Abstract
Arsenic is a widely distributed toxic natural element. Chronic arsenic ingestion causes several cancers, especially skin cancer. Arsenic-induced cancer mechanisms are not well defined, but several studies indicate that mutation is not the driving force and that microRNA expression changes play a role. Chronic low arsenite exposure malignantly transforms immortalized human keratinocytes (HaCaT), serving as a model for arsenic-induced skin carcinogenesis. Early changes in miRNA expression in HaCaT cells chronically exposed to arsenite will reveal early steps in transformation. HaCaT cells were maintained with 0/100 nM NaAsO2 for 3 and 7 weeks. Total RNA was purified. miRNA and mRNA expression was assayed using Affymetrix microarrays. Targets of differentially expressed miRNAs were collected from TargetScan 6.2, intersected with differentially expressed mRNAs using Partek Genomic Suite software, and mapped to their pathways using MetaCore software. MDM2, HMGB1 and TP53 mRNA, and protein levels were assayed by RT-qPCR and Western blot. Numerous miRNAs and mRNAs involved in carcinogenesis pathways in other systems were differentially expressed at 3 and 7 weeks. A TP53 regulatory network including MDM2 and HMGB1 was predicted by the miRNA and mRNA networks. Total TP53 and TP53-S15-phosphorylation were induced. However, TP53-K382-hypoacetylation suggested that the induced TP53 is inactive in arsenic exposed cells. Our data provide strong evidence that early changes in miRNAs and target mRNAs may contribute to arsenic-induced carcinogenesis.
Keywords: arsenic, skin cancer, miRNA, mRNA, transformation, TP53
Arsenic is a toxic metalloid that is naturally prevalent in the earth’s crust and widely distributed in air and water. Ground water as well as certain foods such as seafood and rice are the most common sources of ingested arsenic (Hunt et al., 2014; American Cancer Society, 2014). The toxicity of long-term exposure to arsenic is associated with a broad range of health problems including skin lesions, pigmentary abnormalities, arteriosclerosis, peripheral neuropathy, chronic lung disease, wasting syndrome, and cancer (IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, 2012; Hunt et al., 2014; Mazumder, 2000; Minamoto et al., 2005; Smith and Steinmaus, 2009; States, 2016). Arsenic is a known carcinogen associated with skin, lung, bladder, prostate, and liver cancers (Anetor et al., 2007). Arsenic has been ranked as number one in the U.S. Agency for Toxic Substances and Disease Registry (ATSDR) Priority List of Hazardous Substances since 2001 (Agency for Toxic Substances and Disease Registry, 2014). The upper limit of arsenic concentration recommended in drinking water in the United States is 10 μg/l. However, in countries such as Bangladesh, Taiwan, India, China, and Argentina, and in unregulated private well water in the United States, arsenic water levels often exceed 10 μg/l.
Skin cancer is the most common of all human cancers, and arsenic is second to UV in sunlight as a cause of skin cancer. In multiple epidemiologic studies, arsenical keratosis, hyperpigmentation, and multiple cutaneous malignancies were associated with arsenic exposure (Centeno et al., 2002; Maloney, 1996). The signs of arsenic toxicity appear clearly in the skin as arsenical keratosis in the palms, soles, and trunk (Centeno et al., 2002; Hunt et al., 2014). Chronic arsenic exposure causes both basal cell carcinoma (BCC) and squamous cell carcinoma (SCC) of the skin (IARC, 1987).
Several arsenic toxicity and carcinogenicity mechanisms have been suggested and proposed including abnormal signaling cascades, oxidative stress, and chromosomal aberrations as well as abnormal transcriptional activity and global gene expression (Isokpehi et al., 2012). Moreover, arsenic can induce environmentally driven epigenetic alterations that are known to influence disease development, including differential microRNA (miRNA) expression (Gonzalez et al., 2015; Su et al., 2011).
MiRNAs are a family of small RNAs and an acknowledged component of the epigenome regulating gene expression. miRNAs were discovered in 1993 as small temporal RNAs (stRNAs) that regulate developmental transitions in Caenorhabditis elegans (Lee et al., 1993). MiRNAs target mRNAs by hybridization to complementary sequences in their 3′-untranslated regions (UTRs) and repress translation (Felekkis et al., 2010). Recent studies on some miRNAs showed they also can bind to the 5′-UTR or the open reading frame region (Iorio and Croce, 2012). MiRNAs play a role in all the most important processes in every biologic system (Lagos-Quintana et al., 2001). A single miRNA can target hundreds of mRNAs, and the expression of a gene can be regulated by multiple miRNAs creating complex feedback and feed-forward gene regulatory loops in a cell. The regulatory loops provide checks and balances within and across gene networks (Sato et al., 2011; Tsang et al., 2010).
Several studies have shown that differential miRNA expression can be a hallmark of cancer and miRNAs can function as potential oncogenes or tumor suppressor genes influencing tumor development, progression, and response to therapy (Iorio and Croce, 2012; Kunej et al., 2012). MiRNAs exhibit differential expression in different cancers, including cervical, lung, esophageal, oral, pharyngeal, and tongue SCC (Pi et al., 2008). Furthermore, dysregulation of several miRNAs, including miR-21, miR-200a, miR-141, and let-7c has been reported in HaCaT cells after acute or subchronic exposure to moderate levels of arsenic (Gonzalez et al., 2015; Jiang et al., 2014).
HaCaT cells are a spontaneously immortalized human epithelial cell line established in 1988 from adult human skin (Boukamp et al., 1988). HaCaT cells are an in vitro model commonly used to study epidermal carcinogenesis (Williams et al., 2011). HaCaT cells malignantly transformed by chronic incubation in low concentration of sodium arsenite (Sun et al., 2009) are the only currently available in vitro model to study arsenic-induced skin carcinogenesis (Bruegger et al., 2013; Cha et al., 2014; Chowdhari and Saini, 2014; Jian et al., 2014; Syed et al., 2013; Xu et al., 2012). However, miRNA expression profiling has never been obtained in human keratinocytes chronically exposed to low physiologically relevant arsenic (100 nM). We hypothesize that investigating differential miRNA expression at different times during arsenite-induced transformation of HaCaT cells will show key events in the cellular transformation process. Thus, this report is focused on identifying differentially expressed miRNAs and their target genes in the early stages of arsenic-induced skin cancer using HaCaT cells exposed to 100 nM NaAsO2 for 3 and 7 weeks. For broad detection coverage, hybridization microarrays were used to measure mRNAs and small RNAs differentially expressed at the early stages of arsenic-induced skin cancer using HaCaT cells exposed to 100 nM NaAsO2 for 3 and 7 weeks.
MATERIALS AND METHODS
Cell culture and RNA isolation
HaCaT cells were the kind gift of Dr TaiHao Quan, University of Michigan. HaCaT cells are known to be compound heterozygous mutant in TP53 (H179Y and R282W; Lehman et al., 1993). The cells were cultured in MEM alpha modification media supplemented with 10% fetal bovine serum, 100 units/ml penicillin/100 µg/ml streptomycin) and 2 mM glutamine. Cultures were maintained at 37°C in a humidified 5% CO2 atmosphere. Multiple cultures of cells (4 with and 4 without 100 nM NaAsO2) were maintained separately for 7 weeks providing quadruplicate biological replicates. Cells were passaged twice a week and a million cells were plated per 100 mm dish every time. NaAsO2 (CAS 7784-0698) cell culture media and supplements were obtained from Thermo Fisher Scientific INC., Waltham. MA. Total RNA was purified from the cells (quadruplicate unexposed and exposed cultures) after 3 and 7 weeks using the mirVana RNA Isolation Kit (ThermoFisher Scientific Inc., Waltham, Massachusetts). RNA quality was determined using the Agilent RNA 6000 Pico Kit, Eukaryote, version 2.6 and the Agilent 2100 Bioanalyzer instrument (Agilent Technologies Inc., Santa Clara, California). All samples used had RIN (RNA integrity number) > 9.
