By studying three independent samples of healthy volunteers using fMRI, Chen and Ursini et al. report that polygenic risk scores for schizophrenia predict hippocampal activity during memory encoding, a putative neuroimaging intermediate phenotype. This finding suggests a link between the polygenic architecture of schizophrenia and a component of disease pathophysiology.
Keywords: schizophrenia, genetics, polygenic risk score (PRS), neuroimaging genetics, hippocampal activity
Abstract
The use of polygenic risk scores has become a practical translational approach to investigating the complex genetic architecture of schizophrenia, but the link between polygenic risk scores and pathophysiological components of this disorder has been the subject of limited research. We investigated in healthy volunteers whether schizophrenia polygenic risk score predicts hippocampal activity during simple memory encoding, which has been proposed as a risk-associated intermediate phenotype of schizophrenia. We analysed the relationship between polygenic risk scores and hippocampal activity in a discovery sample of 191 unrelated healthy volunteers from the USA and in two independent replication samples of 76 and 137 healthy unrelated participants from Europe and the USA, respectively. Polygenic risk scores for each individual were calculated as the sum of the imputation probability of reference alleles weighted by the natural log of odds ratio from the recent schizophrenia genome-wide association study. We examined hippocampal activity during simple memory encoding of novel visual stimuli assessed using blood oxygen level-dependent functional MRI. Polygenic risk scores were significantly associated with hippocampal activity in the discovery sample [P = 0.016, family-wise error (FWE) corrected within Anatomical Automatic Labeling (AAL) bilateral hippocampal-parahippocampal mask] and in both replication samples (P = 0.033, FWE corrected within AAL right posterior hippocampal-parahippocampal mask in Bari sample, and P = 0.002 uncorrected in the Duke Neurogenetics Study sample). The relationship between polygenic risk scores and hippocampal activity was consistently negative, i.e. lower hippocampal activity in individuals with higher polygenic risk scores, consistent with previous studies reporting decreased hippocampal-parahippocampal activity during declarative memory tasks in patients with schizophrenia and in their healthy siblings. Polygenic risk scores accounted for more than 8% of variance in hippocampal activity during memory encoding in discovery sample. We conclude that polygenic risk scores derived from the most recent schizophrenia genome-wide association study predict significant variability in hippocampal activity during memory encoding in healthy participants. Our findings validate mnemonic hippocampal activity as a genetic risk associated intermediate phenotype of schizophrenia, indicating that the aggregate neurobiological effect of schizophrenia risk alleles converges on this pattern of neural activity.
Introduction
A major role of inherited genetic variants in the aetiology of schizophrenia is supported by the high heritability of this complex disorder (Sullivan, 2005). In recent years, genome wide association studies (GWAS) with increasing sample sizes have been able to detect increasingly more risk-associated loci (International Schizophrenia Consortium et al., 2009; Schizophrenia Psychiatric Genome-Wide Association Study, 2011; Schizophrenia Working Group of the Psychiatric Genomics, 2014). The most recent state-of-the-art GWAS of schizophrenia conducted by the Psychiatric Genetics Consortium (PGC) identified 108 risk-associated genetic loci achieving genome-wide statistical significance (P < 5 × 10−8), but each locus accounted for only a very small fraction of genetic risk, with odds ratios generally between 0.9 and 1.1 (Schizophrenia Working Group of the Psychiatric Genomics, 2014), which is consistent with the putative polygenic architecture of schizophrenia. This phenomenon is also consistent with previous family studies (Gottesman and Shields, 1967; McGue et al., 1985) showing that the change in relative risk with decreasing relatedness is inconsistent with Mendelian inheritance patterns, and with the presence of a majority of sporadic cases not linked to a family history of schizophrenia (Yang et al., 2010).
The small prediction of risk attributable to an individual GWAS locus has encouraged application of a strategy based on summing genome-wide risk alleles in the form of polygenic risk scores (PRSs) (International Schizophrenia Consortium et al., 2009; Schizophrenia Working Group of the Psychiatric Genomics, 2014). PRSs allow much greater prediction of liability than individual variant genotypes (International Schizophrenia Consortium et al., 2009; Schizophrenia Working Group of the Psychiatric Genomics, 2014). PRSs are calculated by summarizing the genetic effects of an ensemble of independent markers identified through large-scale GWAS (International Schizophrenia Consortium et al., 2009; Schizophrenia Working Group of the Psychiatric Genomics, 2014). The assumptions underlying the PRS approach are that genetic risk variants act collectively, and thousands of individual loci of small effect, with relatively few being GWAS-significant at any given sample size, together account for a substantial proportion of variation in risk (International Schizophrenia Consortium et al., 2009). Based on the weighted sum of reference alleles for single nucleotide polymorphisms (SNPs) with a certain statistical level of association, PRSs provide a parsimonious approach to detect the aggregate genetic effect of markers with small individual effects. In recent years, PRSs have been widely used in investigating the polygenic architecture of schizophrenia (International Schizophrenia Consortium et al., 2009; Schizophrenia Psychiatric Genome-Wide Association Study, 2011; Derks et al., 2012; Schizophrenia Working Group of the Psychiatric Genomics, 2014) and other complex phenotypes (Stahl et al., 2012; Cross-Disorder Group of the Psychiatric Genomics, 2013), such as personality traits and mood disorders (Middeldorp et al., 2011; Luciano et al., 2012), measures of depression and anxiety (Demirkan et al., 2011), creativity (Power et al., 2015), and cancer (Szulkin et al., 2015).
A crucial issue in identifying mechanisms through which genetic factors confer risk for neuropsychiatric disorders is to define whether the risk genes modulate brain functions corresponding to biological phenomena implicated in the disorder and associated with increased genetic risk, so-called intermediate phenotypes (Rasetti and Weinberger, 2011) or endophenotypes. As a heritable quantitative biological trait that is associated with the illness state and with genetic risk, an intermediate phenotype is presumably located in the path from genetic predisposition to psychopathology. Its link to risk for disease can be inferred by showing greater prevalence in unaffected relatives of patients than in the general population (Rasetti and Weinberger, 2011). However, while the presence of these phenotypes in healthy relatives may reflect potentially genetic or shared familial and other environmental factors, they do not prove genetic association. A relationship between polygenic risk scores and intermediate phenotypes is a more direct approach to establishing this genetic association. Moreover, to circumvent the potential effect of confounding factors associated with the illness state, genetic association with intermediate phenotypes is best validated in healthy participants.
