Abstract
Prions are conformationally flexible proteins capable of adopting a native state and a spectrum of alternative states associated with a change in the function of the protein. These alternative states are prone to assemble into amyloid aggregates, which provide a structure for self-replication and transmission of the underlying conformer and thereby the emergence of a new phenotype. Amyloid appearance is a rare event in vivo, regulated by both the aggregation propensity of prion proteins and their cellular environment. How these forces normally intersect to suppress amyloid appearance and the ways in which these restrictions can be bypassed to create protein-only phenotypes remain poorly understood. The most widely studied and perhaps most experimentally tractable system to explore the mechanisms regulating amyloid appearance is the [PIN+] prion of Saccharomyces cerevisiae. [PIN+] is required for the appearance of the amyloid state for both native yeast proteins and for human proteins expressed in yeast. These observations suggest that [PIN+] facilitates the bypass of amyloid regulatory mechanisms by other proteins in vivo. Several models of prion appearance are compatible with current observations, highlighting the complexity of the process and the questions that must be resolved to gain greater insight into the mechanisms regulating these events.
Keywords: prion, chaperone, [PSI+], [PIN+], proteostasis, amyloid
Prion appearance is the gateway to the creation of protein-only phenotypes, and recent studies highlight both the complexity of this process and the unanswered mechanistic questions remaining.
INTRODUCTION
Protein-only traits underlie an increasing cross-section of biology. Outcomes as varied as the regulation of gene expression in budding yeast and the emergence and progression of neurodegenerative disease in humans have now been linked to a protein-only mechanism (Tuite and Serio 2010). A subset of these traits, which are determined by proteins known as prions (Prusiner 1982), are transmissible through either infection or heredity, shattering the long-held belief that information transfer had sole provenance in nucleic acids (Crick 1970). The breakthrough that led to this advance was the uncovering of the distinct nature of the information itself: while nucleic acid-based information is encoded by sequence, protein-based information is encoded in conformation. Thus, when a prion protein adopts a new conformation, its native activity is altered, and/or it acquires a new one, leading to a novel phenotype (Tuite and Serio 2010). Because the transmission of a protein-only trait requires the replication of its determinant in a new individual, prions represent the first in vivo example of an autonomous self-replicating shape (Penrose and Penrose 1957).
In the more than three decades that have elapsed since this breakthrough, much insight into the structure of the self-replicating conformation and the mechanism of self-replication has emerged (Knowles, Vendruscolo and Dobson 2014). Both prion proteins and prionoids, which determine protein-only but non-transmissible traits (Aguzzi 2009), can access an alternative protein folding trajectory, which competes with the pathway leading to the native state (Jahn and Radford 2008). Within this extended energy landscape, monomeric protein has the propensity to self-assemble into amyloid, a filamentous complex characterized by a cross-β structure, where the strands of a continuous β-sheet are arranged along the fiber length (Sunde et al. 1997). At each end of the fiber, an exposed strand acts as a templating surface, allowing the formation of hydrogen bonds between backbone residues and the packing of side chains into a steric zipper (Nelson et al. 2005; Sawaya et al. 2007). This configuration promotes bidirectional growth of the fiber (Goldsbury et al. 1999; Blackley et al. 2000; Scheibel et al. 2001) and concomitantly the depletion of alternative conformers of the same protein (Satpute-Krishnan and Serio 2005; Knowles et al. 2009).
Given the self-replicating nature of amyloid, its appearance is the primary gateway to the emergence of new traits associated with this state. For many proteins, the kinetic threshold for amyloidogenesis appears to be high, primarily due to the need for self-assembly (Gazit 2002; Baldwin et al. 2011). Amyloid formation proceeds via a nucleated process, in which monomers must assemble into an oligomer of defined size to become thermodynamically stable (i.e. the nucleus) (Jarrett and Lansbury 1993). Once this threshold is reached, amyloid accumulation increases both through continued assembly onto this nucleus and through the formation of secondary nuclei by fragmentation of existing fibers to create new ends or by de novo assembly stimulated along the lateral surfaces of fibers (Masel, Jansen and Nowak 1999; Masel and Jansen 2001; Knowles et al. 2009; Gaspar et al. 2017). The kinetic threshold for amyloidogenesis is easily overcome in vitro, where protein concentrations can be readily manipulated. However in vivo, amyloid appearance seems to be regulated beyond the intrinsic aggregation propensity of these proteins even at high concentration. For example, amyloid appearance increases during aging, with the decline of protein-quality control pathways known as the proteostasis network (Powers et al. 2009; Koga, Kaushik and Cuervo 2011) and in the presence of other misfolded proteins (Derkatch et al. 2001; Osherovich and Weissman 2001; Gidalevitz et al. 2006).
To understand the emergence of protein-only traits, we must then uncover not only how the complex energy landscape of protein folding is balanced by the intricate proteostasis network to suppress amyloidogenesis but also where the points of vulnerability in this intersection lie. In this review, I examine the literature on prion appearance in the yeast Saccharomyces cerevisiae, focusing on the interaction between the [PSI+] and [PIN+] prions, in pursuit of this insight.
[PSI+] and [PIN+]
[PSI+] is the protein-only trait determined by the amyloid form of the prion protein Sup35 (Cox 1965; Doel et al. 1994; Ter-Avanesyan et al. 1994; Chernoff et al. 1995; Patino et al. 1996; Paushkin, Kushnirov and Smirnov 1996; Glover et al. 1997; King et al. 1997; Paushkin et al. 1997). [PSI+] arises spontaneously in [psi−] yeast, which have non-prion state Sup35, at a frequency of ∼10−8–10−7/generation (Lancaster et al. 2010), but this ability to acquire [PSI+] is specifically regulated in vivo. [PIN+] yeast strains, which are inducible to [PSI+] either spontaneously or by transient overexpression of Sup35, can be converted to a non-inducible state ([pin−]) by treatment with millimolar concentrations of guanidium HCl (GdnHCl) (Lund and Cox 1981; Derkatch et al. 1997). The [PIN+] trait segregates 4:0 in the meiotic progeny of a diploid strain formed by crossing [PIN+] and [pin−] yeast strains and is eliminated by deletion or overexpression of the molecular chaperone Hsp104 (Chernoff et al. 1995; Derkatch et al. 1997). Together, these observations suggested that the [PIN+] determinant was an epigenetic factor (i.e. reversible) transmitted through the cytoplasm (i.e. inherited in a non-Mendelian pattern), and subsequent studies revealed that [PIN+] was also a protein-only trait (Derkatch and Liebman 2007).
This [PIN+]-dependent ability of Sup35 to bypass the forces that normally restrict its transition to the amyloid state provides a unique experimental tool to gain mechanistic insight into amyloid appearance in vivo. Indeed, the presence of [PIN+] also promotes the accumulation of SDS-resistant aggregates, a hallmark of the amyloid state (Serio et al. 2000), of other proteins when they are overexpressed in the yeast cytosol, including the polyglutamine-expanded forms of the Machado-Joseph disease protein and exon 1 of huntingtin (Osherovich and Weissman 2001; Meriin et al. 2002; Alexandrov et al. 2008; Kochneva-Pervukhova, Alexandrov and Ter-Avanesyan 2012). This generality suggests that the [PSI+]-[PIN+] interplay is a representative example of a broader system of amyloid regulation in vivo.