Microarray analysis
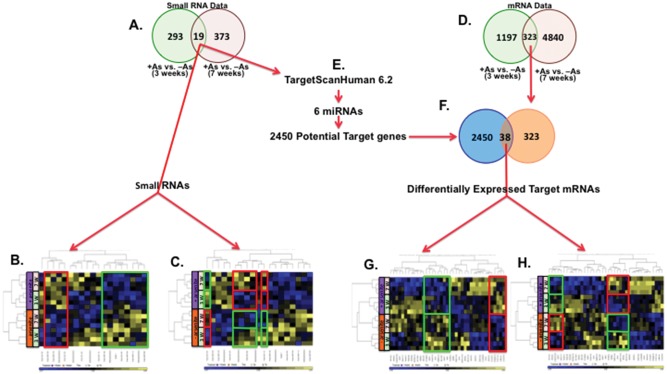
Expression profiles of mRNA and small RNA were obtained for 4 independent cultures of HaCaT cells exposed to 0 or 100 nM NaAsO2 for 3 and 7 weeks using the GeneChip PrimeView Human Gene Expression and GeneChip miRNA 4.0 Affymetrix arrays. The former, used for measuring mRNA expression, contains more than 530 000 probes that detect over 36 000 transcripts and variants, representing over 20 000 genes mapped via RefSeq or via UniGene annotation. The latter arrays probe for small noncoding RNA transcripts: 100% miRBase v20 coverage, 30 424 mature miRNAs (all organisms), 5214 human, mouse, and rat miRNAs, 1996 human small nucleolar RNAs (snoRNA), small Cajal body RNAs (scaRNA) and 3770 probe sets unique to human, mouse, and rat pre-miRNA hairpin sequences including 3 types of small RNAs, snoRNAs, and both stem-loop and mature miRNAs. Biotinylated complementary RNA (cRNA) was prepared according to the standard protocol for Affymetrix 3′ IVT Express Plus Reagent Kit from 250 ng total RNA. Following fragmentation, cRNA was hybridized for 16 h at 45°C to Affymetrix Primeview Human arrays according to the Affymetrix GeneChip 3′ array Hybridization User Manual. GeneChips were scanned using GeneChip Scanner 3000 7G (Affymetrix) and the GeneChip Command Console 4.0 (Affymetrix). For miRNA 4.0 arrays, biotinylated RNAs were prepared according to the standard protocol for Affymetrix FlashTag Biotin HSR RNA labeling Kit from 800 ng total RNA. Biotin-labeled samples were hybridized for 18 h at 48°C to Affymetrix miRNA 4.0 arrays. GeneChips were scanned using GeneChip Scanner 3000 7G (Affymetrix) and the GeneChip Command Console 4.0 (Affymetrix). The CEL files were imported into Partek software Version 6.6 (Partek Inc.) and normalized using Robust Multi-Array normalization. Contrasts of interest were analyzed using a 2-way ANOVA considering treatment and time. Using Partek Genomic Suite, potential targets of 6 of the 19 differentially expressed miRNAs (p-value ≤ .05) at both 3 and 7 weeks were obtained from the TargetScanHuman 6.2 database (2450 genes). Intersecting the target mRNA list with the differentially expressed mRNA lists at both 3 and 7 weeks (323 mRNAs, p-value ≤ 0.05), we obtained a list of differentially expressed mRNAs that are potential targets of differentially expressed miRNAs at both 3 and 7 weeks based on having the same probe annotation number on GeneChip (Figure 1). Data have been deposited in the GEO database, accession numbers GSE97303, GSE97305, and GSE97306.
Figure 1.
Exposure and time-dependent differential expression of small RNAs and mRNAs detected by hybridization microarrays. A, Venn diagrams of differentially expressed small RNAs at both 3 and 7 weeks (p < .05). Unsupervised hierarchical clustering of the 19 small RNAs differentially expressed at both 3 and 7 weeks of exposure to arsenite. B, Arsenite exposure-dependent induced (red boxed) and suppressed (green boxed) small RNAs. C, Time-dependent differentially expressed miRNAs and the expression pattern is flipped by arsenite exposure compared with nonexposed cells, induced (red boxed) and suppressed (green boxed). D, Venn diagram of differentially expressed mRNAs at 3 and 7 weeks. E, Predicted mRNA targets of 6 differentially expressed miRNAs were found in TargetScan Human V6.2 database. F, Venn diagram of intersection of predicted mRNA targets of differentially expressed miRNAs and differentially expressed 323 mRNAs at both 3 and 7 weeks. Unsupervised hierarchical clustering of the 39 mRNAs differentially expressed at both 3 and 7 weeks of exposure to arsenite that are predicted targets of 6 miRNAs differentially expressed at both 3 and 7 weeks. G, Arsenite exposure-dependent induced (red boxed) and suppressed (green boxed) mRNAs. H, Time-dependent differentially expressed mRNAs and the expression pattern is flipped by arsenite exposure compared with nonexposed cells, induced (red boxed) and suppressed (green boxed). Analysis was done using Partek Genomic Suite.
Most of miRNAs lead the RNA-induced silencing complex to the 3′ UTRs of their mRNA targets causing degradation or inhibiting translation of the mRNAs (Macfarlane and Murphy, 2010; Soifer et al., 2007). Using this approach, we obtained lists of differentially expressed target mRNAs at 3 and 7 weeks with expression direction opposite to that of the targeting miRNAs (differentially expressed miRNAs at 3 or 7 weeks, p-value ≤ .05), e.g. induced mRNA targets for suppressed miRNA and vice versa (Supplementary Tables 3 and 4).
Immunoblotting
Total protein was extracted from the cells (quadruplicate unexposed and exposed cultures) after 7 weeks using lysis solution (0.01 M Tris-HCl pH 7.4, 1 mM EDTA, 0.1% SDS, 180 µg/mL PMSF and 1× protease inhibitor cocktail [Complete, Roche, Mannheim, Germany]). Lysates were sonicated and protein concentrations were measured using the BCA assay kit (Sigma-Aldrich, BCA1 and B9643). Proteins were resolved by electrophoresis using Bio-Rad 4%–15% Mini-PROTEAN TGX Precast Protein gels. Gels were electrotransferred onto cellulose nitrate (wet transfer). Membranes were blocked using 5% skim milk in Tris-buffered saline containing 0.1% Tween-20 (TBS-T) for 2 h, then incubated at 4°C overnight with the following antibodies diluted in 5% (w/v) BSA in TBS-T: rabbit mAb anti-HMGB1 (D3E5) (6893S) (Cell Signaling Inc., Danvers, Massachusetts) (1:800), mouse monoclonal anti-MDM2 (Santa Cruz Biotechnology Inc., Dallas, Texas) (1:800), rabbit polyclonal anti-TP53 (no. 9282) (1:650), rabbit polyclonal anti-Phospho-p53 (Ser15) (1:800) and rabbit polyclonal anti-Acetyl-p53 (Lys382) (1:800) (Cell Signaling Inc., Danvers, Massachusetts), followed by an anti-rabbit (for anti-HMG, anti-TP53, anti-Phospho-p53, anti-Acetyl-p53) and anti-mouse (for anti-MDM2) secondary antibodies conjugated with horseradish peroxidase (Cell Signaling, 7074 and 7076, respectively) (1:3000). PageRuler Plus Prestained Protein Ladder no. 26619 (ThermoFisher Scientific) was used as a molecular weight marker. β-Actin was used loading control (Santa CruzBiotechnology Inc., Dalla, TX, SC47778-HRP, 1:2500). Antibody reactive bands were detected using Pierce ECL Plus kit (ThermoScientific, 32132). Imaging and quantitation were done using ImageQuant LAS 4000 biomolecular imager (GE Healthcare Life Sciences, Pittsburgh, Pennsylvania). Two-tailed Student’s t-test was used for statistical analyses.
