Abstract
Either of the first two introns of the Arabidopsis tryptophan pathway gene PAT1 elevates mRNA accumulation from a PAT1:β-glucuronidase (GUS) fusion roughly 5-fold without affecting the rate of PAT1:GUS transcription. To further explore the mechanism of this intron-mediated enhancement of gene expression, we wanted to determine whether splicing or specific intron sequences were necessary. In-frame derivatives of PAT1 intron 1, whose splicing was prevented by a point mutation or large deletions, were able to increase mRNA accumulation from a PAT1:GUS fusion, demonstrating that splicing per se is not required. Furthermore, each of a series of introns containing overlapping deletions that together span PAT1 intron 1 increased PAT1:GUS mRNA accumulation as much as the full-length intron did, indicating that all intron sequences are individually dispensable for this phenomenon. These results eliminate the simple idea that this intron stimulates mRNA accumulation via a unique RNA-stabilizing sequence or through the completed act of splicing. However, they are consistent with a possible role for redundant intron sequence elements or an association of the pre-mRNA with the spliceosome.
Introns are documented in many cases to have a large positive effect on gene expression in a broad range of organisms including nematodes, insects, and mammals (Buchman and Berg, 1988; Chung and Perry, 1989; Meredith and Storti, 1993; Okkema et al., 1993). In plants, the inclusion of one or more introns in a gene construct usually leads to increased accumulation of mRNA and protein relative to similar fusions that lack introns (for review, see Koziel et al., 1996; Simpson and Filipowicz, 1996). This effect has been termed intron-mediated enhancement (IME) of gene expression (Mascarenhas et al., 1990).
Introns known to stimulate expression in monocots include those from the maize Adh1, Sh1, Bz1, Hsp82, actin, and GapA1 genes (Callis et al., 1987; Luehrsen and Walbot, 1991; Maas et al., 1991; Sinibaldi and Mettler, 1992; Donath et al., 1995) and the rice salT, Act1, and tpi genes (McElroy et al., 1990; Xu et al., 1994; Rethmeier et al., 1997). Similarly, dicot introns that elevate expression include those from the petunia rbcS gene SSU301 (Dean et al., 1989), the potato ST-LS1 gene (Leon et al., 1991), and the Arabidopsis UBQ3, UBQ10, PAT1, atpk1, A1 EF-1α, and At eEF-1β genes (Curie et al., 1993; Norris et al., 1993; Zhang et al., 1994; Gidekel et al., 1996; Rose and Last, 1997). The magnitude of IME can be more than 100-fold (Maas et al., 1991; Zhang et al., 1994), but is more commonly in the range of 2- to 10-fold and is typically larger in monocots than dicots. While widespread, the phenomenon of IME is not universal. Plant introns that apparently fail to elevate expression include those from the bean phaseolin gene (Chee et al., 1986) and the maize Hsp81 gene (Sinibaldi and Mettler, 1992), and many genes lack introns.
Very little is known about the mechanism of IME, and different introns might affect expression by different means. However, some common features have emerged from those cases in which the mechanisms have been explored. In plants, the available evidence indicates that introns act post-transcriptionally to increase mRNA accumulation, presumably by facilitating mRNA maturation or enhancing the stability of nascent transcripts. Indirect support for a post-transcriptional role is provided by the findings that introns must be contained within transcribed sequences and in the proper orientation to elevate gene expression (Callis et al., 1987; Mascarenhas et al., 1990; Clancy et al., 1994; Donath et al., 1995), unlike transcriptional enhancers, which are usually position and orientation independent. Furthermore, a nuclear mode of action for IME is suggested in one case by the observation that the rice salT intron elevates cat gene expression but has no effect on the cytoplasmic stability of cat mRNA in cultured maize cells (Rethmeier et al., 1997). More directly, introns were shown to increase mRNA accumulation without significantly affecting the transcription rate of the petunia rbcS gene SSU301 in tobacco (Dean et al., 1989). Similarly, fusions of the β-glucuronidase (GUS) gene to the first or third exon of the Arabidopsis PAT1 gene (which encodes the Trp pathway enzyme phosphoribosylanthranilate transferase) are transcribed at comparable rates, even though only the fusions to the third exon accumulate PAT1:GUS mRNA to appreciable levels (Rose and Last, 1997). Either of the first two PAT1 introns increases GUS activity in transgenic lines containing PAT1:GUS fusions roughly 5-fold relative to an otherwise identical fusion lacking introns (Rose and Last, 1997), strongly suggesting that these introns stimulate mRNA accumulation by a post-transcriptional mechanism.
Another approach that has been taken to help define the mechanism of IME is to ask which features of an intron are required. Several deletion derivatives of introns from the maize Adh1 and Sh1 genes can still enhance expression (Clancy et al., 1994; Luehrsen and Walbot, 1994), suggesting that most intron sequences are dispensable for IME. However, even though these deletions are large, they do not include all parts of the intron, leaving open the possibility that a sequence element needed for IME is present in each of the deletion-containing introns tested.
The role of intron splicing in IME has been investigated using two monocot introns. Large internal deletions of the first intron of the maize Adh1 gene that strongly reduced splicing also impaired the expression-enhancing effect of this intron (Luehrsen and Walbot, 1994). Similarly, when the maize Hsp82 intron was rendered unspliceable by a point mutation at the 5′ or 3′ splice site, the ability of that intron to enhance expression was also lost (Sinibaldi and Mettler, 1992).