Given its epigenetic nature, the determinant of the [PIN+] trait was once proposed to be an intermediate conformation of the non-amyloid form of Sup35. According to this model, the [PIN+], but not [pin−], conformation of Sup35 was competent to self-assemble into amyloid (Derkatch et al. 1997). However, the authors also showed that the [PIN+] phenotype is propagated in and transmissible through a yeast strain deleted for the prion-determining domain of the Sup35 protein (Derkatch et al. 1997), a glutamine and asparagine-rich N-terminal segment of the protein required for prion formation and propagation (Ter-Avanesyan et al. 1994). While these observations did not rule out a role for the non-prion domain of Sup35 in the propagation of [PIN+], other factors were soon implicated. Overexpression of yeast proteins with prion-like QN-rich domains, including New1, Ure2, Lsm4, Ste18, Pin2, Yck1, Nup116 and Cyc8, was capable of inducing [PIN+] in a [pin−] strain, as expected for an amyloid-based trait (Wickner 1994; Derkatch et al. 2001; Osherovich and Weissman 2001). Moreover, deletion of the gene encoding Rnq1, the rich in asparagine (N) and glutamine (Q) protein determinant of the previously identified [RNQ+] yeast prion (Sondheimer and Lindquist 2000), was sufficient to convert a strain from [PIN+] to [pin−] and to block transmission of [PIN+] (Derkatch et al. 2001; Osherovich and Weissman 2001). Strikingly, the spontaneous appearance and GdnHCl-curing of [PIN+] correlated with the appearance and disappearance, respectively, of Rnq1 aggregates (Derkatch et al. 2001). Moreover, in vitro assembled Rnq1 amyloid, but not non-aggregated Rnq1, induced the appearance of [PIN+] when transformed into [pin−] yeast (Patel and Liebman 2007). Thus, [PIN+] is not conferred by an intermediate conformation of Sup35. Rather, it is primarily determined by the amyloid form of the Rnq1 protein in laboratory yeast strains, although the amyloid forms of multiple Q/N-rich proteins can serve in a similar, albeit reduced, capacity (Sondheimer and Lindquist 2000; Derkatch et al. 2001; Osherovich and Weissman 2001).
The genetic and physical interaction of the Sup35 and Rnq1 prion proteins in their amyloid, native and denatured states has been extensively analyzed in vitro and in vivo, with Susan Lindquist, her long-term collaborator Susan Liebman, and many of her former trainees (cited throughout) contributing to our mechanistic understanding of prion appearance in vivo through this body of work. These studies have been organized into multiple models (Fig. 1), which I present below along with a summary of experimental evidence and remaining open questions.
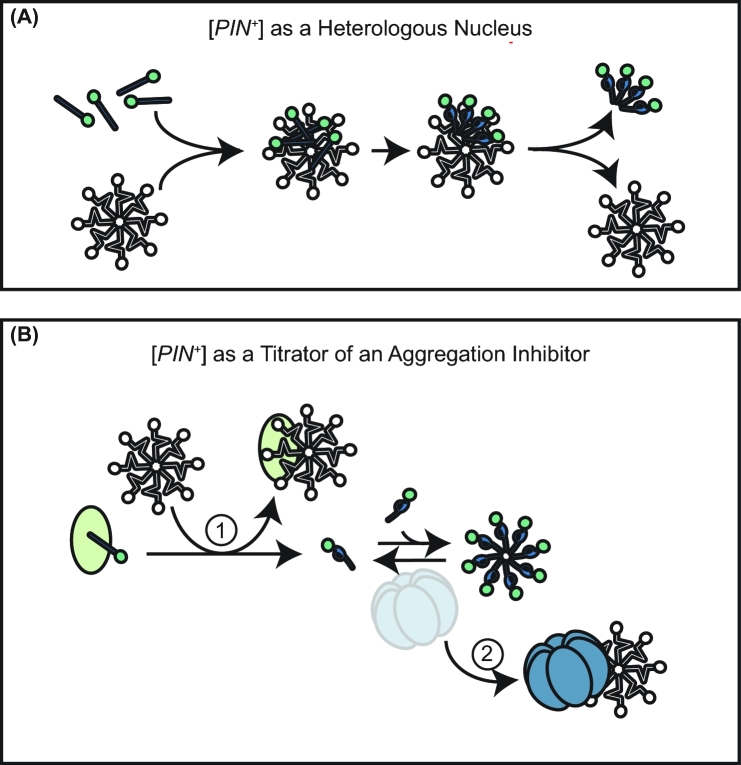
Figure 1.
Models for the role of [PIN+] in [PSI+] appearance. Two models have been proposed to explain the requirement for [PIN+] in the appearance of [PSI+] in vivo. (A) The most widely accepted model for [PIN+] is that of heterologous nucleus (unfilled pinwheel), stimulating the nucleation of Sup35 (green and blue) by direct interaction. Once nucleated, Sup35 and Rnq1 (the determinant of [PIN+] in laboratory yeast strains) amyloids propagate as separate aggregates. (B) [PIN+] may also promote [PSI+] formation by titrating an inhibitor (green oval) that binds to non-prion state Sup35 and blocks its self-association (step 1). As a variation on the inhibitor model, [PIN+] may titrate the fragmentation machinery (blue hexamer) away from spontaneously forming nascent Sup35 aggregates, allowing them to persist and amplify (step 2). See the text for details on each of the models.
Model 1: [PIN+] acts as a heterologous nucleus for Sup35 amyloid formation
What is the role of Rnq1 amyloid in promoting the formation of a stably propagating [PSI+] state? The most widely accepted possibility is the heterologous nucleation model. According to this idea, Rnq1 amyloid templates the formation of Sup35 amyloid through direct interaction, providing a pathway to overcome the kinetic barrier to amyloid appearance by promoting nucleation (Fig. 1A) (Derkatch et al. 1997, 2000; Osherovich and Weissman 2002). Indeed, fusing the Sup35 prion domain to Rnq1 promotes [PSI+] formation in [PIN+] yeast in the absence of Sup35 overexpression, suggesting that the act of artificially bringing together Sup35 monomers is sufficient to bypass a barrier to prion formation in vivo (Choe et al. 2009). The question is: Does [PIN+] play the same role when it is not fused to Sup35? Many studies have been undertaken, both in vitro and in vivo, to directly test this model.
