor lesions revealed that all (5/5) mice injected with vehicle-treated SUM-159 cells formed tumors 6 weeks after injection (Figure 4A). By contrast, in mice injected with SUM-159 cells pretreated with DOA, 20% (1/5) failed to develop tumors and 80% of injected animals (4/5) developed less macroscopically visible tumors. Indeed, the lesions in mice injected with DOA-pretreated cells were 15 times smaller in size than those observed in control animals (76 versus 1176 mm3; Figure 4A). Accordingly, the time required for 50% of animals to develop palpable tumors of either ≥50 or ≥10 mm3 was lengthened by 78% (from 36 days with vehicle-treated cells to 64 days with DOA-pretreated cells) and 119% (from 28 days with vehicle-treated cells to 64 days with DOA-pretreated cells), respectively.
Figure 4.
DOA blocks tumor-initiating capacity of CSC-like cells in vivo. (A) Tumor growth rate was calculated by measuring volumes along several weeks after injection of DOA-pretreated of untreated control cells. Shown are the mean volumes (±SD) for at least 9 weeks. Tumor-free survival in mice bearing SUM-159 xenografts with volumes ≥10 and ≥50 mm3 is shown as a function of time. [*P < 0.01 and **P < 0.001, statistically significant differences from the untreated (control) group; n.s. not statistically significant]. (B) Mammosphere-initiating cells were isolated and exposed to graded concentrations of DOA for 2 h. Viable single cell suspensions (1 × 104 cells) were orthotopically injected into the mammary fat pads of SCID/Beige mice, and tumor growth was monitored for at least 4 months. Tumor-free survival in mice bearing orthotopically implanted SUM-159 CSC-like cells is shown as a function of time.
DOA suppresses the growth of CSC-enriched BC-cell populations in orthotopic implantation model
We next evaluated the ability of DOA to inhibit tumor formation using the orthotopic implantation of low numbers of SUM-159 cells precultured under nonadherent and nondifferentiating conditions for 48 h, an experimental environment selecting for mammosphere-initiating CSC-like cells (Figure 4B). Single cell suspensions (5 × 103) isolated from these cultures were exposed or not to graded concentrations of DOA for 2 h before injection into the mammary fat pads of SCID/Beige mice. Examination of tumor formation revealed that all of the mice (5/5) injected with mammosphere-initiating SUM-159 cells formed tumors 9 weeks after orthotopic injection (Figure 4B). By contrast, more than half of the mice (3/5) injected with mammosphere-initiating SUM-159 cells pretreated with the lowest dose of DOA (5 μmol/L) failed to develop visible tumors at the same time point. Accordingly, the time required for 50% of animals to develop palpable lesions ≥50 mm3 was lengthened by 155%, that is, from 47 days with vehicle-treated cells to 120 days with DOA-treated cells (Figure 4B). Strikingly, all the mice (5/5) that were orthotopically injected with CSC-like SUM-159 cells pretreated with DOA at 10 or 20 μmol/L remained free from tumors more than 2 months after the control arm was ended (Figure 4B).
DOA regulates the expression of genes involved in stem cell fate
We designed secondary assays to gain some insights into the molecular mechanisms through which DOA might functionally deplete CSC-like states without promoting nonspecific cytotoxic effects on heterogeneous BC cell populations. First, we carried out a Stem Cell Transcription Factors Assay (TaqMan® OpenArray® Human Stem Cell Panel) to assess the impact of DOA on the expression of 609 stemness-related genes. A total of 160 genes were found significantly altered in their baseline expression status (arbitrary fold-change cutoff of >2) when monolayer cultures of SUM-159 cells were cultured in the presence of 20 μmol/L DOA for 3 days with daily refeeding of the compound (Supplementary Material, Table S1 and Figure S2, available at Carcinogenesis Online). Supporting the ALDEFLUOR inhibitory effect of DOA, ALDH1A1—the key ALDH isoenzyme-linked CSC function (40)—was among the genes significantly downregulated (3-fold) by DOA. The expression of the CSC marker OCT4 (8,9,41), a homeodomain transcription factor of the POU family, was notably downregulated (4-fold) in the presence of DOA. DOA exposure drastically augmented the expression of WRN (13-fold), which has been shown to promote stem cell differentiation via epigenetic inactivation of OCT4 (42). DOA treatment similarly augmented the expression of several suppressors of the undifferentiated metastatic phenotype (43,44) including SPRED1 (4-fold), SPRED2 (9-fold) and SMURF2 (7-fold).