While the above data are consistent with the hypothesis that IME requires intron splicing, an alternative hypothesis is also possible. If an intron contains sequences such as a stop codon, which would interfere with translation, mRNAs that retain the intron might be destabilized, obscuring the ability of the unspliced intron to stimulate mRNA accumulation. Transcripts containing premature stop codons are often rapidly degraded by the process of nonsense-mediated RNA decay (Sullivan and Green, 1993). Therefore, the need for splicing in IME can still be considered unresolved.
To explore potential mechanisms of IME in dicots, we characterized the effects of introns on the expression of a PAT1:GUS reporter gene in transgenic Arabidopsis plants. Here we show that either of the first two PAT1 introns elevates steady-state levels of PAT1:GUS mRNA without significantly affecting the rate of transcription, clearly establishing that these introns increase expression by a post-transcriptional mechanism. The intron features required for this enhancement were further investigated using the first PAT1 intron. To eliminate the possibility that nonsense-mediated RNA decay could influence the results, this intron was modified to maintain the reading frame of the adjacent exons of a PAT1:GUS fusion so that translation could proceed in the absence of splicing. Using mutated derivatives of this in-frame intron, we found that neither splicing nor unique intron sequences are required to increase mRNA accumulation.
MATERIALS AND METHODS
Creating Sequence Changes
All sequence changes were generated by PCR amplification using primers containing the desired alterations. Each amplified DNA was cloned and sequenced to confirm the introduction of only the intended mutations. First, a PstI site was created immediately upstream of PAT1 intron 1 and the reading frame of the intron was adjusted. This in-frame intron was subsequently used as the template for creating all other mutant introns. A PstI site into which modified introns could be inserted was introduced at the PAT1 exon 1:exon 2 junction in a 309-bp HindIII-XbaI fragment, which was then replaced into the larger context of a PAT1:GUS fusion by conventional cloning (Rose and Last, 1997).
Transformations
All PAT1:GUS fusions were inserted as KpnI fragments into the binary vector pEND4K (Klee et al., 1985) in the same orientation as pAR209 (Rose and Last, 1997). The resulting plasmids were transformed first into Agrobacterium tumefaciens strain GV3101 (pMP90) (Koncz and Schell, 1986) by electroporation and then into Arabidopsis ecotype Columbia by vacuum transformation (Bechtold et al., 1993). Transformed T1 seedlings were selected on medium containing 50 mg/L kanamycin. Blots of genomic DNA from T2 plants were probed with the GUS gene to identify lines that contain a single transgene insertion. The T3 progeny of several self-pollinated individuals from each single-copy line were screened for kanamycin resistance to identify homozygotes.
Expression Analysis
RNA gel blots of steady-state mRNA levels in leaf tissue of 3-week-old homozygous T3 lines were prepared and probed as described by Radwanski et al. (1995) with a 2.2-kb GUS fragment and a 1.0-kb PAT1 cDNA fragment spanning exons 4 through 9. Nuclear run-on transcription assays were performed as described previously (Rose and Last, 1997). All quantitation was with a phosphor imager (Storm model 860, Molecular Dynamics, Sunnyvale, CA) using ImageQuant software (Molecular Dynamics).
Splicing Efficiency
Total RNA isolated from transgenic lines was digested with RQ1 RNAse-free DNAse1 (Promega, Madison, WI). Reverse transcription (RT) using primers specific for GUS RNA was performed with a RT system from Promega. The resulting cDNA was used as the template for PCR using a PAT1 exon 1 primer and a primer that anneals to GUS upstream of the RT primers. For comparison, the splicing efficiency of the first intron from the endogenous PAT1 mRNA was estimated by reverse-transcribing and PCR-amplifying RNA from untransformed plants using PAT1 primers. The amplification was for 25 cycles of 94°C for 1 min, 50°C for 1 min, 72°C for 2 min, which preliminary tests indicated was within the linear range (not shown). The relative quantity of spliced and unspliced RNA was estimated by probing blots of 3% (w/v) agarose gels (NuSieve, FMC, Rockland, ME) containing the PCR reactions with an equimolar mixture of 167-bp HindIII-PstI exon 1 and 142-bp PstI-XbaI exon 2 probes. RT-PCR reactions from representative lines containing each construct were amplified for 35 cycles as above, and the products were sequenced directly.
RESULTS
PAT1 Introns Increase Expression Post-Transcriptionally
The GUS activity in transgenic lines containing PAT1:GUS fusions is increased approximately 5-fold by either of the first two PAT1 introns (Rose and Last, 1997). To directly test the hypothesis that introns affect expression by a post-transcriptional mechanism and to rule out the possibility that the differences in GUS activity between lines were due to position effects or variations in transgene copy number, transcription rates and mRNA accumulation were measured in several homozygous, single-copy transgenic lines containing fusions that differed only in the presence or absence of an intron.
