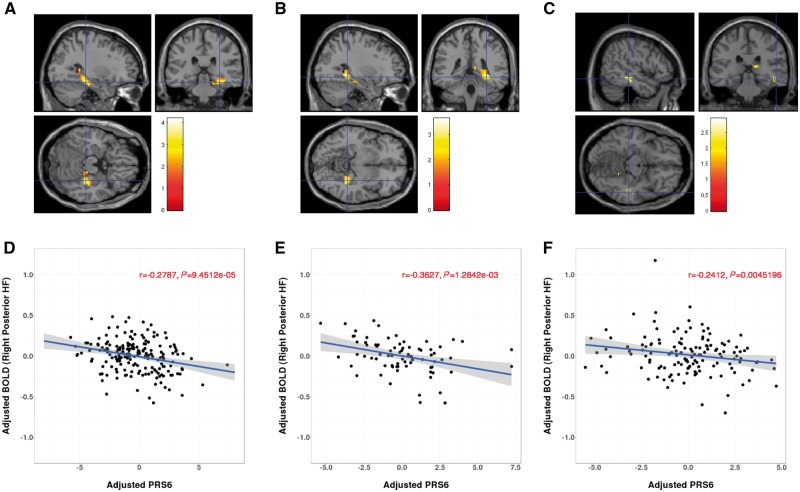
Figure 1.
Association between PRS6 and hippocampus activation during memory encoding in the Lieber discovery sample (A and D), and in Bari (B and E) and DNS (C and F) replication samples. (A–C) Section views (sagittal, coronal and transversal views) of the relationship between PRS6 and functional MRI hippocampal activity during neutral encoding in Lieber sample (A), Bari sample (B), and DNS sample (C), respectively. Genetic risk for schizophrenia is associated with right hippocampal activation (P < 0.05, uncorrected for illustration). (D–F) Scatter plot of the relationship between PRS6 and right posterior hippocampal activity, showing that subjects with greater genetic risk have lower hippocampal activation. In Lieber sample (D), peak activation is at [30, −30, −18] in right posterior hippocampal-parahippocampal region with Z = 4.10, P < 0.001 uncorrected (P = 0.016, FWE corrected within AAL bilateral hippo-parahippocampal mask). In the Bari sample (E), peak of association is at [30, −40, −8], with Z = 3.47, P < 0.001 uncorrected (P = 0.033, FWE corrected within AAL right posterior hippo-parahippocampal mask). In the DNS sample (F), peak of association is at [51, −29, −13], with Z = 2.89, P = 0.002 uncorrected. PRS6 accounted for approximately 8.23% of the variability in hippocampal activity in Lieber sample, and 12.82% and 5.71% for the Bari and DNS samples, respectively.