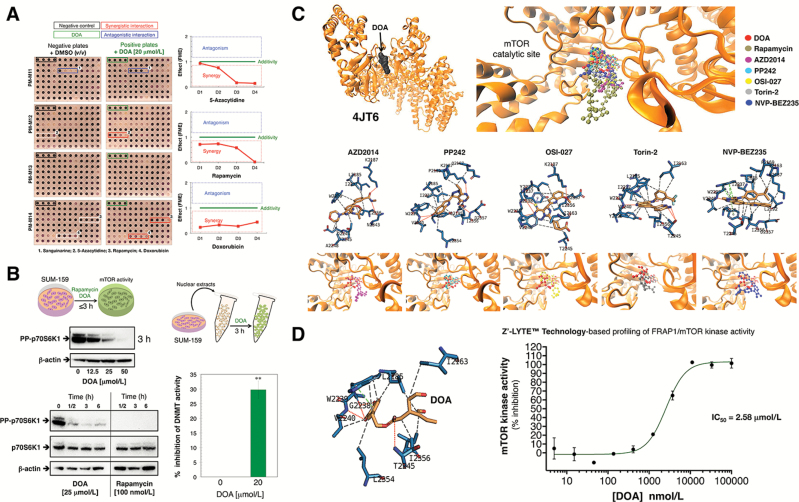
DOA synergistically interacts with the mTOR inhibitor rapamycin and the DNMT inhibitor 5-azacytidine
To further characterize the molecular targets through which DOA might exert its antistemness properties in cancer cell populations, we utilized commercially available Biolog Phenotype MicroArrays (PM) for mammalian cells. This platform is commonly employed to analyze components of pathways that might be involved in resistance phenotypes, but its major role is for drug development by inference of the mechanism of action of a given compound (45–47). We simultaneously profiled SUM-159 cells in four microplates (termed PM-M11, PM-M12, PM-M13 and PM-M14) whose wells had been coated with antibiotics and other growth inhibitors to create 92 unique culture conditions. The PM assay was conducted over 2 days in the absence (negative plates) or presence (positive plates) of 20 μmol/L DOA, an optimal concentration that fully impeded mammosphere formation but without cytotoxic effects against SUM-159 cells grown under adherent conditions (Figure 5A). DOA treatment appeared to strongly protect SUM-159 cells from the cytotoxic effects of the multifaceted alkaloid sanguinarine (48,49). Conversely, DOA was found to synergistically interact, in a dose-dependent manner, with the allosteric mTOR inhibitor rapamycin and the DNA methyltransferase (DNMT) inhibitor 5-azacytidine and in a dose-independent manner with the chemotherapeutic agent doxorubicin (Figure 5A). We therefore chose to further investigate the metabolo-epigenetic nature of DOA in terms of its capacity to target and inhibit mTOR and DNMT.
Figure 5.
(A) DOA synergistically interacts with mTOR and DNMT inhibitors. Shown are representative microphotographs of PM arrays (microplates PM-M11–PM-M14) from two biological replicates. The different interactions were defined as described in Supplementary Material, available at Carcinogenesis Online. (B) DOA decreases mTOR and DNMT activities in cultured cells. Left. Representative western blotting analyses of phospho-p70 S6 Kinase (Thr389)/phospho-p85 S6 Kinase (Thr412) in SUM-159 cells cultured in the absence of presence of DOA or rapamycin for up to 6 h, as specified. Right. Nuclear extracts of SUM-159 were exposed to 20 μmol/L DOA for 2 h before assessing DNMT activity using the DNMT activity/Inhibition Assay Kit of Active Motif as per manufacturer’s instructions. The results are expressed as percentages of the means (columns) ± SD (bars); three technical replicates per n; n = 2 biological replicates. (**P < 0.001 versus DNMT activity in untreated nuclear extracts). (C) DOA binds the ATP-binding pocket in mTOR kinase domain and inhibits mTOR kinase activity. Overall structures and views of the interaction between DOA (red) and well-known TORKinhibs with the ATP-dependent catalytic pocket of mTOR (PDB code 4JT6). Figure shows in sticks all the interaction residues involved in the binding of DOA/TORKinhibs using PLIP. Hydrogen bond interactions are represented by orange dashed lines; salt bridges are represented by yellow dashed lines and charge centers by yellow spheres; Cation-π interactions are represented by blue dashed lines and white spheres for the center of the aromatic ring. (D) DOA directly inhibits the ATP-dependent catalytic activity of mTOR. A dose-response curve of ATP-dependent mTOR kinase activity was created by plotting FRET signal of the Z′-LYTE Kinase assay as the function of DOA concentration.
We first tested the ability of DOA to alter the phosphorylation status of the mTOR pathway kinase p70S6K1, a commonly used readout of mTOR signaling (50). The phosphorylation of p70S6K1 on Thr389 was suppressed by graded concentrations of DOA in a time- and dose-dependent manner (Figure 5B, left panels). Moreover, DOA decreased the cellular activity of the mTOR kinase as early as 30 min after treatment, although to a lesser extent than that achieved by rapamycin. When total DNMT activity in nuclear extracts of SUM-159 cells pretreated with DOA for 2 h was assessed using an ELISA-based DNMT activity/inhibition assay, a ~30% reduction in DNMT activity was found in the presence of DOA (Figure 5B, right panels).
DOA is an ATP-competitive mTOR inhibitor
Biophenols structurally related to DOA such as quercetin have been suggested to inhibit the enzymatic activity of mTOR by bonding to its ATP-binding catalytic pocket (51). Given this information, we performed computational studies using several cocrystal structures of a complex of mTOR with an ATP transition state mimic and with several ATP-site inhibitors: 4JT6 (in complex with PI-103), 4JSX (in complex with Torin 2), 4JT5 (in complex with PP242) and 4JSV/4JSP (in complex with ATPgamma-Mg2+). Results of in silico binding experiments using rigid-docking calculation, which were run twice to avoid false positives, showed that DOA bound all the above crystal structures with energies ranging from −6.0 kcal/mol for 4JSV to −7.1 kcal/mol for 4JT6, which was selected for further analysis (Supplementary Material, Table S2, available at Carcinogenesis Online). To add protein flexibility to the analysis and to test the stability of the selected DOA-mTOR complex, we carried out short MD simulations of 1 ns and applied MM/GBSA calculations to estimate a −26.8226 kcal/mol free energy of the binding of DOA to 4JT6 (Supplementary Material, Table S2, available at Carcinogenesis Online).