RESULTS
Arsenite-Dependent Small RNA and mRNA Differential Expression
Samples of RNA purified from quadruplicate cultures of arsenite-exposed and unexposed HaCaT cells after 3 and 7 weeks of culture were analyzed by hybridization to miRNA 4.0 Affymetrix microarrays. The results showed that 293 and 373 small RNAs were differentially expressed, respectively, after 3 and 7 weeks exposure to 100 nM sodium arsenite (p-value ≤ .05) (Figure 1A). Nineteen small RNAs were differentially expressed at both time points and included 8 snoRNAs, 5 stem-loop miRNAs and 6 mature miRNAs (Table 1; Figure 1A). Unsupervised hierarchical clustering of these 19 small RNAs showed exposure-dependent differential expression of 12 small RNAs with 4 increased and 8 decreased compared with unexposed cells at both time points (Figure 1B). Time-dependent differential expression was observed for 6 small RNAs and this differential expression was reversed by arsenite exposure (Figure 1C). Analysis of mRNA expression revealed that 1197 and 4840 mRNAs were differentially expressed after 3 and 7 weeks exposure to 100 nM sodium arsenite (p-value ≤ .05), respectively, and 323 mRNAs were differentially expressed at both time points (Figure 1D).
Table 1.
Differentially Expressed Small RNAs at Both 3 and 7 Weeks of Exposure to Arsenite
| No. | Transcript ID (Array Design) | Sequence Type | 3 Weeks |
7 Weeks |
||
|---|---|---|---|---|---|---|
| Fold-Change | p-Value | Fold-Change | p-Value | |||
| 1. | 14qII-1 | CDBox (subtype of snoRNA) | −1.197 | 0.04363 | −1.296 | 0.00691 |
| 2. | ENSG00000212338 | snoRNA | −1.246 | 0.02753 | 1.241 | 0.02979 |
| 3. | ENSG00000221345 | snoRNA | −1.132 | 0.01756 | 1.148 | 0.00968 |
| 4. | ENSG00000221496 | snoRNA | 1.187 | 0.00091 | 1.109 | 0.02166 |
| 5. | ENSG00000238611 | snoRNA | −1.070 | 0.02915 | −1.121 | 0.00132 |
| 6. | ENSG00000238807 | snoRNA | −1.351 | 0.00779 | 1.307 | 0.01494 |
| 7. | ENSG00000239188 | snoRNA | −1.562 | 0.00005 | −1.235 | 0.01288 |
| 8. | ENSG00000252290 | snoRNA | −1.241 | 0.02864 | 1.237 | 0.03033 |
| 9. | hsa-miR-339 | stem-loop | 1.289 | 0.03267 | 1.312 | 0.02403 |
| 10. | hsa-miR-1228a | stem-loop | −1.167 | 0.02415 | 1.160 | 0.02888 |
| 11. | hsa-miR-4309a | stem-loop | 1.448 | 0.00026 | −1.172 | 0.04897 |
| 12. | hsa-miR-4692 | stem-loop | −1.298 | 0.03144 | 1.284 | 0.03749 |
| 13. | hsa-miR-548au | stem-loop | −1.234 | 0.02995 | −1.292 | 0.01113 |
| 14. | hsa-miR-548a-3pa | miRNA | −2.388 | 0.02041 | −2.564 | 0.01362 |
| 15. | hsa-miR-645a | miRNA | 1.194 | 0.03779 | 1.204 | 0.03067 |
| 16. | hsa-miR-1254a | miRNA | −1.671 | 0.02557 | −1.657 | 0.02756 |
| 17. | hsa-miR-2682-5p | miRNA | 1.189 | 0.03746 | 1.316 | 0.00298 |
| 18. | hsa-miR-3618a | miRNA | −1.183 | 0.03511 | −1.194 | 0.02762 |
| 19. | hsa-miR-8083 | miRNA | −1.270 | 0.03868 | −1.287 | 0.03050 |
miRNAs with available predicted targets in the TargetScanHuman 6.2 database.
Cancer-Associated Pathways of Arsenite-Dependent mRNA Differential Expression (Targets of Differentially Expressed Small RNAs)
In order to learn about the potential impact of the 11 differential expressed miRNAs (Table 1), potential targets of differentially expressed miRNAS were sought. Only 6 of the 11 miRNAs had predicted targets listed (Figure 1E). There were 2450 potential targets of these 6 miRNAs listed in TargetScanHuman 6.2 database. By intersecting the list of predicted target mRNAs (2450 mRNAs) with the 323 mRNAs differentially expressed at both 3 and 7 weeks of arsenite exposure (Figure 1D;Supplementary Table 1), 38 predicted target mRNAs were found differentially expressed in arsenic-exposed cells at both time points (p-value ≤ .05) (Supplementary Table 2; Figure 1F). Unsupervised hierarchical clustering of the 38 mRNAs showed exposure-dependent differential expression of 13 mRNAs with 5 increased and 8 decreased compared with unexposed cells at both time points (Figure 1G). Time-dependent differential expression was also observed for some of the 38 mRNAs. In comparison to 3 weeks, 15 mRNAs were induced and 15 were suppressed at 7 weeks. Expression of 12 mRNAs was reversed by arsenite exposure (Figure 1H).
In order to gain an understanding of the potential impact of these differentially expressed genes, we performed pathway analysis using Metacore software to place the differentially expressed target mRNAs on pathways. The top 10 pathways are listed in Table 2. Three of these pathways, cytoskeleton remodeling (predicted to be induced), WNT signaling (predicted to be induced), and signal transduction PTEN (predicted to be suppressed), are all known to play a role in carcinogenesis, and have a single mRNA in common. This mRNA, TCF7L2, was induced at 3 and 7 weeks and is a predicted target for miR-548a-3p, which is significantly suppressed at both 3 and 7 weeks (Table 1). TCF7L2 is a transcription factor that participates in the WNT signaling pathway (among others) and promotes c-MYC expression (Shao et al., 2013). TCF7L2 has a known role in colon cancer (Chen et al., 2013; Rosales-Reynoso et al., 2016). These results suggest that the chronic arsenite exposure at early times decreases the expression of miR-548a-3p, relieving suppression of TCF7L2 expression. The increase in TCF7L2 expression then promotes these oncogenic pathways.
Table 2.