A factor acting as a heterologous nucleus for amyloid formation by another protein should accelerate the assembly of the latter both in vitro and in vivo in a manner that increases with the concentration of the former templates. While the prion-determining domain of Sup35 formed amyloid in vitro in the absence of Rnq1 amyloid (Glover et al. 1997; King et al. 1997), the addition of Rnq1 amyloid accelerated its formation, as assessed by thioflavin T fluorescence (Derkatch et al. 2004; Vitrenko et al. 2007; Sharma and Liebman 2013a). However, this stimulation did not recapitulate in vivo observations of conformation-specific genetic interactions between Rnq1 and Sup35 (Sharma and Liebman 2013a), was only weakly dependent on Rnq1 concentration (Derkatch et al. 2004) and required Rnq1 amyloid potentially in excess of the Rnq1:Sup35 ratios observed in vivo (Ghaemmaghami et al. 2003; Kulak et al. 2014). These points raise the possibility of non-specific effects. For example, Rnq1 amyloid at high concentration may increase molecular crowding and thereby Sup35 assembly (Lansbury 1999; Minton 2005; Huang et al. 2015). This alternative possibility also provides an explanation for the increased stimulation of Sup35 amyloid formation upon sonication of Rnq1 fibers (Sharma and Liebman 2013a), an effect that is smaller in magnitude than would be predicted for end-dependent polymerization (Serio et al. 2000) but consistent with an increased efficiency of crowding expected at lower viscosity and with a smaller crowder (Ellis and Minton 2006; Bokvist and Gröbner 2007). Intriguingly, the stimulation of Sup35 assembly in vitro is not specific to Rnq1 fibers, which was cited as evidence of specificity (Derkatch et al. 2004). However, another possibility remains. The proteins that are capable of this activity (immunoglobulin, insulin and Rnq1) have isoelectric points close to neutrality, while those that are incapable of doing so (α-synuclein, lysozyme and transthyretin) have highly acidic or basic isoelectric points. If crowding is indeed the mechanism of stimulation in vitro, the charge of the crowder could be an important component of the effect, as has been previously suggested (Minton 1983).
In vivo, the relationship between [PSI+] induction and Rnq1 amyloid concentration is similarly confounding in the context of the heterologous nucleation model. [PIN+] can exist as a spectrum of variants, each of which corresponds to a different conformation of Rnq1 amyloid and varies in its frequency of [PSI+] induction (Bradley et al. 2002; Sharma and Liebman 2013a; Stein and True 2014). Notably, the frequencies of [PSI+] induction associated with these variants correspond neither to the number of heritable Rnq1 aggregates nor to the accumulation of aggregated Rnq1, as would be predicted for a heterologous nucleus (Bradley et al. 2002; Bardill and True 2009; Kalastavadi and True 2010; Sharma and Liebman 2013a, 2013b). Thus, studies of the effects of Rnq1 amyloid concentration on Sup35 amyloidogenesis in vitro and in vivo cannot currently provide strong support for the heterologous nucleation model.
The heterologous nucleation model also predicts a direct interaction between Rnq1 amyloid and Sup35 protein. Several lines of evidence support this prediction. First, some mutations in Rnq1 retain the ability to propagate [PIN+], as assessed by Rnq1 aggregation and transmissibility, but have a reduced ability to support [PSI+] induction (Bardill and True 2009; Stein and True 2014). The reduced [PSI+]-inducibility of one mutant can be suppressed by a mutation in Sup35, providing support for molecular specificity in this process, although not necessarily through a direct Rnq1-Sup35 interaction (Keefer, Stein and True 2017). Second, overexpressed Sup35 and Rnq1 co-localize to cytoplasmic ring and dot structures, as detected by immunofluorescence or tagging with fluorescent proteins (Derkatch et al. 2004; Kimura et al. 2004; Tyedmers et al. 2010; Du and Li 2014; Arslan et al. 2015), and these structures have been previously linked to [PSI+] appearance (Zhou, Derkatch and Liebman 2001). Third, Sup35 and Rnq1 have been demonstrated to physically interact by immunoprecipitation/immunocapture from yeast lysates, the capture of Rnq1 from yeast lysates on a Sup35-affinity resin and in vitro cross-linking of purified proteins (Salnikova et al. 2005; Tyedmers et al. 2010; Sharma and Liebman 2013a; Keefer, Stein and True 2017). Together, these observations provide support for an interaction between Rnq1 and Sup35, a necessary component of the heterologous nucleation model.
Conceptually, the idea of an unrelated protein serving as a heterologous nucleus can be seen as counter to the known specificity of amyloid copolymerization, which requires a high degree of sequence identity (Krebs et al. 2004). For example, the Sup35 homologs from the closely related species S. paradoxus and S. bayanus and from the more distantly related Pichia methanolica access a prion state, as assessed by loss of Sup35 native activity and GdnHCl reversibility, when overexpressed in the S. cerevisiae cytosol. However, these proteins are unable to support [PSI+] propagation upon deletion of the S. cerevisiae SUP35 gene, demonstrating the sequence specificity required for heterologous nucleation (Chernoff et al. 2000; Chen, Newnam and Chernoff 2007). Indeed, a single amino-acid change in Sup35 (S17R) disrupts the ability of preformed Sup35 amyloid fibers to accelerate the assembly of soluble wild-type Sup35 in vitro, although both wild-type and mutant Sup35s retain the ability to form amyloid on their own (DePace et al. 1998). More extensive studies have revealed that exact homology in short stretches of Sup35 is required for copolymerization, prion induction and prion propagation (Santoso et al. 2000; Resende et al. 2002), and these sequences mediate direct contacts between monomers that likely nucleate distinct Sup35 conformations (Chien et al. 2003; Krishnan and Lindquist 2005; Tessier and Lindquist 2007). Such examples of high-sequence specificity for amyloidogenesis among Sup35 proteins must necessarily lead to skepticism about [PIN+] acting as a heterologous nucleus, especially given the large number of QN-rich proteins capable of promoting [PSI+] appearance when overexpressed in vivo (Derkatch et al. 2001; Osherovich and Weissman 2001).
Nonetheless, the specificity described above reflects end-dependent copolymerization as a mechanism of nucleation (Derkatch and Liebman 2007), and there have been relatively few examples of such heteropolymeric amyloids identified to date (Sarell, Stockley and Radford 2013). Consistent with this reality, Sup35 and Rnq1 form separate SDS-resistant aggregates in vivo (Bagriantsev and Liebman 2004). Moreover, overexpression of Sup35 homologs promotes [PSI+] appearance by S. cerevisiae Sup35 without efficiently adopting an SDS-resistant state, indicating that they have not formed amyloid themselves and would therefore be incapable of promoting [PSI+] appearance by amyloid-based cross-seeding (Chen, Newnam and Chernoff 2007; Vishveshwara and Liebman 2009). Thus, other mechanisms must be rigorously considered in any attempt to understand a role for [PIN+] as a heterologous nucleus.
An expanding repertoire of possibilities has been reported for other pairs of amyloidogenic proteins. For example, the sequence-specific binding of Aβ to tau promotes phosphorylation of the latter, which in turn reduces the affinity between the two proteins and potentially promotes their aggregation (Guo et al. 2006). New1 induces ATP-dependent fragmentation of Sup35 fibers in vitro to create new ends (Inoue et al. 2011), and an N-terminally truncated variant of β2-microglobulin induces a conformational change in the wild-type protein to promote amyloidogenesis (Eichner et al. 2011). Sickle hemoglobin polymerization is believed to include a heterogeneous nucleation step mediated along the lateral surface of the polymer through sequence-specific contacts (Ferrone et al. 1980; Ferrone, Hofrichter and Eaton 1985; Rotter et al. 2005), a mechanism proposed to explain the kinetics of amyloidogenesis in vitro for other proteins (Knowles et al. 2009; Gaspar et al. 2017) and the [PIN+] activity of Lsm4 in vivo (Oishi et al. 2013).
