Abstract
Advances in the methods for detecting protein–DNA interactions have played a key role in determining the directions of research into the mechanisms of transcriptional regulation. The most recent major technological transformation happened a decade ago, with the move from using tiling arrays [chromatin immunoprecipitation (ChIP)-on-Chip] to high-throughput sequencing (ChIP-seq) as a readout for ChIP assays. In addition to the numerous other ways in which it is superior to arrays, by eliminating the need to design and manufacture them, sequencing also opened the door to carrying out comparative analyses of genome-wide transcription factor occupancy across species and studying chromatin biology in previously less accessible model and nonmodel organisms, thus allowing us to understand the evolution and diversity of regulatory mechanisms in unprecedented detail. Here, we review the biological insights obtained from such studies in recent years and discuss anticipated future developments in the field.
Keywords: ChIP-seq, evolution, chromatin, transcription factors, eukaryotes
Introduction
The locations of transcription factor occupancy and the distribution of chromatin marks along the genome constitute indispensable information for understanding the mechanisms of gene expression and its regulation. Chromatin immunoprecipitation (ChIP) has been the main tool for their characterization since its invention in the 1980s [1–3]. ChIP and its numerous variants rely on the chemical cross-linking (typically using formaldehyde) of proteins to DNA, the subsequent enrichment of cross-linked fragments bound to the protein of interest, followed by reversal of cross-links and measurement of the purified DNA. Improvements in the tools for carrying out the latter task has made ChIP a progressively more powerful assay over the years. Initially, qPCR was used to assay enrichment over a small number of predefined sites [4]. Later, ChIP was coupled with microarrays (ChIP-on-Chip/ChIP-Chip) [5–10], which allowed a large number of targets to be probed. ChIP-Chip assays could in principle encompass the whole genome if tiling arrays were used, but in practice, this was relatively straightforward to accomplish only for small genomes such as those of yeast, Arabidopsis [11], flies [12] or worms [13], and most array studies focused on promoters and subsets of noncoding regions. In addition to usually falling short of fully genome-wide coverage, arrays also had numerous other issues having to do with noise levels, detailed resolution, signal range, interoperability of different platforms and others.
Early efforts to overcome these limitations by moving the ChIP readout from arrays to direct sequencing in the form of ChIP-PET (paired-end tagging) [14, 15] also suffered from difficulties related to low throughput, cumbersome library construction protocols and the short read tags being generated by these methods.
The advent of high-throughput sequencing in the mid-00s overcame most of these limitations, leading to the development of ChIP-seq [16–19] in 2006–07, which provided truly genome-wide coverage (with the exception of highly repetitive areas of the genome), with unprecedented sensitivity and resolution. In the decade since, ChIP-seq has become the workhorse of functional genomic studies of gene regulation. The genomic occupancy of hundreds of human, mouse, worm and fly transcription factors and the genomic distribution of histone marks in hundreds of human cell types have been systematically mapped using ChIP-seq by large-scale efforts such as the ENCODE [20], mouseENCODE [21, 22], modENCODE [13, 12], Roadmap Epigenomics [23], BLUEPRINT Epigenome and International Human Epigenome Consortium [24] and others, in addition to the method having been used in thousands of other scientific publications.
But ChIP-seq did not just resolve the technical limitations preventing the comprehensive high-resolution characterization of protein occupancy in the genomes of model organisms, it also opened the door to doing the same in any species for which a genome sequence is available. It did so by removing the major initial barrier of designing and manufacturing arrays, a slow and expensive process that was out of reach for many labs in the past. This enabled several lines of research that were not viable in the prior years. Studies of comparative analysis of transcription factor occupancy across species have now begun to unravel the biochemical and evolutionary factors behind its conservation and divergence. The direct mapping of regulatory elements in nonmodel species is helping us understand morphological changes during embryonic development. Finally, we have the tools to study transcriptional and regulatory biology across the whole tree of life, allowing us to track the deep evolutionary roots of its logic.
While these studies are still largely in their early stages in groups other than human, mouse, Drosophila and Caenorhabditiselegans, we review some of the most important insights they have delivered so far and discuss future directions for work in the field.
The evolution of transcription factors occupancy
Gene expression in the vast majority of eukaryotes is regulated largely by the orchestrated action of sequence-specific transcription factors binding to proximal and distal cis-regulatory elements. Knowledge of the mechanisms of evolution of transcription factor binding is therefore vital for a complete understanding of gene regulatory networks (GRNs), as they exist and function in extant organisms.
An intuitive expectation following from the importance of gene regulation for the proper functioning of the cell is that cis-regulatory elements would be highly conserved in evolution. This is indeed generally true. The sequencing of the human, mouse and other vertebrate genomes revealed that only not much >∼2% of their sequence consists of the exons of protein coding and noncoding genes [25], but whole-genome comparisons between these genomes identify a considerably higher fraction of conserved sequence, up to 10% [26, 27], with conserved noncoding elements (the bulk of which are thought to likely be cis-regulatory elements) making up the difference. However, the absence of conservation does not necessarily imply absence of function, and overall sequence conservation does not necessarily mean strict functional conservation (i.e. the same transcription factors need not be associated with a given conserved element in different species), meaning that the question to what extent regulatory networks themselves are conserved or divergent between species cannot be answered based on sequence alone.
Patterns of regulatory element evolution
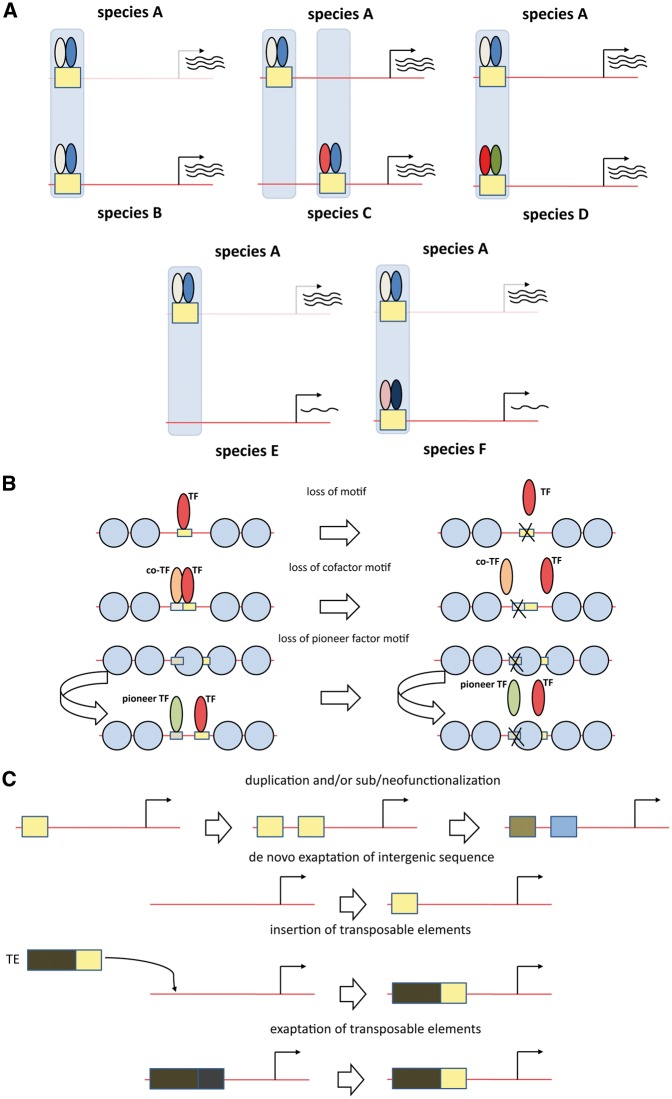
In the late 1990s and early 2000s, initial approaches toward answering these questions used sequence comparisons between well-characterized loci in different species, tracking the conservation of individual transcription factor-binding sites, and experimental in vivo examination of expression patterns driven by orthologous or synthetic cis-regulatory elements. Application of these techniques to the endo16 gene in the sea urchins Strongylocentrotus purpuratus and Strongylocentrotus droebachiensis [28], to even skipped enhancers in multiple Drosophila and other fly species [29–31] and to promoters of mammalian genes [32] revealed that functional conservation can be maintained despite considerable divergence at the sequence level, by means of compensatory mutations and of turnover of nonconserved regulatory elements (Figure 1A).
Figure 1.
Patterns and mechanisms of regulatory element, transcription factor occupancy and gene expression evolution. (A) Patterns of conservation and divergence. Functional conservation of gene expression (top row) can be achieved through conservation of both regulatory elements and transcription factor occupancy (left), but it can also be maintained in the presence of significant turnover, both of occupancy and at the sequence level, either through replacement of individual transcription factors occupying an orthologous regulatory region (right) or by the evolution of nonorthologous regulatory regions (middle). Loss of regulatory elements and alterations in transcription factor occupancy can also lead to changes in gene expression (bottom row). (B and C) Mechanisms of transcription factor occupancy and regulatory element evolution. (B) Mechanisms of loss of transcription factor binding: loss of cognate motif (top), loss of cofactor binding (middle), loss of pioneer factor binding (bottom); (C) Mechanisms of gain of transcription factor binding and de novo evolution of regulatory elements: duplication and/or sub/neofunctionalization, direct de novo exaptation of existing intergenic space, insertion of TEs, exaptation of ancestral TE sequences.
Yet to what extent such observations generalize could only be answered using genomic techniques. Pioneering studies addressing the question first appeared in 2007 [33], using ChIP-Chip over a set of orthologous human and mouse putative regulatory regions to compare the occupancy of the FOXA2, HNF1A, HNF4A and HNF6 transcription factors in hepatocytes of the two species. Strikingly, depending on the factor, between 41 and 89% of binding events appeared to be species-specific, and conserved occupancy around promoters was frequently observed in the absence of direct orthologous conservation of binding events. In a different study, only 20% of E2F4 sites were found to be conserved between human and mouse [34].
ChIP-Chip was also used to map the occupancy of Ste12 and Tec1 in the yeasts Saccharomyces cerevisiae, Saccharomycesmikatae and Saccharomycesbayanus, revealing only 20–21% conservation across all three species [35]; large turnover was also observed for Mcm1 in S. cerevisiae, Kluyveromyces lactis and Candida albicans using the same approach [36].
The advent of ChIP-seq eventually allowed truly genome-wide corroboration of these initial results in addition to much more detailed insights into the mechanisms driving conservation and divergence of occupancy. Mapping of transcription factor binding in the livers of five vertebrates (human, mouse, dog, opossum and chicken, separated by as much as 300 My of evolution) [37] showed conservation levels of 10–22% between placental mammals and of 6% between humans and opossum for CEBPa and HNF4A, and only 2% between humans and chicken for CEBPa. Half of lineage-specific losses of binding are associated with lineage-specific occupancy events in the nearby genomic vicinity, suggesting widespread regulatory element turnover through the evolution of compensatory binding, with novel nonorthologous regulatory elements taking over the function of the ones having been lost.
At the opposite extreme of phylogenetic distances, HNF4A, CEBPA and FOXA1 were profiled in livers from rat and five closely related mouse strains and species [38]. Again, significant divergence of occupancy was observed, with 30% conservation of FOXA1 binding events for the 20 My phylogenetic distance between mouse and rat, 40% within 6 My and 70% for divergence within 1 Mya.
Large-scale divergence of occupancy has been reported by all other studies in mammals. A comparative study of HNF4A, CEBPA, ONECUT1 and FOXA1 in human, macaque, mouse, rat and dog found 21 and 37% conservation between human and macaque, 21–31% between mouse and rat and as little as 7% for more distal relationships [39]. Comparison of PPARγ ChIP-seq profiles between human and mouse adipocytes also revealed that only ∼20% of occupancy sites are shared [40]. The mouseENCODE project carried out comparative analysis of several dozen factors between mouse and human [22, 41, 42]. In this analysis, while up to half of identified occupied sites were not alignable to the other species at the sequence level, an even smaller fraction (varying between different factors but generally between 15 and 33%) of conserved occupancy was observed, underscoring the larger extent of sequence conservation relative to occupancy conservation.
Widespread turnover has also been observed even for polymerase (Pol) III occupancy. RNA Pol III primarily transcribes transfer RNAs (tRNAs) plus a few other classes of small RNAs; accordingly, it associates with localized genomic regions around these genes. Of note, many copies of genes for the same tRNA isotype can be found in mammalian genomes. Comparison of Pol III binding between six mammals demonstrated that a large fraction of occupancy sites are species-specific, but overall binding at the tRNA isotype level was largely conserved [43].
Mechanisms of regulatory element evolution
Comparative ChIP-seq studies have thus at this point firmly established that regulatory element turnover is a fundamental feature of the functional evolution of mammalian genomes. Gene expression regulation in functionally equivalent cell types is frequently conserved, even though substantial fractions of the regulatory elements driving it have diverged. But what exactly are the mechanisms through which individual factor occupancy and larger regulatory elements are gained and lost?