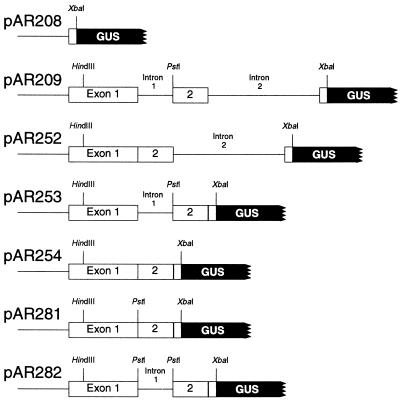
All of the constructs shown in Figure 1 are translational fusions of GUS to the same position in PAT1 exon 3 except pAR208, in which GUS is fused to exon 1. The plasmids pAR209, pAR252, pAR253, and pAR254 differ only in the introns they contain; each encodes an identical protein, they all have the same 2.4-kb PAT1 promoter, and each intron is in its natural location. The construction of these fusions has been described (Rose and Last, 1997), and single-copy transformants were identified by genomic DNA blots from among the previously reported lines.
Figure 1.
Structural details of PAT1:GUS fusions. Thin lines show the PAT1 promoter and introns, white boxes are the PAT1 exons, and the black box is the GUS gene. All other fusions containing modified introns have the structure shown for pAR282.
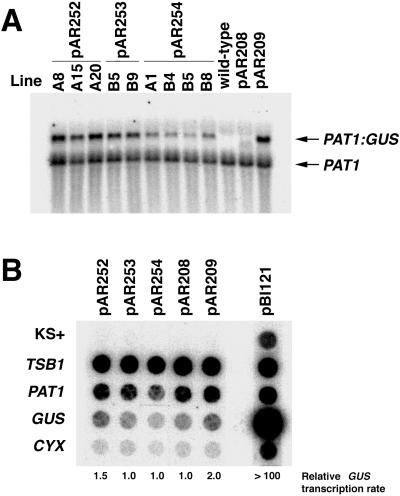
Constructs containing either the first or second PAT1 intron (pAR253 and pAR252, respectively) yielded three to five times more steady-state PAT1:GUS mRNA in homozygous single-copy lines than the comparable fusion lacking introns (pAR254, Fig. 2A), which is consistent with the previously observed 5-fold difference in GUS activity between these lines (Rose and Last, 1997). Fusions containing both introns 1 and 2 (pAR209) gave rise to slightly more PAT1:GUS mRNA than the constructs containing a single intron, although this effect was less than additive. The nuclear run-on transcription assays shown in Figure 2B indicate that the transgene is transcribed at virtually identical rates in lines containing pAR253, pAR254, and pAR208. (Compared with the other lines, pAR254 gave a proportionately weaker signal with all of the genes tested, indicating that less radiolabeled RNA was used in that hybridization.) While fusions containing intron 2 (pAR252 and pAR209) are apparently transcribed at modestly higher rates, these differences are insufficient to explain the differences in PAT1:GUS mRNA accumulation in these lines. Therefore, either of the first two PAT1 introns is capable of increasing mRNA accumulation by a post-transcriptional mechanism.
Figure 2.
Effect of PAT1 introns 1 and 2 on mRNA accumulation and transcription rate. A, RNA gel blot hybridized with GUS and a PAT1 cDNA fragment spanning exons 4 through 9. Each lane contains RNA from an independent single-copy transgenic line containing the PAT1:GUS fusion indicated. B, Nuclear run-on transcription assay. Radiolabeled RNA synthesized by nuclei isolated from the lines indicated along the top (pAR252 line A8, pAR253 line B9, and pAR254 line B4) was hybridized to filter strips on which the DNAs listed along the left had been affixed by dot blot. The relative GUS transcription rate shown is based on the radioactivity hybridized to the GUS DNA, relative to that in the pAR208 line, corrected for variations in RNA probe intensity using the PAT1 signal. The pBI121 positive control line contains the GUS gene driven by the strong cauliflower mosaic virus 35S promoter.
Modifying PAT1 Intron 1
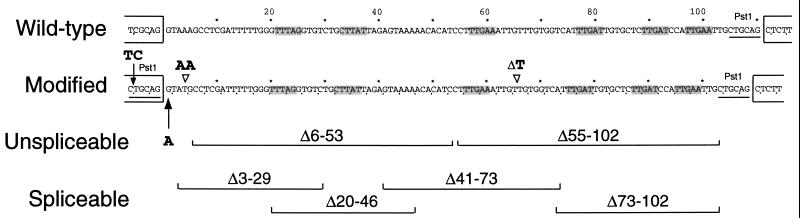
To further explore the intron features required to enhance mRNA accumulation, the first PAT1 intron was selected because it is small (110 nt) and easy to manipulate, yet stimulates expression as much as the larger intron 2. Three minor sequence modifications were made to facilitate the use of this intron in this and other expression studies. The first enables the isolation of the intron as a PstI restriction fragment. As shown in Figure 3, the last six nucleotides at the 3′ end of the intron form a natural PstI restriction enzyme site (CTGCAG), and the final six bases at the 3′ end of exon 1 differ from a PstI site only in the order of the first two nucleotides (TCGCAG). By reversing the order of these two nucleotides, a second PstI site was created immediately upstream of intron 1. This new site allowed the intron to be isolated as a PstI fragment, and provides a site in an intronless PAT1:GUS fusion into which modified introns can be returned to test their effect on expression in a virtually normal context. The PAT1:GUS constructs containing this new PstI site without or with intron 1 are pAR281 and pAR282, respectively (Fig. 1).