Whether the mechanism of heterologous nucleation proceeds via end-dependent polymerization or another pathway, a significant gap in proof for the model remains: a definitive demonstration that the Sup35-Rnq1 interaction is required for [PSI+] induction. For example, while the Sup35-Rnq1 interaction in yeast lysates is [PIN+]-dependent, the extent of the interaction does not correlate with the distinct [PSI+]-induction frequencies of [PIN+] variants (Sharma and Liebman 2013a). Moreover, most mutations in Rnq1 that reduce [PSI+]-inducibility, and the Sup35 mutation that suppresses this effect for one mutant, have not been assessed for corresponding changes in interaction (Bardill and True 2009; Stein and True 2014; Keefer, Stein and True 2017). In the one case where this analysis has been undertaken, both the [PSI+]-induction defective Rnq1(N297S) (Bardill and True 2009) and wild-type Rnq1 are immunocaptured from yeast lysates with the Sup35 prion-determining domain to the same extent (Sharma and Liebman 2013a). In the absence of evidence linking a reduction in [PSI+] appearance to a decrease in Rnq1-Sup35 association, effects on unknown events downstream of the Rnq1-Sup35 association and/or indirect effects independent of this association cannot be eliminated from consideration (Sharma and Liebman 2013b).
Model 2: [PIN+] titrates an aggregation inhibitor
[PIN+] has also been proposed to function as a factor that titrates an inhibitor of aggregation (Fig. 1B—step 1) (Derkatch et al. 2001; Osherovich and Weissman 2001, 2002; Vitrenko et al. 2007). This model is not mutually exclusive with the heterologous template model because, by definition, it describes the in vivo regulation of protein misfolding. Consistent with this idea, growth under conditions of stress led to an increase in the spontaneous frequency of [PSI+] appearance in a strain expressing a mutant form of Sup35 that more readily converts to the prion state (Liu and Lindquist 1999; Tyedmers, Madariaga and Lindquist 2008). Thus, the threshold for amyloidogenesis likely changes in distinct proteostatic niches, suggesting the potential for regulation.
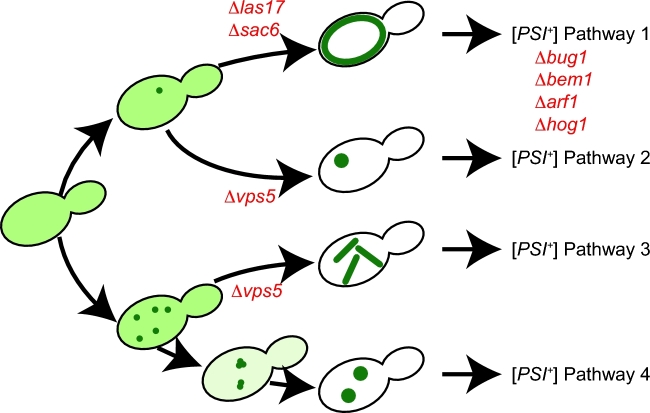
As was the case for the heterologous nucleation model, a more nuanced consideration of potential mechanisms of inhibition is warranted. Several lines of evidence suggest that prion appearance in vivo is a multistep process. First, genes, whose deletion reduce [PSI+] induction in [PIN+] strains, fall into two classes: those that interfere with the formation of cytoplasmic rings of GFP fused to the Sup35 prion-determining domain, a hallmark of [PSI+] induction in wild-type strains (Derkatch et al. 1997), and those that do not (Manogaran et al. 2011) (Fig. 2). Thus, there are at least two genetically separable events. Second, fluorescently detectible structures, once appearing, dynamically evolve into different forms, and this evolution appears to be genetically regulated (Sharma et al. 2017; Wisniewski et al. 2018) (Fig. 2). Third, SDS-resistant oligomers of the Sup35 prion-determining domain fused to GFP appear prior to microscopically visible structures (Sharma et al. 2017). Fourth, [PSI+] induction frequencies increase with less stringent selection, suggesting that aggregate appearance occurs more commonly than standard [PSI+]-selection conditions capture (Tyedmers, Madariaga and Lindquist 2008; McGlinchey, Kryndushkin and Wickner 2011; Gorkovskiy et al. 2017). Thus, the initial aggregation of Sup35 and the transition of these nascent aggregates into a stable [PSI+] state appear to be separable events, raising the possibility that they are independently regulated (Manogaran et al. 2011).
Figure 2.
Genetic regulation of early steps of [PSI+] appearance (adapted from Sharma et al. 2017). Overexpression of the prion-determining domain of Sup35 fused to GFP in [PIN+] yeast cells leads to the appearance and evolution of microscopically visible protein aggregates of distinct types: rings (pathway 1), single dots (pathway 2), lines (pathway 3) and multiple dots (pathway 4), which all lead to [PSI+] appearance (Sharma et al. 2017). Deletion mutants (red), which reduce [PSI+] appearance, differentially impact the appearance and/or accumulation of these visible aggregates (e.g. Δlas17, Δvps5 or Δsac6) or appear to act downstream of these events (e.g. Δbug1, Δbem1, Δarf1 or Δhog1) (Manogaran et al. 2011; Sharma et al. 2017; Wisniewski et al. 2018).
Sup35 polymerization in vivo, as in vitro, is a nucleated event: GFP-tagged Sup35, which is newly synthesized or introduced by mating into a [PSI+] strain expressing untagged Sup35, localized to cytoplasmic foci, reflecting conversion onto existing aggregates (Patino et al. 1996; Satpute-Krishnan and Serio 2005), and in vitro assembled Sup35 amyloid transformed into a [psi−] strain induces [PSI+] appearance (Tanaka et al. 2004). Thus, the barriers to [PSI+] appearance likely involve the formation of an initial nucleus, its survival and its amplification. [PIN+] is dispensable for [PSI+] induction following transformation of Sup35 amyloid (Tanaka et al. 2004; King, Wang and Chang 2006) and for the propagation of existing [PSI+] (Derkatch et al. 2000). Because each of these [PIN+]-independent processes require continued amplification of Sup35 amyloid, [PIN+] would most likely exert its effects on nucleus formation and/or survival in this model.
Factors interfering with nucleus formation would be expected to interact with non-prion state Sup35 or with a prion-competent intermediate form of the protein, and overexpression of Sup35 alone might be expected to overcome this mechanism of inhibition by exceeding the concentration of the inhibitor. While overexpression of Sup35 to a high level does not induce [PSI+] in a [pin−] strain (Derkatch et al. 1997), its inability to do so could simply reflect differential affinities of Rnq1 amyloid and Sup35 for the titration target. Indeed, the idea of an aggregation inhibitor is supported by the observation Sup35 amyloid formation in vitro is inhibited by the addition of yeast lysates (Uptain et al. 2001). Unfortunately, this inhibition was not assessed for dependence on [PIN+]; thus, the relationship of this inhibitory activity to [PIN+] remains an open question.