Top 10 Pathways of the Differentially Expressed 38 mRNAs at Both 3 and 7 Weeks That Are Predicted Targets of miRNAs Differentially Expressed at Both 3 and 7 Weeks
| No. | Pathway | FDR |
|---|---|---|
| 1. | Development WNT signaling pathway.Part 1. Degradation of beta-catenin in the absence WNT signaling | 1.231E−02 |
| 2. | Action of GSK3 β in bipolar disorder | 1.231E−02 |
| 3. | Cytoskeleton remodeling TGF, WNT, and cytoskeletal remodeling | 1.231E−02 |
| 4. | Signal transduction PTEN pathway | 2.498E−02 |
| 5. | Immune response NFAT in immune response | 2.498E−02 |
| 6. | Development WNT signaling pathway. Part 2 | 2.50E−02 |
| 7. | PGE2 pathways in cancer | 2.50E−02 |
| 8. | Immune response CD28 signaling | 2.50E−02 |
| 9. | Ligand-independent activation of androgen receptor in prostate cancer | 2.92E−02 |
| 10. | Signal transduction mTORC2 downstream signaling | 2.92E−02 |
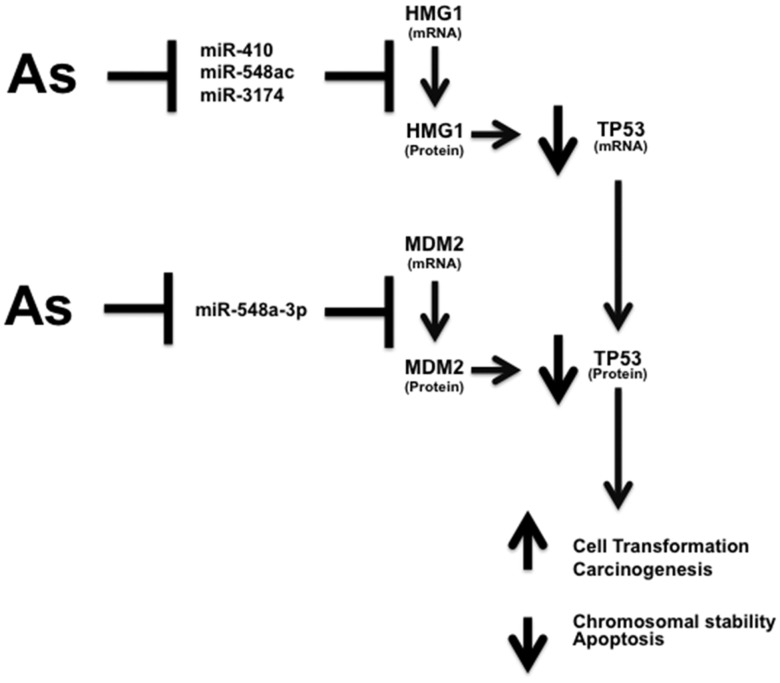
In addition to TCF7L2, MDM2 (mouse double minute 2 homolog) also is among the 38 target mRNAs and also is a predicted target of miR-548a-3p (Figure 3). MDM2 is a well-established suppressor of TP53 expression (Shangary and Wang, 2008). MDM2 mRNA was induced at 3 weeks and suppressed at 7 weeks as assayed by microarray hybridization. Suppression of miR-548a-3p would increase MDM2 levels, which then can act to suppress TP53 (Figure 3) at 3 weeks. High mobility group box 1 protein (HMGB1), a target of multiple differentially expressed miRNAs (Figure 3), also was among the differentially expressed target mRNAs at 3 and 7 weeks by microarray hybridization. HMGB1 is a predicted target of 3 differentially expressed miRNAs: miR-410, miR-548ac, and miR-3174 that were all suppressed at either 3 or 7 weeks (Figure 3). HMGB1 acts to suppress TP53 transcription (Štros et al., 2002). Thus, there appears to be a temporal switching of suppression of miRNAs that target HMGB1, with miR-410 and miR-548ac suppressed at 3 weeks (Supplementary Table 3) and miR-3174 suppressed at 7 weeks (Supplementary Table 4). This change in expression is likely an adaptive response to elevated TP53.
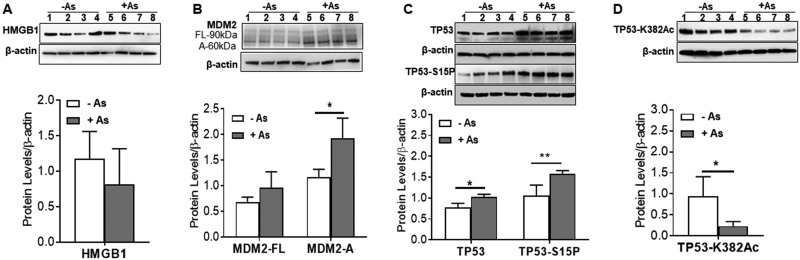
Figure 3.
Arsenite exposure effects on HMGB1, MDM2, and TP53 expression. After 7 weeks chronic exposure (A) Western blot and quantitation showing no change in HMGB1. B, Western blot and quantitation showing increased expression of MDM2 isoform A (MDM2-A, 60 kDa), but not isoform MDM2-FL. C, Western blot and quantitation showing both TP53 and TP53 phosphorylated at serine 15 (TP53-S15P) were induced with arsenic exposure. D, Western blot and quantitation showing TP53 hypoacetylated at lysine 382 (TP53-K382Ac) in arsenite exposed cells. Proteins levels were normalized to β-actin levels and means ± SD are plotted. Student’s t-test was used for statistical analyses. *p-value ≤ .05. Samples from 4 independent cultures incubated without arsenite for 7 weeks (lanes 1–4) were compared with samples from 4 independent cultures incubated in parallel with 100 nM NaAsO2 for 7 weeks (lanes 5–8). After probing for query proteins (HMGB1, MDM2, TP53), blots were stripped and probed for β-actin.
HMGB1, MDM2, and TP53 protein levels were examined at 7 weeks (Figure 3). The levels of HMGB1 were induced, yet the induction was not statistically significant (Figure 3A). MDM2 is an oncoprotein that has multiple isoforms and few of which can bind TP53 (Bartel et al., 2002; Schuster et al., 2007). Several MDM2 isoforms (multiple bands) were detected in exposed and unexposed cells (Figure 3B). Although levels of MDM2 isoforms increased in arsenite-exposed cells, only the increase of MDM2-A (∼60 kDa) was statistically significant. The levels of HMGB1 were induced, yet the induction was not statistically significant (Figure 3A). Total TP53 and phosphorylated TP53 (TP53-S15P) were induced (Figure 3C). In contrast, acetylated TP53 (TP53-K382Ac) was suppressed with arsenite exposure (Figure 3D).
Additional data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.3q26p.
DISCUSSION
The mechanism of arsenic-induced skin cancer is not well understood, but several studies indicate that mutation is not the driving force as it is for sunlight-induced skin cancers (Castren et al., 1998; Hsu et al., 1999; States, 2015). Different mechanisms of induction and progression of arsenic-induced carcinogenesis have been proposed including miRNA expression dysregulation (States, 2015). The current study focuses on early changes in gene expression linked to arsenic-induced changes in mRNA and small RNA expression in the HaCaT-cell chronic arsenic exposure model using Affymetrix hybridization microarrays. The results showed that arsenite exposure changes the expression pattern of small RNAs, including miRNAs, after 3 and 7 weeks exposure to 100 nM arsenite (Figure 1). This concentration of arsenite is physiologically relevant in that it reflects in vivo arsenic concentration in people exposed to moderately high arsenic in drinking water (Pi et al., 2008).
A major strength of the current study is that unlike previous reports investigating arsenic transformation of HaCaT cells, we have used four independent cultures of exposed and four independent cultures of exposed cells. Because transformation events are stochastic by nature, this approach is more realistic than reporting on a single culture, although it risks some variability in response. In spite of the variability, we were able to infer information on early events in arsenic transformation of keratinocytes in this model system.
The results identified 3 of the 6 miRNAs with predicted targets that were differentially expressed after 3 and 7 weeks (miR-1228, miR-1254, miR-645). These miRNAs were associated with cancer in previous studies (Foss et al., 2011; Lin et al., 2015; Sun et al., 2015). MiR-1228 (suppressed at 3 weeks, induced at 7 weeks) was reported to be induced in breast cancer tissues and cell lines and regulates the levels of mRNA and protein of SCAI (suppressor of cancer cell invasion) (Lin et al., 2015). MiR-1254 (suppressed at 3 and 7 weeks) was reported to be induced in the sera of nonsmall cell lung cancer patients (Foss et al., 2011). MiR-645 (induced at 3 and 7 weeks) was induced in head and neck SCC and its induction was found to promote cell invasion and metastasis (Sun et al., 2015).
The results also showed that arsenite dysregulated (mostly suppressed) several snoRNAs. SnoRNAs are 60–300 nucleotide small RNAs that are concentrated in the nucleolus (Williams and Farzaneh, 2012). SnoRNAs are involved in guiding premature ribosomal and other spliceosomal RNAs for nucleoside post-transcriptional modifications, crucial for accurate ribosomes (Williams and Farzaneh, 2012). SnoRNAs have two main classes: C/D box and H/ACA box snoRNAs, both reported to be dysregulated in several diseases including cancer (Dong et al., 2009; Williams and Farzaneh, 2012). These results suggest that dysregulation of snoRNAs may play a role in arsenic-induced carcinogenesis.