Divergence of transcription factor sequence specificity does not appear to be the answer. Although orthologous transcription factors can on occasions evolve distinct sequence preferences [44], large-scale characterization of transcription factor sequence specificity using methods such as HT-SELEX [45] has shown that it is usually conserved across deep evolutionary distances, going back even as far as the dawn of metazoan evolution.
Genomic alterations in cis are therefore the main drivers of regulatory evolution, a conclusion supported by the observation that human chromosome 21 transplanted in mice preserves human-specific gene expression and transcription factor occupancy profiles [46].
The most obvious mechanism for losing occupancy is loss-of-function mutations in the corresponding transcription factor motif. One of the surprising trends to emerge from comparative ChIP-seq studies, both in mammals and in flies, is that such changes often do not account for even a majority of lineage-specific occupancy losses [37, 38, 47, 48]. Instead, the explanation for a significant portion of such cases appears to be the loss of motifs of cofactors or pioneer factors (Figure 1B). This conclusion (substantial binding differences without obvious sequence changes) is also corroborated by studies of the effects of within-population genetic variation on transcription factor occupancy [49–51], as well as by observed patterns of binding conservation, such as the finding that cobinding factors frequently lose or gain occupancy in concert in different species [38, 41].
Some of the key known mechanisms of regulatory element gain include the duplication in cis of existing elements followed by the subfunctionalization of the two new copies or the neofunctionalization of one or both, exaptation from ancestral DNA and derivation from either recent or more ancient transposable element (TE) insertions, as TEs often contain functional transcription factor-binding sites and active promoter/enhancer elements (Figure 1C). Comparative ChIP-seq analyses have helped clarify the relative roles of these processes in evolution.
Exaptation of TEs turns out to be particularly important for certain kinds of regulatory elements and factors, a conclusion that emerged early in the history of global transcription factor mapping studies [52]. For example, CTCF binding sites often arise thanks to insertions of short interspersed nuclear elements (SINEs), which contain sequences with high affinity for CTCF [53]. Other important examples involve the neuronal-gene repressor NRSF/REST, which frequently evolves new binding sites through the exaptation of low-affinity sequences found in TEs such as ERV1 [54], and the contribution of TEs to the rewiring of the core regulatory network in mammalian embryonic stem cells [55]. Of note, TE exaptation seems to be most important for factors with long recognition motifs (to which group both CTCF and NRSF/REST belong), as binding sites for such factors are naturally more difficult to evolve through random drift from ancestral sequence.
Several efforts have compared regulatory elements as a whole (rather than individual transcription factors, using ChIP-seq against the enhancer-associated H3K27ac and the promoter-associated H3K4me3 histone marks) in multiple mammalian species [22, 41, 56–59], which has allowed first, a comparison of the rates of evolution between different types of elements, and second, an assessment of the relative contribution of the different mechanisms for their gain. Remarkably, promoters are much more conserved than enhancers; most promoters are highly conserved between any two mammals, while in contrast enhancer elements turn over rapidly. Exapted TEs (long terminal repeats and SINEs) are enriched in newly evolved promoters, both in promoters arising from recent and from ancestral DNA regions, with the former constituting the majority [57]. In contrast, most new enhancers appear to evolve from exaptation of ancestral DNA sequences, and TEs are highly enriched only in enhancers evolving from phylogenetically recent sequences [57, 59].
The conservation of regulatory elements unsurprisingly appears to be correlated to functional constraints. That such constraints acting on promoters are higher than those applying to any individual enhancer is fairly obvious, thus it is not surprising that promoters are more highly conserved. Probably related to such constraints is the higher conservation of individual transcription factor occupancy in promoters and promoter-proximal regulatory regions than in distal ones, observed both in mammals and insects [41, 48, 60], as is the observation by the ENCODE and mouseENCODE consortia that regulatory elements active in multiple tissues and likely having pleiotropic effects exhibit higher degrees of occupancy conservation, as do transcription factors that co-associate with many other factors.
To what extent these principles of regulatory conservation apply to other phylogenetic groups is of considerable interest, and remains to be answered by future expansions of the range of lineages covered by comparative functional genomic studies.
Evolutionary dynamics of regulatory element turnover
Data for lineages other than mammals are also of key importance for answering the other major question regarding regulatory evolution: What are the evolutionary forces driving it, i.e. what are the relative contributions of mutation, selection and genetic drift to the observed patterns of conservation and divergence?
The only other group in which extensive comparative ChIP-seq studies have been carried out is flies, and the picture painted by these studies has generally been rather different from what is seen in mammals, mostly returning much higher estimates of occupancy conservation. An early analysis of six developmental transcription factors in Drosophila melanogaster and Drosophila yakuba reported that fewer than 1–5% (depending on the factor) of ChIP-seq peaks observed in one species were clearly absent or displaced in the other [61]. Mapping of Twist occupancy in six Drosophila species [48] estimated a 60% conservation of binding events between D. melanogaster and Drosophilapseudoobscura, which are estimated (according to substitutions per neutral site) to be as equally diverged as humans and chickens. On the other hand, comparative analysis of four transcription factors (BCD, GT, HB and KR) involved in early anterior–posterior patterning in D. melanogaster, D. yakuba, D. pseudoobscura and Drosophilavirilis returned lower conservation estimates, with 15–38% of regions bound in D. melanogaster also bound in D. yakuba and D. pseudoobscura [60].
The one notable difference between flies and mammals clearly going in the other direction in terms of conservation concerns the insulator protein CTCF. CTCF occupancy is generally the most conserved of any transcription factor in mammals [53, 62, 63], likely reflecting its critical role in partitioning the spatial organization of the genome but has been reported to diverge rapidly in Drosophila [64]. This is possibly because flies contain multiple insulator proteins (see further discussion below), of which CTCF is not the most important, relaxing the constraints on the evolution of its occupancy.
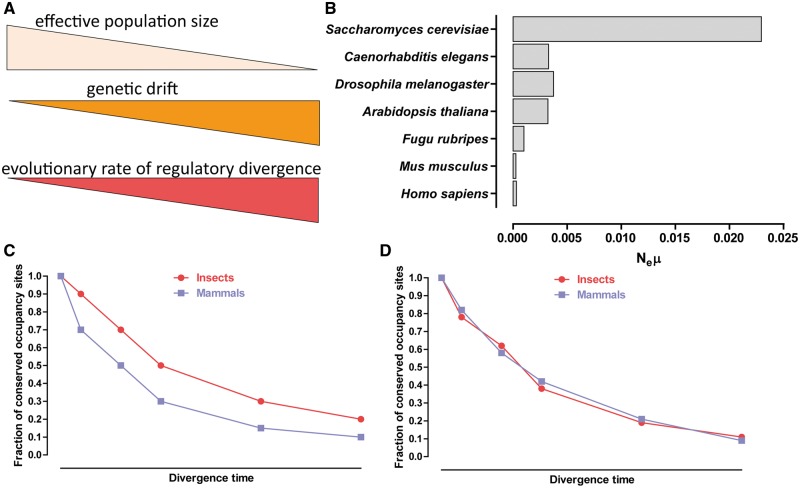
How are the observed patterns of conservation and divergence to be interpreted? Transcription factor-binding sites and regulatory element evolution have attracted considerable attention from theoretical geneticists in the past [65, 66, 67]. Based on population genetic considerations and available data on key population genetic parameters across eukaryotes [28, 29, 67], the following model, centered on the balance between selection and genetic drift, has been proposed. The power of natural selection to eliminate maladaptive alleles and fix adaptive ones is limited by the effective population size (Ne) of real-life populations. More specifically, when , where s is the selection coefficient associated with them, alleles behave effectively neutrally. Thus, in populations with large values of Ne, selection is highly efficient, and even small selective differences are ‘visible’ to it, and vice versa—in populations with small Ne, slightly maladaptive genomic changes can drift to fixation because of the relaxed strength of selection. Most mutations that eventually lead to regulatory turnover are expected to be maladaptive to some extent because they can lead to misregulation of gene expression. It is thus natural to expect lineages with small effective population size to exhibit increased rates of regulatory turnover (Figure 2A). More specifically, Ne is highest in prokaryotes, and then generally decreases from prokaryotes to unicellular eukaryotes to smaller invertebrates, with large-bodied complex multicellular organisms having the smallest effective population sizes (Figure 2B). Following this line of reasoning, faster cis-regulatory evolution would be expected in vertebrates than in flies because of the significantly lower Ne of the former (Figure 2C).
Figure 2.
Theoretical expectations regarding the evolutionary dynamics of transcription factor occupancy and interpretations of existing data sets. (A). Population genetic theoretical considerations lead to an expectation of faster turnover of regulatory sites in organisms with low effective population sizes (Ne) than in species with large Ne. (B) Distribution of effective population size values in some of the main model systems [68]. Shown is the product of Ne and the mutation rate μ, which can be most directly estimated empirically, unlike Ne alone [68, 69]. (C) The compilation of individual studies summarized in [47] suggested higher rates of regulatory site turnover in mammals than in flies. (D). In contrast, a reanalysis of several data sets in flies and mammals using a uniform data processing pipeline found similar rates of regulatory divergence within the two groups [70].
Does available data confirm these theoretical expectations? A review of published comparative studies appeared in 2014 [47], and it found concordance with the model presented above, with reported transcription factor occupancy conservation being higher in flies than what is observed in mammals (Figure 2C). However, it was based on compiling estimates of the fraction of conserved occupied sites directly taken from individual studies, which used different analysis methods and therefore might not have been directly comparable with each other. A reanalysis of a subset of several factors was published later in 2015 [70]. It used uniform data processing and analysis procedures across flies and mammals and reported that in fact the two lineages exhibit similar rates of regulatory evolution (Figure 2D). However, while it did apply an uniform analytical pipeline, it did not fully control for all potential relevant variables (such as, for example, organismal generation times), and it also focused on only a small set of transcription factors.
The issue of how empirically observed rates of regulatory divergence fit with theoretical considerations will have to be resolved in the future by a much expanded compendium of transcription factors, the application of uniform data processing pipelines and, most importantly, the inclusion of a much wider coverage of lineages, preferably with different population genetic environments (such as, for example, unicellular eukaryotes with high effective population sizes [71]), and covering a wide spectrum of evolutionary distances. Such studies are beginning to emerge in other metazoan systems. For example, ChIP-seq was recently used to compare occupancy by the developmental transcription factor Tbrain (Tbr) in the sea star Patiria miniata and the sea urchin S.purpuratus [44], which diverged ∼450–500 Mya; ∼10% of sites appear to be conserved in this case. Much more data will certainly become available in the future, but more systematic approaches to experimental design and data collection will undoubtedly also be helpful for its integration into a coherent picture of evolutionary dynamics.
Mapping the rewiring of gene regulatory networks
The development of multicellular organisms is driven by GRNs, primarily composed of transcription factors acting in concert to specify developmental patterning. GRNs are understood to have an ultimately hierarchical structure, beginning with the fertilized zygote. Each developmental phase specifies the events happening in the subsequent stages through the activation or repression of certain GRN sub-circuits in defined regions of the embryo, with individual terminally differentiated cell types residing at the bottom of these hierarchies [72, 73].
The key to understanding the evolution of organismal morphology and development therefore lies in large part in studying the rewiring of GRNs over evolutionary time. As a well-known example, the pelvic apparatus of the threespine stickleback fish (Gasterosteus aculeatus) usually contains prominent spines, but in multiple freshwater stickleback populations, these structures have been partially or entirely lost (possibly because of the absence of predatory fish, nutrient limitations or other factors). Such losses can be tracked to the recurrent loss of an enhancer for the Pitx1 gene [74], which encodes a homeodomain transcription factor and is involved in the specification of hindlimb development in vertebrates.
Painstaking work over many years aimed at picking apart the details of the cis-regulation of individual developmental genes has resulted in detailed GRNs maps in several systems, with the one of early sea urchin embryonic development being perhaps the most famous example [75]. Such GRNs have often been presented in the form of network circuit diagrams, in which different factors have explicit binary relationships with each other. The conceptual clarity of depictions of this sort is highly attractive, and finding a way for regulatory relationships to be comprehensively mapped onto such diagrams over the whole genome is the ultimate goal/holy grail for the field.