Nonetheless, if this model is correct, several predictions can be made. First, deletion or mutation of the inhibitor should promote [PSI+] induction. Second, overexpression of the inhibitor should reduce [PSI+] induction but not propagation, if its action is specific to nucleation or nucleus persistance. Third, binding of the inhibitor to Sup35 should be enriched in [psi−] strains, if it is directly blocking conversion to the prion state. And fourth, [PIN+] should play a role in titrating such an inhibitory factor away from its interaction with Sup35. To date, no candidate genes meeting these criteria have been identified despite the initial promise of some factors. For example, deletion of the cotranslationally acting Hsp70 homologs in yeast, SSB1 and SSB2, promotes [PSI+] appearance, both spontaneously and following transient overexpression of Sup35, by a factor of 10 (Chernoff et al. 1999). While the effect of Ssb1 overexpression on [PSI+] appearance was not analyzed, overexpression of Ssb1 was subsequently shown to promote loss of existing [PSI+] (Chernoff et al. 1999; Kushnirov et al. 2000; Chacinska et al. 2001; Allen et al. 2005), and Ssb1 binds to Sup35 in both [psi−] and [PSI+] strains by immunocapture (Allen et al. 2005; Holmes et al. 2014). Another once promising candidate is Sup45, the functional partner of Sup35 in translation termination (Dever and Green 2012). Overexpression of Sup45 reduces [PSI+] induction by Sup35 overexpression and does not interfere with [PSI+] propagation (Derkatch, Bradley and Liebman 1998), but Sup45 binding to Sup35 persists in [PSI+] strains (Pezza et al. 2014; Arslan et al. 2015). Finally, deletion of either of the genes encoding the small heat shock proteins (sHsp) Hsp26 or Hsp42 mildly increases, while overexpression of either factor mildly reduces, [PSI+] appearance following Sup35 overexpression, but the latter is likely due to an effect on existing aggregates, as overexpression of sHsps promotes loss of existing [PSI+] in vivo (Duennwald, Echeverria and Shorter 2012). More generally, EMS mutagenesis, undertaken to inactivate the putative aggregation inhibitor, did not identify a factor capable of increasing the frequency of [PSI+] appearance (Derkatch et al. 2001), but redundancy in and essentiality of genes necessarily complicate the search for putative aggregation inhibitors via this approach. Thus, inhibition of Sup35 nucleation and a role for [PIN+] in promoting the bypass of this regulation remain theoretical possibilities to explore as new candidate genes are identified.
Whether a nucleus is formed spontaneously without regulation or following the bypass of inhibitory processes, it must persist and be amplified to establish a stable, transmissible prion state in vivo (Pezza and Serio 2007). Several lines of evidence suggest that persistence and amplification are related processes, reflecting a balance between growth and fragmentation of existing aggregates. For [PSI+], amplification requires the fragmentation of existing Sup35 amyloid by the chaperone machinery, specifically the AAA + ATPase Hsp104 and its co-chaperones Hsp70 (Ssa1) and Hsp40 (Sis1) (Chernoff et al. 1995; Song et al. 2005; Satpute-Krishnan, Langseth and Serio 2007; Higurashi et al. 2008; Tipton, Verges and Weissman 2008). In a balanced system, where wild-type factors are expressed at native levels and growth occurs in the absence of stress, introduction of a single, preformed Sup35 aggregate is theoretically sufficient to induce a stable [PSI+] state (Tanaka et al. 2004). However, a Sup35 mutant that reduces the kinetic stability of its amyloid state or the upregulation of molecular chaperones in response to a sublethal heat shock create proteostatic niches in which existing amyloid is cleared through the process of Hsp104-dependent fragmentation (DiSalvo et al. 2011; Klaips et al. 2014; Pei et al. 2017). Importantly, these niches occur in compartments characterized by an elevated chaperone:amyloid ratio due to the asymmetric inheritance of factors during yeast cell division (Derdowski et al. 2010; Klaips et al. 2014; Pei et al. 2017).
A similar situation likely exists during nucleation, where nascent amyloid appears at low abundance, and this reality raises the possibility of a different type of aggregation inhibitor—one that promotes disassembly of existing aggregates (Fig. 1B—step 2) (Derkatch et al. 2001; Osherovich and Weissman 2001, 2002; Vitrenko et al. 2007; Davis and Sindi 2016). Indeed, the importance of balance in the amplification and clearance pathways during [PSI+] appearance is supported by studies of Sup35 and Hsp104 mutants. [PSI+] can be induced in a strain expressing a fragmentation-defective deletion mutant of Sup35 (Δ22–69) only if Hsp104 levels are elevated (Borchsenius et al. 2001). Likewise, [PSI+] variants induced in a strain overexpressing Hsp104 or in one expressing an Hsp104 mutant that is incapable of promoting [PSI+] loss when overexpressed (T160M) are eliminated when Hsp104 is returned to its wild-type state (Borchsenius et al. 2006; Gorkovskiy et al. 2017).
Does [PIN+] act to tip this balance in favor of amyloid persistence and amplification? The concept of titration of cellular factors by one amyloidogenic protein from another is supported by the surprising observation that the proteins that most efficiently confer the [PIN+] phenotype when overexpressed (Pin4C, Cyc8C, Yck1, Ste18) also induce loss of existing [PSI+] at the same elevated levels (Yang et al. 2013). In the case of Pin4C, overexpression leads to an increase in the size of Sup35 aggregates, as assessed by GFP-tagging/microscopy and by gel-based analysis of SDS-resistant aggregates from yeast lysates, and to a decrease in their mobility in some cells (Yang et al. 2013). These observations are consistent with a defect in Hsp104-mediated fragmentation and loss of [PSI+] by the failure to transmit Sup35 aggregates during cell division (Satpute-Krishnan, Langseth and Serio 2007; Kawai-Noma et al. 2009). Consistent with this hypothesis, both Hsp104-GFP and Sis1-GFP co-localize with RFP-tagged Pin4C under these conditions, and [PSI+] loss is ameliorated by overexpression of Hsp104 (T160M) or by overexpression of Sis1 (Hung and Masison 2006; Yang et al. 2013). However, the interplay between amyloid and the proteostasis network is complex, as the opposite scenario also appears to promote [PSI+] formation: overexpression of Cyc8C elevates the levels of Hsp104 (significantly) and Sis1 (modestly) (Yang et al. 2013). Nonetheless, the impact of Pin4C overexpression provides clear support for the idea that overexpression of an amyloidogenic protein can titrate the fragmentation machinery away from nascent Sup35 aggregates, allowing them to persist (Yang et al. 2013).
The question then becomes, can the [PIN+] phenotype, conferred by a self-replicating conformation of a prion protein expressed at its native level, be similarly linked to titration of the fragmentation machinery? Intriguingly, the presence of [PIN+] can induce [PSI+] loss in some cases (Bradley and Liebman 2003; Mathur, Hong and Liebman 2009; Westergard and True 2014), an observation that could reflect competition for cellular factors. However, the presence of [PIN+] does not appear to alter the size of SDS-resistant aggregates of Sup35 (Bagriantsev and Liebman 2004), an observation that is at odds with a titration model or one that at a minimum suggests that titration is slight. Indeed, variants of [PIN+] do not appear to be characterized by differences in their binding to chaperones (Sharma and Liebman 2013a; Stein and True 2014), despite their distinct [PSI+]-induction frequencies (Bradley et al. 2002). Thus, strong support for the [PIN+]-dependent titration of factors promoting the disassembly of nascent Sup35 aggregates is currently lacking.