Arsenite exposure also induced dramatic changes in differential mRNA expression at 3 weeks (1197 mRNAs) and 7 weeks (4840 mRNAs) (Figure 1). The current report is focused on the 38 mRNAs differentially expressed at both time points that also were predicted targets of miRNAs differentially expressed at both time points to gain insight on plausible mechanisms by which the changes in miRNA expression would contribute to carcinogenesis.
Several carcinogenesis-associated pathways such as cytoskeleton remodeling, WNT signaling, and signal transduction PTEN pathways were populated by these 38 differentially expressed target mRNAs. These results support the hypothesis that the differential expression of miRNAs at early stages of arsenite exposure is contributing to the process of transformation to a cancerous phenotype. A mutually differentially expressed mRNA in these pathways was TCF7L2 (Figure 2). TCF7L2 is the most studied member of the mammalian TCF/LEF family of nuclear factors. TCF7L2 polymorphisms have been associated with type-2 diabetes susceptibility in several studies (Cadigan and Peifer, 2009; Hrckulak et al., 2016; Jin, 2016) and dysregulation of TCF7L2 may be related to the association of arsenic exposure and diabetes (Murea et al., 2012). The WNT signaling pathway is one of the main signaling mechanisms during embryogenesis and cancer development (Cadigan and Peifer, 2009; Hrckulak et al., 2016; Jin, 2016). The β-catenin/TCF (including TCF7L2) complex is the key effector in the WNT pathway, which leads to the activation of downstream targets and increased cell differentiation, proliferation, migration, and insulin sensitivity (Cadigan and Peifer, 2009; Hrckulak et al., 2016; Jin, 2016). Both β-catenin and TCF7L2 were induced at 7 weeks (1.41- and 1.13-fold, respectively). However, only TCF7L2 was induced at 3 weeks, suggesting that there is a progression in the gene expression changes with continued arsenite exposure. Analysis of gene expression changes with longer exposure times will be needed to determine if these early changes are maintained and perhaps enhanced as the cells become transformed.
Figure 2.
miRNA and target genes expression target TP53. Arsenite suppressed the expression miR-410, -548ac, and -548a-3p. MDM2 mRNA expression levels were induced at 3 weeks of arsenite exposure which might lead to the induction of MDM2 protein levels. HMGB1 mRNA expression levels were induced at 3 and 7 weeks of arsenite exposure which might lead to the induction in HMGB1 protein levels. Arsenite suppression of miR-410 and -548ac might induce MDM2 and HMGB1 at 3 weeks leading to the decrease in the levels of TP53.
E-cadherin is a calcium-dependent cell-cell adhesion protein and a key gene in the epithelial-to-mesenchymal transition (EMT, which is WNT signaling related) (Cadigan and Peifer, 2009; Hrckulak et al., 2016; Jin, 2016). Low E-cadherin expression is associated with the induction of EMT in cervical SCC and breast cancer (Cappellesso et al., 2015; Ma et al., 2015). E-cadherin mRNA expression was suppressed at 7 weeks of arsenite exposure (1.46-fold). Enhanced cell migration is an indicator of EMT. However, the migration capability of unexposed and arsenite exposed cells at 3 weeks did not differ (data not shown). These results suggest that at these early times of exposure, the changes in the mRNA levels of extracellular matrix components are not yet reflected in cellular phenotype. As for the progression observed with changes in mRNA expression, full development of the EMT phenotype may require longer exposures.
Of the 38 mRNAs differentially expressed after 3 and 7 weeks chronic exposure, MDM2 mRNA was induced at 3 weeks and suppressed after 7 weeks. MDM2 is a potential target of several miRNAs including miR-548a-3p that was suppressed (> 2.4-fold; in early exposure). Moreover, HMGB1 mRNA was induced in early exposure. HMGB1 is a potential target for 3 miRNAs found suppressed in early exposure. Both MDM2 and HMGB1 can regulate TP53 expression. The induction in MDM2 and HMGB1 expression was investigated along with MDM2 and HMGB1 effect on TP53 protein levels.
The MDM2 is a TP53-specific E3 ubiquitin ligase and the main cellular antagonist of TP53 (Moll and Petrenko, 2003). MDM2 has multiple isoforms, a few of which can bind TP53, such as MDM2-FL (Bartel et al., 2002; Schuster et al., 2007). The protein levels of some MDM2 isoforms were changed at 7 weeks. MDM2 splice variant expression has been observed in several types of cancer including bladder cancer (Sigalas et al., 1996), cancerous and normal breast tissues (Lukas et al., 2001; Matsumoto et al., 1998), soft tissue sarcomas (Bartel et al., 2001; Tamborini et al., 2001), and giant cell tumors of the bone (Evdokiou et al., 2001). MDM2-FL is the MDM2 isoform responsible for the ubiquitination of p53 for proteasomal degradation because it contains TP53 binding domain (Bartel et al., 2002; Honda et al., 1997; Schuster et al., 2007). MDM2-FL levels did not change significantly (Figure 3B). MDM2-A is a less common isoform of MDM2 and along with MDM2-B it binds to MDM2-FL and the interaction was found to prevent MDM2-mediated TP53 degradation (Zheng et al., 2013). MDM2-A (∼60 kDa) was expressed in higher levels with arsenite exposure, consistent with the observed decrease in expression of miR-548a-3p. Therefore, the induction in MDM2-A expression may be preventing MDM-FL from regulating TP53. The elevated levels of the MDM2 isoform MDM2-A (Figure 3B) also likely contribute to TP53 stabilization by preventing its ubiquitinylation (Shieh et al., 1997) resulting in the higher TP53 levels observed.
HMGB1 is a well-known active chromatin-associated protein implicated in autophagy (Tang et al., 2010). HMG proteins can modulate transcriptional activity of several receptors and protein complexes such as steroid hormone receptors, NF-KB, TP53 and TP73 transcriptional complexes, and homeobox containing proteins (TBP), and also facilitates V(D)J recombination (Sharma et al., 2008). HMGB1 is overexpressed in several tumors and tumor cells including chemically induced SCC in mice, human bladder cancer tissue, and melanoma cells (Boukamp et al., 1988; Poser et al., 2003; Sharma et al., 2008; Wang et al., 2013). Although HMGB1 mRNA levels were dysregulated, HMGB1 protein levels were variable and did not significantly change after 7 weeks chronic arsenic exposure (Figure 3A). Thus, it is unlikely that HMGB1 plays a role in regulating TP53 at this time point.
In sunlight-induced skin cancer, unrepaired DNA damage (UV photoproducts) leads to multiple mutations, especially in the TP53. UV signature mutations in TP53 appear in all sunlight-induced SCC (Bodak et al., 1999; Brash et al., 1991) but not in arsenic-induced skin cancer (Castren et al., 1998; Hsu et al., 1999). Moreover, preliminary studies found no mutations in TP53 exons 5–9 from FFPE samples of arsenic-induced SCC, BCC, or hyperkeratoses (J. C. States, unpublished data). Thus, TP53 mutations are rare in arsenic-induced skin cancer. Because dominant-negative TP53 mutations can result in stabilized TP53, it was thought that TP53 mutations were present in arsenic-induced skin cancers due to the overexpression of TP53 in keratinocytes exposed to arsenic (Chang et al., 1998). However, later studies showed that arsenic-induced skin lesions and carcinomas lack TP53 mutations and the gene is functional (Castren et al., 1998; Hsu et al., 1999). Investigators have also reported that arsenite exposure (0.1–5 μM) in several cell lines can induce TP53 (Huang et al., 2008; Sandoval et al., 2007). Hybridization microarray data showed that TP53 mRNA was suppressed in early chronic exposure. However, TP53 protein levels are regulated principally at the post-translational level and were induced along with TP53 phosphorylated at Ser-15 (Figure 3C). Phosphorylation of TP53 at Ser-15 prevents MDM2 from binding leading to the stabilization of TP53 (Sakaguchi et al., 1998). TP53 at Ser-15 phosphorylation also activates ATM-mediated DNA-damage response (Sakaguchi et al., 1998). However, mRNAs for DNA repair genes that are known TP53 targets such as DDB2, XPC, and DNA polymerase beta were not induced in the array data, suggesting the DNA damage response is being dysregulated. It should be noted that HaCaT cells are compound heterozygous mutant in TP53 and both mutations, H179Y and R282W, are gain-of-function mutations (Martynova et al., 2012). Thus, the TP53 proteins produced in HaCaT cells appear to retain transcription factor activity, although the battery of genes targeted is expanded. Overexpression of H179Y induces cyclin A and CDK4 (Yang et al., 2007). However, neither of these genes were induced in the microarray data. Thus, the induced TP53 appears not to be active in these cells (Luo et al., 2004).