One of the putative intermediate phenotypes identified in our previous studies is altered hippocampal physiological engagement in patients with schizophrenia and in their healthy siblings, as measured with functional MRI during a simple incidental declarative memory task (Di Giorgio et al., 2013; Rasetti et al., 2014). Converging evidence from many lines of research, including post-mortem (Harrison and Weinberger, 2005), animal (Lipska and Weinberger, 2000; Lipska, 2004; Tseng et al., 2009), and human cognition and neuroimaging studies (Bertolino et al., 1996; Achim and Lepage, 2005), implicates abnormal hippocampal function as a consistent pathological feature of schizophrenia. These studies and many others in diverse mammalian species implicate the hippocampus as a key structure in declarative memory, a cognitive domain that is consistently impaired in patients with schizophrenia (Heinrichs and Zakzanis, 1998). Further, declarative memory deficits in patients with schizophrenia are moderately heritable, share apparent genetic overlap with this disease in twin samples (Owens et al., 2011), and are thought to be due primarily to deficient memory encoding that likely reflects hippocampal dysfunction (Dickerson and Eichenbaum, 2010; Rasetti et al., 2014). Consistently, hippocampal-parahippocampal activation during memory encoding has been associated with impaired memory (Allen et al., 2011; Di Giorgio et al., 2013; Rasetti et al., 2014), and loss of hippocampal activation over time has been linked with declining memory and with clinical decline (O'Brien et al., 2010; Persson et al., 2012).
While many confounding factors associated with the experience of the disorder may contribute to declarative memory dysfunction in patients with schizophrenia, an impairment of hippocampal recruitment during a declarative memory task measured with functional MRI has also been reported in healthy first-degree biological relatives of patients (Di Giorgio et al., 2013; Rasetti et al., 2014), thus supporting the notion of altered hippocampal function as a risk-associated intermediate phenotype. However, since findings in siblings and high-risk individuals may reflect other familial and environmental factors and not genetic risk, per se, investigating the association of hippocampal activity during memory encoding with schizophrenia PRS in healthy participants may provide a compelling validation of this intermediate phenotype as being related to genomic risk.
The aim of this study was thus to examine whether PRS constructed based on risk loci from the most recent schizophrenia GWAS (Schizophrenia Working Group of the Psychiatric Genomics, 2014) are associated with hippocampal activity during memory encoding as a putative neuroimaging intermediate phenotype (Di Giorgio et al., 2013; Rasetti et al., 2014). To that end, we investigated the relationship between PRS and hippocampal activity during simple encoding of novel visual stimuli while participants underwent blood oxygenation level-dependent functional MRI (BOLD functional MRI). We found that the PRS that has the highest prediction accuracy for diagnostic status (i.e. so-called ‘PRS6,’ see below) is associated with decreased hippocampal activity in a discovery sample as well as in two independent replication samples of healthy participants. Critically, the directionality of the association is consistent with decreased hippocampal activity reported in patients with schizophrenia and in their healthy siblings. These results support the conclusion that the cumulative effect of schizophrenia risk alleles may converge on a pattern of mnemonic hippocampal activity associated with vulnerability for the disorder.
Materials and methods
Discovery sample
A total of 191 unrelated and extensively evaluated healthy volunteers (73 males; mean age 30.3 ± 8.7 years) with high quality genetic and neuroimaging data were included in the discovery analysis of association between PRS and hippocampal activity (Table 1). The participants were all Caucasians of European ancestry and were selected from the Clinical Brain Disorders Branch Sibling Study of schizophrenia at the National Institute of Mental Health (Daniel R. Weinberger, Principal Investigator). The study was approved by the Institutional Review Board of the Intramural Program of the National Institute of Mental Health. All participants were assessed in person with a Structured Clinical Interview for DSM-IV (American Psychiatric Association, 1994). Exclusion criteria for healthy participants included: presence of a psychiatric disorder at the time of the study and by history; having a first-degree relative with a psychiatric disorder; IQ < 80; recent drug or alcohol abuse (within 1 year) or >5 years of previous abuse; substantial medical or neurological conditions; and current psychotropic pharmacological treatment. Details of exclusion criteria can be found in the Supplementary material. Only right-handed participants [Edinburgh Handedness Index (EHI) > 80] with retrieval accuracy above chance (>50%) were included in this study, to analyse the relationship between genetic risk and hippocampal activity, reducing the potential confounding effect related to handedness, and to differences in successful and unsuccessful encoding. After a complete description of the study to the participants, written informed consent was obtained.
Table 1.
Demographic and performance data of the discovery sample, with statistics of the relationship between each variable and PRS
| Sex (M/F) | Age, years (SD) | WAIS score (SD) | d′ (SD) | |
|---|---|---|---|---|
| Mean (SE) | 73/118 | 30.3 (8.7) | 110.4 (9.5) | 2.6 (0.8) |
| Association with PRS | ||||
| Test | Two-sample t-test | Correlation | Correlation | Correlation |
| PRS1 | T = −0.5088 | r = −0.0088 | r = 0.1139 | r = −0.0916 |
| P = 0.6115 | P = 0.9038 | P = 0.1166 | P = 0.2077 | |
| PRS2 | T = −1.0861 | r = −0.0138 | r = 0.1131 | r = −0.0575 |
| P = 0.2790 | P = 0.8497 | P = 0.1194 | P = 0.4297 | |
| PRS3 | T = −1.6182 | r = −0.1023 | r = 0.0066 | r = −0.0020 |
| P = 0.1079 | P = 0.1589 | P = 0.9279 | P = 0.9785 | |
| PRS4 | T = −1.5084 | r = −0.1088 | r = −0.0593 | r = 0.0482 |
| P = 0.1338 | P = 0.1341 | P = 0.4150 | P = 0.5080 | |
| PRS5 | T = −2.1009 | r = −0.1661 | r = −0.1105 | r = 0.0421 |
| P = 0.0373 | P = 0.0217 | P = 0.1282 | P = 0.5632 | |
| PRS6 | T = −2.5751 | r = −0.1818 | r = −0.1679 | r = 0.0500 |
| P = 0.0109 | P = 0.0118 | P = 0.0203 | P = 0.4919 | |
| PRS7 | T = −1.8513 | r = −0.1864 | r = −0.2030 | r = 0.0015 |
| P = 0.0659 | P = 0.0098 | P = 0.0049 | P = 0.9834 | |
| PRS8 | T = −2.2100 | r = −0.1774 | r = −0.1973 | r = 0.0147 |
| P = 0.0285 | P = 0.0141 | P = 0.0062 | P = 0.8397 | |
| PRS9 | T = −1.8050 | r = −0.1997 | r = −0.2110 | r = 0.0065 |
| P = 0.0729 | P = 0.0056 | P = 0.0034 | P = 0.9289 | |
| PRS10 | T = −2.0090 | r = −0.2004 | r = −0.2019 | r = 0.0075 |
| P = 0.0461 | P = 0.0054 | P = 0.0051 | P = 0.9183 | |
F = female; M = male; SE = standard error; WAIS = Wechsler Adult Intelligence Scale.