Figure 3.
DNA sequence of PAT1 intron 1 and derivatives. The sequence of the Columbia wild-type PAT1 intron 1 is shown, and the modifications to introduce a PstI site and to maintain reading frame are indicated. Adjacent exon sequences are in open boxes, PstI sites are underlined, codons are delineated by ticks, and putative branchpoint sequences are shaded. The 5′ splice site point mutation and the extent of the various deletions tested are indicated.
Two additional modifications were introduced to maintain the PAT1 reading frame so that retention of the intron in an unspliced mRNA would not prevent translation of the GUS reporter. Intron nucleotides 4 and 5 were replaced with a single T residue, and one of the three T residues that form bases 66 to 68 was deleted (see Fig. 3). These two minor changes destroyed one stop codon, placed the remaining seven stop codons in the intron out of frame, and shortened the intron to 108 nucleotides (precisely 36 codons). This in-frame intron was inserted into the PstI site of pAR281 to make pAR284, which is otherwise identical in structure to pAR282 (Fig. 1). The in-frame intron was the starting material for all of the derivatives described below, which were also inserted into pAR281 as PstI fragments.
For all of the constructs, differences in expression due to transgene copy number were eliminated by identifying lines homozygous for a single-copy transgene (data not shown). Expression was measured as the PAT1:GUS mRNA accumulation relative to that in the intronless fusion pAR254 line B4, the line with expression closest to the mean of the four single-copy pAR254 lines isolated. Splicing efficiency was estimated from the relative amounts of RT-PCR product derived from spliced and unspliced RNA. The results for all lines are summarized in Table I, and representative blots are shown in Figure 4.
Table I.
Summary of transgene expression and intron splicing
| PAT1:GUS Fusion | No. of Linesa | Relative PAT1:GUS mRNA Accumulationb
|
Percent Splicedc
|
||
|---|---|---|---|---|---|
| Mean | Range | Mean | Range | ||
| Intronless fusions | |||||
| pAR254 | 4 | 1.2 | 0.8–1.5 | ||
| pAR281 (+ PstI site) | 2 | 1.6 | 0.9–2.2 | ||
| With PAT1 intron 1 | |||||
| pAR253 | 2 | 4.6 | 4.2–4.8 | 94.9 | 94.4–95.3 |
| pAR282 (+ PstI site) | 1 | 5.6 | 95.6 | ||
| pAR284 (In-frame) | 2 | 4.0 | 3.9–4.1 | 93.2 | 93.1–93.3 |
| Spliceable introns | |||||
| Δ3-29 | 3 | 3.4 | 3.0–4.0 | 40.1 | 31.6–48.6 |
| Δ20-46 | 6 | 5.1 | 3.8–6.6 | 84.2 | 80.2–90.0 |
| Δ41-73 | 4 | 5.9 | 4.5–8.1 | 88.0 | 86.7–89.7 |
| Δ73-102 | 2 | 4.4 | 4.0–4.9 | 85.7 | 82.8–88.6 |
| Unspliceable introns | |||||
| Reverse | 2 | 0.6 | Both 0.6 | <2 | Both <2 |
| 5′ Splice site | 4 | 3.0 | 2.6–3.3 | <2 | All <2 |
| Δ6-53 | 1 | 2.6 | 38.5d | ||
| Δ55-102 | 4 | 4.5 | 3.5–6.0 | <2 | <2–2.5 |
The number of independent, homozygous, single-copy lines tested for each transgene.
The average amount of GUS-hybridizing mRNA in each line relative to pAR254 line B4, corrected for variations in loading using the endogenous PAT1 mRNA signal.
The average proportion of RT-PCR product derived from spliced mRNA.
Aberrantly spliced.
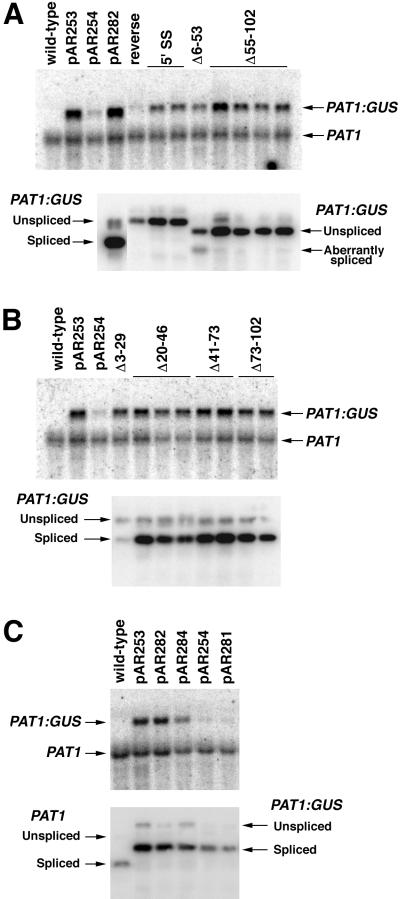
Figure 4.