Acetylation is required for TP53 transcriptional activity. Therefore, acetylation at the key acetylation site Lys-382 (Gu and Roeder, 1997) was quantified and found lower in exposed cells (Figure 3D). TP53 hypoacetylation supports the hypothesis that the induced TP53 is transcriptionally inactive. These results suggest that inactive TP53 may contribute to increased clastogenesis that arsenic is known to cause contributing to genomic instability (Salazar and Ostrosky-Wegman, 2016) by not properly activating the DNA damage response. At 100 nM sodium arsenite exposure, the HaCaT cells do not have high apoptosis indexes, further suggesting the induced TP53 is not be active in these cells (Luo et al., 2004). K382 acetylation is crucial for the proapoptotic functions (Yamaguchi et al., 2009). Thus, hypoacetylation not only reduces transcriptional activity but also reduces binding of BAX and induction of apoptosis. Reduction of apoptosis also may contribute to carcinogenesis.
Taken together the results described in this study suggest that the early changes in miRNA profiles and their target genes in human keratinocytes contribute to arsenic-induced carcinogenesis. Therefore, studies evaluating gene expression at longer exposure times will be needed to reveal the role miRNA and mRNA expression changes play as arsenic-induced carcinogenesis progresses. Testing the hypotheses by looking at protein levels and posttranslational modifications indicate that TP53 was induced, but inactive, stressing the importance of testing predictions based on the RNA data.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
National Institute of Environmental Health Sciences (T35 ES014559, R21 ES023627, R01 ES027778); UofL Genomics Facility, which is supported by National Institute of General Medical Sciences (P20 RR016481 [KY IDeA Networks of Biomedical Research Excellence], P30 GM106396 [UofL J. G. Brown Cancer Center Phase III CoBRE]), the J. G. Brown Foundation, and user fees.
Supplementary Material
ACKNOWLEDGMENT
This work was submitted in partial fulfillment of the requirements for the Doctor of Philosophy at the University of Louisville by L.A.
The authors declare no conflict of interest.
REFERENCES
- Agency for Toxic Substances and Disease Registry. (2014). Priority List of Hazardous Substances. Available at: http://www.atsdr.cdc.gov/spl/index.html. Accessed June 2016. [Google Scholar]
- American Cancer Society. (2014). Arsenic. Available at: http://www.cancer.org/cancer/cancercauses/othercarcinogens/intheworkplace/arsenic. Accessed June 2016. [Google Scholar]
- Anetor J. I., Wanibuchi H., Fukushima S. (2007). Arsenic exposure and its health effects and risk of cancer in developing countries: micronutrients as host defence. Asian Pac J Cancer Prev 8, 13–23. [PubMed] [Google Scholar]
- Bartel F., Meye A., Wurl P., Kappler M., Bache M., Lautenschlager C., Grunbaum U., Schmidt H., Taubert H. (2001). Amplification of the MDM2 gene, but not expression of splice variants of MDM2 MRNA, is associated with prognosis in soft tissue sarcoma. Int. J. Cancer 95, 168–175. [DOI] [PubMed] [Google Scholar]
- Bartel F., Taubert H., Harris L. C. (2002). Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell 2, 9–15.http://dx.doi.org/10.1016/S1535-6108(02)00091-0 [DOI] [PubMed] [Google Scholar]
- Bodak N., Queille S., Avril M. F., Bouadjar B., Drougard C., Sarasin A., Daya-Grosjean L. (1999). High levels of patched gene mutations in basal-cell carcinomas from patients with xeroderma pigmentosum. Proc. Natl. Acad. Sci. USA. 96, 5117–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. (1988). Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106, 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brash D. E., Rudolph J. A., Simon J. A., Lin A., McKenna G. J., Baden H. P., Halperin A. J., Ponten J. (1991). A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl. Acad. Sci. USA. 88, 10124–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruegger C., Kempf W., Spoerri I., Arnold A. W., Itin P. H., Burger B. (2013). MicroRNA expression differs in cutaneous squamous cell carcinomas and healthy skin of immunocompetent individuals. Exp. Dermatol. 22, 426–428.http://dx.doi.org/10.1111/exd.12153 [DOI] [PubMed] [Google Scholar]
- Cadigan K. M., Peifer M. (2009). Wnt signaling from development to disease: Insights from model systems. Cold Spring Harb. Perspect. Biol. 1, a002881.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellesso R., Marioni G., Crescenzi M., Giacomelli L., Guzzardo V., Mussato A., Staffieri A., Martini A., Blandamura S., Fassina A. (2015). The prognostic role of the epithelial-mesenchymal transition markers E-cadherin and Slug in laryngeal squamous cell carcinoma. Histopathology 67, 491–500. [DOI] [PubMed] [Google Scholar]
- Castren K., Ranki A., Welsh J. A., Vahakangas K. H. (1998). Infrequent p53 mutations in arsenic-related skin lesions. Oncol Res. 10, 475–482. [PubMed] [Google Scholar]
- Centeno J. A., Mullick F. G., Martinez L., Page N. P., Gibb H., Longfellow D., Thompson C., Ladich E. R. (2002). Pathology related to chronic arsenic exposure. Environ. Health Perspect. 110, 883–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha H. A., Kim O.-Y., Lee G. T., Lee K. S., Lee J. H., Park I.-C., Lee S.-J., Kim Y. R., Ahn K. J., An I.-S. et al. , (2014). Identification of ultraviolet B radiationinduced microRNAs in normal human dermal papilla cells. Mol. Med. Rep. 10, 1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. H., Tsai R. K., Chen G. S., Yu H. S., Chai C. Y. (1998). Expression of bcl-2, p53 and Ki-67 in arsenical skin cancers. J. Cutan. Pathol. 25, 457–462. [DOI] [PubMed] [Google Scholar]
- Chen J., Yuan T., Liu M., Chen P. (2013). Association between TCF7L2 gene polymorphism and cancer risk: a meta-analysis. PLoS One 8, e71730.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhari S., Saini N. (2014). hsa-miR-4516 mediated downregulation of STAT3/CDK6/UBE2N plays a role in PUVA induced apoptosis in keratinocytes. J. Cell Physiol. 229, 1630–1638.http://dx.doi.org/10.1002/jcp.24608 [DOI] [PubMed] [Google Scholar]
- Dong X. Y., Guo P., Boyd J., Sun X., Li Q., Zhou W., Dong J. T. (2009). Implication of snoRNA U50 in human breast cancer. J. Genet. Genomics 36, 447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokiou A., Atkins G. J., Bouralexis S., Hay S., Raggatt L. J., Cowled P. A., Graves S. E., Clayer M., Findlay D. M. (2001). Expression of alternatively-spliced MDM2 transcripts in giant cell tumours of bone. Int. J. Oncol. 19, 625–632. [DOI] [PubMed] [Google Scholar]