To characterize the mechanism of action of DOA against mTOR, we selected five different second-generation ATP-competitive mTOR inhibitors (TORKinhibs): AZD2014 (vistusertib), PP242 (torkinib), OSI-027, Torin 2 and NBP-BEZ235 (dactolisib) to perform rigid docking, MD simulations and MM/GBSA calculations (Figure 5C; Supplementary Material, Table S3, available at Carcinogenesis Online). The binding mode of DOA shared key amino acid residues of the binding modes of TORKinhibs and apparently mimicked the binding behavior of PP242 and Torin 2 to the ATP-binding catalytic pocket of mTOR, although the presence of more aromatic rings in the DOA molecule resulted in a slightly different binding strength from that of PP242 and Torin 2. Specifically, rigid-docking calculations revealed that π–π stacking appears to occur between the aromatic ring of DOA and the Trp2239 residue (or Tyr2225 upon conformational changes of either DOA or the pocket itself) in the catalytic site of mTOR. A stronger stabilization of the DOA binding appeared to involve Gly2238 and Val2240, which not only might stabilize the Trp2239-DOA stacking but also establish direct electrostatic interactions with DOA (Supplementary Material, Table S4, available at Carcinogenesis Online). When we added the solvation effect and the dynamic nature of the interaction in the DOA–mTOR complex, the generated trajectories by MD simulations confirmed the main occurrence of the π-π stacking with Trp2239 (and a more fluctuating interaction with Tyr2225), as well as a significant number of additional residues performing key electrostatic interactions depending on the conformation adopted by the DOA–mTOR complex (Supplementary Material, Table S4, available at Carcinogenesis Online).
We then used the FRET-based Z-LYTE™ Kinase Assay to test the ability of DOA to inhibit mTOR activity. Ten concentrations of DOA spanning over five logarithmic decades were selected. To validate the bioassay procedure, we also measured the inhibitory profile of the TORKinhibs PP242 and Torin 2. Figure 5D shows the mTOR activity rate as a function of DOA concentration. DOA inhibited mTOR activity with an IC50 of 2.6 μmol/L, whereas the measured IC50 values for PP242 and Torin 2 were 18.3 and 1.3 nmol/L, respectively (Supplementary Material, Figure S3, available at Carcinogenesis Online).
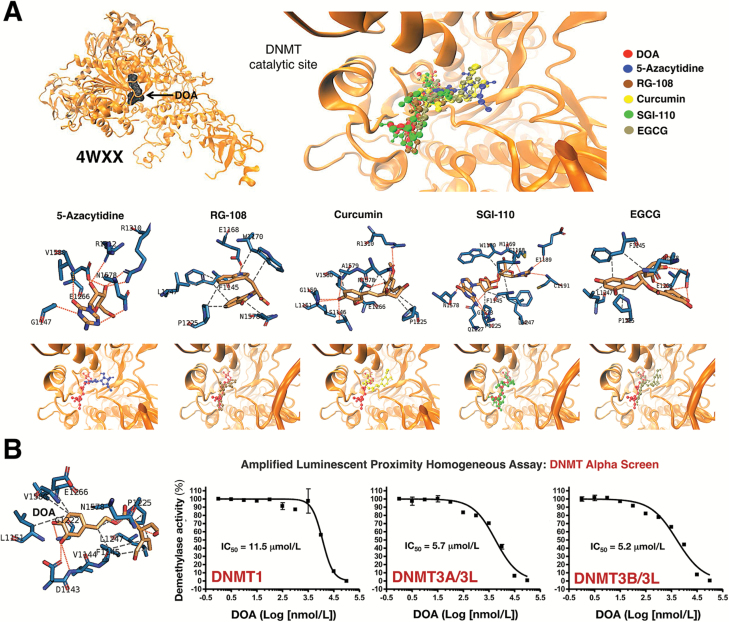
DOA is an SAM competitive inhibitor of DNMTs
We next performed computational studies using the crystal structure of human DNMT (351–1600) 4WXX (Figure 6A). In silico binding experiments employing rigid-docking calculations confirmed the ability of DOA to bind the DNMT/4WXX crystal structure with an energy of −7.8 kcal/mol. Short MD simulations and MM/GBSA estimated a −30.567 kcal/mol free energy of the binding of DOA to DNMT/4WXX (Supplementary Material, Table S2, available at Carcinogenesis Online). To characterize the mechanism of action of DOA against DNMT, we selected five DNMT inhibitors: 5-azacytidine, RG108, curcumin, SGI-110 and epigallocatechin gallate (Figure 6). Similarly to what happened with the binding mode of DOA to mTOR, the binding mode of DOA appeared to share that of all the chosen DNMT inhibitors with the presence of one or two more aromatic rings being responsible for the differences in the free energy binding, which ranged from −20.4119 kcal/mol for RG108 to −40.1785 kcal/mol for epigallocatechin gallate (Supplementary Material, Table S3, available at Carcinogenesis Online). The DOA pattern of spatial orientation more closely resembles that of 5-azacytidine, SGI-110 and curcumin. Rigid-docking calculations and MD simulations suggested that the main residues involved in the stabilization of the DOA-DNMT complex were Ser1146, Pro1125, Asp1143, Phe1145, Gly1150, Leu1151, Asn1158, Val1580 and Gly1223, along with a significant number of additional residues performing key electrostatic interactions depending on the conformation adopted by the DOA–DNMT complex (Supplementary Material, Table S4, available at Carcinogenesis Online).