Bari replication sample
To determine the reliability of our results, we analysed the relationship between PRS and hippocampal activity during the identical incidental memory encoding task in an independent sample of 76 healthy unrelated participants (33 males; mean age 27.8 ± 6.1 years) collected at the University of Bari, Italy. All participants were Caucasians of European ancestry from the Apulia region of Italy. These individuals underwent the same screening and evaluations as participants in the discovery sample, and all were right-handed (EHI > 50) with retrieval accuracy above chance (>50%). Details of exclusion criteria are in the Supplementary material. All participants provided written informed consent after complete description of the study protocol as approved by the Institutional Review Board of the University of Bari.
Duke Neurogenetics Study replication sample
We further evaluated the reliability of the association between PRS and hippocampal activity during a similar memory encoding task in a second replication sample of 137 (63 males; mean age: 19.8 ± 1.2) non-Hispanic Caucasian university students of European ancestry who had successfully completed the Duke Neurogenetics Study (DNS) (Supplementary Table 2). Ancestry was determined by self-report and confirmed using multidimensional scaling (MDS) of genome-wide SNP genotypes [i.e. no individuals were ±6 standard deviations (SDs) from the mean on the top 10 components (Purcell et al., 2007)]. All participants were in good general health and free of the following study conditions: (i) medical diagnoses of cancer, stroke, head injury with loss of consciousness, untreated migraine headaches, diabetes requiring insulin treatment, chronic kidney or liver disease; (ii) use of psychotropic, glucocorticoid, or hypolipidaemic medication; and (iii) conditions affecting cerebral blood flow and metabolism (e.g. hypertension). All participants provided informed consent in accordance with Duke University Medical Center Institutional Review Board guidelines prior to participation.
Genotyping and imputation
DNA of the discovery and Bari replication samples was extracted from blood using standard procedures, and genotyping was done using variate Illumina Bead Chips including 510K/610K/660K/2.5M. We divided samples into two groups according to genotyping chips. One group included samples genotyped with low-resolution BeadChips (510K/610K/660K), and the other included samples genotyped with high-resolution BeadChips (2.5 M), and imputation was performed separately for these two groups. To control for the use of two different imputations, we included genotyping batch label as a covariate in the statistical analysis.
Quality control was performed using PLINK (version 1.07; http://pngu.mgh.harvard.edu/purcell/plink/) (Purcell et al., 2007), as reported by the Psychiatric Genomic Consortium (Schizophrenia Working Group of the Psychiatric Genomics, 2014). Pre-phasing was done before imputation with SHAPEIT (Delaneau et al., 2012), and imputation was done with IMPUTE2 (Howie et al., 2009) using 1000 genome phase 1 as reference panel (Howie et al., 2011; Delaneau et al., 2012). To control for population stratification in the association analysis, the first 10 principal components of the whole genome data were calculated using EIGENSOFT v5.01 (EIGENSOFT, http://www.hsph.harvard.edu/alkes-price/software/).
In the DNS replication sample, DNA was isolated from saliva derived from Oragene DNA self-collection kits (DNA Genotek) customized for 23andMe (www.23andme.com). DNA extraction and genotyping were performed by the National Genetics Institute (NGI), a CLIA-certified clinical laboratory and subsidiary of Laboratory Corporation of America. One of two different Illumina arrays with custom content was used to provide genome-wide SNP data, the HumanOmniExpress or HumanOmniExpress-24. Genotype imputation was run separately for all DNS participants with genome-wide chip data using the pre-phasing/imputation stepwise approach implemented in SHAPEIT and IMPUTE2 using only biallelic SNPs and the default value for effective size of the population (20 000), and chunk sizes of 3 Mb and 5 Mb for the respective arrays.
For discovery and replication samples, within each array batch, genotyped SNPs used for imputation were required to have missing rate < 0.02, Hardy-Weinberg equilibrium P > 10−6, and minor allele frequency (MAF) > 0.01. After imputation, imputed dosage data for each SNP with imputation quality (INFO) > 0.1 were used for polygenic risk scores calculation.
Polygenic risk scores
PRSs for schizophrenia were calculated for each individual in each sample, as a measure of genomic risk for schizophrenia, based on the most recent GWAS (Schizophrenia Working Group of the Psychiatric Genomics, 2014). We obtained odds ratios of 102 217 index SNPs from a meta-analysis of PGC2 GWAS using datasets excluding the discovery sample (PGC 2014, non-Lieber sample PGC2 GWAS) (Schizophrenia Working Group of the Psychiatric Genomics, 2014). These 102K SNPs are linkage disequilibrium-independent (R2 < 0.1) and span across the whole genome. We then calculated a weighted sum of risk alleles for schizophrenia, by summing the imputation probability for the reference allele of the index SNP, weighted by the natural log of the odds ratio of association with schizophrenia, at each independent locus across the whole genome, as described elsewhere (International Schizophrenia Consortium et al., 2009; Schizophrenia Working Group of the Psychiatric Genomics, 2014). Consistent with the original approach taken by the Psychiatric Genomics Consortium in the GWAS study (Schizophrenia Working Group of the Psychiatric Genomics, 2014), 10 PRSs (PRS1-PRS10) were calculated using subsets of the 102K SNPs under different thresholds of the PGC2 GWAS P-values of association with schizophrenia: 5 × 10−8, 10−6, 10−4, 0.001, 0.01, 0.05, 0.1, 0.2, 0.5, and 1. SNPs in sets with lower P-values are also in sets with higher P-values (e.g. SNPs in PRS1 are included in PRS2, SNPs in PRS2 are included in PRS3, and so on). Details of number of SNPs for each PRS level can be found in Supplementary Table 3.
We focused our analyses on PRS6 (PPGC2 < 0.05), which was constructed using SNPs with GWAS association P-value < 0.05, since this PRS has been shown to have the highest prediction accuracy for diagnostic status in multiple independent samples (Schizophrenia Working Group of the Psychiatric Genomics, 2014). It has been assumed that SNPs at this threshold represent most, if not all, true schizophrenia risk-associated genomic loci, even though most of the individual loci are not GWAS significant. To address specificity of observed associations with PRS6, exploratory analyses using the other PRSs are reported in the Supplementary material.