The ability of PAT1 intron 1 and derivatives to stimulate PAT1:GUS mRNA accumulation and to be spliced. Total RNA was subjected to RNA gel-blot hybridization (top panels of A–C) and RT-PCR (bottom panels). Each lane represents an independent single-copy transgenic line, with the RNA and RT-PCR blots for each aligned. The RNA blots were probed with GUS and a PAT1 cDNA spanning exons 4 through 9. The lower panels show gel blots of the RT-PCR products hybridized with an equal mixture of PAT1 exon 1 and exon 2 probes. A, PAT1:GUS fusions containing unspliceable introns. The introns were inverted (reverse), had a point mutation at the 5′ splice site (5′ SS), or were too small to be spliced (Δ6-53 and Δ55-102). B, PAT1:GUS fusions containing spliceable introns. See Figure 3 for deletion details. C, Control PAT1:GUS fusions. The structure of each fusion is shown in Figure 1.
Splicing Is Not Required for IME
To determine if an intron could enhance expression in the absence of splicing, three mutations that abolish splicing were made in the in-frame PAT1 intron 1. In the first, the G of the conserved GU dinucleotide at the 5′ splice site was converted to an A (Unspliceable, Fig. 3). Similar mutations are known to eliminate intron splicing in several Arabidopsis genes (Brown, 1996). The other two mutations (Δ6-53 and Δ55-102) are large deletions of the 5′ or 3′ half that each reduce the size of the intron to 60 nucleotides, below the lower size limit of 70 to 73 nucleotides needed for intron splicing in plants (Goodall and Filipowicz, 1990).
RT-PCR analysis confirmed that these mutations prevent splicing. No RT-PCR product derived from properly spliced mRNA was observed from transgenic lines containing any of these mutated introns (Fig. 4A, bottom). However, in lines containing the 5′ intron deletion (Δ6-53), roughly one-third of the RT-PCR product was derived from aberrantly spliced RNA. This product is a result of splicing between the proper 5′ splice site and a cryptic 3′ splice site in exon 2, 19 bp downstream of the normal splice acceptor.
All of the “introns” rendered unspliceable by mutation were able to increase PAT1:GUS mRNA accumulation relative to the constructs lacking introns (Fig. 4A, top), although the magnitude of the enhancement is generally less than that mediated by the wild-type or in-frame intron (Table I). The wild-type intron is not spliced when inserted backwards in the PAT1:GUS fusion (reverse, Fig. 4A), presumably because it lacks a 5′ splice site in this orientation. In contrast to the other unspliced introns, this reversed intron not only failed to elevate PAT1:GUS mRNA accumulation, it reduced expression 2-fold relative to a construct lacking introns (Fig. 4A, top). The wild-type intron in this orientation would certainly abolish translation in the absence of splicing because it contains multiple stop codons and would cause a frame shift. Therefore, splicing is not necessary for PAT1 intron 1 to elevate mRNA accumulation, but this effect can only be seen when the unspliced sequences allow translation to proceed.
Intron Sequences Are Dispensable for IME
To determine whether specific intron sequences are required for PAT1 intron 1 to mediate an increase in mRNA accumulation, four small overlapping in-frame deletions that together spanned the intron were generated (Spliceable, Fig. 3). The introns containing deletions of nucleotides 20 to 46, 41 to 73, and 73 to 102 are each spliced nearly as efficiently as the wild-type intron. The splicing of the intron lacking nucleotides 3 to 29 was reduced so that the intron was retained in one-half to two-thirds of the PAT1:GUS mRNA (Fig. 4B, bottom), perhaps because the deletion creates a poor match to the 5′ splice site consensus sequence at the +4 and +5 positions (Brown et al., 1996). However, all four of these introns elevate PAT1:GUS mRNA accumulation as much as does the wild-type intron (Fig. 4B, top; Table I). The result that individual intron sequences between residues 3 and 102 are entirely dispensable for IME clearly demonstrates that if any intron sequences are involved in elevating mRNA accumulation, they must be redundant or nonspecific.
The Modifications Do Not Affect Splicing or IME
In addition to the deletions or point mutation designed to test the need for individual sequences and splicing, each of the above introns also contains the modifications to introduce a PstI site and to render the intron in-frame. To test the possibility that the ability of an intron to be spliced or to elevate mRNA accumulation is influenced by these minor modifications, the expression and intron splicing efficiency of several control fusions were compared. The wild-type intron flanked by PstI sites and the in-frame intron (fusions pAR282 and pAR284, respectively) both stimulate PAT1:GUS mRNA accumulation 4-fold or more relative to a fusion lacking introns, an effect very similar to that mediated by the wild-type intron in pAR253 (Fig. 4C, top). Furthermore, intronless PAT1:GUS fusions with or without a PstI site at the exon 1:exon 2 junction (pAR281 and pAR254, respectively) yield comparable amounts of PAT1:GUS mRNA (Fig. 4C, top). These controls demonstrate that the introduced PstI site does not significantly influence the expression of the PAT1:GUS reporter in the presence or absence of an intron, and that the modifications to eliminate stop codons have little or no effect on the ability of the intron to elevate mRNA accumulation.