Based on a naive understanding of the relationship between transcription factor occupancy and gene expression, ChIP-seq promised to provide maps of these circuits on a genome-wide scale, and across lineages, something especially attractive given that many striking developmental and morphological innovations in metazoans are to be found outside of the traditional model systems. The reality has turned out to be significantly more complicated though. First, transcription factor occupancy exhibits a continuum, with many more weaker sites than strong ones, and without a clear delineation between true binding events and background [76, 77]. Second, while there is some evidence that low-level occupancy sites are more frequently nonfunctional [78], there is no absolute relationship between ChIP signal strength and functional effects on nearby genes or a clear way to clarify that relationship on a per-site basis from ChIP data alone. Third, the combinatorial complexity of transcription factor binding has turned out to be high, and it is not always clear which of all the factors associated with a given putative regulatory element are of critical importance for its functioning. Similar redundancy seems to be also widespread at the level of the set of regulatory elements associated with a given gene [79]. Finally, and perhaps most important, most putative regulatory sites in metazoans with large genomes are distally located from promoters. Consequently, identifying which putative enhancers regulate which promoter is not a trivial task without performing additional experiments.
These inherent complexities of transcription regulation in eukaryotes are perhaps the reason why while traditional circuit diagrams are by no means obsolete, deriving them from functional genomic data alone has been difficult, and why mapping their rewiring on a genome-wide across evolution has not been widely done yet. However, ChIP-seq provides highly useful lists of candidate regulatory elements, which can be subsequently characterized in more depth, especially with the now widespread CRISPR-based tools for genome and epigenome editing [80, 81] provided that the organism studied is amenable to such manipulations. ChIP-seq is also informative about global trends at the genomic level. Several studies using it to track regulatory changes during evolution and/or development of nonmodel species have been published over the past few years.
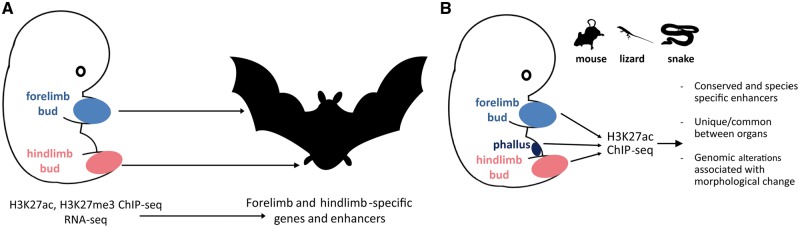
An example is the mapping of epigenomic and transcriptomic difference between forelimbs and hindlimbs in bat development using Miniopterus natalensis as a model system [82] (Figure 3A). Unlike most tetrapods, bat forelimbs and hindlimbs develop into morphologically different from each other structures. The forelimbs become elongated webbed wings, while the hindlimbs develop into fairly short legs. The integrated analysis of H3K27ac and H3K27me3 ChIP-seq, transcriptomic and comparative genomic data identified pathways involved in the differential patterning of the two structures, including the Wnt/β-catenin, Wnt-PCP and BMP signaling pathways as well as several ribosomal proteins. Increased forelimb mesenchymal condensation apparently driven by diminished β-catenin signaling seems to be particularly important for forelimb growth and patterning.
Figure 3.
Examples of using ChIP-seq to map GRNs in development and evolution. (A). Cataloging enhancers involved in patterning the morphology of bat limbs. The forelimb bud of bats develops into elongated webbed wings, while the hindlimb bud produces much shorter legs. H3K27ac and H3K27me3 ChIP-seq and RNA-seq were applied to developing limb buds to chart the developmental enhancer landscape involved in the specification of these structures [82]. (B) Understanding the role of limb-specific enhancers in snake evolution. Snakes lack legs, but H3K27ac ChIP-seq in mouse and lizard embryos and comparative genomics reveal that a substantial portion of limb enhancers are in fact conserved in snakes, one major reason for which is their role during phallus development, where a similar developmental program is deployed [83].
ChIP-seq has also provided insights into vertebrate limb development in a different context. Mapping of H3K27ac enriched regions in mouse and lizard developing embryos and comparative genomic analysis relative to available snake genomes has revealed that numerous limb-specific enhancers are retained in snakes, even though snakes are limbless and have been so for nearly 100 million years [83] (Figure 3B). A major reason for this relatively unexpected conservation appears to be that many limb enhancers are also used during the development of genital structures, which, unlike limbs, are not reduced in snakes. Detailed functional testing of an enhancer of the Tbx4 gene (which is important for hindlimb development) showed loss of its hindlimb activity in snakes as well as its importance for both hindlimb and urogenital development in mice.
The application of functional genomic approaches to developmental questions in nonmodel systems is still in its early days, but we can expect its future expansion to shed light on the genomic foundations of developmental gene regulation, including in metazoan lineages outside vertebrates such as arthropods and echinoderms. Insects in particular provide an endless diversity of morphological innovations. With large numbers of sequenced genomes expected to come online through efforts such as the i5K Consortium [84], the infrastructure for research in that direction will become available in the near future. Epigenomic maps were already used to annotate the regulatory genome in the context of head development in the butterfly Heliconius erato [85], and many more studies of this type will surely be carried out moving forward.
Charting the evolution of gene regulatory mechanisms
Transcription and transcriptional regulation in mammals, in which the relevant processes have been studied in greatest detail, is driven by the interplay of certain classes of regulatory elements—chiefly promoters, enhancers acting at a distance and insulators blocking long-range interactions and the spread in cis of epigenetic states. These mechanisms of regulation fit well with the overall architecture of large-sized vertebrate genomes, with their sparsely distributed coding regions, and large expanses of intergenic and intronic space. Different classes of regulatory elements are marked by specific subsets of chromatin-binding proteins and histone marks, and so are genomic regions associated with active transcription, heterochromatin, etc., defining characteristic chromatin states [86]. Well-known examples include the association of H3K27ac and H3K4me1 with enhancer elements [87–89], of H3K4me3 with active promoters of genes [90, 91], of H3K36me3 with active transcription [92, 93], of H3K27me3 and H3K9me3 with repressed facultative and constitutive heterochromatin [94–96] and the demarcation of insulator elements by CTCF and cohesin [97, 98]. More generally, specific highly choreographed arrays of dynamic histone modifications are associated with all aspects of chromatin biology, including transcription [99], splicing [100], replication and mitosis [101], DNA repair [102] and others. Histone modifications serve as platforms for the recruitment of effector proteins containing corresponding recognition domains, and thus constitute what is often referred to as the ‘histone code’ [103]. The complexity of histone marks is dizzying [104]; hundreds of them have been identified in mammalian cells by proteomics studies, but so far only a fraction have been carefully studied and are well understood in mechanistic detail.
A remarkable feature of histone proteins is their extreme conservation across eukaryotes, especially at sites carrying key modifications involved in the processes listed above [105, 106]. This implies an accordingly strong general (but not necessarily absolute) conservation of the corresponding biochemical processes these residues are associated with, and that at the least a core set of histone marks involved in the transcriptional cycle, heterochromatin formation and a few other areas date back to the last common eukaryotic ancestor (LECA). It also makes the biology of the few organisms that represent major exceptions from the usual rules all the more interesting.
LECA may have had a small and compact genome, as did the more recent ancestors of metazoans, with the baroque mammalian genome and transcription regulation architecture evolving later in the process of metazoan diversification. What the deep evolutionary factors behind these developments are and what their relationship to the phenotypic complexity of mammals is, i.e. is the regulation-from-a-distance, enhancers/promoters/insulators model a prerequisite for building a highly complex multicellular organism such as us, remains an open question.
As we discuss below, understanding of chromatin biology across the eukaryotic phylogeny will be of crucial importance for answering these questions.
The known eukaryotic diversity in the 2010s
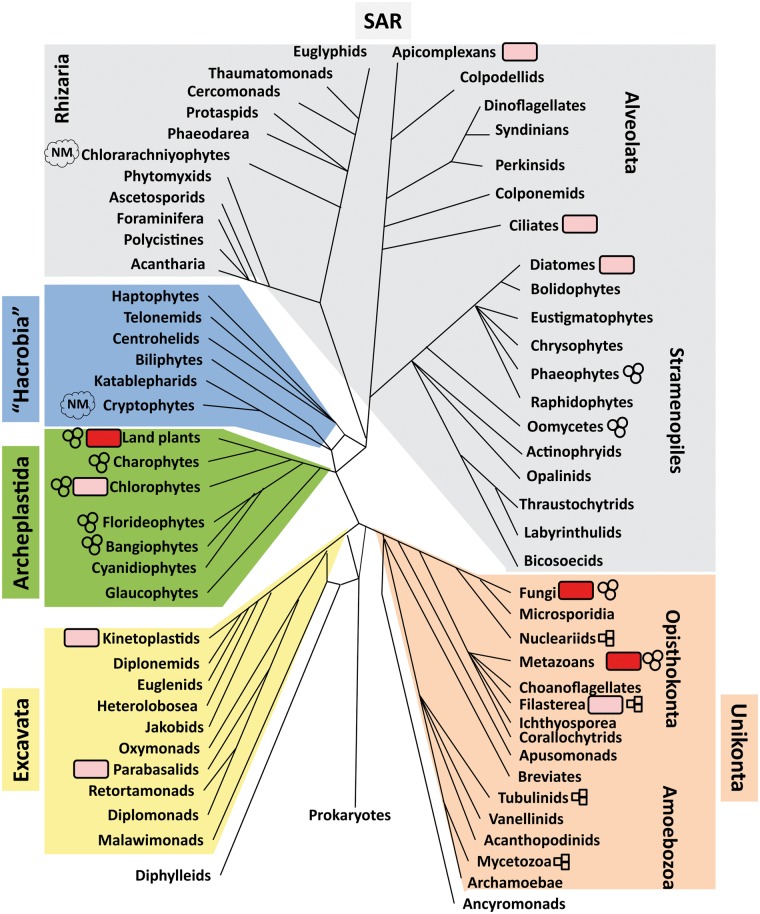
Traditional presentations of eukaryotic diversity have focused on the three main lineages where complex multicellularity evolved—land plants, fungi and metazoans—with other eukaryotes lumped into the overlapping categories of ‘algae’ and ‘protists’ (depending on whether they photosynthesize). The phylogenomic era has radically transformed this simplistic understanding. We now know of in the neighborhood of a hundred distinct eukaryotic lineages that are as deeply divergent (and should be thought of as equivalent in rank clades) as metazoans [107] (see Figure 4 for a partial overview), and new lineages of major phylogenetic importance are still being regularly discovered [108, 109]. These are tentatively grouped in several more or less phylogenetically coherent groups—Opisthokonta, Amoebozoa, Excavata, Archaeplastida, Hacrobia, Rhizaria, Alveolata and Stramenopiles. Opisthokonta and Amoebozoans are clearly grouped together in the Unikonts; Stramenopiles, Alveolates and Rhizaria are tentatively grouped in the SAR (Stramenopiles-Alveolates-Rhizaria) superclade; and some poorly studied lineages are still difficult to confidently link with any of the major groups. Where exactly the root of the eukaryotic tree lies is still unclear; different topologies have been proposed by various studies [110, 111]. The root might lie between Excavata and the rest of eukaryoties, or it may even be located within the current Excavata assemblage (making it paraphyletic), with the Discobeans (the clade containing the Jakobids and Kinteoplastids; Figure 4) on one branch and all other eukaryotes on the other [110].
Figure 4.
Major eukaryotic clades, their epigenomic characterization and the origins of multicellularity. The tree shown follows previously published topologies [112], but it should be noted that the precise deep branching is still to be fully resolved by future phylogenomic studies. Red rounded rectangles are placed next to the lineages in which chromatin and transcriptional biology have been studied in considerable detail. Pink rounded rectangles are placed next to clades for some representatives of which initial epigenomic studies have been carried out in some detail. Clades in which multicellularity has evolved are indicated with three circles where multicellularity results from cell division, and by three triangles where multicellularity is aggregative [113]. The lineages containing nucleomorphs (the chlorarachniyophytes and the cryptophytes) are indicated with ‘NM’.
The Opistokonts feature both metazoans and fungi, and the model systems in which the bulk of cellular and molecular biology research is being carried out. In addition, Monosiga brevicolis and Capsaspora owczarzaki are emerging as model systems in the two phylogenetically closest to metazoans groups, Choanoflagellata and Filasterea, respectively. Among the Amoebozoans, the mycetozoan Dictyostelium has long been studied as a model for the origins of multicellularity. The Archaeplastida assemblage features all groups with a primary plastid (i.e. derived from the original endosymbiotic event back in the Precambrian), including land plants (Arabidopsis being the main model system for plants) and green algae (with Chlamydomonas and Volvox being popular model organisms). The Excavates include several clades featuring important disease agents, which have accordingly attracted significant attention from researchers such as Trypanosoma and Leishmania in Kinetoplastida, Trichomonas in Parabasalia, Giardia among the diplomonads and Naegleria in Heterolobosea. The Alveolates include the Apicomplexans, the major parasitic lineage featuring the malaria plasmodium, Toxoplasma and a number of other pathogens and the free-living ciliates, among which Tetrahymena and Paramecium have a long history of being model organisms. Finally, model systems are emerging in the diatomes and the brown algae (Phaeophyta) among the Stramenopiles.