Figure 6.
DOA binds the DNA-binding pocket in DNMT methylation domain and inhibits SAM-dependent DNA methylation activity. (A) Overall structures and views of the interaction between DOA (red) and well-known DNMT inhibitors with the DNA-binding catalytic pocket of DNMT1 (PDB code 4WXX). Figure shows in sticks all the interaction residues involved in the binding of DOA/DNMT inhibitors as described in Figure 5. (B) Dose-response curves of SAM-dependent methylation activities of DNMT1, DNMT3A/3L and DNMT3B/3L were created by plotting AlphaScreen signals as the function of DOA concentration.
We then used an amplified luminescent proximity homogeneous assay (AlphaScreen) to in vitro test the ability of DOA to inhibit DNMT activity. As before, 10 concentrations of DOA over 4 logarithmic decades were selected and tested in the presence of 20 μmol/L S-adenosyl-l-methionine (SAM), a catalytic substrate of DNMTs. To validate the bioassay procedure, we also measured the inhibitory profile of the natural inhibitor of DNMT activity, S-adenosyl-l-homocysteine (SAH). Figure 6B shows the activity rates of DNMT1, DNMT3A/3L and DNMT3B/3L as a function of DOA concentration. DOA inhibited the methyltransferase activity of DNMT1, 3A/3L and 3B/3L with an IC50 of 11.5, 5.7 and 5.2 μmol/L, respectively (Figure 6, bottom panels), whereas the measured IC50 values for SAH were 9.9, 5.8 and 2.8 μmol/L, respectively (Supplementary Material, Figure S4, available at Carcinogenesis Online).
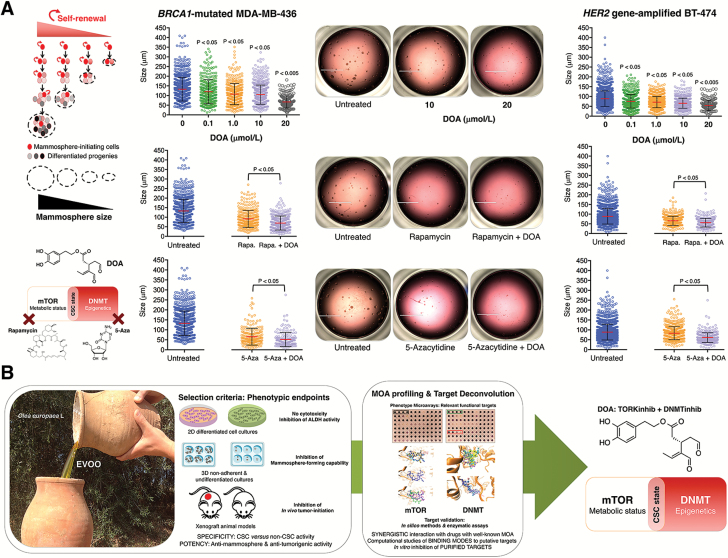
DOA synergistically inhibits self-renewal capacity of CSC-like cells in combination with rapamycin and 5-azacytidine
Given that mammosphere size reflects the self-renewal capacity of each CSC-like cell, we used the recently developed Cell2Sphere™ Kit to evaluate the impact of DOA for self-renewal in combination with either rapamycin or 5-azacytidine. We used two in vitro models for this analysis: the MDA-MB-436 cell line is an M-like BC model that harbors the deleterious mutation 5396 + 1G>A in the splice donor site of exon 20 of the BRCA1 gene and the BT-474 cell line is a luminal-like BC model with an amplification of the HER2 oncogene. The size of MDA-MB-436 and BT-474 mammospheres decreased significantly in the presence of graded concentrations of DOA (Figure 7A). Moreover, the concurrent supplementation with a suboptimal concentration of DOA significantly enhanced the ability of rapamycin and 5-azacytidine to limit the size of MDA-MB-436 and BT-474 mammospheres, indicating a reduction in self-renewal capacity. Under these conditions, mammospheres, when found, were considerably smaller than those generated from untreated control cultures, and there were no mammospheres >50 μm in size (Figure 7A).
Figure 7.