Functional MRI paradigm in the discovery and Bari replication samples
The functional MRI paradigm in discovery and Bari replication samples is a simple declarative memory task, which has been used in previous studies (Di Giorgio et al., 2013; Rasetti et al., 2014). The task included blocks of incidental encoding and retrieval of neutral and aversive images selected from the International Affective Picture System (Lang et al., 1997; Ito et al., 1998). During the encoding run, four blocks of neutral scenes (six scenes for 3 s per scene) and four blocks of aversive scenes were continuously presented, alternating with nine blocks of rest (18 s of fixation). The order of the presentation of blocks (neutral and aversive) was counterbalanced across individuals. To make the encoding incidental, participants were not informed about the subsequent recognition/retrieval phase before scanning and thus were not aware that they were engaged in a memory task. During the encoding session, participants determined whether each picture represented an ‘indoor’ or ‘outdoor’ scene, and responded via a button press (left button for ‘indoor’ and right button for ‘outdoor’). The presentation order of ‘indoor’ and ‘outdoor’ scenes was randomized. A subsequent retrieval session started ∼2 min after the encoding session. During retrieval, participants determined whether the scene presented was seen during the encoding session, and responded via a button press (left button for ‘new’ and right button for ‘old’). The presentation order of ‘new’ and ‘old’ scenes was also randomized. During retrieval, half of the scenes were old (i.e. presented during encoding), and half were new (i.e. not presented during encoding). As with the encoding session, there were eight blocks (four neutral and four aversive) interleaved with nine rest blocks during retrieval, and the order of neutral and aversive blocks was counterbalanced across participants.
In the current study, we focused on hippocampal activity during encoding, because retrieval is likely a mixture of encoding and retrieval processes as half of the scenes shown during retrieval are novel. Furthermore, we limited our analyses to neutral scenes in order to specifically compare our findings with the previously described intermediate phenotype (Di Giorgio et al., 2013; Rasetti et al., 2014). Both accuracy and reaction time were extracted from log files of task performance after scanning.
Functional MRI paradigm in the Duke Neurogenetics Study replication sample
The functional MRI paradigm in the DNS replication sample consisted of the encoding and subsequent recall of novel neutral face-name pairs. A distractor task (odd/even number identification) is interleaved between encoding and recall blocks to prevent maintenance of information in working memory. During each of four encoding blocks, participants view six novel face-name pairs for 3.5 s each. During each of four recall blocks, participants view six faces each presented for 2 s and immediately followed by an incomplete name fragment for 1 s during which they are required by forced-choice to determine if the fragment is correct or incorrect. A 1-s intertrial interval is used during recall blocks. During each of four distractor blocks, participants view six different numbers for 3.5 s each and are required to determine if the numbers are odd or even (Zeineh et al., 2003). Total task length is 324 s. As with the discovery and Bari replication samples, analyses in the DNS sample were similarly focused on hippocampal activity during encoding blocks in comparison with distractor blocks.
Functional MRI acquisition and processing in the discovery and Bari replication samples
Data acquisition, quality control, preprocessing and first level analysis were performed in the same way in the discovery and in the Bari replication samples. Each subject underwent BOLD functional MRI on a GE Signa 3 T scanner using gradient echo BOLD-EPI pulse sequence (repetition time/echo time = 2000/28 ms, flip angle = 900, field of view = 24 cm, 64 × 64 matrix). One hundred and seventy whole brain images comprising 24 (4-mm thick, 1 mm gap) axial slices covering the entire cerebrum and most of the cerebellum were acquired for each subject while they performed the simple declarative memory task.
The images were preprocessed in SPM5 (http://www.fil.ion.ucl.ac.uk/spm) using standard procedures as detailed in previous reports (Rasetti et al., 2014). Briefly, images were realigned to the first image of the scan run using INRIalign, spatially normalized to a 3 × 3 × 3 mm3 voxel size into a standard stereotactic space (MNI template) using affine and non-linear transformation, and smoothed using an 8-mm full-width at half-maximum isotropic 3D Gaussian kernel. Datasets were individually examined to ensure head motion was <2 mm translation and <1.5° rotation.
All functional MRI data were individually examined and carefully screened for data quality using a variety of procedures including visual inspection for image artefacts, estimating indices for ghosting artefacts, signal-to-noise ratio across the time series, and head motion (data from participants with head motion >2 mm translation and/or head rotation >1.5° were excluded), as stated previously (Di Giorgio et al., 2013; Rasetti et al., 2014). In the first-level analysis, encoding condition and control condition (visual fixation) were modelled using a box car convolved with the haemodynamic response function at each voxel. Head movement parameters obtained from the realignment procedure were included in the model as covariates, taking into account the effects of subject head motion. A contrast map was generated for each individual using the beta value of encoding of neutral visual stimulus versus fixation cross-hair at each voxel.
Functional MRI acquisition and processing in the Duke Neurogenetics Study replication sample
In the DNS sample, each participant was scanned using one of two identical research-dedicated GE MR750 3 T scanners equipped with high-power high-duty-cycle 50-mT/m gradients at 200 T/m/s slew rate, and an eight-channel head coil for parallel imaging at high bandwidth up to 1 MHz at the Duke-UNC Brain Imaging and Analysis Center. The AFNI program 3dREMLfit (Cox, 1996) was used to fit a general linear model for first-level functional MRI data analyses. Following preprocessing, linear contrasts employing canonical haemodynamic response functions were used to estimate effects of condition (Encoding > Distractor) for each individual (see Supplementary material for more detail on data acquisition, preprocessing, and quality assurance).
Demographic and performance data analysis
We evaluated the relationship of each PRS with demographic and performance data, i.e. sex, age, WAIS score, and d′ (measure of retrieval accuracy; see Supplementary material for details), in all samples using R. To examine the difference of PRS between genders, we used a two-sample t-test. In examining the relationship between PRS and other variables, we used Pearson correlation.
Association between polygenic risk score and hippocampal activity
To evaluate the association between brain activation and PRS, individual contrast images were used in second-level random effects models in SPM12 accounting for participant-to-participant variability. Based on the primary hypothesis of this study and prior studies showing hippocampal activation during performance of a declarative memory task (Di Giorgio et al., 2013; Rasetti et al., 2014), the association between PRS and memory encoding hippocampal activity was examined within the hippocampal formation region using a bilateral hippocampal-parahippocampal mask from the Anatomical Automatic Labeling (AAL) atlas (Tzourio-Mazoyer et al., 2002). Sex and age were included as covariates of no interest to adjust for potential effects of these variables on hippocampal activity. Before the association analysis, 10 principle components of whole genome data and genotyping batch indicator (batch) for genotyping chip types were projected out from the original PRS using linear regression in R (details in Supplementary material). The distribution of adjusted PRS is shown in Supplementary Fig. 1.