- Felekkis K., Touvana E., Stefanou C., Deltas C. (2010). microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia 14, 236–240. [PMC free article] [PubMed] [Google Scholar]
- Foss K. M., Sima C., Ugolini D., Neri M., Allen K. E., Weiss G. J. (2011). miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J. Thorac. Oncol. 6, 482–488. [DOI] [PubMed] [Google Scholar]
- Gonzalez H., Lema C., Kirken R. A., Maldonado R. A., Varela-Ramirez A., Aguilera R. J. (2015). Arsenic-exposed keratinocytes exhibit differential microRNAs expression profile; potential implication of miR-21, miR-200a and miR-141 in melanoma pathway. Clin. Cancer Drugs 2, 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Roeder R. G. (1997). Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606.http://dx.doi.org/10.1016/S0092-8674(00)80521-8 [DOI] [PubMed] [Google Scholar]
- Honda R., Tanaka H., Yasuda H. (1997). Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27.http://dx.doi.org/10.1016/S0014-5793(97)01480-4 [DOI] [PubMed] [Google Scholar]
- Hrckulak D., Kolar M., Strnad H., Korinek V. (2016). TCF/LEF transcription factors: An update from the internet resources. Cancers 8, 70.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. H., Yang S. A., Wang J. Y., Yu H. S., Lin S. R. (1999). Mutational spectrum of p53 gene in arsenic-related skin cancers from the blackfoot disease endemic area of Taiwan. Br. J. Cancer 80, 1080–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Zhang J., McHenry K. T., Kim M. M., Zeng W., Lopez-Pajares V., Dibble C. C., Mizgerd J. P., Yuan Z. M. (2008). Induction of cytoplasmic accumulation of p53: a mechanism for low levels of arsenic exposure to predispose cells for malignant transformation. Cancer Res. 68, 9131–9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. (2012). Arsenic, metals, fibres, and dusts. IARC Monogr. Eval. Carcinog. Risks Hum. 100(Pt C), 11–465. [PMC free article] [PubMed] [Google Scholar]
- Hunt K. M., Srivastava R. K., Elmets C. A., Athar M. (2014). The mechanistic basis of arsenicosis: Pathogenesis of skin cancer. Cancer Lett. 354, 211–219.http://dx.doi.org/10.1016/j.canlet.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC (1987). Overall Evaluations of Carcinogenicity: An Updating of IARC Monographs Volumes 1 to 42: This Publication Represents the Views and Expert Opinions of an IARC Ad-Hoc Working Group on the Evaluation of Carcinogenic Risks to Humans, Which Met in Lyon, 10-18 March 1987. World Health Organization; Distributed by the World Health Organization, Distribution and Sales Service, Lyon, France; Geneva, Switzerland.
- Iorio M. V., Croce C. M. (2012). MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 4, 143–159.http://dx.doi.org/10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isokpehi R. D., Udensi U. K., Anyanwu M. N., Mbah A. N., Johnson M. O., Edusei K., Bauer M. A., Hall R. A., Awofolu O. R. (2012). Knowledge building insights on biomarkers of arsenic toxicity to keratinocytes and melanocytes. Biomarker Insights 7, 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian Q., An Q., Zhu D., Hui K., Liu Y., Chi S., Li C. (2014). MicroRNA 340 is involved in UVB-induced dendrite formation through the regulation of RhoA expression in melanocytes. Mol. Cell. Biol. 34, 3407–3420.http://dx.doi.org/10.1128/MCB.00106-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R., Li Y., Zhang A., Wang B., Xu Y., Xu W., Zhao Y., Luo F., Liu Q. (2014). The acquisition of cancer stem cell-like properties and neoplastic transformation of human keratinocytes induced by arsenite involves epigenetic silencing of let-7c via Ras/NF-κB. Toxicol. Lett. 227, 91–98. [DOI] [PubMed] [Google Scholar]
- Jin T. (2016). Current understanding and dispute on the function of the Wnt signaling pathway effector TCF7L2 in hepatic gluconeogenesis. Genes Dis. 3, 48–55.http://dx.doi.org/10.1016/j.gendis.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunej T., Godnic I., Horvat S., Zorc M., Calin G. A. (2012). Cross talk between microRNA and coding cancer genes. Cancer J. 18, 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854.http://dx.doi.org/10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Lehman T. A., Modali R., Boukamp P., Stanek J., Bennett W. P., Welsh J. A., Metcalf R. A., Stampfer M. R., Fusenig N., Rogan E. M. et al. , (1993). p53 mutations in human immortalized epithelial cell lines. Carcinogenesis 14, 833–839. [DOI] [PubMed] [Google Scholar]
- Lin L., Liu D., Liang H., Xue L., Su C., Liu M. (2015). MiR-1228 promotes breast cancer cell growth and metastasis through targeting SCAI protein. Int. J. Clin. Exp. Pathol. 8, 6646–6655. [PMC free article] [PubMed] [Google Scholar]
- Lukas J., Gao D. Q., Keshmeshian M., Wen W. H., Tsao-Wei D., Rosenberg S., Press M. F. (2001). Alternative and aberrant messenger RNA splicing of the mdm2 oncogene in invasive breast cancer. Cancer Res. 61, 3212–3219. [PubMed] [Google Scholar]
- Luo J., Li M., Tang Y., Laszkowska M., Roeder R. G., Gu W. (2004). Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 101, 2259–2264.http://dx.doi.org/10.1073/pnas.0308762101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zheng X., Zhou J., Zhang Y., Chen K. (2015). ZEB1 promotes the progression and metastasis of cervical squamous cell carcinoma via the promotion of epithelial-mesenchymal transition. Int. J. Clin. Exp. Pathol. 8, 11258–11267. [PMC free article] [PubMed] [Google Scholar]
- Macfarlane L. A., Murphy P. R. (2010). MicroRNA: Biogenesis, function and role in cancer. Curr. Genomics 11, 537–561.http://dx.doi.org/10.2174/138920210793175895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney M. E. (1996). Arsenic in dermatology. Dermatol. Surg. 22, 301–304.http://dx.doi.org/10.1111/j.1524-4725.1996.tb00322.x [DOI] [PubMed] [Google Scholar]
- Martynova E., Pozzi S., Basile V., Dolfini D., Zambelli F., Imbriano C., Pavesi G., Mantovani R. (2012). Gain-of-function p53 mutants have widespread genomic locations partially overlapping with p63. Oncotarget 3, 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto R., Tada M., Nozaki M., Zhang C. L., Sawamura Y., Abe H. (1998). Short alternative splice transcripts of the mdm2 oncogene correlate to malignancy in human astrocytic neoplasms. Cancer Res. 58, 609–613. [PubMed] [Google Scholar]
- Mazumder D. N. G. (2000). Diagnosis and treatment of chronic arsenic poisoning, http://www.who.int/water_sanitation_health/dwq/arsenicun4.pdf. Accessed January 2018.