(A) DOA augments the capacity of mTOR inhibitor rapamycin and DNMT inhibitor 5-azacytidine to inhibit BC mammosphere formation activity. Cell2Sphere™ assays using BRCA1-deficient MDA-MB-436 (left) and HER2-overexpressing BT-474 cell (right) were performed as per the manufacturer’s instructions. Drugs were added to sextuplicate sets of wells on days 1 and 4 without replenishing the medium. ImageJ was used to quantify the size (central lines indicate mean values) of 6-day-old mammospheres. Size bar = 2000 μm. (B) EVOO-derived oleoside DOA is a metabolo-epigenetic suppressor of CSC cellular states: A phenotypic drug discovery approach. EVOO is the juice from fruits of olive trees (Olea europaea L.) obtained solely by mechanical means and consumed without further refining processes other than washing, filtration, decantation and centrifugation. Whereas well-known secoiridoids such as the glucoside oleuropein are characteristic and abundant constituents easily accessible from the drupes and other organs (leaves) of O. europaea L., DOA is an oleuropein-dialdehyde derivative confirmed only in EVOO in highly variable concentrations. We performed a holistic approach of phenotypic drug discovery coupled to mechanism-of-action (MOA) functional profiling and target deconvolution that identified DOA as an unforeseen EVOO bioactive phytochemical that operates as a dual TORKinhib/DNMTinhib molecule capable of specifically and potently suppressing the functional traits of CSC irrespective of the mutational landscape of BC populations.
Discussion
Advancement in cancer prevention and treatment may derive from the identification of molecules capable of eliminating tumor- and metastasis-initiating chemoresistant CSC. A potential approach to eliminate the polycausal and multidimensional genotype–phenotype interactions that drive CSC cellular states may involve the usage of multifaceted drugs capable of interfering with the biologic functioning of cancer stemness itself. Plant evolution has produced a rich source of refined, multitargeting phytochemicals to overcome environmental stresses, including protection against fungi, insects and predators. Not surprisingly, plant-derived phytochemicals targeting the multiple molecular machineries that support CSC cellular states are emerging as valuable tools for therapy. Here, we aimed to characterize the health benefits chemically encrypted in secoiridoids that are unique to oleaceous plants in terms of their bioactivity against CSC. Our systematic in vitro, in vivo and in silico approach (Figure 7B) reveals for the first time that EVOO contains a phenol-conjugated oleosidic secoiridoid (DOA) capable of functionally depleting the CSC-like states responsible for maintaining tumor-initiating properties within genetically diverse types of BC populations.
In contrast to the target-based strategies that have been widely employed in the pharmaceutical cancer industry in the past decades, including those aimed to discover new anti-CSC drugs, we chose to uncover novel EVOO-derived anti-CSC biophenols by employing a cell-based phenotypic drug discovery strategy coupled to mechanism-of-action profiling and target deconvolution (52,53). Without prior knowledge of the phenolic compound target(s), we first screened and selected a lead compound from the EVOO phenolic fraction based on quantifiable phenotypic endpoints such as in vitro ALDH activity and mammosphere formation, and in vivo tumor initiation. This experimental approach identified DOA as a hitherto unrecognized anti-CSC phytochemical that, when used as single-agent at low micromolar concentrations, is sufficient to fully prevent the ability of CSC-like cellular states to survive and proliferate as floating mammospheres under nonadherent and nondifferentiating conditions. It is believed that the size of tumorspheres reflects self-renewal rate of stem cells. Consequently, the observed effects of DOA on both the number and size of mammospheres point to its ability to not only target the tumor-initiating traits possessed by CSC-like states but also to reduce their self-renewal capabilities. DOA has a null inhibitory effect on BC cell growth and viability under two-dimensional differentiated growth conditions, thus confirming that the DOA-responsive phenotype is exclusively manifested under three-dimensional stem cell culture conditions. Indeed, the substantial decrease in tumor initiation rates upon pre-exposure of BC cells to DOA prior to implantation in mice strongly supports the notion that DOA is particularly efficient at specifically removing CSC and preventing the occurrence of CSC-like cellular states from the bulk population of nontumorigenic cancer cells. Accordingly, treatment with noncytotoxic concentrations of DOA for only 2 h successfully impaired anoikis-resistant cancer cells with stem-like characteristics from forming tumors in mammary fat pads even though the compound is presumably not present 4 months after injection. These findings lend experimental support to mathematical models proposing that CSC-targeted therapies are a better choice for long-term maintenance therapy via suppression of CSC-driven tumor relapse in vivo, even when such therapies, as it is the case for DOA, might not exert cytotoxic effects in bulk cancer cell populations (54).
Despite the diversity of genetic changes and molecular aberrations driving the different molecular subtypes of BC (e.g., luminal, HER2-enriched, basal-like, claudin-low), it is known that CSC-like states within these distinct BC subtypes have similar gene expression patterns, suggesting common, shared regulatory signaling pathways in CSC subpopulations across the molecular subtypes of BC (13,33). DOA treatment induced specific and potent suppression of E- and M-CSC cellular states irrespective of the mutational landscape of the starting BC population, strongly suggesting that its mechanism of action might efficiently interfere with the biologic functioning of cancer stemness per se. To couple the phenotypic drug discovery of DOAgly anti-CSC activity and target deconvolution, we used mechanism-of-action profiling with PM-based screenings, which indicated synergistic interactions of DOA with the mTOR inhibitor rapamycin and the DNMT inhibitor 5-azacytidine. The statistically significant reduction in the number and size of mammospheres generated with DOA plus rapamycin or 5-azacytidine would indicate that DOA functions synergistically to decrease the magnitude of the tumor-initiating CSC-like populations as well as their self-renewal capacity by, in part, targeting the rapamycin and 5-azacytidine molecular targets, mTOR and DNMT, respectively. Indeed, in silico computational approaches suggested that DOA binds and inhibits the ATP-binding kinase domain site in mTOR and the SAM cofactor-binding pocket in DNMTs. Consistent with this notion, FRET-based Z-LYTE™ and AlphaScreen-based in vitro assays confirmed its capacity to function as an ATP-competitive inhibitor that directly targets the mTOR catalytic site and to block the SAM-dependent methylation activity of DNMTs.