In Bari sample and DNS sample, the association between PRSs and memory encoding activity were specifically examined within the hippo-parahippocampal region. A right posterior hippo-parahippocampal mask from the AAL atlas was used for small volume correction in the analysis of the replication samples.
Variance of hippocampal activation explained by polygenic risk score
To estimate the variance of hippocampal activation explained by PRSs, the BOLD responses within the cluster including peak activation in the right posterior hippocampal-parahippocampal region of interest under threshold P < 0.05 were extracted without correction. The relationship between the averaged extracted BOLD response and PRSs was evaluated by linear regression in R with sex and age as non-interest covariates. The variance explained by PRSs was estimated using the difference of goodness of fit (R2) of the full model and the reduced model. Details can be found in the Supplementary material. Variance of hippocampal activation explained by sex, age, IQ, and d′ was also evaluated.
Polygenic risk score and IQ effects on hippocampal activation
We extensively evaluated hippocampal activation associated with PRS6 and IQ, in separate and combined regression models. Furthermore, a potential IQ effect on hippocampal activation was investigated in different ranges of PRS6 (high, middle, and low). Different regression models were set up in SPM12 and corresponding plots were generated in R.
Results
Demographics and behavioural results
Demographic and behavioural data for the discovery and replication samples are shown in Table 1, Supplementary Table 1 and Supplementary Table 2, respectively. In the discovery sample, no significant correlation was found between PRSs and retrieval accuracy. PRS5-PRS10 showed significant correlations with sex and age, so in the following analyses we included sex and age as non-interest covariates to remove their confounding effects. PRS6-PRS10 also showed significant correlations with IQ. We therefore analysed whether the relationship between PRS6 and hippocampal activation was affected by the addition of IQ as a covariate, and whether the relationship between IQ and hippocampal activation was affected by the addition of PRS6 as a covariate. Corresponding results are reported in Supplementary Figs 5 and 6.
Association between polygenic risk score and hippocampal activity
Analyses in our discovery sample revealed that PRS6 was significantly negatively associated with hippocampal activity as shown in Fig. 1 (P < 0.001, uncorrected for illustration). The negative association indicates that higher genomic risk (PRS) for schizophrenia was associated with reduced hippocampal activity during encoding, which is consistent with the directionality of the previously reported intermediate phenotype association of reduced hippocampal activity in patients and siblings compared with healthy volunteers, during declarative memory tasks (Di Giorgio et al., 2013; Rasetti et al., 2014). The peak voxel for the association between PRS6 and brain activity was located in the right posterior hippocampal-parahippocampal region, which is the same location of our previous neuroimaging intermediate phenotype study (Rasetti et al., 2014). The peak is located at 30, −30, −18 (x, y, z, MNI coordinates), with Z = 4.10 and P = 0.016 family-wise error (FWE) correction within bilateral hippocampal-parahippocampal mask. The peaks, statistics of peaks and number of voxels of all PRSs are shown in Supplementary Table 4.
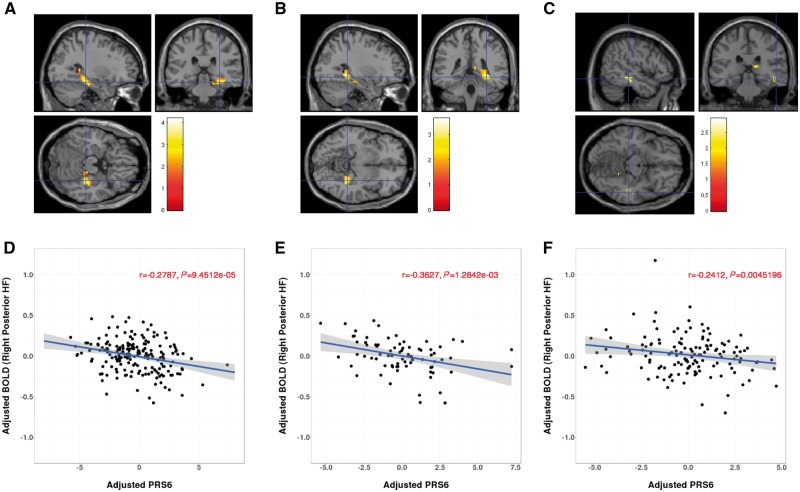
Figure 1.
Association between PRS6 and hippocampus activation during memory encoding in the Lieber discovery sample (A and D), and in Bari (B and E) and DNS (C and F) replication samples. (A–C) Section views (sagittal, coronal and transversal views) of the relationship between PRS6 and functional MRI hippocampal activity during neutral encoding in Lieber sample (A), Bari sample (B), and DNS sample (C), respectively. Genetic risk for schizophrenia is associated with right hippocampal activation (P < 0.05, uncorrected for illustration). (D–F) Scatter plot of the relationship between PRS6 and right posterior hippocampal activity, showing that subjects with greater genetic risk have lower hippocampal activation. In Lieber sample (D), peak activation is at [30, −30, −18] in right posterior hippocampal-parahippocampal region with Z = 4.10, P < 0.001 uncorrected (P = 0.016, FWE corrected within AAL bilateral hippo-parahippocampal mask). In the Bari sample (E), peak of association is at [30, −40, −8], with Z = 3.47, P < 0.001 uncorrected (P = 0.033, FWE corrected within AAL right posterior hippo-parahippocampal mask). In the DNS sample (F), peak of association is at [51, −29, −13], with Z = 2.89, P = 0.002 uncorrected. PRS6 accounted for approximately 8.23% of the variability in hippocampal activity in Lieber sample, and 12.82% and 5.71% for the Bari and DNS samples, respectively.
Region of interest analysis revealed that PRS6 had a significant negative relationship with the averaged BOLD responses within the right posterior hippocampus (r = −0.2787, P = 9.45 × 10−5) (Fig. 1D). Regression analysis showed PRS6 explained 8.23% of the variance in right hippocampal BOLD signal change.
When analysing the relationship between other PRSs and right hippocampal BOLD signal change, we found that PRS6 had the strongest association with hippocampal activation (Supplementary Table 4 and Supplementary Fig. 2), which is analogous to the findings of PRS6 having the strongest prediction accuracy of diagnosis in GWAS studies (Schizophrenia Working Group of the Psychiatric Genomics, 2014). Indeed, the stepwise increase in the strength of association of PRS from level one to level six closely parallels the pattern seen in prediction of case-control status (Schizophrenia Working Group of the Psychiatric Genomics, 2014).
Replication
We replicated the negative association between PRS6 and memory encoding activity within the right posterior hippocampal-parahippocampal AAL mask in both the Bari sample (peak voxel: 30, −40, −8; Z = 3.47; P = 0.033, FWE corrected within mask, Fig. 1B) and the DNS sample (peak voxel: 51, −29, −13; Z = 2.89, P = 0.002 uncorrected, Fig. 1C). PRS6 scores accounted for approximately 12.82% and 5.71% of the variability in hippocampal activity for the Bari and DNS samples, respectively (Fig. 1E and F).