However, the vast majority of the eukaryote diversity is still extremely poorly studied, and the discipline of evolutionary cell and molecular biology [114], which aims to understand not only how exactly cellular processes function in different organisms but also why they came to be organized in the way they are, is still in its infancy because of the limited amount of data available.
The evolution of eukaryotic chromatin organization
The ancestral eukaryote genome, the appearance of complex genomes and the origins of morphological complexity
As outlined above, the organization of chromatin and the logic of gene expression regulation in metazoans are generally well known. But while rapid advances are being made in understanding the detailed mechanistic workings of this system, how and why it came to be remain open questions. Also not understood at present is the relationship between the enhancer/promoter/insulator model and the emergence of complex multicellularity. The genomes of unicellular yeasts, which have served as a model system for studying transcriptional regulation for many decades, are highly compact, with little intergenic space, and appear to use primarily promoter-proximal mechanisms of regulating transcription. While the transcriptional biology of most of them has not been studied in detail, the genomes of many (but by no means all) other unicellular eukaryotes are also similarly compact. In contrast, animal genomes typically contain large expanses of intergenic space, and long introns, as do the genomes of land plants, the lineage that has achieved the second highest level of morphological multicellular complexity as meausured by the number of distinct cell types in the organism [115].
The expansion of the genomes of multicellular eukaryotes is usually explained as a consequence of the lower efficiency of natural selection in lineages with decreased effective population size [69], which in turn is a natural consequence of the increased physical size of multicellular organisms. In such a population genetic environment, noncoding DNA and TEs, whose presence is usually nonadaptive and which are efficiently eliminated by natural selection in microbial lineages, can proliferate leading to large genomes full of repeats, introns and other noncoding DNA. As discussed above, these conditions have also been proposed to accelerate GRN evolution through duplication and sub/neofunctionalization of regulatory elements [67]. In turn, this might have enabled morphological complexification by facilitating the appearance of new cell types. The utilization of different distal regulatory elements to drive the expression of the same gene in different contexts may have been particularly useful for that purpose, which fits well with the on average a dozen or more putative such elements per gene that have been observed in mammalian genomes by large-scale candidate regulatory elements mapping efforts such as ENCODE and mouseENCODE.
One plausible view of how complex multicellularity developed that emerges based on these considerations sees regulatory complexification, genome expansion and organismal complexification as going hand in hand in a feed-forward loop relationship. More complex organisms tend to be physically larger and with lower effective population sizes, which allows for proliferation of noncoding DNA and the appearance of more complex GRNs, which in turn enables further organismal complexification. Gene regulatory mechanisms centered around distal regulatory elements and working in 3D space play a central role in this model. The diversity of gene expression programs in which an individual gene may participate is most likely substantially more limited if the gene is to be regulated exclusively through promoter-proximal means. But if regulatory elements are physically decoupled from the gene’s promoter, the constraints on their number and subfunctionalization are lifted, and they can more freely evolve in ways allowing the gene to be expressed in different developmental contexts and cell types.
A prediction this model makes is that long-range gene regulation and large genomes should be prevalent in large-bodied organisms with complex multicellularity. Such level of complexity has been achieved in metazoans and plants, on more than one occasion in fungi, in red and in brown algae, and it is indeed frequently accompanied by expanded genomes. To what extent long-range gene regulation is also common will be elaborated in more detail further below.
Before that, we will note that this model for the origin of complex gene regulation (or its alternatives) does not necessarily explain when exactly it evolved and what its molecular foundations are. Phylogenomic studies over the past couple decades have converged on the view that eukaryotes arose from within archaea. Most likely this happened as the direct result of an endosymbiotic event with a member of the α-proteobacteria, from which the modern mitochondrion derives [116, 117]. Thus, the ancestral eukaryotic genome probably looked much like the genomes of modern archaea, i.e. following the typical prokaryote organization of genes into operons and being small, compact and free of spliceosomal introns. It is tempting to think that the LECA genome was also streamlined, and this would be the more parsimonious explanation given the distribution of genome sizes across modern eukaryotes (most of which are unicellular species with compact genomes).
However, LECA seems to have been an essentially modern eukaryote, with all core characteristics that define the eukaryotic cell and sharply separate it from prokaryotes, and no clear intermediates, stemming from the period before the radiation of the known modern eukaryote supergroups, have ever been found. Thus, some time seems to have passed between eukaryogenesis and LECA. One thing we can say with some confidence about the LECA genome is that it was intron-rich, as comparisons of intron positions across eukaryotes have shown that many introns are ancestrally shared by all modern groups and must therefore date back to the time before LECA [118]. Combined with the clear relationship between eukaryotic spliceosomal introns and prokaryotic Group II self-splicing introns, and with the observation that the latter are phylogenetically restricted to bacteria but absent from archaea, an attractive model has been proposed, according to which spliceosomal introns originated as a result of the invasion of the archaeal genome by Group II self-splicing introns from the α-proteobacterial endosymbiont [119, 120]. Spliceosomal introns evolved later as a result of the loss by individual Group II introns of the ability to autonomously splice in cis and the transfer of that function in trans to the spliceosome. As Group II introns are rather large ribozymes, that would imply that the original eukaryote lineage might have had a larger genome than what is seen in modern eukaryotes with compact genomes. Whether that was also the case for LECA or the genome had already shrunk considerably by that time cannot be known with certainty at the moment though. In addition, the presence of large introns does not necessarily imply the existence of large intergenic spaces.
Still, the core of the machinery mediating long-range enhancer-promoter interactions seems to have been present in LECA. The Mediator complex, which plays an important role in the process of establishing such loops [121], is ancestral to all eukaryotes, as is the cohesin complex [122] (even if its primary ancestral role might have been chromatin cohesion during mitosis and meiosis).
The main multicellular lineages, however, have still apparently all emerged from ancestors with compact genomes, such as green algae, and the various holozoan opisthokont lineages sister to metazoans. On the other hand, large genomes are not unique to multicellular eukaryotes. The famous 750 Gb genome size estimates for some amoebas [123] are possibly artifacts of polyploidy or methodology, but a number of protists do indeed have large genomes. Dinoflagellate genomes are notorious for their enormous sizes, with the smallest ones being ∼1.5 Gb (for example, Symbiodinium [124]) and most being considerably larger (for example, the 100 Gb genome of the dinoflagellate Prorocentrum micans [125]). Giant genomes much larger than those of mammals have also been reported in diatoms [126] and in other protozoan lineages. Unfortunately, but for understandable reasons, genome sequencing efforts have so far targeted representative of each lineage species with small genomes. As a result, little is known about the organization of large protist genomes. Similar constraints have also applied to genome sequencing projects in fungi. Hundreds of genomes have been sequenced, but no sequences are available for the largest ones, which approach 1 Gb [127]. Yet, based on genome size estimates, it seems that increases in genome size relative to a more compact ancestral state can be found in all multicellular lineages: red algae genomes range between 100 Mb and 2.8 Gb [128], and those of brown algae range between 200 Mb and 3.6 Gb [129].
Several major questions thus emerge. Is a sparse genomic organization with distal regulatory elements tightly phylogenetically linked to the emergence of multicellularity? If yes, are the two causally related? If it evolved independently on multiple occasions did it evolve through similar modifications of the same preexisting components, and did it converge onto similar mechanisms everywhere or to divergent states operating under somewhat different principles? Are the identity and characteristics of distal regulatory elements the same in all eukaryotes? What can such similarities/differences tell us about the logic of mammalian gene regulation and its origins?
A separate set of similar questions concerns the conservation and divergence of chromatin biology. As mentioned above, histone proteins are generally extremely highly conserved across eukaryotes, with the key residues on which the best studied histone modifications are deposited being ancestrally present in all lineages [105]. The core histone marks themselves also appear to be ancestral as evident by their presence in both metazoans, plants and ciliates, as do some histone variants such as H2A.Z [130]. However, conservation of histone marks does not necessarily mean conservation of associated biochemical processes, much less conservation of combinatorial sets of histone marks and resulting chromatin states. What exactly the ancestral state of the histone code was, how it came it to be and how conserved and constrained all of its aspects are among eukaryotes is still not entirely clear.
These questions can only be fully answered through comprehensive functional genomic studies of eukaryotic lineages throughout the tree of life. We are far from achieving that goal, but initial insights have already been obtained from a number of clades.
The three most important multicellular lineages all contain well-studied model systems; thus, most information for such comparisons is naturally available from those species.
Functional genomic studies in non-yeast fungi
In the case of the fungal lineage, all model systems are yeasts or molds with compact genomes, but the wealth of sequenced genomes includes a number of complex multicellular species, which appear to have compact genomes too (for example, the Amanita muscaria mushroom has a ∼40 Mb genomes with ∼18 000 genes [131]), and are likely using primarily promoter-proximal regulatory elements. Of note, unicellular yeasts lack H3K27me3, the developmentally regulated heterochromatin histone modification, while that modification is present in the mold Neurospora [132]. A curious feature of Neurospora biology that has been revealed by functional genomics is that it appears to use a unique mechanism for organizing its chromatin in 3 D space. Unlike the CTCF/cohesin-driven looping used in metazoan genomes, studies using ChIP-seq and 3 D genome structure mapping techniques such as Hi-C have shown that heterochromatin regions marked by H3K9me3 and H3K27me3 organize most 3 D interactions in Neurospora nuclei [132, 133].
Functional genomics and the chromatin biology of land plants
It is remarkable given their economic importance, but the study of enhancers in plants is still in its infancy. Plants do have enhancers, but only a small number of individual enhancer elements have been characterized in any depth [134, 135], and functional genomic studies have so far painted a rather inconclusive picture regarding their properties. DNAse hypersensitivity maps have identified thousands of intergenic and intronic open chromatin regions in rice, in Arabidopsis and in Brachyopodium [136–139]. Similarly, comparative genomics efforts have identified large numbers of conserved noncoding sequences, exhibiting enhancer-like characteristics [140, 141]. Typical features of eukaryotic chromatin, such as the association of H3K4me3 with promoters, of H3K9me3 and of Polycomb-deposited H3K27me3 with heterochromatin, are also present in plants.
However, despite numerous studies using ChIP-seq to map histone marks and chromatin states in the main plant systems [142–146] as well as the moss Physcomitrella patens [147], clear chromatin signatures of active enhancers have not yet emerged [132, 133]. H3K27ac appears to correlate with active enhancers [148], as it does in metazoans. However, the association is weaker, and to what extent the additional features that seem to distinguish mammalian enhancers (H3K4me1, eRNA transcription, poised enhancers being marked by H3K4me1 alone but not by H3K27ac and others) are shared by their plant counterparts is also not clear. Part of the challenge is that there are few gold-standard enhancer elements in plants around which detailed studies are to be built. Another is that ChIP experiments are usually done on whole parts of plants, containing a mixture of cell types, thus producing a convolution of signals and decreasing the signal-to-noise ratio. Methods for the specific isolation of pure cell populations will be needed to resolve these issues [149].
How enhancer–promoter interactions in 3 D space are regulated in plants is at present also not understood and remains to be elucidated in the future. A remarkable feature of Arabidopsis chromatin, in stark contrast to what is observed in most metazoans, is the absence of topologically associated domains (TADs; regions of increased 3 D interaction frequency, which are also generally not crossed by promoter–enhancer interactions) [150–152]. On the other hand, the existence of insulator elements in plants has been postulated [153]. Of note Arabidopsis, with its fairly compact genome (27 655 genes in ∼ 135 Mb [154]), may not be the ideal system for such studies. Other plants with sparser genomes, such as rice and maize, are potentially more informative. Indeed, a more recent study [155] found that about a quarter of the rice genome is organized into TADs, but data from a number of other lineages will be needed to fully understand plant chromatin organization.
No epigenomic data providing information about the regulatory organization of multicellular red algae and brown algae are available at the moment, but their future study will be highly informative regarding the origin and phylogenetic distribution of distal regulatory elements and its relationship to multicellularity, as will be the study of the closest algal relatives of land plants.
A number of protozoan lineages have, however, been examined in some detail, and these efforts have revealed some of the extent of conservation and divergence of chromatin biology in eukaryotes.