RT-PCR was used to determine whether these minor sequence changes affected intron splicing. More than 93% of the RT-PCR product from lines containing the modified introns (pAR282 and pAR284) is derived from spliced mRNAs (Fig. 4C, bottom; Table I). A similar splicing efficiency was observed for the wild-type PAT1 intron 1 in either a PAT1:GUS fusion (94.9%, pAR253) or the endogenous PAT1 mRNA in wild-type plants (94.3%, Fig. 4C, bottom). Furthermore, sequencing of RT-PCR products revealed that these modified introns are precisely spliced using the normal donor and acceptor sites. Therefore, none of the mutations that alter exon sequences just upstream of the intron and render the intron in-frame affect either intron splicing or IME. This means that the deletions and splice site mutation must be solely responsible for any of the observed differences between the other introns tested.
DISCUSSION
We have shown that either of the first two PAT1 introns act post-transcriptionally to increase mRNA accumulation from a PAT1:GUS fusion in stably transformed Arabidopsis, and that neither specific intron 1 sequences nor splicing per se is required for this effect. While it is likely that many other introns will share these properties with PAT1 intron 1, the possibility cannot be ruled out that more than one mechanism of IME exists.
After copy number differences were eliminated, the expression of each PAT1:GUS fusion in independent lines was remarkably similar. For all of the fusions listed in Table I except one, the most highly expressing line accumulated less than twice as much PAT1:GUS mRNA as the least active line containing the same construct. This indicates that PAT1:GUS fusions are insensitive to position effects, as previously noted (Rose and Last, 1997); therefore, reliable results can be obtained from a relatively small number of independent single-copy lines.
Specific Sequences and Splicing Are Dispensable
The finding that individual PAT1 intron sequences between residues 3 and 102 are entirely dispensable for IME is consistent with previous findings that the first introns from the maize Adh1 and Sh1 genes can tolerate large deletions and still enhance expression (Clancy et al., 1994; Luehrsen and Walbot, 1994). If any PAT1 intron 1 sequences are involved in increasing mRNA accumulation, they must be non-specific or redundant. In light of this sequence plasticity, it is not surprising that splicing and IME are unaffected by the minor sequence alterations to introduce a PstI site adjacent to PAT1 intron 1 and to render this intron in-frame.
In contrast, the result that splicing of PAT1 intron 1 is unnecessary for IME was unexpected, because unspliceable derivatives of the first introns from the maize Adh1 (Luehrsen and Walbot, 1994) and Hsp82 (Sinibaldi and Mettler, 1992) genes fail to enhance expression. This apparent discrepancy can be resolved by the hypothesis that translation-blocking sequences in unspliced introns could destabilize mRNA in addition to preventing translation of the reporter whose expression is being monitored. Nonsense-mediated RNA decay might have caused the reduction in PAT1:GUS mRNA accumulation (relative to intron-lacking fusions) observed for the reversed wild-type PAT1 intron because the intron is not spliced and would abolish translation in this orientation. Similarly, the unspliceable deletion-containing derivatives of maize Adh1 intron 1 contain stop codons in all three reading frames (Dennis et al., 1984; Luehrsen and Walbot, 1994). Because the mutated introns were located near the 5′ end of the coding sequences, the stops they encode might be recognized as premature and trigger nonsense-mediated RNA decay. Even though the unspliceable Hsp82 introns were placed in the 5′ untranslated region of the gene (Sinibaldi and Mettler, 1992), they might interfere with translation of the downstream reporter because they contain at least one short open reading frame. Thus, the mRNA stabilizing effects of introns might be obscured by the destabilizing effects of early stop codons if any are present in the unspliced introns.
Possible Mechanisms of IME
Our results eliminate the simple models that PAT1 intron 1 contains a transcriptional enhancer element or that IME requires unique intron sequences or completed splicing. However, two general mechanisms for IME remain consistent with the ability of all of the mutated introns tested in this report to promote mRNA accumulation. One is that redundant sequence elements in introns could directly stabilize nascent transcripts, either by adopting a stable secondary structure or by providing targets for protective RNA binding factors such as oligo-U-binding proteins (Gniadkowski et al., 1996) or heterogenous nuclear ribonucleoproteins (Krecic and Swanson, 1999). An alternative model is that spliceosome assembly onto the introns in pre-mRNA could facilitate an association with enzymes involved in other aspects of RNA maturation known to stabilize mRNA.
Capping, polyadenylation, and export from the nucleus to the cytoplasm all increase mRNA stability, and examples from several species suggest an interconnection between these processes and splicing (Huang and Gorman, 1990; Niwa et al., 1990; Izaurralde et al., 1994, 1995; Lou et al., 1996; Minvielle-Sebastia and Keller, 1999), probably involving an interaction with RNA polymerase (for review, see Bentley, 1999). In this scenario, each of the mutated introns must retain the necessary features (such as U-richness and splice sites) to be recognized as an intron, even in the absence of splicing. The observation that unspliceable introns elevate PAT1:GUS expression to a lesser degree than the spliceable ones could indicate that splicing itself helps to stabilize mRNA or that the small size or altered 5′ splice site of the unspliceable introns reduces their resemblance to a functional intron.