Discussion
In the present work, we found that the PRS derived from the most recent schizophrenia GWAS is associated with decreased hippocampal activity during memory encoding in three independent samples of healthy participants. We believe these results serve to advance our understanding of both schizophrenia PRS and schizophrenia-associated neuroimaging intermediate phenotypes. While previous studies have indicated a role of the additive effect of risk alleles in schizophrenia susceptibility (Schizophrenia Working Group of the Psychiatric Genomics, 2014; Agerbo et al., 2015; Power et al., 2015), they have offered little insight into potential effects of this cumulative risk at the level of brain mechanisms. Our main finding of the association between PRS and reduced hippocampal activity during memory encoding in healthy participants suggests, firstly, an aggregate neurobiological effect of schizophrenia risk alleles on hippocampal function and, second, that this effect is a potential neural mechanism mediating genetic risk. More specifically, our findings indicate that the cumulative effect of risk alleles detected by the most recent schizophrenia GWAS may converge on a pattern of neural activity that has been associated with schizophrenia diagnosis, with familial risk for schizophrenia (Rasetti et al., 2014) and with the putative prodromal state (Allen et al., 2011). Since we found that higher PRS, i.e. higher genomic risk for schizophrenia, is associated with decreased hippocampal activity, our results are consistent with previous studies reporting decreased hippocampal-parahippocampal recruitment during declarative memory tasks in patients with schizophrenia (Achim and Lepage, 2005; Rasetti et al., 2014) and in their healthy siblings (Rasetti et al., 2014), and in individuals at high clinical risk for schizophrenia (Allen et al., 2011).
Importantly, while earlier findings of a putative intermediate phenotype in individuals of increased genetic risk suggested that it might be a genetic risk-associated characteristic, this prior evidence alone did not establish a genetic basis for their origin above and beyond the shared familial and other environmental factors, even if the phenotype is heritable. To our knowledge, no functional MRI study has assessed the heritability of hippocampal activity during memory encoding; however, there is evidence that the behavioural aspects of the task are heritable (Glahn et al., 2007; Owens et al., 2011). We found that genomic risk for schizophrenia predicts a schizophrenia risk-associated pattern of hippocampal activity in healthy participants, thus avoiding the potential role of confounding factors associated with the experience/familiarity and treatment of the disorder. We, therefore, propose that hippocampal activity during memory encoding is a genetically-mediated intermediate phenotype of schizophrenia risk, which is linked to the polygenic architecture of the clinical aspects of the disorder.
It is noteworthy that PRS6 explained about 8% of variance in hippocampal activity. Logically, while most variance of hippocampal activity during memory encoding is not related to PRS6, as other genetic and environmental factors also likely contribute to hippocampal memory function, the degree of prediction by schizophrenia genomic risk is substantial, considering that the same risk profile predicts ∼7% of liability for the clinical disorder (Schizophrenia Working Group of the Psychiatric Genomics, 2014). It is also likely that in patients with schizophrenia, due to the diverse epiphenomena of the illness state (e.g. medication, ongoing symptoms), a smaller fraction of variance in hippocampal function would be found related to genetic risk in ill subjects. Nevertheless, our results showing that a component of mnemonic hippocampal activity is related to schizophrenia genomic risk in three independent healthy samples instantiates that at least a component of the genetically determined pathophysiological architecture of schizophrenia is based on altered hippocampal function.
Our report is not the first to explore the relationship between schizophrenia PRS and functional MRI-based physiology. Previous studies have detected a relationship between polygenic scores for schizophrenia and prefrontal cortex activation during working memory (Walton et al., 2013, 2014; Kauppi et al., 2015), another putative intermediate phenotype associated with schizophrenia. However, these studies have adopted a measure of polygenic risk for schizophrenia that was based on SNPs selected from the literature (Walton et al., 2013) or on smaller GWAS samples (Walton et al., 2014; Kauppi et al., 2015) in comparison with the basis of PRS in our report, which is the most recent and largest schizophrenia GWAS (Schizophrenia Working Group of the Psychiatric Genomics, 2014). Moreover, these studies found a relationship between PRS and prefrontal activation in combined samples of healthy volunteers and patients with schizophrenia (Walton et al., 2013, 2014; Kauppi et al., 2015), which makes it impossible to exclude potential effects on brain function of factors related to the active illness state (e.g. treatment, symptoms, smoking, and general health issues). A recent study (Erk et al., 2017) analysed the relationship between PRS1—which is calculated from a small number of GWAS-significant alleles (e.g. 106 in our data)—and brain activation during working memory, reward processing, episodic memory, social cognition and emotion processing. This study reported association of PRS1 only with perigenual anterior cingulate cortex activation during the memory recognition phase of an associative episodic memory task. These effects were not replicated and the authors concluded that their finding has limited validity for intermediate phenotype characterization. To note, we also did not find a significant consistent association between PRS1 and hippocampal activity during memory encoding (Supplementary Table 4 and Supplementary Fig. 2), in contrast to the significant association that we found with PRS6. It is noteworthy that PRS1 alone also shows limited prediction of case status (Schizophrenia Working Group of the Psychiatric Genomics, 2014).
Further, we found that the different PRSs, constructed from alleles with schizophrenia association at different thresholds of significance, show different prediction accuracy of hippocampal activity during memory encoding (Supplementary Figs 2–4). The results of variance analysis in the Lieber and Bari samples are remarkably consistent with previous evidence highlighting PRS6 as the risk score with the highest prediction accuracy (Schizophrenia Working Group of the Psychiatric Genomics, 2014) of schizophrenia case status and showing that PRS derived from a smaller number of SNPs selected based on a more significant threshold of association with schizophrenia (PRS1–5) have a lower prediction accuracy compared with PRS6. Above PRS6, the PRSs constructed with a greater number of SNPs (PRS7 to 10) do not explain more variance in hippocampal activity compared with PRS6, echoing evidence showing that PRS7–10 also do not explain more variance in schizophrenia liability, compared with PRS6 (Schizophrenia Working Group of the Psychiatric Genomics, 2014). These results also support the assumption that the loci represented in the PRS6 threshold contain most if not all true schizophrenia risk genes, even though most of these loci individually do not achieve GWAS significance with current sample sizes. On the other hand, in the DNS sample, the variance of hippocampal activation explained by the different PRSs showed a different pattern. This may be due to different characteristics of the population (e.g. the age range) and suggests that in different samples the cumulative effects of PRSs on hippocampal activity may be driven by different SNPs.