Rigorous analysis of this model, moreover, is not a straightforward endeavor for a number of reasons. First, given the low frequency of [PSI+] appearance, the highest probability of detecting such an activity will occur in strains containing incompatible prions, which by definition induce each other's loss. Second, incompatibility between different prions is quite specific and is impacted not only by the constellation of prions present but also by their conformations, further restricting the experimental bandwidth in which to assess these effects (Bradley and Liebman 2003; Du and Li 2014). Third, the propagation of different prions and even different variants of the same prion are differentially sensitive to chaperone levels (Kushnirov et al. 2000; Wegrzyn et al. 2001; Kryndushkin et al. 2002; Fan et al. 2007; Tipton, Verges and Weissman 2008; Mathur, Hong and Liebman 2009; Hines et al. 2011a,b; DeSantis and Shorter 2012; Dulle and True 2013; Lancaster, Dobson and Rachubinski 2013; Dulle, Stein and True 2014; Harris et al. 2014; Stein and True 2014), raising the possibility of distinct titration targets. Fourth, the fragmentation machinery, if the target, would be required for both clearance and amplification of nascent Sup35 aggregates, and reductions in the expression of or mutations in these factors would be expected to lead to prion loss at additional points in the prion cycle beyond appearance. Fifth, chaperone-substrate interactions are notoriously transient. Given the constellation of intriguing observations compatible with this model, these challenges must be overcome to fully explore the pathway of prion appearance in vivo.
CONCLUSION
The predominant models for the role of [PIN+] in [PSI+] appearance are not mutually exclusive. Rather, the complexity and dynamics of the system, at the intersection between overlapping protein folding trajectories and proteostasis, and the large number of factors identified with the capacity to provide [PIN+] activity in their amyloid state suggest a multitude of pathways to prion appearance in vivo. Indeed, the requirement for [PIN+] can be completely bypassed if the prion-determining domain of Sup35 is fused to a random C-terminal extension (RVDLQACKLMIQYQRK), suggesting another route to overcome the in vivo barriers to prion appearance (Derkatch et al. 1997, 2000). Recent studies have embraced this possibility, and the extensive body of work contributed over the past two decades provides a strong foundation of genetic and physical interactions to guide future inquiry. To move forward, we must develop new approaches beyond the endpoint readout of assessing the appearance of a stable [PSI+] state. The numerous questions remaining on prion appearance underscore the fact that mechanistic insight will only be gleaned through deconvolution of the interconnected processes of amyloid nucleation, persistence and amplification in vivo.
Acknowledgements
This review is dedicated to the memory of Susan Lindquist, whose creativity, intellect, passion and drive were and continue to be a source of personal inspiration for me. Sue had the confidence necessary to engage with the most complex of biological problems, the clarity of thought to identify a viable path to pursue and the talent to actually succeed. She is truly missed. I also thank the members of my research group for comments on this manuscript and the community of researchers, who have contributed to this fascinating area of prion appearance.
FUNDING
This work was supported by the National Institute of General Medical Sciences at the National Institutes of Health [grant number R35 GM118042].
Conflict of interest. None declared.
REFERENCES
- Aguzzi A. Beyond the prion principle. Nature 2009;459:924–5. [DOI] [PubMed] [Google Scholar]
- Alexandrov IM, Vishnevskaya AB, Ter-Avanesyan MD et al. Appearance and propagation of Polyglutamine-based amyloids in yeast. J Biol Chem 2008;283:15185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen KD, Wegrzyn RD, Chernova TA et al. Hsp70 chaperones as modulators of prion life cycle. Genetics 2005;169:1227–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan F, Hong JY, Kanneganti V et al. Heterologous aggregates promote de novo prion appearance via more than one mechanism. PLoS Genet 2015;11:e1004814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem 2004;279:51042–8. [DOI] [PubMed] [Google Scholar]
- Baldwin AJ, Knowles TPJ, Tartaglia GG et al. Metastability of native proteins and the phenomenon of amyloid formation. J Am Chem Soc 2011;133:14160–3. [DOI] [PubMed] [Google Scholar]
- Bardill JP, True HL. Heterologous prion interactions are altered by mutations in the prion protein Rnq1p. J Mol Biol 2009;388:583–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackley HK, Sanders GH, Davies MC et al. In-situ atomic force microscopy study of beta-amyloid fibrillization. J Mol Biol 2000;298:833–40. [DOI] [PubMed] [Google Scholar]
- Bokvist M, Gröbner G. Misfolding of amyloidogenic proteins at membrane surfaces: the impact of macromolecular crowding. J Am Chem Soc 2007;129:14848–9. [DOI] [PubMed] [Google Scholar]
- Borchsenius AS, Muller S, Newnam GP et al. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr Genet 2006;49:21–29. [DOI] [PubMed] [Google Scholar]
- Borchsenius AS, Wegrzyn RD, Newnam GP et al. Yeast prion protein derivative defective in aggregate shearing and production of new seeds'. EMBO J 2001;20:6683–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Edskes HK, Hong JY et al. Interactions among prions and prion "strains" in yeast. P Natl Acad Sci USA 2002;99 Suppl 4:16392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Liebman SW. Destabilizing interactions among [PSI+] and [PIN+] yeast prion variants. Genetics 2003;165:1675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Szczesniak B, Kochneva-Pervukhova NV et al. Ssb1 chaperone is a [PSI+] prion-curing factor. Curr Genet 2001;39:62–67. [DOI] [PubMed] [Google Scholar]
- Chen B, Newnam GP, Chernoff YO. Prion species barrier between the closely related yeast proteins is detected despite coaggregation. P Natl Acad Sci USA 2007;104:2791–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Galkin AP, Lewitin E et al. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol 2000;35:865–76. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B et al. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+]. Science 1995;268:880–4. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Newnam GP, Kumar J et al. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol 1999;19:8103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, DePace AH, Collins SR et al. Generation of prion transmission barriers by mutational control of amyloid conformations. Nature 2003;424:948–51. [DOI] [PubMed] [Google Scholar]
- Choe Y-J, Ryu Y, Kim H-J et al. Increased [PSI+] appearance by fusion of Rnq1 with the prion domain of Sup35 in Saccharomyces cerevisiae. Eukaryot Cell 2009;8:968–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BS. [PSI], a cytoplasmic suppressor of super-suppression in yeast. Heredity 1965;20:505–21. [Google Scholar]
- Crick F. Central dogma of molecular biology. Nature 1970;227:561–3. [DOI] [PubMed] [Google Scholar]
- Davis JK, Sindi SS. A mathematical model of the dynamics of prion aggregates with chaperone-mediated fragmentation. J Math Biol 2016;72:1555–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePace AH, Santoso A, Hillner P et al. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 1998;93:1241–52. [DOI] [PubMed] [Google Scholar]