Even though the GU to AU mutation at the PAT1 intron 5′ splice site completely eliminated the formation of spliced product, it may still allow an association of the pre-mRNA with the spliceosome. The first committed step of intron splicing is the binding of the U1 small nuclear ribonucleoprotein to the pre-mRNA via base pairing of the U1 small nuclear RNA with the 5′ splice site (Simpson and Filipowicz, 1996). In examples from Arabidopsis (Liu and Filipowicz, 1996), yeast (Newman et al., 1985), and mammals (Aebi et al., 1986), introns containing GU to AU mutations can undergo the first splicing reaction (cleavage at the 5′ splice site and intron lariat formation) but not the second (3′ splice site cleavage and exon joining). In these cases, point mutation of the 5′ terminal G must not prevent spliceosome assembly onto the pre-mRNA, perhaps because sufficient homology remains to allow annealing of the U1 small nuclear RNA with the unaffected 5′ splice site sequences. Partially spliced PAT1:GUS products were not detected in lines containing the 5′ splice site mutation, and at least some RNA uncleaved at the 5′ splice site must accumulate to give the unspliced RT-PCR product seen in Figure 4A. However, this does not rule out the presence of cleaved RNA because splicing intermediates might co-migrate with either the PAT1 or PAT1:GUS mRNAs and would not be amplified by RT-PCR.
Cloning Introns as PstI Fragments
The technique of flanking an intron with PstI sites demonstrated here with PAT1 intron 1 could have broad applications. The introduction of a PstI site greatly facilitated the manipulation of this intron without reducing its ability to be spliced or to elevate mRNA accumulation. Now PAT1 intron 1 can be inserted into a PstI site in any gene, thereby precisely introducing a complete natural intron with no extraneous sequences and without altering the reading frame. This may provide a means to increase the expression in plants of a transgene that otherwise lacks introns, such as a cDNA or bacterial gene, with potential scientific or agricultural benefit. It is also likely that other introns could be modified in a similar way to contain PstI sites at both ends without affecting splicing because the PstI recognition sequence (CTGCAG) is a perfect match with the consensus DNA sequences at both the 3′ splice site (NTGCAG) and the exon sequences adjacent to the 5′ splice site (NNNC/A AG, Brown et al., 1996). This approach is being used to further explore the intron requirements for IME, and may prove helpful in determining what proportion of plant genes need introns for full expression.
ACKNOWLEDGMENTS
We thank Linda Fritts for technical assistance, Dr. Irwin Segel for sharing laboratory space and equipment, and Drs. Judy Callis and Lesilee Rose for helpful comments on the manuscript.
Footnotes
This work was supported by the U.S. Department of Agriculture-National Research Initiatives Competitive Grants Program (grant no. 97353014392).
LITERATURE CITED
- Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986;47:555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser III Sci Vie. 1993;316:1194–1199. [Google Scholar]
- Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- Brown JWS. Arabidopsis intron mutations and pre-mRNA splicing. Plant J. 1996;10:771–780. doi: 10.1046/j.1365-313x.1996.10050771.x. [DOI] [PubMed] [Google Scholar]
- Brown JWS, Smith P, Simpson CG. Arabidopsis consensus intron sequences. Plant Mol Biol. 1996;32:531–535. doi: 10.1007/BF00019105. [DOI] [PubMed] [Google Scholar]
- Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- Chee PP, Klassy RC, Slightom JL. Expression of a bean storage protein ‘phaseolin minigene’ in foreign plant tissues. Gene. 1986;41:47–57. doi: 10.1016/0378-1119(86)90266-0. [DOI] [PubMed] [Google Scholar]
- Chung S, Perry R. Importance of introns for expression of mouse ribosomal protein gene rpL32. Mol Cell Biol. 1989;9:2075–2082. doi: 10.1128/mcb.9.5.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy M, Vasil V, Hannah LC, Vasil IK. Maize Shrunken-1 intron and exon regions increase gene expression in maize protoplasts. Plant Sci. 1994;98:151–161. [Google Scholar]
- Curie C, Axelos M, Bardet C, Atanassova R, Chaubet N, Lescure B. Modular organization and developmental activity of an Arabidopsis thaliana EF-1α gene promoter. Mol Gen Genet. 1993;238:428–436. doi: 10.1007/BF00292002. [DOI] [PubMed] [Google Scholar]
- Dean C, Favreau M, Bond-Nutter D, Bedbrook J, Dunsmuir P. Sequences downstream of translation start regulate quantitative expression of two petunia rbcS genes. Plant Cell. 1989;1:201–208. doi: 10.1105/tpc.1.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Gerlach WL, Pryor AJ, Bennetzen JL, Inglis A, Llewellyn D, Sachs MM, Ferl RJ, Peacock WJ. Molecular analysis of the alcohol dehydrogenase (Adh1) gene of maize. Nucleic Acids Res. 1984;12:3983–4000. doi: 10.1093/nar/12.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donath M, Mendel R, Cerff R, Martin W. Intron-dependent transient expression of the maize GapA1 gene. Plant Mol Biol. 1995;28:667–676. doi: 10.1007/BF00021192. [DOI] [PubMed] [Google Scholar]
- Gidekel M, Jimenez B, Herrera-Estrella L. The first intron of the Arabidopsis thaliana gene coding for elongation factor 1β contains an enhancer-like element. Gene. 1996;170:201–206. doi: 10.1016/0378-1119(95)00837-3. [DOI] [PubMed] [Google Scholar]