One of the limitations of the current study is the blocked design of the functional MRI tasks, which limits the ability to discriminate the activity associated with successful encoding from that related to unsuccessful encoding. Future studies using an event-related design may help differentiate these two processes in greater detail. For this reason, we included only participants with retrieval accuracy above 50%, so that the potential confounding effect because of differences in successful and unsuccessful encoding was reduced. We believe this study design is appropriate to evaluate variance of hippocampal activity contributed by genetic risk. Concerning the relationship between performance and hippocampal activity, we detected a negative association between retrieval accuracy (d′) and hippocampal activation (Supplementary Fig. 7). The small variance in performance in this sample and the inclusion of only individuals with high behavioural performance requires caution in interpreting these results. However, we believe that they are consistent with the possibility that, in controls, compensatory mechanisms exist, so that individuals with lower activation and high genetic risk are still able to perform well.
It is also necessary to mention that, while the intermediate phenotype that we studied is known to be essentially related to memory encoding (Stern et al., 1996; Hariri et al., 2003; Bertolino et al., 2006, 2008; Di Giorgio et al., 2013; Rasetti et al., 2014), it is likely more heterogeneous, since we cannot exclude potential confounding effects of other brain functions related to working memory and decision-making, especially in the second replication sample, given the characteristic of the distractor task. In this regard, while from one point of view the use of different functional MRI paradigms represents another potential limitation of this study, we also believe that the consistent association between genetic risk for schizophrenia and hippocampal activation during two different task variants of the same basic declarative memory function strengthens our conclusion that risk alleles for schizophrenia may cumulatively affect the physiology of this brain region during this form of information processing. Another limitation is that PRSs were calculated based on the weighted linear combination of independent SNPs, which does not consider gene-by-gene interaction (epistasis) or even more complicated gene network architectures. Further studies using non-linear approaches may be critical to examine how multiple SNPs can interact to affect hippocampal function. Finally, the genes influenced by the SNPs included in PRS may not be directly related to hippocampal function. We cannot exclude whether the relationship between PRS and hippocampal activity may be mediated by other genetic factors and brain regions. The study of gene networks and brain connectivity networks related to mnemonic hippocampal function may help to answer this question.
In conclusion, we found a replicable negative association between PRS, derived from the most recent and largest schizophrenia GWAS, and hippocampal activity during memory encoding in healthy participants, thus arguing that the cumulative effect of polygenic risk alleles converges on this pattern of neural activity associated with schizophrenia risk and vulnerability. It is tempting to speculate that the use of GWAS-derived polygenic scores and hippocampal intermediate phenotypes could aid in identifying subgroups of individuals with high risk for schizophrenia who could benefit from treatments specifically designed to improve hippocampal-associated memory function.
Supplementary Material
Acknowledgements
We are grateful to the Lieber and Maltz families for their visionary support that funded the analytic work of this project. We thank all the participants in the study and their families. We thank Annabella Di Giorgio, Leonardo Fazio, Roberta Rasetti, Paolo Taurisano, for help with data acquisition. We thank the Psychiatry Genomics Consortium for providing the statistics for PRS calculation.
Funding
The collection of the genetic data for the American samples was supported by direct funding from the Intramural Research Program of the NIMH to the Clinical Brain Disorders Branch (DR Weinberger, PI, protocol 95-M-0150, NCT00001486, annual report number: ZIA MH002942-03 CTNB). The analytic work was funded by the Lieber Institute for Brain Development.
The Duke Neurogenetics Study received support from Duke University as well as US-National Institutes of Health grants R01DA033369 and R01DA031579. ARK and ARH received further support from US-National Institutes of Health grant R01AG049789.
Analysis of European data was supported by a “Capitale Umano ad Alta Qualificazione” grant by Fondazione Con Il Sud and by the “Ricerca Finalizzata” (grant number: PE-2011-02347951) awarded to Alessandro Bertolino. Additionally, this project has received funding from the European Union Seventh Framework Programme for research, technological development and demonstration under grant agreement no. 602450. This paper reflects only the author's views and the European Union is not liable for any use that may be made of the information contained therein.
Supplementary material
Supplementary material is available at Brain online.
Glossary
Abbreviations
- BOLD
blood oxygenation level-dependent
- DNS
Duke Neurogenetics Study
- GWAS
genome-wide association study
- PGC
Psychiatric Genetics Consortium
- PRS
polygenic risk score
- SNP
single nucleotide polymorphism
References
- Achim AM, Lepage M. Episodic memory-related activation in schizophrenia: meta-analysis. Br J Psychiatry 2005; 187: 500–9. [DOI] [PubMed] [Google Scholar]
- Agerbo E, Sullivan PF, Vilhjalmsson BJ, Pedersen CB, Mors O, Borglum AD, et al. Polygenic risk score, parental socioeconomic status, family history of psychiatric disorders, and the risk for schizophrenia: a danish population-based study and meta-analysis. JAMA Psychiatry 2015; 72: 635–41. [DOI] [PubMed] [Google Scholar]
- Allen P, Seal ML, Valli I, Fusar-Poli P, Perlini C, Day F, et al. Altered prefrontal and hippocampal function during verbal encoding and recognition in people with prodromal symptoms of psychosis. Schizophr Bull 2011; 37: 746–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association Diagnostic and statistical manual of mental disorders: DSM-IV-TR. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- Bertolino A, Di Giorgio A, Blasi G, Sambataro F, Caforio G, Sinibaldi L, et al. Epistasis between dopamine regulating genes identifies a nonlinear response of the human hippocampus during memory tasks. Biol Psychiatry 2008; 64: 226–34. [DOI] [PubMed] [Google Scholar]
- Bertolino A, Nawroz S, Mattay VS, Barnett AS, Duyn JH, Moonen CT, et al. Regionally specific pattern of neurochemical pathology in schizophrenia as assessed by multislice proton magnetic resonance spectroscopic imaging. Am J Psychiatry 1996; 153: 1554–63. [DOI] [PubMed] [Google Scholar]
- Bertolino A, Rubino V, Sambataro F, Blasi G, Latorre V, Fazio L, et al. Prefrontal-hippocampal coupling during memory processing is modulated by COMT val158met genotype. Biol Psychiatry 2006; 60: 1250–8. [DOI] [PubMed] [Google Scholar]
- Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 1996; 29: 162–73. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381: 1371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods 2012; 9: 179–81. [DOI] [PubMed] [Google Scholar]
- Demirkan A, Penninx BW, Hek K, Wray NR, Amin N, Aulchenko YS, et al. Genetic risk profiles for depression and anxiety in adult and elderly cohorts. Mol Psychiatry 2011; 16: 773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derks EM, Vorstman JA, Ripke S, Kahn RS, Schizophrenia Psychiatric Genomic Consortium, Ophoff RA. Investigation of the genetic association between quantitative measures of psychosis and schizophrenia: a polygenic risk score analysis. PloS One 2012; 7: e37852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio A, Gelao B, Caforio G, Romano R, Andriola I, D'Ambrosio E, et al. Evidence that hippocampal-parahippocampal dysfunction is related to genetic risk for schizophrenia. Psychol Med 2013; 43: 1661–71. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Eichenbaum H. The episodic memory system: neurocircuitry and disorders. Neuropsychopharmacology 2010; 35: 86–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erk S, Mohnke S, Ripke S, Lett TA, Veer IM, Wackerhagen C, et al. Functional neuroimaging effects of recently discovered genetic risk loci for schizophrenia and polygenic risk profile in five RDoC subdomains. Transl Psychiatry 2017; 7: e997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Almasy L, Blangero J, Burk GM, Estrada J, Peralta JM, et al. Adjudicating neurocognitive endophenotypes for schizophrenia. Am J Med Genet B Neuropsychiatr Genet 2007; 144B: 242–9. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Shields J. A polygenic theory of schizophrenia. Proc Natl Acad Sci USA 1967; 58: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, et al. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci 2003; 23: 6690–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry 2005; 10: 40–68; image 5. [DOI] [PubMed] [Google Scholar]
- Heinrichs RW, Zakzanis KK. Neurocognitive deficit in schizophrenia: a quantitative review of the evidence. Neuropsychology 1998; 12: 426–45. [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011; 1: 457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium, Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Cacioppo JT, Lang PJ. Eliciting affect using the international affective picture system: trajectories through evaluative space. Pers Soc Psychol Bull 1998; 24: 855–79. [Google Scholar]
- Kauppi K, Westlye LT, Tesli M, Bettella F, Brandt CL, Mattingsdal M, et al. Polygenic risk for schizophrenia associated with working memory-related prefrontal brain activation in patients with schizophrenia and healthy controls. Schizophr Bull 2015; 41: 736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang PJ, Bradley MM, Cuthbert BN. International Affective Picture System (IAPS): Technical Manual and Affective Ratings. Gainesville, FL: NIMH Center for the Study of Emotion and Attention, University of Florida; 1997. [Google Scholar]
- Lipska BK. Using animal models to test a neurodevelopmental hypothesis of schizophrenia. J Psychiatry Neurosci 2004; 29: 282–6. [PMC free article] [PubMed] [Google Scholar]
- Lipska BK, Weinberger DR. To model a psychiatric disorder in animals: schizophrenia as a reality test. Neuropsychopharmacology 2000; 23: 223–39. [DOI] [PubMed] [Google Scholar]
- Luciano M, Huffman JE, Arias-Vasquez A, Vinkhuyzen AA, Middeldorp CM, Giegling I, et al. Genome-wide association uncovers shared genetic effects among personality traits and mood states. Am J Med Genet B Neuropsychiatr Genet 2012; 159B: 684–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Gottesman II, Rao DC. Resolving genetic models for the transmission of schizophrenia. Genet Epidemiol 1985; 2: 99–110. [DOI] [PubMed] [Google Scholar]
- Middeldorp CM, de Moor MH, McGrath LM, Gordon SD, Blackwood DH, Costa PT, et al. The genetic association between personality and major depression or bipolar disorder: a polygenic score analysis using genome-wide association data. Transl Psychiatry 2011; 1: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien JL, O'Keefe KM, LaViolette PS, DeLuca AN, Blacker D, Dickerson BC, et al. Longitudinal fMRI in elderly reveals loss of hippocampal activation with clinical decline. Neurology 2010; 74: 1969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens SF, Picchioni MM, Rijsdijk FV, Stahl D, Vassos E, Rodger AK, et al. Genetic overlap between episodic memory deficits and schizophrenia: results from the Maudsley Twin Study. Psychol Med 2011; 41: 521–32. [DOI] [PubMed] [Google Scholar]
- Persson J, Pudas S, Lind J, Kauppi K, Nilsson LG, Nyberg L. Longitudinal structure-function correlates in elderly reveal MTL dysfunction with cognitive decline. Cereb Cortex 2012; 22: 2297–304. [DOI] [PubMed] [Google Scholar]
- Power RA, Steinberg S, Bjornsdottir G, Rietveld CA, Abdellaoui A, Nivard MM, et al. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Meurosci 2015; 18: 953–5. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasetti R, Mattay VS, White MG, Sambataro F, Podell JE, Zoltick B, et al. Altered hippocampal-parahippocampal function during stimulus encoding: a potential indicator of genetic liability for schizophrenia. JAMA Psychiatry 2014; 71: 236–47. [DOI] [PubMed] [Google Scholar]
- Rasetti R, Weinberger DR. Intermediate phenotypes in psychiatric disorders. Curr Opin Genet Dev 2011; 21: 340–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet 2011; 43: 969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Wegmann D, Trynka G, Gutierrez-Achury J, Do R, Voight BF, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet 2012; 44: 483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern CE, Corkin S, Gonzalez RG, Guimaraes AR, Baker JR, Jennings PJ, et al. The hippocampal formation participates in novel picture encoding: evidence from functional magnetic resonance imaging. Proc Natl Acad Sci USA 1996; 93: 8660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF. The genetics of schizophrenia. PLoS Med 2005; 2: e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulkin R, Whitington T, Eklund M, Aly M, Eeles RA, Easton D, et al. Prediction of individual genetic risk to prostate cancer using a polygenic score. Prostate 2015; 75: 1467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng KY, Chambers RA, Lipska BK. The neonatal ventral hippocampal lesion as a heuristic neurodevelopmental model of schizophrenia. Behav Brain Res 2009; 204: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002; 15: 273–89. [DOI] [PubMed] [Google Scholar]
- Walton E, Geisler D, Lee PH, Hass J, Turner JA, Liu J, et al. Prefrontal inefficiency is associated with polygenic risk for schizophrenia. Schizophr Bull 2014; 40: 1263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton E, Turner J, Gollub RL, Manoach DS, Yendiki A, Ho BC, et al. Cumulative genetic risk and prefrontal activity in patients with schizophrenia. Schizophr Bull 2013; 39: 703–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Visscher PM, Wray NR. Sporadic cases are the norm for complex disease. Eur J Hum Genet 2010; 18: 1039–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineh MM, Engel SA, Thompson PM, Bookheimer SY. Dynamics of the hippocampus during encoding and retrieval of face-name pairs. Science 2003; 299: 577–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.