Functional genomics and the chromatin biology of apicomplexans
A relative wealth of information has become available in recent years from the apicomplexan Plasmodium falciparum [156–158], whose epigenome exhibits some curious features. Unlike most other eukaryotes, in which H3K4me3 is localized around promoters, as is the histone variant H2A.Z, in Plasmodium H3K4me3 and H2A.Z, together with the apicomplexan-specific histone variant H2B.Z, demarcate intergenic regions in their entirety. In addition, the H3.3 variant, which is typically deposited around sites of active transcription, has been suggested to also associate with subtelomeric regions in Plasmodium [159]. Additional intriguing properties of Plasmodium chromatin include strong nucleosome positioning over splice sites and transcription termination sites (TTSs), but an absence of the typical [160] tightly positioned nucleosomes immediately around transcription start sites (TSSs). It is possible that intergenic regions are occupied by a different type of nucleosomes because of the extreme AT-richness of the Plasmodium, which reaches and sometimes even exceeds 90% in intergenic regions.
Functional genomics and the chromatin biology of kinetoplastids
Another interesting group that has attracted attention because of its medical relevance is kinetoplastids. As mentioned above, it is possible that the discobeans as a whole are the deepest diverging eukaryotic clade [110], which may or may not be related to kinetoplastids exhibiting a number of divergent features. The most remarkable among them is the apparently complete loss of gene regulation at the transcriptional level [161]. Instead, genes are organized into long constitutively expressed polycistronic units, pre-mRNAs are trans-spliced with splice leader sequences to produce mature mRNAs and gene expression regulation happens primarily at the posttranscriptional level through mechanisms regulating RNA stability and translation.
Kinetoplastids also have more divergent than usual histone tails, missing a number of key histone code residues [162, 163], such as H3K9. The kinetoplastid epigenome is far from comprehensively characterized, but multiple ChIP-seq studies have revealed some aspects of its organization. It appears that the boundaries of polycistronic transcription units in trypanosomatids are demarcated by nucleosomes containing four histone variants, H2AZ, and the unique to kinetoplastids H2BV, H3V and H4V [164] (reminiscent in some ways to what is observed in apicomplexans) as well as by histone acetylation [165]. Kinetoplastid genomes are also remarkable for containing a unique elaborate DNA modification, glucosylated hydroxymethyluracil, also known as base J. Base J has been mapped genome-wide and appears to be a marker for transcriptional termination at the end of polycistronic units [166, 167].
How this strikingly divergent genomic organization came to be is an open question. Parasitism is a defining theme of kinetoplastid biology (although free-living kinetoplastids do exist), and parasitic lineages are often highly derived. However, the sequencing of the genome of Bodo saltans, one of the free-living kinetoplastids, suggested that polycistronic organization is ancestral to the groups [168]. How much deeper this feature extends within the Discobea clade is not known, as little data exist for the other lineages in the group, except for the Heteroloboseans/Percolozoa, where a couple of Naegleria genomes have been sequenced [169, 170] and appear to have a conventional organization. However, not much is known about their functioning beyond that, and even less is known about other related lineages, even though the fascinating biological questions regarding them are plenty, such as the organization of permanently condensed chromosomes in euglenids [171].
Functional genomic studies of other protozoan lineages
Within excavates, limited epigenome mapping has also been carried out in Trichomonas vaginalis (Parabasalia) [172], targeting only two histone marks, H3K4me3 and H3K27ac, and revealing the usual enrichment around TSSs of active genes. Interestingly, chromatin in the diplomonad Giardia lamblia has been reported to contain an HU-like protein (HU-type proteins are histone-like nucleoid-associated nonspecific DNA-binding proteins typically found in bacteria, although they can also bind to RNA [173, 174]) in addition to the linker and the four core histones [175]. In addition, its promoters have been suggested to produce highly abundant transcripts in both orientations, unlike in other eukaryotes [176], but detailed studies using more modern functional genomic tools are lacking.
More comprehensive histone mark profiling has been carried out in two algal lineages. Five histone marks (H3K4me3, H3K27ac, H3K9me3, H3K36me3 and H3K27me3) were mapped along the compact genome of the classic chlorophyte model species Chlamydomonas reinhardtii [177]. Surprisingly, H3K9me3, which is generally strongly associated with heterochromatin, was reported to associate with the promoters of most genes, forming a bivalent signature with the H3K4me3 and H3K27ac marks, while H3K27me3, also a heterochromatin mark, was found to form bivalent domains along gene bodies with the transcription elongation mark H3K36me3. Both of these are unusual and unexpected chromatin states not found in other eukaryotes.
In the diatom Phaeodactylum tricornutum, H3K4me2, H3K9me2, H3K9me3, H3K9/K14Ac and H3K27me3 were profiled [178], providing the first glimpse into the epigenome of a stramenopile and revealing largely shared features with what is observed in most other eukaryotes.
No comprehensive epigenomic characterization has been carried out so far for any lineage of the Hacrobians, the Amoebozoans or the Rhizarians.
Functional genomics and the chromosomal biology of ciliates
Within the alveolates, aside from apicomplexans, it is the ciliates that include several established and emerging model systems (Paramecium, Tetrahymena, Oxytricha, Euplotes, Stentor). Except for MNAse-seq-based nucleosome positioning studies in Tetrahymena [179, 180], epigenomes have not yet been mapped in ciliates, but ChIP-seq has proven highly useful in understanding the unique biology of their chromosomes. One of the defining features of ciliates is their nuclear dualism. Ciliates have a somatic macronucleus (MAC), which arises from a germline micronucleus (MIC) through the elimination of internal eliminated segments (IESs; largely containing repetitive elements) from the MIC chromosomes. Macronuclear destined segments (MDSs) in the developing MAC are retained and stitched together into somatic chromosomes. In addition to the differences in sequence content, the MAC is also highly polyploid, with tens to thousands of copies of each chromosome. How this transformation is accomplished varies between different ciliate lineages, but common between all of them is the major role that small RNAs play in the process.
In Paramecium and Tetrahymena, the MIC genome is fully transcribed from both strands, generating double-stranded RNA (dsRNA), from which small RNAs (called scnRNAs) are produced by Dicer proteins, loaded onto Piwi-family Argonaute proteins and exported to the old MAC. There they are ‘scanned’ against transcripts generated from the rearranged chromosomes present in it. If matches to scnRNAs are found, they are degraded, leaving only scnRNAs targeting IESs, which are then exported to the developing MAC, where they guide the excision of IESs. Once initial IESs are excised, a secondary population of small RNAs is produced, as a feed-forward loop to ensure the complete elimination of IESs from the developing MAC. In Paramecium, where IES excision is mostly precise, so-called iesRNAs are generated through the formation of circular DNA segments from the excised IESs, which are then transcribed bidirectionally resulting in dsRNAs further processed into iesRNAs [181]. But in Tetrahymena, where IESs are mostly in intergenic regions and excisions can be imprecise, the mechanism is chromatin-based. Early scnRNAs guide the establishment of heterochromatin (marked by H3K9me3 and H3K27me3) and recruitment of HP1 (heterochromatin protein 1 [182, 183]) homologs along IESs; heterochromatin formation spreads in cis and results in the production of late scnRNAs. Mechanisms for restricting its spreading into MDSs must exist, however, and ChIP-seq maps have helped establish that this is accomplished through the binding of a different HP1-like protein, Coi6p, together with several interaction partners and histone demethylases to MDSs [184, 185]. This mechanism appears to be analogous to how insulator elements block the spreading of heterochromatin in other eukaryotes.
Genome rearrangement is most striking in Oxytricha and related spirotricheans. In these organisms, MDS segments in the MIC are not collinear and have to be unscrambled during MAC formation (a process apparently guided by long RNA templates from the old MAC). Remarkably, the end result of unscrambling is the formation of nanochromosomes just a few kilobases long, typically containing only a single gene [186, 187]. The structure of nanochromosomes poses a number of interesting questions regarding the organization of chromatin in these nuclei, as they contain extremely short (only a few tens of bases) stretches of DNA between TSSs and TTTs and telomeres. How transcription factor binding around promoters, nucleosome positioning and histone marks in Oxytricha deviate from the typical eukaryotic norms remains to be studied in the future. Of note, nanochromosomes seem to have evolved independently on multiple occasions within the ciliates [188, 189], but their features have not been studied outside of Oxytricha.
Poorly studied groups with radically different chromatin organization
It is the third major alveolate lineage, the dinoflagellates, where the most striking nuclear organization among all eukaryotes that have been studied in any detail is to be found, so divergent in fact, that decades ago it was thought that they may represent an intermediate state between prokaryotes and eukaryotes [190]. The packaging of chromatin by nucleosomes is a universal eukaryotic feature except for special cases such as mammalian sperm cells, during the development of which histones are largely replaced by protamines [191]. However, in dinoflagellates, chromatin appears to not be packaged by histones, and chromosomes are permanently condensed throughout the life cycle, existing in a liquid crystalline state [192, 193]. Dinoflagellates do, however, possess histone genes, although they are highly divergent in sequence, and they do also have a number of traditional chromatin modification enzymes and remodeling factors, including the FACT histone chaperone, which plays a major role in cotranscriptional nucleosome disassembly in all eukaryotes [106]. Thus, histones likely are of some importance for dinoflagellate chromatin biology, and transcription through nucleosomal arrays is probably happening, but nothing is known about the group beyond that. Research on dinoflagellates has been hampered by the massive size of their genomes (usually much bigger than those of humans), which has until recently precluded genome assembly efforts. But sequences for several Symbiodinium species, whose genomes are relatively small, have recently become available [194, 195], and the mysteries of these nuclei should be resolved in the future through the application of functional genomic tools.
Another curious case of likely highly divergent chromatin structure is provided by nucleomorphs. Secondary endosymbiosis, i.e. the engulfment of one photosynthetic eukaryote by another, has happened on numerous occasions in evolution [113], and it usually leads to the eventual disappearance of the endosymbiont’s nucleus, leaving only the plastid. However, on two independent occasions, in the chlorarachniophytes and the cryptophytes (Figure 4), the nucleus has been retained in the form of a nucleomorph, with chlorarachniophyte nucleomorphs being of green algal and cryptophyte ones of red algal origin. Nucleomorph genomes are extremely small (a few hundred kilobases in length) and highly compact, and exhibit a strikingly convergent organization in both lineages [196]. Interestingly, nucleomorph histone tails are divergent in chlorarachniophyte nucleomorphs (and more conserved in cryptophytes) [197]. Key histone modifications ancestrally involved in the transcriptional cycle in eukaryotes have been lost, presenting numerous questions regarding the conservation of the corresponding biochemical processes in these nuclei.
The deep evolution of chromatin organization in the context of current knowledge
While the precise rooting of the eukaryotic tree of life and functional genomic studies of early diverging lineages, particularly in excavates, may change the picture significantly, we can, based on current knowledge, draw some conclusions about LECA’s chromatin and about the evolution of chromatin biology following the eukaryotic radiation. A core set of histone marks and associated processes is likely ancestral to all eukaryotes. These include the association of histone acetylation with open chromatin (which may follow simply from the resulting relaxation of the electric charge of histone molecules and not be position-specific), the demarcation of promoters by H3K4me3 and the H2A.Z histone variant, the transcription-associated deposition of H3K36 methylation and the use of H3K9 and H3K27 methylation for gene repression and heterochromatinization. Given the close connection between heterochromatin and small RNA-mediated mechanisms for TE suppression [198–200], found throughout diverse eukaryotes, it seems plausible that the latter were part of the solution to the problem of keeping TEs in check in LECA too. It is likely that numerous other histone marks also date back to LECA, but currently available data encompassing a sufficient diversity of lineages are still limited to only a few modifications. It is also likely that different eukaryotes possess diverse repertoires of lineage-specific marks, especially on the faster evolving H2A, H2B and H1 histones, remaining to be understood in depth in the future.
The conservation of histone marks and their functions does not, however, appear to imply strict conservation of chromatin states, and divergent modes of partitioning the genome have evolved, as shown by the examples of Trypanosomatids, Plasmodium, Chlamydomonas and others. Some evolutionary plasticity with respect to chromatin organization appears to exist, considerable in some lineages, with a common set of chromatin states (as defined by a limited set of histone modifications) shared by most eukaryotes, and additional lineage-specific states appearing during evolution, through novel combinations of histone marks, addition of novel histone variants and other chromatin-associated proteins and even DNA modifications The diversity and functional significance of these variations remains to be characterized in detail.
Also not yet fully resolved at present is the question of the relationship between organismal complexity and regulatory architecture. As discussed above, enhancers in plants are still poorly understood, and no data are available for other complex multicellular lineages such as red and brown algae. Relevant data do, however, exist regarding metazoans and some of their closest relatives.
The origins and evolution of metazoan regulatory architecture
The holozoan lineage comprises metazoans, and their closes unicellular relatives, which are, in order of phylogenetic proximity, the choanoflagellates, the filastereans, the ichtyosporeans and corallochytrids (Figures 4 and 5). The ancestor of metazoans was most likely a colonial organism similar to choanoflagellates. The striking resemblance between choanoflagellates and the choanocyte cells of sponges was noticed as far back as the mid-19th century [212] and has been extensively confirmed by modern genomics [213].