- Gniadkowski M, Hemmings-Mieszczak M, Klahre U, Liu H-X, Filipowicz W. Characterisation of intronic uridine-rich sequence elements acting as possible targets for nuclear proteins during pre-mRNA splicing in Nicotiana plumbaginifolia. Nucleic Acids Res. 1996;24:619–627. doi: 10.1093/nar/24.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall GJ, Filipowicz W. The minimum functional length of pre-mRNA introns in monocots and dicots. Plant Mol Biol. 1990;14:727–733. doi: 10.1007/BF00016505. [DOI] [PubMed] [Google Scholar]
- Huang MTF, Gorman CM. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzyn-kiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Klee HJ, Yanofsky MF, Nester EW. Vectors for transformation of higher plants. Biotechnology. 1985;3:637–642. [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Koziel MG, Carozzi NB, Desai N. Optimizing expression of transgenes with an emphasis on post-transcriptional events. Plant Mol Biol. 1996;32:393–405. doi: 10.1007/BF00039392. [DOI] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- Leon P, Planckaert F, Walbot V. Transient gene expression in protoplasts of Phaseolus vulgaris isolated from a cell suspension culture. Plant Physiol. 1991;95:968–972. doi: 10.1104/pp.95.3.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H-X, Filipowicz W. Mapping of branchpoint nucleotides in mutant pre-mRNAs expressed in plant cells. Plant J. 1996;9:381–389. doi: 10.1046/j.1365-313x.1996.09030381.x. [DOI] [PubMed] [Google Scholar]
- Lou H, Gagel RF, Berget SM. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- Luehrsen KR, Walbot V. Intron enhancement of gene expression and the splicing efficiency of introns in maize cells. Mol Gen Genet. 1991;225:81–93. doi: 10.1007/BF00282645. [DOI] [PubMed] [Google Scholar]
- Luehrsen KR, Walbot V. Addition of A- and U-rich sequence increases the splicing efficiency of a deleted form of a maize intron. Plant Mol Biol. 1994;24:449–463. doi: 10.1007/BF00024113. [DOI] [PubMed] [Google Scholar]
- Maas C, Laufs J, Grant S, Korfhage C, Werr W. The combination of a novel stimulatory element in the first exon of the maize Shrunken-1 gene with the following intron 1 enhances reporter gene expression up to 1000-fold. Plant Mol Biol. 1991;16:199–207. doi: 10.1007/BF00020552. [DOI] [PubMed] [Google Scholar]
- Mascarenhas D, Mettler IJ, Pierce DA, Lowe HW. Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol Biol. 1990;15:913–920. doi: 10.1007/BF00039430. [DOI] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell. 1990;2:163–171. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith J, Storti RV. Developmental regulation of the Drosophila tropomyosin II gene in different muscles is controlled by muscle-type-specific intron enhancer elements and distal and proximal promoter control elements. Dev Biol. 1993;159:500–512. doi: 10.1006/dbio.1993.1259. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- Newman AJ, Lin R-J, Cheng S-C, Abelson J. Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell. 1985;42:335–344. doi: 10.1016/s0092-8674(85)80129-x. [DOI] [PubMed] [Google Scholar]
- Niwa M, Rose SD, Berget SM. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- Norris SR, Meyer SE, Callis J. The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol Biol. 1993;21:895–906. doi: 10.1007/BF00027120. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwanski ER, Zhao J, Last RL. Arabidopsis thaliana tryptophan synthase alpha: gene cloning, expression, and subunit interaction. Mol Gen Genet. 1995;248:657–667. doi: 10.1007/BF02191705. [DOI] [PubMed] [Google Scholar]
- Rethmeier N, Seurinck J, Van Montagu M, Cornelissen M. Intron-mediated enhancement of transgene expression in maize is a nuclear, gene-dependent process. Plant J. 1997;12:895–899. doi: 10.1046/j.1365-313x.1997.12040895.x. [DOI] [PubMed] [Google Scholar]
- Rose AB, Last RL. Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J. 1997;11:455–464. doi: 10.1046/j.1365-313x.1997.11030455.x. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Filipowicz W. Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol Biol. 1996;32:1–41. doi: 10.1007/BF00039375. [DOI] [PubMed] [Google Scholar]
- Sinibaldi RM, Mettler IJ. Intron splicing and intron-mediated enhanced expression in monocots. In: Cohn WE, Moldave K, editors. Progress in Nucleic Acid Research and Molecular Biology. Vol. 42. New York: Academic Press; 1992. pp. 229–257. [DOI] [PubMed] [Google Scholar]
- Sullivan ML, Green PJ. Post-transcriptional regulation of nuclear-encoded genes in higher plants: the roles of mRNA stability and translation. Plant Mol Biol. 1993;23:1091–1104. doi: 10.1007/BF00042344. [DOI] [PubMed] [Google Scholar]
- Xu Y, Yu H, Hall TC. Rice triosephosphate isomerase gene 5′ sequence directs β-glucuronidase activity in transgenic tobacco but requires an intron for expression in rice. Plant Physiol. 1994;106:459–467. doi: 10.1104/pp.106.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S-H, Lawton MA, Hunter T, Lamb CJ. atpk1, a novel ribosomal protein kinase gene from Arabidopsis: I. Isolation, characterization, and expression. J Biol Chem. 1994;269:17586–17592. [PubMed] [Google Scholar]