The dual mTOR/DNMT inhibitory nature of DOA appears to target a highly relevant metabolo-epigenetic dimension that drives the functional traits of tumor-initiating CSC-like cells. Against this background, the translational landscape of mTOR signaling is beginning to be viewed as a key component of the protein synthesis-addicted metabolic profile of tumor-initiating CSC-like cells (55). Accordingly, administration of rapamycin efficiently suppresses the tumorsphere-forming potential of mammosphere formation of BCSC (56). Moreover, DNMT1 is essential for the self-renewal and maintenance of CSC, and thus 5-azacytidine treatment during premalignant stages efficiently blocks mammary tumorigenesis and reduces tumorsphere-forming potential of CSC (57). Our findings suggest that the DOA’s ability to strongly and negatively impact the tumorigenic and self-renewal nature of CSC occurs through DNMT-related epigenetic regulation in addition to the inhibition of the mTOR-related anabolic phenotype of CSC. DOA turned off the expression of pivotal factors that positively regulate the activation of the CSC phenotype such as the stemness transcription factor OCT4 and the telomerase catalytic subunit TERT, while concomitantly augmented the expression of genes contributing to cell differentiation (e.g., FOS, CASP3) rather than execution of cell death programs (58–60). Therefore, it appears reasonable to suggest that DOA might functionally deplete tumor-initiating CSC-like states that sustain tumorigenicity by impacting fundamental controllers of cell fate choice, a metabolo-epigenetic mechanism involving both the downregulation of stemness-driving transcription factors and the re-activation of epigenetically suppressed differentiation programs.
Our discovery that EVOO, a unique functional food with a major contribution to the health-promoting effects of the Mediterranean diet (24–29), contains a natural inhibitor of mTOR and DNMTs capable of specifically and potently suppressing the functional traits of CSC within heterogeneous cancer cell populations, might open new avenues for introducing innovations in CSC-targeted therapy (61) based on the molecular bridge that connects metabolism and epigenetics with the state of stemness.
Supplementary material
Supplementary Material, Table S1–S4 and Figure S1–S3, can be found at Carcinogenesis online.
Funding
This work was supported by grants from the Ministerio de Ciencia e Innovación (Grant SAF2016-80639-P to J.A.M.), Plan Nacional de I+D+I, Spain, the Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR; Grant 2014 SGR229 to J.A.M.), Departament d’Economia i Coneixement, Catalonia, Spain, the Andalusian Regional Government Council of Innovation and Science (Grant P11-CTS-7625 to A.S.-C.), the Ministerio de Economía, Industria y Competitividad, Spain (Grants AGL2015-67995-C2-3-R and AGL2015-67995-C3-1-R to A.S.-C. and V.M.) and Conselleria d’Educació, Investigació, Cultura I Esport, Generalitat Valenciana, Spain (Grant PROMETEO/2016/006 to V.M). E.C. is supported by the Sara Borrell post doctoral contract (CD15/00033) from the Ministerio de Sanidad y Consumo, Fondo de Investigación Sanitaria (FIS), Spain. We are grateful to Custodio Borrego for giving us free use of the photograph he took of EVOO and olive trees in Granada (Spain), which have been included in Figure 7. This work has been awarded with the IV Premio Internacional Castillo de Canena de Investigación Oleícola ‘LUIS VAÑÓ’(IV Edition of Castillo de Canena LUIS VAÑÓ Award for Research on Olive Cultivation and Olive Oil; UC Davis Olive Center, Castillo de Canena, and Universidad de Jaén).
Acknowledgements
The authors would like to thank Dr Kenneth McCreath for editorial support. We are greatly indebted to Prof Robert A. Weinberg (Whitehead Institute for Biomedical Research, Cambridge, MA) for providing the HMLERshCntrol/HMLERshEcad cells used in this work.
Conflict of Interest Statement: Stock ownership: Á.G.M., StemTek Therapeutics (CEO). All other authors have no competing interests to declare.
Abbreviations
- ALDH
aldehyde dehydrogenase
- BC
breast cancer
- CSC
cancer stem cells
- DNMT
DNA methyltransferase
- DOA
decarboxymethyl oleuropein aglycone
- EVOO
extra virgin olive oil
- FBS
fetal bovine serum
- MD
molecular dynamics
- MM/GBSA
mechanics/generalized Borne surface area
- MSFE
mammosphere-forming efficiency
- OA
oleuropein aglycone
- OD
optical density
- PM
phenotype microarrays
- SAM
S-adenosyl-l-methionine.