- Derdowski A, Sindi SS, Klaips CL et al. A size threshold limits prion transmission and establishes phenotypic diversity. Science 2010;330:680–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY et al. Prions affect the appearance of other prions. Cell 2001;106:171–82. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Liebman SW. Overexpression of the SUP45 gene encoding a Sup35p-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. P Natl Acad Sci USA 1998;95:2400–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Masse SV et al. Dependence and independence of [PSI+] and [PIN+]: a two-prion system in yeast? EMBO J 2000;19:1942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P et al. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997;147:507–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Liebman SW. Prion-prion interactions. Prion 2007;1:161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Uptain SM, Outeiro TF et al. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. P Natl Acad Sci USA 2004;101:12934–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis ME, Shorter J. Hsp104 drives “protein-only” positive selection of Sup35 prion strains encoding strong [PSI+]. Chem Biol 2012;19:1400–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. CSH Persp Biol 2012;4:a013706–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSalvo S, Derdowski A, Pezza JA et al. Dominant prion mutants induce curing through pathways that promote chaperone-mediated disaggregation. Nat Struct Mol Biol 2011;18:486–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doel SM, McCready SJ, Nierras CR et al. The dominant PNM2- mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics 1994;137:659–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Li L. Investigating the Interactions of Yeast Prions: [SWI+], [PSI+], and [PIN+]. Genetics 2014;197:685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Echeverria A, Shorter J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol 2012;10:e1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulle JE, Stein KC, True HL. Regulation of the Hsp104 middle domain activity is critical for yeast prion propagation. PLoS One 2014;9:e87521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulle JE, True HL. Low activity of select Hsp104 mutants is sufficient to propagate unstable prion variants. Prion 2013;7:394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner T, Kalverda AP, Thompson GS et al. Conformational conversion during amyloid formation at atomic resolution. Mol Cell 2011;41:161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ, Minton AP. Protein aggregation in crowded environments. Biol Chem 2006;387:485–97. [DOI] [PubMed] [Google Scholar]
- Fan Q, Park K-W, Du Z et al. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics 2007;177:1583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrone FA, Hofrichter J, Eaton WA. Kinetics of sickle hemoglobin polymerization. J Mol Biol 1985;183:611–31. [DOI] [PubMed] [Google Scholar]
- Ferrone FA, Hofrichter J, Sunshine HR et al. Kinetic studies on photolysis-induced gelation of sickle cell hemoglobin suggest a new mechanism. Biophys J 1980;32:361–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar R, Meisl G, Buell AK et al. Secondary nucleation of monomers on fibril surface dominates α-synuclein aggregation and provides autocatalytic amyloid amplification. Quart Rev Biophys 2017;50:e6. [DOI] [PubMed] [Google Scholar]
- Gazit E. The “Correctly Folded” state of proteins: is it a metastable state? Angew Chem Int Ed 2002;41:257–9. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K et al. Global analysis of protein expression in yeast. Nature 2003;425:737–41. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH et al. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 2006;311:1471–4. [DOI] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 1997;89:811–9. [DOI] [PubMed] [Google Scholar]
- Goldsbury C, Kistler J, Aebi U et al. Watching amyloid fibrils grow by time-lapse atomic force microscopy. J Mol Biol 1999;285:33–39. [DOI] [PubMed] [Google Scholar]
- Gorkovskiy A, Reidy M, Masison DC et al. Hsp104 disaggregase at normal levels cures many [PSI+] prion variants in a process promoted by Sti1p, Hsp90, and Sis1p. P Natl Acad Sci USA 2017;114:E4193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J-P, Arai T, Miklossy J et al. Abeta and tau form soluble complexes that may promote self aggregation of both into the insoluble forms observed in Alzheimer's disease. P Natl Acad Sci USA 2006;103:1953–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JM, Nguyen PP, Patel MJ et al. Functional diversification of hsp40: distinct j-protein functional requirements for two prions allow for chaperone-dependent prion selection. PLoS Genet 2014;10:e1004510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi T, Hines JK, Sahi C et al. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. P Natl Acad Sci USA 2008;105:16596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines JK, Higurashi T, Srinivasan M et al. Influence of prion variant and yeast strain variation on prion-molecular chaperone requirements. Prion 2011;5:238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines JK, Li X, Du Z et al. [SWI], the prion formed by the chromatin remodeling factor Swi1, is highly sensitive to alterations in Hsp70 chaperone system activity. PLoS Genet 2011b;7:e1001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WM, Mannakee BK, Gutenkunst RN et al. Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nat Commun 2014;5:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Liu X, Cheng B et al. How our bodies fight amyloidosis: effects of physiological factors on pathogenic aggregation of amyloidogenic proteins. Arch Biochem Biophys 2015;568:46–55. [DOI] [PubMed] [Google Scholar]
- Hung G-C, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 2006;173:611–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Kawai-Noma S, Koike-Takeshita A et al. Yeast prion protein New1 can break Sup35 amyloid fibrils into fragments in an ATP-dependent manner. Genes Cell 2011;16:545–56. [DOI] [PubMed] [Google Scholar]
- Jahn TR, Radford SE. Folding versus aggregation: polypeptide conformations on competing pathways. Arch Biochem Biophys 2008;469:100–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett JT, Lansbury PT. Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell 1993;73:1055–8. [DOI] [PubMed] [Google Scholar]
- Kalastavadi T, True HL. Analysis of the [RNQ+] prion reveals stability of amyloid fibers as the key determinant of yeast prion variant propagation. J Biol Chem 2010;285:20748–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai-Noma S, Pack CG, Tsuji T et al. Single mother-daughter pair analysis to clarify the diffusion properties of yeast prion Sup35 in guanidine-HCl-treated [PSI] cells. Gene Cells 2009;14:1045–54. [DOI] [PubMed] [Google Scholar]
- Keefer KM, Stein KC, True HL. Heterologous prion-forming proteins interact to cross-seed aggregation in Saccharomyces cerevisiae. Sci Rep 2017;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Koitabashi S, Kakizuka A et al. The role of pre-existing aggregates in Hsp104-dependent polyglutamine aggregate formation and epigenetic change of yeast prions. Gene Cells 2004;9:685–96. [DOI] [PubMed] [Google Scholar]
- King CY, Tittmann P, Gross H et al. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. P Natl Acad Sci USA 1997;94:6618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C-Y, Wang H-L, Chang H-Y. Transformation of yeast by infectious prion particles. Methods 2006;39:68–71. [DOI] [PubMed] [Google Scholar]
- Klaips CL, Hochstrasser ML, Langlois CR et al. Spatial quality control bypasses cell-based limitations on proteostasis to promote prion curing. Elife 2014;3:1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TPJ, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Bio 2014;15:384–96. [DOI] [PubMed] [Google Scholar]
- Knowles TPJ, Waudby CA, Devlin GL et al. An analytical solution to the kinetics of breakable filament assembly. Science 2009;326:1533–7. [DOI] [PubMed] [Google Scholar]
- Kochneva-Pervukhova NV, Alexandrov AI, Ter-Avanesyan MD. Amyloid-mediated sequestration of essential proteins contributes to mutant huntingtin toxicity in yeast. PLoS One 2012;7:e29832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: the importance of exquisite quality control. Ageing Res Rev 2011;10:205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs MRH, Morozova-Roche LA, Daniel K et al. Observation of sequence specificity in the seeding of protein amyloid fibrils. Protein Sci 2004;13:1933–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 2005;435:765–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Smirnov VN, Ter-Avanesyan MD et al. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J Biol Chem 2002;277:23702–8. [DOI] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I et al. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 2014;11:319–24. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV, Kryndushkin DS, Boguta M et al. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol 2000;10:1443–6. [DOI] [PubMed] [Google Scholar]
- Lancaster AK, Bardill JP, True HL et al. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics 2010;184:393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster DL, Dobson CM, Rachubinski RA. Chaperone proteins select and maintain [PIN+] prion conformations in Saccharomyces cerevisiae. J Biol Chem 2013;288:1266–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury PT. Evolution of amyloid: what normal protein folding may tell us about fibrillogenesis and disease. P Natl Acad Sci USA 1999;96:3342–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate “protein-only” inheritance in yeast. Nature 1999;400:573–6. [DOI] [PubMed] [Google Scholar]