Figure 5.
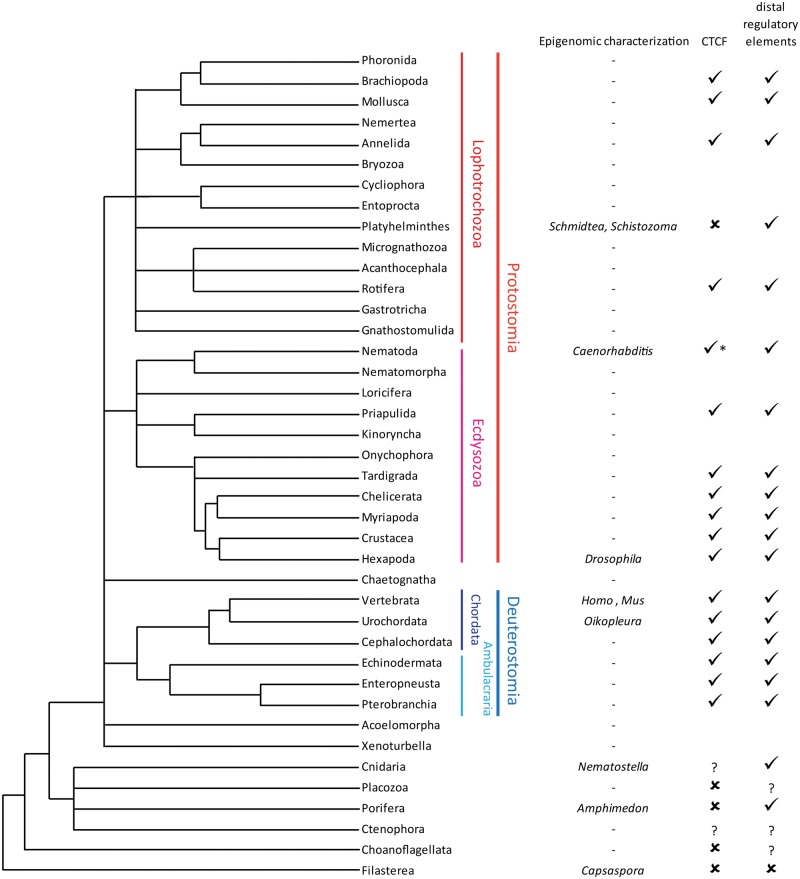
Relationships between metazoan phyla and their closest relatives and the extent of their epigenomic characterization. Taxons for which the correct phylogenetic relationships is still to be conclusively established (such as the position of Ctenophora relative to other metazoans [201–205], the monophyly of poriferans and the overall relationships between non-bilaterian clades [206] and the placement of Chaetognatha [207], Acoelomorpha [208, 209], Xenoturbellida [209, 210] and others) are incorporated as unresolved branches on the cladogram, with Orthonectida and Dicyemida (tentatively placed in Lophotrochozoa [211]) being omitted. Phyla in which at least some epigenomic studies have been carried out are indicated on the right-hand side with the corresponding genus names for the main taxa studied. Lineages without published sequenced genomes have been left blank. Note that CTCF has been lost in some nematodes such as C. elegans but is present in other members of the phylum.
The availability of genome sequences from these groups has allowed the tracing of the evolution of the metazoan transcription factor toolkit, which appears to have been assembled gradually but beginning before the appearance of multicellular animals, with many TF families already present in unicellular holozoans [214]. Their genomes are fairly compact (but not as compact as those in yeasts such as S.cerevisiae) leaving open the question when exactly enhancers and other conserved features of metazoan genomes appeared in the evolution of animals.
A comprehensive mapping of histone marks and open chromatin regions was carried out in C.owczarzaki, the best studied filasterean [215]. It did not find apparent distant regulatory elements, with potential such regions being promoter-proximal. Remarkably, putative filasterean regulatory elements appear to smaller in size than in metazoans, indicating that during metazoan evolution, both their combinatorial and individual complexity increased. Curiously, the repressive H3K9me3 and H3K27me3 modifications were not observed in Capsaspora.
The early non-bilaterian metazoan groups include the Placozoa, sponges/Porifera, Cnidaria and Ctenophora. Owing to their deep and probably rapid divergence, it has been difficult to conclusively determine their relationships [201–206]. Different studies have suggested strongly conflicting topologies: rootings of the metazoan tree between Ctenophora and all other phyla, between Porifera and all other groups, within Porifera and others have all been proposed.
Recently, ChIP-seq has been used to map histone modifications in the sponge Amphimedon queenslandica [216] and the chidarian Nematostella vectensis [217]. These studies revealed the presence of distal regulatory elements and an overall typically metazoan regulatory architecture. Therefore, metazoan enhancer elements seem to have originated somewhere between filastereans and poriferans/cnidarians, but when and how exactly remain to be answered by analysis of other non-bilaterians and in choanoflagellates, and by the hoped for future firm establishing of the correct topology of the metazoan tree.
Interestingly, while they do seem to have enhancers, it is not clear whether cnidarians and sponges have typical insulator elements. The phylogenetic distribution of CTCF, the classic insulator protein in metazoans, appears to be restricted to bilaterians, with some possibility for the existence of distant homologs in Cnidaria and Ctenophora [218, 219] (Figure 5).
CTCF is also missing, as a result of secondary losses, in some nematodes, including C.elegans, as well as in flatworms. Caenorhabditiselegans does possess distal enhancers [220], but it appears to, remarkably, lack TADs on its chromosomes, perhaps as a consequence of losing CTCF, except, even more remarkably, for chrX. On chrX the dosage compensation complex, a condensin complex that localizes to sequence-specific recruitment elements, plays a role analogous to the CTCF/cohesin combination in other bilaterians. How exactly enhancers are restricted to their cognate promoters on autosomes remains to be fully understood, as is any potential relationship between the loss of TADs and the organization of many C. elegans genes into polycistronic operons.
In the evolution of insects, on the other hand, the array of insulator proteins has expanded [221]. Six such factors in addition to CTCF operate in Drosophila. The mechanisms of metazoan insulation thus seem to exhibit a considerable degree of plasticity, but to fully understand the functional constraints on them and their relationship with 3 D nuclear organization, they need to be studied in representative species from a much wider sampling of animal phyla (Figure 5) than the limited data that exist at present includes.
In addition to vertebrates, insects, nematodes, Amphimedon and Nematostella, epigenomic studies have been carried out in a couple flatworms and in the urochordate Oikopleura dioica.
An important result to emerge from histone marks profiling in the parasitic trematode Schistosoma mansoni [222] and in the planarian Schmidtea mediterranea [223] is the existence of bivalent promoters, simultaneously marked by H3K4me3 and H3K27me3 in flatworm stem cells. These bivalent structures were first identified in mammalian embryonic stem cells [224], where they serve to poise the expression of developmentally regulated genes, and are resolved toward an active or repressed state with the progression of embryonic development. The presence of such domains in flatworms suggests deep conservation of this feature of animal stem cells.
The highly derived Oikopleura genome is intriguing for being extremely compact, at only 70 Mb, compared with the genomes of other chordates, which are at least an order of magnitude larger, and for genes being organized into operons. However, the profiling of 19 histone modifications and CTCF in Oikopleura revealed generally conserved epigenomic organization.Distal enhancers and insulators and typical chromatin states are observed, the main difference being the restricted extent of heterochromatin (because of the compacted nature of the genome) and sex-specific chromatin states on the Y chromosome [225].
Overall, the properties of metazoan chromatin present a pattern of general conservation, but, as is the case with the deeper evolutionary splits between protist groups, some notable divergence cases are observed too. The most comprehensive comparison of chromatin organization in metazoans was carried out by the ENCODE and modENCODE consortia using hundreds of human, fly and worm data sets [12, 13, 226]. It revealed an overall conservation of histone modification patterns around regulatory elements but also some differences. Examples of the latter include the enrichment of H3K23ac around expressed promoters in C. elegans, but across both active and inactive gene bodies in Drosophila, and the association of H4K20me1 with both active and silent genes in humans but only with expressed genes in fly and worm. Perhaps, the most significant such difference is the colocalization of the heterochromatin marks H3K27me3 and H3K9me3 in worms, in contrast to the distinct domains that they tend to form in mammals and flies [226].
How widespread such differences are between different metazoans and whether they are restricted to these marks awaits further research. The systematic functional genomic characterization of species from all phyla using comprehensive arrays of histone mark profiles should provide a much fuller and detailed picture of the evolution of gene regulation in metazoans.
Remaining technical challenges
While ChIP-seq has been successfully adapted to a wide diversity of lineages in the past decade, a number of technical challenges remain to be overcome to fully unlock its potential for understanding their biology, with many of the issues being specific to particular lineages.
For example, a major limitation to functional genomic studies of chromatin in clades such as the apicomplexans, ciliates, Dictyostelium and others is the extremes of GC-composition biases that can be found outside mammals. Such genomes can often approach or exceed an AT content of 80%. On its own this would not be an enormous problem; however, GC content is not uniform across the genome, with exons of protein-coding genes typically being relatively GC-rich (because they are under stronger purifying selection at the codon level) in contrast to the even more extremely AT-rich intronic and intergenic regions. Under such conditions, polymerase chain reaction (PCR)-based methods for sequencing library generation produce strong biases toward exons, making data generated in such ways difficult to interpret, as the true ChIP signal is often almost completely overwhelmed by the PCR bias. For example, it has been proposed that nucleosome positioning in the MAC of Tetrahymena is strongly driven by sequence features, in particular exons and GC content [179], but such a result is also what would be expected if the observed effect is because of PCR biases, and PCR is not even the only problematic step in a ChIP protocol, as cross-link reversal, typically carried out at 65°C, can bias against extremely AT-rich sequences, which become denatured even at such relatively low temperatures and are therefore underrepresented by library building protocols dependent on double-stranded DNA ligation steps. Linear amplification methods that resolve these issues have been developed [227–230], but they have not been widely adopted yet.
Another significant lineage-specific issue concerns background and artifactual signals in poorly studied (and often, not well assembled) genomes of emerging model and nonmodel species. ChIP-seq data sets in traditional model systems are now understood to often contain artifacts of several sources, such as a bias toward open chromatin regions [231–234] and the presence of strong but artifactual enrichment over so-called ‘blacklist’ regions [235]. Historically, it took several years for these sources of bias to be understood, classified and corrected for, and for a set of standard practices to be compiled [76]. Some sources of artifacts (such as the bias toward open chromatin) are likely to be common to all species, but nevertheless such accumulated experience is not available for nonmodel systems, and new genomes can always present unexpected surprises in terms of sources of artifacts [236]. It is not clear at present to what extent published results in insufficiently well-characterized species are affected by such factors.
It is also not clear how generally applicable analysis tools are, as evidenced by the discrepancy between the published metaanalyses of transcription factor occupancy divergence in metazoans. ChIP-seq processing and analysis pipelines have been designed primarily with mammalian genomes in mind, with their particular repeat and background structure, but most genomes have different properties. Strong transcription factor peaks are probably successfully captured regardless of these issues, but the impacts of the convolution of algorithms and genome and background properties on the overall sets of identified occupancy sites are currently not well understood.
A more general challenge is the lack of purified uniform cell lines for most nonmodel species. As discussed above regarding plants, this confounds interpretation, especially of histone modification data sets, because of the mixing of different cell types together in the same ChIP reactions. Such issues may exist even for single-celled organisms if substantial heterogeneity of cell states exists within the experimental population. In the light of these considerations and the ones regarding experimental artifacts outlined above, results reporting unexpected combinations of histone marks such as bivalency between H3K9me3/H3K27me3 and marks usually associated with active transcription may have to be taken with a grain of salt until independently verified. Of note, even H3K4me3/H3K27me3 bivalent domains in mammalian embryonic stem cells were not truly proven not to be the result of heterogeneity between cells or between individual chromosomes within the same cells until methods for directly mapping the combinatorial modification states of individual nucleosomes emerged recently [237]. Such approaches should prove invaluable in resolving chromatin state conundrums in other species.
Finally, reagent availability remains a major remaining challenge for ChIP-seq studies even in human and mouse cells, and it is even more of an issue in other organisms. High-quality reliable antibodies for most human transcription factors are not available, which has necessitated the application of epitope tagging methods toward the mapping of their occupancy sites. CRISPR/Cas9-based genome editing has eventually allowed such tagging to be carried out as knock-ins into the native locus [238], mitigating against possible overexpression artifacts. ChIP-grade antibodies are even less accessible for nonmodel organisms; thus, epitope tagging is likely to be the approach of choice in such cases, with its success depending on the particular details of the system in question.