- Minamoto K., Mascie-Taylor C. G., Moji K., Karim E., Rahman M. (2005). Arsenic-contaminated water and extent of acute childhood malnutrition (wasting) in rural Bangladesh. Environ. Sci. 12, 283–292. [PubMed] [Google Scholar]
- Moll U. M., Petrenko O. (2003). The MDM2-p53 interaction. Mol. Cancer Res. 1, 1001–1008. [PubMed] [Google Scholar]
- Murea M., Ma L., Freedman B. I. (2012). Genetic and environmental factors associated with type 2 diabetes and diabetic vascular complications. Rev. Diabet. Stud. 9, 6–22.http://dx.doi.org/10.1900/RDS.2012.9.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi J., Diwan B. A., Sun Y., Liu J., Qu W., He Y., Styblo M., Waalkes M. P. (2008). Arsenic-induced malignant transformation of human keratinocytes: Involvement of Nrf2. Free Radic. Biol. Med. 45, 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser I., Golob M., Buettner R., Bosserhoff A. K. (2003). Upregulation of HMG1 leads to melanoma inhibitory activity expression in malignant melanoma cells and contributes to their malignancy phenotype. Mol. Cell. Biol. 23, 2991–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agency for Toxic Substances and Disease Registry. (2014). Priority List of Hazardous Substances. Available at: http://www.atsdr.cdc.gov/SPL/index.html [Google Scholar]
- Rosales-Reynoso M. A., Arredondo-Valdez A. R., Juarez-Vazquez C. I., Wence-Chavez L. I., Barros-Nunez P., Gallegos-Arreola M. P., Flores-Martinez S. E., Moran-Moguel M. C., Sanchez-Corona J. (2016). TCF7L2 and CCND1 polymorphisms and its association with colorectal cancer in Mexican patients. Cell. Mol. Biol. 62, 13–20. [PubMed] [Google Scholar]
- Sakaguchi K., Herrera J. E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C. W., Appella E. (1998). DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar A. M., Ostrosky-Wegman P. (2016). Genotoxicity States JC, ed. Arsenic: Exposure Sources, Health Risks, and Mechanisms of Toxicity. John Wiley & Sons Inc, Hoboken, NY. [Google Scholar]
- Sandoval M., Morales M., Tapia R., del Carmen Alarcon L., Sordo M., Ostrosky-Wegman P., Ortega A., Lopez-Bayghen E. (2007). p53 response to arsenic exposure in epithelial cells: Protein kinase B/Akt involvement. Toxicol. Sci. 99, 126–140. [DOI] [PubMed] [Google Scholar]
- Sato F., Tsuchiya S., Meltzer S. J., Shimizu K. (2011). MicroRNAs and epigenetics. FEBS J. 278, 1598–1609.http://dx.doi.org/10.1111/j.1742-4658.2011.08089.x [DOI] [PubMed] [Google Scholar]
- Schuster K., Fan L., Harris L. C. (2007). MDM2 splice variants predominantly localize to the nucleoplasm mediated by a COOH-terminal nuclear localization signal. Mol. Cancer Res. 5, 403–412.http://dx.doi.org/10.1158/1541-7786.MCR-06-0146 [DOI] [PubMed] [Google Scholar]
- Shangary S., Wang S. (2008). Targeting the MDM2-p53 interaction for cancer therapy. Clin. Cancer Res. 14, 5318–5324.http://dx.doi.org/10.1158/1078-0432.CCR-07-5136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W., Wang D., Chiang Y. T., Ip W., Zhu L., Xu F., Columbus J., Belsham D. D., Irwin D. M., Zhang H. (2013). The Wnt signaling pathway effector TCF7L2 controls gut and brain proglucagon gene expression and glucose homeostasis. Diabetes 62, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Ray R., Rajeswari M. R. (2008). Overexpression of high mobility group (HMG) B1 and B2 proteins directly correlates with the progression of squamous cell carcinoma in skin. Cancer Invest. 26, 843–851.http://dx.doi.org/10.1080/07357900801954210 [DOI] [PubMed] [Google Scholar]
- Shieh S. Y., Ikeda M., Taya Y., Prives C. (1997). DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91, 325–334.http://dx.doi.org/10.1016/S0092-8674(00)80416-X [DOI] [PubMed] [Google Scholar]
- Sigalas I., Calvert A. H., Anderson J. J., Neal D. E., Lunec J. (1996). Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: Transforming ability and frequent detection in human cancer. Nat. Med. 2, 912–917.http://dx.doi.org/10.1038/nm0896-912 [DOI] [PubMed] [Google Scholar]
- Smith A. H., Steinmaus C. M. (2009). Health effects of arsenic and chromium in drinking water: recent human findings. Annu. Rev. Public Health 30, 107–122.http://dx.doi.org/10.1146/annurev.publhealth.031308.100143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soifer H. S., Rossi J. J., Sætrom P. (2007). MicroRNAs in disease and potential therapeutic applications. Mol. Ther. 15, 2070–2079. [DOI] [PubMed] [Google Scholar]
- States J. C. (2016). Arsenic: Exposure Sources, Health Risks, and Mechanisms of Toxicity. Hoboken, NJ: John Wiley & Sons, Inc. [Google Scholar]
- States J. C. (2015). Disruption of mitotic progression by arsenic. Biol. Trace Elem. Res. 166, 34–40.http://dx.doi.org/10.1007/s12011-015-0306-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Štros M., Ozaki T., Bačı´ková A., Kageyama H., Nakagawara A. (2002). HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J. Biol. Chem. 277, 7157–7164. [DOI] [PubMed] [Google Scholar]
- Su L. J., Mahabir S., Ellison G. L., McGuinn L. A., Reid B. C. (2011). Epigenetic contributions to the relationship between cancer and dietary intake of nutrients, bioactive food components, and environmental toxicants. Front. Genet. 2, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Chen S., Zhao X., Yan M., Fang Z., Wang H., Zhao J., Sun M., Han X., Chen W. et al. , (2015). Dysregulated miR-645 affects the proliferation and invasion of head and neck cancer cell. Cancer Cell Int. 15, 87.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Pi J., Wang X., Tokar E. J., Liu J., Waalkes M. P. (2009). Aberrant cytokeratin expression during arsenic-induced acquired malignant phenotype in human HaCaT keratinocytes consistent with epidermal carcinogenesis. Toxicology 262, 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed D. N., Khan M. I., Shabbir M., Mukhtar H. (2013). MicroRNAs in skin response to UV radiation. Curr. Drug Targets 14, 1128–1134.http://dx.doi.org/10.2174/13894501113149990184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborini E., Della Torre G., Lavarino C., Azzarelli A., Carpinelli P., Pierotti M. A., Pilotti S. (2001). Analysis of the molecular species generated by MDM2 gene amplification in liposarcomas. Int. J. Cancer 92, 790–796. [DOI] [PubMed] [Google Scholar]
- Tang D., Kang R., Zeh H. J. III, Lotze M. T. (2010). High-mobility group box 1 and cancer. Biochim. Biophys. Acta 1799, 131–140.http://dx.doi.org/10.1016/j.bbagrm.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J. S., Ebert M. S., van Oudenaarden A. (2010). Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol. Cell 38, 140–153.http://dx.doi.org/10.1016/j.molcel.2010.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Jiang H., Zhu H., Zhang H., Gong J., Zhang L., Ding Q. (2013). Overexpression of high mobility group box 1 and 2 is associated with the progression and angiogenesis of human bladder carcinoma. Oncol. Lett. 5, 884–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. T., Farzaneh F. (2012). Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 12, 84–88. [DOI] [PubMed] [Google Scholar]
- Williams K. A., Kolappaswamy K., Detolla L. J., Vucenik I. (2011). Protective effect of inositol hexaphosphate against UVB damage in HaCaT cells and skin carcinogenesis in SKH1 hairless mice. Comp. Med. 61, 39–44. [PMC free article] [PubMed] [Google Scholar]
- Xu X. M., Qian J. C., Deng Z. L., Cai Z., Tang T., Wang P., Zhang K. H., Cai J. P. (2012). Expression of miR-21, miR-31, miR-96 and miR-135b is correlated with the clinical parameters of colorectal cancer. Oncol. Lett. 4, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Woods N. T., Piluso L. G., Lee H. H., Chen J., Bhalla K. N., Monteiro A., Liu X., Hung M. C., Wang H. G. (2009). p53 acetylation is crucial for its transcription-independent proapoptotic functions. J. Biol. Chem. 284, 11171–11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Qi Y., Chen Q., Wang Z., Jin X., Gao J., Fu J., Xiao X., Zhou Z. (2007). The over-expression of p53 H179Y residue mutation causes the increase of cyclin A1 and Cdk4 expression in HELF cells. Mol. Cell Biochem. 304, 219–226. [DOI] [PubMed] [Google Scholar]
- Zheng T., Wang J., Zhao Y., Zhang C., Lin M., Wang X., Yu H., Liu L., Feng Z., Hu W. (2013). Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat. Commun. 4, 2996.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.