References
- 1. Wicha M.S., et al. (2006)Cancer stem cells: an old idea—a paradigm shift. Cancer Res., 66,1883–1889. [DOI] [PubMed] [Google Scholar]
- 2. Kakarala M., et al. (2008)Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J. Clin. Oncol., 26, 2813–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Creighton C.J., et al. (2009)Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. USA, 106, 13820–13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Magee J.A., et al. (2012)Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell, 21, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaffer C.L., et al. (2015)How does multistep tumorigenesis really proceed?Cancer Discov., 5, 22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Menendez J.A. (2015)The metaboloepigenetic dimension of cancer stem cells: evaluating the market potential for new metabostemness-targeting oncology drugs. Curr. Pharm. Des., 21, 3644–3653. [DOI] [PubMed] [Google Scholar]
- 7. Hu X., et al. (2017)Cancer stem cells therapeutic target database: the first comprehensive database for therapeutic targets of cancer stem cells. Stem Cells Transl. Med., 6, 331–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tai M.H., et al. (2005)Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis, 26, 495–502. [DOI] [PubMed] [Google Scholar]
- 9. Trosko J.E. (2006)From adult stem cells to cancer stem cells: oct-4 gene, cell-cell communication, and hormones during tumor promotion. Ann. N. Y. Acad. Sci., 1089, 36–58. [DOI] [PubMed] [Google Scholar]
- 10. Mani S.A., et al. (2008)The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell, 133, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chaffer C.L., et al. (2011)Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. USA, 108, 7950–7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Friedmann-Morvinski D., et al. (2014)Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep., 15, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brooks M.D., et al. (2015)Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell, 17, 260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sai K., et al. (2000)Prevention of the down-regulation of gap junctional intercellular communication by green tea in the liver of mice fed pentachlorophenol. Carcinogenesis, 21, 1671–1676. [DOI] [PubMed] [Google Scholar]
- 15. Nakamura Y., et al. (2005)Augmentation of differentiation and gap junction function by kaempferol in partially differentiated colon cancer cells. Carcinogenesis, 26, 665–671. [DOI] [PubMed] [Google Scholar]
- 16. Li Y., et al. (2011)Implications of cancer stem cell theory for cancer chemoprevention by natural dietary compounds. J. Nutr. Biochem., 22, 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leone A., et al. (2012)The chemopreventive role of dietary phytochemicals through gap junctional intercellular communication. Phytochem. Rev., 11, 285–307. [Google Scholar]
- 18. Kim Y.S., et al. (2012)Cancer stem cells: potential target for bioactive food components. J. Nutr. Biochem., 23, 691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y., et al. (2014)Targeting cancer stem cells by curcumin and clinical applications. Cancer Lett., 346, 197–205. [DOI] [PubMed] [Google Scholar]
- 20. Li Y., et al. (2010)Sulforaphane, a dietary component of broccoli/broccoli sprouts, inhibits breast cancer stem cells. Clin. Cancer Res., 16, 2580–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pandey P.R., et al. (2011)Resveratrol suppresses growth of cancer stem-like cells by inhibiting fatty acid synthase. Breast Cancer Res. Treat., 130, 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsieh C.Y., et al. (1999)Stem cell differentiation and reduction as a potential mechanism for chemoprevention of breast cancer. Chinese Pharm. J., 51, 15–30. [Google Scholar]
- 23. Mineva N.D., et al. (2013)Epigallocatechin-3-gallate inhibits stem-like inflammatory breast cancer cells. PLoS One, 8, e73464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Colomer R., et al. (2006)Mediterranean diet, olive oil and cancer. Clin. Transl. Oncol., 8, 15–21. [DOI] [PubMed] [Google Scholar]
- 25. Menendez J.A., et al. (2006)Mediterranean dietary traditions for the molecular treatment of human cancer: anti-oncogenic actions of the main olive oil’s monounsaturated fatty acid oleic acid (18:1n-9). Curr. Pharm. Biotechnol., 7, 495–502. [DOI] [PubMed] [Google Scholar]
- 26. Escrich E., et al. (2011)Olive oil, an essential component of the Mediterranean diet, and breast cancer. Public Health Nutr., 14, 2323–2332. [DOI] [PubMed] [Google Scholar]
- 27. Visioli F., et al. (2011)Extra virgin olive oil’s polyphenols: biological activities. Curr. Pharm. Des., 17, 786–804. [DOI] [PubMed] [Google Scholar]
- 28. Casaburi I., et al. (2013)Potential of olive oil phenols as chemopreventive and therapeutic agents against cancer: a review of in vitro studies. Mol. Nutr. Food Res., 57, 71–83. [DOI] [PubMed] [Google Scholar]
- 29. Menendez J.A., et al. (2013)Xenohormetic and anti-aging activity of secoiridoid polyphenols present in extra virgin olive oil: a new family of gerosuppressant agents. Cell Cycle, 12, 555–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Obied H.K., et al. (2008)Biosynthesis and biotransformations of phenol-conjugated oleosidic secoiridoids from Olea europaea L. Nat. Prod. Rep., 25, 1167–1179. [DOI] [PubMed] [Google Scholar]