Epitope tagging is a viable solution for transcription factors and other proteins, but perhaps even more substantial is the challenge of obtaining reliable histone modification antibodies. Most such antibodies have been polyclonal (and therefore nonrenewable), although monoclonal ones are becoming increasingly available, and of varying quality [239, 240]. Histone proteins are some of the most conserved in all eukaryotes, but some minor differences in histone tails nevertheless exist, and, as discussed above, in some of the most interesting groups, histones are in fact fairly divergent, making existing antibodies either inapplicable or unreliable. New reagents may have to be generated and extensively characterized in such cases; developing reliable systems for the large-scale screening and validation of reagents would prove highly useful toward that goal.
Conclusions
It may be overused at this point, but the saying that nothing in biology makes sense except in the light of evolution remains as true as ever, and it also applies just as strongly to transcriptional regulation and chromatin biology. If we are to truly grasp these aspects of our biology, we need to understand how and why they evolved to their current state. High-throughput sequencing has been a true game changer in this quest, by making accessible every organisms on the planet to functional genomic techniques. The past decade has seen many exciting insights and advances resulting from its application, with many more certain to come in the coming years, as we make our way around the branches of the eukaryotic tree.
Key Points
Comparative analyses of transcription factor occupancy across species illuminate the mechanisms of and the driving forces behind the evolution of gene regulatory elements
Access to nonmodel and emerging model organisms enables the mapping of GRN rewiring during evolution
Epigenomic studies across the tree of life reveal the conservation and divergence of general regulatory mechanisms.
Much of the known eukaryotic diversity is still to be sampled, including some of the most intriguing groups; comparative transcription factor occupancy studies have also been limited in scope so far.
Unresolved technical challenges remain to be overcome to fully empower research in the field.
Funding
This material is based on work supported by National Institutes of Health awards (grant numbers 1DP2OD022870-01, 1UM1HG009436 and 1U01HG009431).
Georgi K. Marinov is a postdoctoral scholar at the Department of Genetics, Stanford School of Medicine. His research focuses on the application of functional genomic tools to understand the mechanisms and evolution of gene regulation in eukaryotes.
Anshul Kundaje is an assistant professor of Genetics and Computer Science at Stanford University. His primary research interests are computational biology and applied machine learning with a focus on large-scale computational regulatory genomics.
References
- 1. Gilmour DS, Lis JT.. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol Cell Biol 1985;5(8):2009–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gilmour DS, Lis JT.. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proc Natl Acad Sci USA 1984;81(14):4275–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Solomon MJ, Larsen PL, Varshavsky A.. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 1988;53(6):937–47. [DOI] [PubMed] [Google Scholar]
- 4. Hecht A, Strahl-Bolsinger S, Grunstein M.. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 1996;383(6595):92–6. [DOI] [PubMed] [Google Scholar]
- 5. Ren B, Robert F, Wyrick JJ, et al. Genome-wide location and function of DNA binding proteins. Science 2000;290(5500):2306–9. [DOI] [PubMed] [Google Scholar]
- 6. Lieb JD, Liu X, Botstein D, Brown PO.. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 2001;28:327–34. [DOI] [PubMed] [Google Scholar]
- 7. Iyer VR, Horak CE, Scafe CS, et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 2001;409:533–8. [DOI] [PubMed] [Google Scholar]
- 8. Horak CE, Snyder M.. ChIP-chip: a genomic approach for identifying transcription factor binding sites. Methods Enzymol 2002;350:469–83. [DOI] [PubMed] [Google Scholar]
- 9. Weinmann AS, Yan PS, Oberley MJ, et al. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev 2002;16:235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee TI, Rinaldi NJ, Robert F, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 2002;298(5594):799–804. [DOI] [PubMed] [Google Scholar]
- 11. Zhang X, Clarenz O, Cokus S, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 2007;5(5):e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. modENCODE Consortium, Roy S, Ernst J, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 2010;330(6012):1787–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerstein MB, Lu ZJ, Van Nostrand EL, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 2010;330(6012):1775–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei CL, Wu Q, Vega VB, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 2006;124(1):207–19. [DOI] [PubMed] [Google Scholar]
- 15. Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 2006;38(4):431–40. [DOI] [PubMed] [Google Scholar]
- 16. Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007;129(4):823–37. [DOI] [PubMed] [Google Scholar]
- 17. Johnson DS, Mortazavi A, Myers RM, Wold B.. Genome-wide mapping of in vivo protein-DNA interactions. Science 2007;316(5830):1497–502. [DOI] [PubMed] [Google Scholar]
- 18. Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007;448(7153):553–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robertson G, Hirst M, Bainbridge M, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 2007;4(8):651–7. [DOI] [PubMed] [Google Scholar]
- 20. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mouse ENCODE Consortium, Stamatoyannopoulos JA, Snyder M, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 2012;13(8):418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yue F, Cheng Y, Breschi A, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 2014;515(7527):355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518(7539):317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stunnenberg HG. International Human Epigenome Consortium Hirst M. The International Human Epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell 2016;167(5):1145–9. [DOI] [PubMed] [Google Scholar]
- 25. Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012;22(9):1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lindblad-Toh K, Garber M, Zuk O, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 2011;478(7370):476–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meader S, Ponting CP, Lunter G.. Massive turnover of functional sequence in human and other mammalian genomes. Genome Res 2010;20(10):1335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Balhoff JP, Wray GA.. Evolutionary analysis of the well characterized endo16 promoter reveals substantial variation within functional sites. Proc Natl Acad Sci USA 2005;102(24):8591–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ludwig MZ, Bergman C, Patel NH, Kreitman M.. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 2000;403(6769):564–7. [DOI] [PubMed] [Google Scholar]
- 30. Ludwig MZ, Palsson A, Alekseeva E, et al. Functional evolution of a cis-regulatory module. PLoS Biol 2005;3(4):e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hare EE, Peterson BK, Iyer VN, et al. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet 2008;4(6):e1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dermitzakis ET, Clark AG.. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol 2002;19(7):1114–21. [DOI] [PubMed] [Google Scholar]
- 33. Odom DT, Dowell RD, Jacobsen ES, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 2007;39(6):730–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Conboy CM, Spyrou C, Thorne NP, et al. Cell cycle genes are the evolutionarily conserved targets of the E2F4 transcription factor. PLoS One 2007;2(10):e1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Borneman AR, Gianoulis TA, Zhang ZD, et al. Divergence of transcription factor binding sites across related yeast species. Science 2007;317(5839):815–19. [DOI] [PubMed] [Google Scholar]
- 36. Tuch BB, Galgoczy DJ, Hernday AD, et al. The evolution of combinatorial gene regulation in fungi. PLoS Biol 2008;6(2):e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmidt D, Wilson MD, Ballester B, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 2010;328(5981):1036–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stefflova K, Thybert D, Wilson MD, et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell 2013;154(3):530–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ballester B, Medina-Rivera A, Schmidt D, et al. Multi-species, multi-transcription factor binding highlights conserved control of tissue-specific biological pathways. Elife 2014;3:e02626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mikkelsen TS, Xu Z, Zhang X, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010;143(1):156–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheng Y, Ma Z, Kim BH, et al. Principles of regulatory information conservation between mouse and human. Nature 2014;515(7527):371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Denas O, Sandstrom R, Cheng Y, et al. Genome-wide comparative analysis reveals human-mouse regulatory landscape and evolution. BMC Genomics 2015;16:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kutter C, Brown GD, Gonçalves A, et al. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat Genet 2011;43(10):948–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cary GA, Cheatle Jarvela AM, Francolini RD, et al. Genome-wide use of high- and low-affinity Tbrain transcription factor binding sites during echinoderm development. Proc Natl Acad Sci USA 2017;114(23):5854–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nitta KR, Jolma A, Yin Y, et al. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. Elife 2015;4:e04837. doi: 10.7554/eLife.04837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wilson MD, Barbosa-Morais NL, Schmidt D, et al. Species-specific transcription in mice carrying human chromosome 21. Science 2008;322(5900):434–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Villar D, Flicek P, Odom DT.. Evolution of transcription factor binding in metazoans—mechanisms and functional implications. Nat Rev Genet 2014;15(4):221–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. He Q, Bardet AF, Patton B, et al. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet 2011;43(5):414–20. [DOI] [PubMed] [Google Scholar]
- 49. Zheng W, Zhao H, Mancera E, et al. Genetic analysis of variation in transcription factor binding in yeast. Nature 2010;464(7292):1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kasowski M, Grubert F, Heffelfinger C, et al. Variation in transcription factor binding among humans. Science 2010;328(5975):232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reddy TE, Gertz J, Pauli F, et al. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res 2012;22(5):860–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bourque G, Leong B, Vega VB, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 2008;18(11):1752–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schmidt D, Schwalie PC, Wilson MD, et al. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 2012;148(1–2):335–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Johnson R, Gamblin RJ, Ooi L, et al. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res 2006;34(14):3862–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kunarso G, Chia NY, Jeyakani J, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 2010;42(7):631–4. [DOI] [PubMed] [Google Scholar]
- 56. Cotney J, Leng J, Yin J, et al. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell 2013;154(1):185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Villar D, Berthelot C, Aldridge S, et al. Enhancer evolution across 20 mammalian species. Cell 2015;160(3):554–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Reilly SK, Yin J, Ayoub AE, et al. Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 2015;347(6226):1155–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Emera D, Yin J, Reilly SK, et al. Origin and evolution of developmental enhancers in the mammalian neocortex. Proc Natl Acad Sci USA 2016;113(19):E2617–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Paris M, Kaplan T, Li XY, et al. Extensive divergence of transcription factor binding in Drosophila embryos with highly conserved gene expression. PLoS Genet 2013;9(9):e1003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bradley RK, Li XY, Trapnell C, et al. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol 2010;8(3):e1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Martin D, Pantoja C, Fernández Miñán A, et al. Genome-wide CTCF distribution in vertebrates defines equivalent sites that aid the identification of disease-associated genes. Nat Struct Mol Biol 2011;18(6):708–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vietri Rudan M, Barrington C, Henderson S, et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep 2015;10(8):1297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ni X, Zhang YE, Nègre N, et al. Adaptive evolution and the birth of CTCF binding sites in the Drosophila genome. PLoS Biol 2012;10(11):e1001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lynch M, Hagner K.. Evolutionary meandering of intermolecular interactions along the drift barrier. Proc Natl Acad Sci USA 2015;112(1):E30–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stewart AJ, Hannenhalli S, Plotkin JB.. Why transcription factor binding sites are ten nucleotides long. Genetics 2012;192(3):973–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet 2007;8(10):803–13. [DOI] [PubMed] [Google Scholar]
- 68. Lynch M, Conery JS.. The origins of genome complexity. Science 2003;302(5649):1401–4. [DOI] [PubMed] [Google Scholar]
- 69. Lynch M. The Origins of Genome Architecture. Sunderland, MA: Sinauer Associates, 2007. [Google Scholar]
- 70. Carvunis AR, Wang T, Skola D, et al. Evidence for a common evolutionary rate in metazoan transcriptional networks. Elife 2015;4:e11615. pii: e11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sung W, Tucker AE, Doak TG, et al. Extraordinary genome stability in the ciliate Paramecium tetraurelia. Proc Natl Acad Sci USA 2012;109(47):19339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Davidson EH, 2006. The Regulatory Genome. Gene Regulatory Networks in Development and Evolution. San Diego, CA: Academic Press/Elsevier. [Google Scholar]
- 73. Peter IS, Davidson EH.. Evolution of gene regulatory networks controlling body plan development. Cell 2011;144(6):970–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chan YF, Marks ME, Jones FC, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 2010;327(5963):302–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Davidson EH, Rast JP, Oliveri P, et al. A genomic regulatory network for development. Science 2002;295(5560):1669–78. [DOI] [PubMed] [Google Scholar]