- Lund PM, Cox BS. Reversion analysis of [psi-] mutations in Saccharomyces cerevisiae. Genet Res 1981;37:173–82. [DOI] [PubMed] [Google Scholar]
- McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. P Natl Acad Sci USA 2011;108:5337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manogaran AL, Hong JY, Hufana J et al. Prion formation and polyglutamine aggregation are controlled by two classes of genes. PLoS Genet 2011;7:e1001386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J, Jansen VA. The measured level of prion infectivity varies in a predictable way according to the aggregation state of the infectious agent. Biochim Biophys Acta 2001;1535:164–73. [DOI] [PubMed] [Google Scholar]
- Masel J, Jansen VA, Nowak MA. Quantifying the kinetic parameters of prion replication. Biophys Chem 1999;77:139–52. [DOI] [PubMed] [Google Scholar]
- Mathur V, Hong JY, Liebman SW. Ssa1 overexpression and [PIN+] variants cure [PSI+] by dilution of aggregates. J Mol Biol 2009;390:155–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriin AB, Zhang X, He X et al. Huntingtin toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol 2002;157:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton AP. The effect of volume occupancy upon the thermodynamic activity of proteins: some biochemical consequences. Mol Cell Biochem 1983;55:119–40. [DOI] [PubMed] [Google Scholar]
- Minton AP. Influence of macromolecular crowding upon the stability and state of association of proteins: predictions and observations. J Pharm Sci 2005;94:1668–75. [DOI] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature 2005;435:773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Kurahashi H, Pack C-G et al. A bipolar functionality of Q/N-rich proteins: Lsm4 amyloid causes clearance of yeast prions. MicrobiologyOpen 2013;2:415–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell 2001;106:183–94. [DOI] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. The utility of prions. Dev Cell 2002;2:143–51. [DOI] [PubMed] [Google Scholar]
- Patel BK, Liebman SW. “Prion-proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132-405) induces [PIN+]. J Mol Biol 2007;365:773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR et al. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 1996;273:622–6. [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN. Propagation of the yeast prion-like [PSI+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 1996;15:3127–34. [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN et al. In vitro propagation of the prion-like state of yeast Sup35 protein. Science NY 1997;277:381–3. [DOI] [PubMed] [Google Scholar]
- Pei F, DiSalvo S, Sindi SS et al. A dominant-negative mutant inhibits multiple prion variants through a common mechanism. PLoS Genet 2017;13:e1007085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrose LS, Penrose R. A Self-reproducing Analogue. Nature 1957;179:1183-. [Google Scholar]
- Pezza JA, Serio TR. Prion propagation: the role of protein dynamics. Prion 2007;1:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezza JA, Villali J, Sindi SS et al. Amyloid-associated activity contributes to the severity and toxicity of a prion phenotype. Nat Commun 2014;5:4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A et al. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 2009;78:959–91. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982;216:136–44. [DOI] [PubMed] [Google Scholar]
- Resende C, Parham SN, Tinsley C et al. The Candida albicans Sup35p protein (CaSup35p): function, prion-like behaviour and an associated polyglutamine length polymorphism. Microbiology 2002;148:1049–60. [DOI] [PubMed] [Google Scholar]
- Rotter MA, Kwong S, Briehl RW et al. Heterogeneous nucleation in sickle hemoglobin: experimental validation of a structural mechanism. Biophys J 2005;89:2677–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikova AB, Kryndushkin DS, Smirnov VN et al. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem 2005;280:8808–12. [DOI] [PubMed] [Google Scholar]
- Santoso A, Chien P, Osherovich LZ et al. Molecular basis of a yeast prion species barrier. Cell 2000;100:277–88. [DOI] [PubMed] [Google Scholar]
- Sarell CJ, Stockley PG, Radford SE. Assessing the causes and consequences of co-polymerization in amyloid formation. Prion 2013;7:359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P, Langseth SX, Serio TR. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol 2007;5:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P, Serio TR. Prion protein remodelling confers an immediate phenotypic switch. Nature 2005;437:262–5. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 2007;447:453–7. [DOI] [PubMed] [Google Scholar]
- Scheibel T, Kowal AS, Bloom JD et al. Bidirectional amyloid fiber growth for a yeast prion determinant. Curr Biol 2001;11:366–9. [DOI] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 2000;289:1317–21. [DOI] [PubMed] [Google Scholar]
- Sharma J, Liebman SW. Exploring the basis of [PIN+] variant differences in [PSI+] induction. J Mol Biol 2013. a425:3046–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J, Liebman SW. Variant-specific prion interactions. Cellular Logistics 2013b;3:e25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J, Wisniewski BT, Paulson E et al. De novo [PSI +] prion formation involves multiple pathways to form infectious oligomers. Sci Rep 2017;7:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1. Mol Cell 2000;5:163–72. [DOI] [PubMed] [Google Scholar]
- Song Y, Wu Y-X, Jung G et al. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot Cell 2005;4:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein KC, True HL. Extensive diversity of prion strains is defined by differential chaperone interactions and distinct amyloidogenic regions. PLoS Genet 2014;10:e1004337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde M, Serpell LC, Bartlam M et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol 1997;273:729–39. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N et al. Conformational variations in an infectious protein determine prion strain differences. Nature 2004;428:323–8. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV et al. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [PSI+] in the yeast Saccharomyces cerevisiae. Genetics 1994;137:671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature 2007;447:556–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell 2008;32:584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite MF, Serio TR. The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Bio 2010;11:823–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol 2008;6:e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Treusch S, Dong J et al. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. P Natl Acad Sci USA 2010;107:8633–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain SM, Sawicki GJ, Caughey B et al. Strains of [PSI+] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J 2001;20:6236–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishveshwara N, Liebman SW. Heterologous cross-seeding mimics cross-species prion conversion in a yeast model. BMC Biol 2009;7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitrenko YA, Gracheva EO, Richmond JE et al. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem 2007;282:1779–87. [DOI] [PubMed] [Google Scholar]
- Wegrzyn RD, Bapat K, Newnam GP et al. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol 2001;21:4656–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergard L, True HL. Extracellular environment modulates the formation and propagation of particular amyloid structures. Mol Microbiol 2014;92:698–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 1994;264:566–9. [DOI] [PubMed] [Google Scholar]
- Wisniewski BT, Sharma J, Legan ER et al. Toxicity and infectivity: insights from de novo prion formation. Curr Genet 2018;64:117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Hong JY, Derkatch IL et al. Heterologous gln/asn-rich proteins impede the propagation of yeast prions by altering chaperone availability. PLoS Genet 2013;9:e1003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI+] and [PIN+]. Mol Microbiol 2001;39:37–46. [DOI] [PubMed] [Google Scholar]