- 31. Gupta P.B., et al. (2009)Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell, 138, 645–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manuel Iglesias J., et al. (2013)Mammosphere formation in breast carcinoma cell lines depends upon expression of E-cadherin. PLoS One, 8, e77281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martin-Castillo B., et al. (2015)Cancer stem cell-driven efficacy of trastuzumab (Herceptin): towards a reclassification of clinically HER2-positive breast carcinomas. Oncotarget, 6, 32317–32338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lozano-Sánchez J., et al. (2010)Prediction of extra virgin olive oil varieties through their phenolic profile. Potential cytotoxic activity against human breast cancer cells. J. Agric. Food Chem., 58, 9942–9955. [DOI] [PubMed] [Google Scholar]
- 35. Vazquez-Martin A., et al. (2012)Phenolic secoiridoids in extra virgin olive oil impede fibrogenic and oncogenic epithelial-to-mesenchymal transition: extra virgin olive oil as a source of novel antiaging phytochemicals. Rejuvenation Res., 15, 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Charafe-Jauffret E., et al. (2009)Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res., 69, 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Korkaya H., et al. (2008)HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene, 27, 6120–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duru N., et al. (2012)HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clin. Cancer Res., 18, 6634–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Corominas-Faja B., et al. (2014)Chemical inhibition of acetyl-CoA carboxylase suppresses self-renewal growth of cancer stem cells. Oncotarget, 5, 8306–8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tomita H., et al. (2016)Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget, 7, 11018–11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jung J.W., et al. (2011)Metformin represses self-renewal of the human breast carcinoma stem cells via inhibition of estrogen receptor-mediated OCT4 expression. PLoS One, 6, e28068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith J.A., et al. (2010)A role for the Werner syndrome protein in epigenetic inactivation of the pluripotency factor Oct4. Aging Cell, 9, 580–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoshida T., et al. (2006)Spreds, inhibitors of the Ras/ERK signal transduction, are dysregulated in human hepatocellular carcinoma and linked to the malignant phenotype of tumors. Oncogene, 25, 6056–6066. [DOI] [PubMed] [Google Scholar]
- 44. Chandhoke A.S., et al. (2016)The ubiquitin ligase Smurf2 suppresses TGFβ-induced epithelial-mesenchymal transition in a sumoylation-regulated manner. Cell Death Differ., 23, 876–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bochner B.R., et al. (2001)Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res., 11, 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bourne C.R., et al. (2012)Classifying compound mechanism of action for linking whole cell phenotypes to molecular targets. J. Mol. Recognit., 25, 216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mackie A.M., et al. (2014)Biolog phenotype microarrays for phenotypic characterization of microbial cells. Methods Mol. Biol., 1096, 123–130. [DOI] [PubMed] [Google Scholar]
- 48. Choi J., et al. (2011)Sanguinarine is an allosteric activator of AMP-activated protein kinase. Biochem. Biophys. Res. Commun., 413, 259–263. [DOI] [PubMed] [Google Scholar]
- 49. Kalogris C., et al. (2014)Sanguinarine suppresses basal-like breast cancer growth through dihydrofolate reductase inhibition. Biochem. Pharmacol., 90, 226–234. [DOI] [PubMed] [Google Scholar]
- 50. Dennis P.B., et al. (1996)The principal rapamycin-sensitive p70(s6k) phosphorylation sites, T-229 and T-389, are differentially regulated by rapamycin-insensitive kinase kinases. Mol. Cell. Biol., 16, 6242–6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Khanfar M.A., et al. (2015)Olive oil-derived oleocanthal as potent inhibitor of mammalian target of rapamycin: biological evaluation and molecular modeling studies. Phytother. Res., 29, 1776–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moffat J.G., et al. (2014)Phenotypic screening in cancer drug discovery—past, present and future. Nat. Rev. Drug Discov., 13, 588–602. [DOI] [PubMed] [Google Scholar]
- 53. Moffat J.G., et al. (2017)Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov., 16, 531–543. [DOI] [PubMed] [Google Scholar]
- 54. Liu X., et al. (2013)Nonlinear growth kinetics of breast cancer stem cells: implications for cancer stem cell targeted therapy. Sci. Rep., 3, 2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hsieh A.C., et al. (2012)The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature, 485, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lamb R., et al. (2015)Targeting tumor-initiating cells: eliminating anabolic cancer stem cells with inhibitors of protein synthesis or by mimicking caloric restriction. Oncotarget, 6, 4585–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pathania R., et al. (2015)DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nat. Commun., 6, 6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fujita J., et al. (2008)Caspase activity mediates the differentiation of embryonic stem cells. Cell Stem Cell, 2, 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wolter F., et al. (2003)Resveratrol-induced modification of polyamine metabolism is accompanied by induction of c-Fos. Carcinogenesis, 24, 469–474. [DOI] [PubMed] [Google Scholar]
- 60. Dogan F., et al. (2018)Correlation between telomerase and mTOR pathway in cancer stem cells. Gene, 641, 235–239. [DOI] [PubMed] [Google Scholar]
- 61. Menendez J.A. (2015)The metaboloepigenetic dimension of cancer stem cells: evaluating the market potential for new metabostemness-targeting oncology drugs. Curr. Pharm. Des., 21, 3644–3653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.