- 76. Landt SG, Marinov GK, Kundaje A, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 2012;22(9):1813–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kellis M, Wold B, Snyder MP, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci USA 2014;111(17):6131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fisher WW, Li JJ, Hammonds AS, et al. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc Natl Acad Sci USA 2012;109(52):21330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hong JW, Hendrix DA, Levine MS.. Shadow enhancers as a source of evolutionary novelty. Science 2008;321(5894):1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Thakore PI, D'Ippolito AM, Song L, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods 2015;12(12):1143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hilton IB, D'Ippolito AM, Vockley CM, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 2015;33(5):510–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Eckalbar WL, Schlebusch SA, Mason MK, et al. Transcriptomic and epigenomic characterization of the developing bat wing. Nat Genet 2016;48(5):528–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Infante CR, Mihala AG, Park S, et al. Shared enhancer activity in the limbs and phallus and functional divergence of a limb-genital cis-regulatory element in snakes. Dev Cell 2015;35(1):107–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. i5K Consortium. The i5K Initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J Hered 2013;104(5):595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lewis JJ, van der Burg KR, Mazo-Vargas A, Reed RD.. ChIP-seq-annotated Heliconius erato genome highlights patterns of cis-regulatory evolution in lepidoptera. Cell Rep 2016;16(11):2855–63. [DOI] [PubMed] [Google Scholar]
- 86. Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011;473(7345):43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009;459(7243):108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rada-Iglesias A, Bajpai R, Swigut T, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011;470(7333):279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Creyghton MP, Cheng AW, Welstead GG, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 2010;107(50):21931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bernstein BE, Humphrey EL, Erlich RL, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA 2002;99(13):8695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ruthenburg AJ, Allis CD, Wysocka J.. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 2007;25(1):15–30. [DOI] [PubMed] [Google Scholar]
- 92. Li J, Moazed D, Gygi SP.. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem 2002;277(51):49383–8. [DOI] [PubMed] [Google Scholar]
- 93. Bannister AJ, Schneider R, Myers FA, et al. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 2005;280(18):17732–6. [DOI] [PubMed] [Google Scholar]
- 94. Lachner M, O’Carroll D, Rea S, et al. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001;410(6824):116–20. [DOI] [PubMed] [Google Scholar]
- 95. Bannister AJ, Zegerman P, Partridge JF, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001;410(6824):120–4. [DOI] [PubMed] [Google Scholar]
- 96. Schotta G, Lachner M, Sarma K, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 2004;18(11):1251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bell AC, West AG, Felsenfeld G.. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 1999;98(3):387–96. [DOI] [PubMed] [Google Scholar]
- 98. Hnisz D, Day DS, Young RA.. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell 2016;167(5):1188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Barth TK, Imhof A.. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem Sci 2010;35(11):618–26. [DOI] [PubMed] [Google Scholar]
- 100. Luco RF, Pan Q, Tominaga K, et al. Regulation of alternative splicing by histone modifications. Science 2010;327(5968):996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang F, Higgins JM.. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol 2013;23(4):175–84. [DOI] [PubMed] [Google Scholar]
- 102. Uckelmann M, Sixma TK.. Histone ubiquitination in the DNA damage response. DNA Repair 2017;56:92–101. [DOI] [PubMed] [Google Scholar]
- 103. Jenuwein T, Allis CD.. Translating the histone code. Science 2001;293(5532):1074–80. [DOI] [PubMed] [Google Scholar]
- 104. Huang H, Sabari BR, Garcia BA, et al. SnapShot: histone modifications. Cell 2014;159(2):458–58.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Postberg J, Forcob S, Chang WJ, Lipps HJ.. The evolutionary history of histone H3 suggests a deep eukaryotic root of chromatin modifying mechanisms. BMC Evol Biol 2010;10:259.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Marinov GK, Lynch M.. Diversity and divergence of dinoflagellate histone proteins. G3 2015;6(2):397–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Adl SM, Simpson AG, Lane CE, et al. The revised classification of eukaryotes. J Eukaryot Microbiol 2012;59(5):429–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Moore RB, Oborník M, Janouskovec J, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature 2008;451(7181):959–63. [DOI] [PubMed] [Google Scholar]
- 109. Janouškovec J, Tikhonenkov DV, Burki F, et al. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci USA 2015;112(33):10200–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. He D, Fiz-Palacios O, Fu CJ, et al. An alternative root for the eukaryote tree of life. Curr Biol 2014;24(4):465–70. [DOI] [PubMed] [Google Scholar]
- 111. Burki F. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol 2014;6(5):a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Keeling PJ. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol 2013;64:583–607. [DOI] [PubMed] [Google Scholar]
- 113. Sharpe SC, Eme L, Brown MW, Roger AJ.. Timing the origins of multicellular eukaryotes through phylogenomics and relaxed molecular clock analyses In Ruiz-Trillo I, Nedelcu AM (eds). Evolutionary Transitions to Multicellular Life, Advances in Marine Genomics, Vol. 2 Dordrecht: Springer, 2015, 3–29. [Google Scholar]
- 114. Lynch M, Field MC, Goodson HV, et al. Evolutionary cell biology: two origins, one objective. Proc Natl Acad Sci USA 2014;111(48):16990–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Bell G, Mooers AO.. Size and complexity among multicellular organisms. Biol J Linn Soc 1997;60(3):345–63. [Google Scholar]
- 116. Koonin EV, Yutin N.. The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harb Perspect Biol 2014;6(4):a016188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Archibald JM. Endosymbiosis and eukaryotic cell evolution. Curr Biol 2015;25(19):R911–21. [DOI] [PubMed] [Google Scholar]
- 118. Rogozin IB, Carmel L, Csuros M, Koonin EV.. Origin and evolution of spliceosomal introns. Biol Direct 2012;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Martin W, Koonin EV.. Introns and the origin of nucleus-cytosol compartmentalization. Nature 2006;440(7080):41–45. [DOI] [PubMed] [Google Scholar]
- 120. Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct 2006;1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Allen BL, Taatjes DJ.. The mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 2015;16(3):155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Cobbe N, Heck MM.. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol Biol Evol 2004;21(2):332–347. [DOI] [PubMed] [Google Scholar]
- 123. Friz CT. The biochemical composition of the free-living amoebae Chaos chaos, Amoeba dubia and Amoeba proteus. Comp Biochem Physiol 1968;26(1):81–90. [DOI] [PubMed] [Google Scholar]
- 124. LaJeunesse TC, Lambert G, Andersen RA, et al. Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. J Phycology 2005;41(4):880–6. [Google Scholar]
- 125. Veldhuis MJW, Cucci TL, Sieracki ME.. Cellular DNA content of marine phytoplankton using two new fluorochromes: taxonomic and ecological implications. J Phycol 1997;33(3):527–41. [Google Scholar]
- 126. Shuter BJ, Thomas JE, Taylor WD, Zimmerman AM.. Phenotypic correlates of genomic DNA content in unicellular eukaryotes and other cells. Am Nat 1983;122(1):26–44. [Google Scholar]
- 127. Kullman B, Tamm H, Kullman K, 2005. Fungal genome size database. http://www.zbi.ee/fungal-genomesize.
- 128. Kapraun DF, Freshwater DW.. Estimates of nuclear DNA content in red algal lineages. AoB Plants 2012;2012:pls005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Phillips N, Kapraun DF, Gómez Garreta A, et al. Estimates of nuclear DNA content in 98 species of brown algae (Phaeophyta). AoB Plants 2011;2011:plr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Talbert PB, Ahmad K, Almouzni G, et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin 2012;5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kohler A, Kuo A, Nagy LG, et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet 2015;47(4):410–15. [DOI] [PubMed] [Google Scholar]
- 132. Jamieson K, Rountree MR, Lewis ZA, et al. Regional control of histone H3 lysine 27 methylation in Neurospora. Proc Natl Acad Sci USA 2013;110(15):6027–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Galazka JM, Klocko AD, Uesaka M, et al. Neurospora chromosomes are organized by blocks of importin alpha-dependent heterochromatin that are largely independent of H3K9me3. Genome Res 2016;26(8):1069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Weber B, Zicola J, Oka R, Stam M.. Plant enhancers: a call for discovery. Trends Plant Sci 2016;21(11):974–87. [DOI] [PubMed] [Google Scholar]
- 135. Marand AP, Zhang T, Zhu B, Jiang J.. Towards genome-wide prediction and characterization of enhancers in plants. Biochim Biophys Acta 2017;1860(1):131–9. [DOI] [PubMed] [Google Scholar]
- 136. Zhang W, Wu Y, Schnable JC, et al. High-resolution mapping of open chromatin in the rice genome. Genome Res 2012;22(1):151–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Sullivan AM, Arsovski AA, Lempe J, et al. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep 2014;8(6):2015–30. [DOI] [PubMed] [Google Scholar]
- 138. Zhang T, Marand AP, Jiang J.. PlantDHS: a database for DNase I hypersensitive sites in plants. Nucleic Acids Res 2016;44(D1):D1148–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Oka R, Zicola J, Weber B, et al. Genome-wide mapping of transcriptional enhancer candidates using DNA and chromatin features in maize. Genome Biol 2017;18(1):137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Haudry A, Platts AE, Vello E, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet 2013;45(8):891–98. [DOI] [PubMed] [Google Scholar]
- 141. Burgess D, Freeling M.. The most deeply conserved noncoding sequences in plants serve similar functions to those in vertebrates despite large differences in evolutionary rates. Plant Cell 2014;26(3):946–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Sequeira-Mendes J, Aragüez I, Peiró R, et al. The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 2014;26(6):2351–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Roudier F, Ahmed I, Bérard C, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 2011;30(10):1928–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Luo C, Sidote DJ, Zhang Y, et al. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J 2013;73(1):77–90. [DOI] [PubMed] [Google Scholar]
- 145. Zhang W, Garcia N, Feng Y, et al. Genome-wide histone acetylation correlates with active transcription in maize. Genomics 2015;106(4):214–20. [DOI] [PubMed] [Google Scholar]
- 146. Du Z, Li H, Wei Q, et al. Genome-wide analysis of histone modifications: H3K4me2, H3K4me3, H3K9ac, and H3K27ac in Oryza sativa L. Japonica. Mol Plant 2013;6(5):1463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Widiez T, Symeonidi A, Luo C, et al. The chromatin landscape of the moss Physcomitrella patens and its dynamics during development and drought stress. Plant J 2014;79(1):67–81. [DOI] [PubMed] [Google Scholar]
- 148. Zhu B, Zhang W, Zhang T, et al. Genome-wide prediction and validation of intergenic enhancers in Arabidopsis using open chromatin signatures. Plant Cell 2015;27(9):2415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Deal RB, Henikoff S.. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc 2011;6(1):56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Dekker J, Heard E.. Structural and functional diversity of topologically associating domains. FEBS Lett 2015;589(20PartA):2877–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Feng S, Cokus SJ, Schubert V, et al. Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol Cell 2014;55(5):694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Grob S, Schmid MW, Grossniklaus U.. Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol Cell 2014;55(5):678–93. [DOI] [PubMed] [Google Scholar]
- 153. Singer SD, Cox KD, Liu Z.. Enhancer-promoter interference and its prevention in transgenic plants. Plant Cell Rep 2011;30(5):723–31. [DOI] [PubMed] [Google Scholar]
- 154. Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000;408(6814):796–815. [DOI] [PubMed] [Google Scholar]
- 155. Liu C, Cheng YJ, Wang JW, Weigel D.. Prominent topologically associated domains differentiate global chromatin packing in rice from Arabidopsis. Nat Plants 2017;3(9):742–8. [DOI] [PubMed] [Google Scholar]
- 156. Gupta AP, Chin WH, Zhu L, et al. Dynamic epigenetic regulation of gene expression during the life cycle of malaria parasite Plasmodium falciparum. PLoS Pathog 2013;9(2):e1003170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Karmodiya K, Pradhan SJ, Joshi B, et al. A comprehensive epigenome map of Plasmodium falciparum reveals unique mechanisms of transcriptional regulation and identifies H3K36me2 as a global mark of gene suppression. Epigenetics Chromatin 2015;8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Ay F, Bunnik EM, Varoquaux N, et al. Multiple dimensions of epigenetic gene regulation in the malaria parasite Plasmodium falciparum: gene regulation via histone modifications, nucleosome positioning and nuclear architecture in P. falciparum. Bioessays 2015;37(2):182–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Fraschka SA, Henderson RW, Bártfai R.. H3.3 demarcates GC-rich coding and subtelomeric regions and serves as potential memory mark for virulence gene expression in Plasmodium falciparum. Sci Rep 2016;6:31965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Lieleg C, Krietenstein N, Walker M, Korber P.. Nucleosome positioning in yeasts: methods, maps, and mechanisms. Chromosoma 2015;124(2):131–51. [DOI] [PubMed] [Google Scholar]
- 161. Clayton CE. Gene expression in Kinetoplastids. Curr Opin Microbiol 2016;32:46–51. [DOI] [PubMed] [Google Scholar]
- 162. Soto M, Requena JM, Morales G, Alonso C.. The Leishmania infantum histone H3 possesses an extremely divergent N-terminal domain. Biochim Biophys Acta 1994;1219(2):533–5. [DOI] [PubMed] [Google Scholar]
- 163. Lukes J, Maslov DA.. Unexpectedly high variability of the histone H4 gene in Leishmania. Parasitol Res 2000;86(3):259–61. [DOI] [PubMed] [Google Scholar]
- 164. Siegel TN, Hekstra DR, Kemp LE, et al. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev 2009;23(9):1063–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Thomas S, Green A, Sturm NR, et al. Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genomics 2009;10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Reynolds D, Cliffe L, Förstner KU, et al. Regulation of transcription termination by glucosylated hydroxymethyluracil, base J, in Leishmania major and Trypanosoma brucei. Nucleic Acids Res 2014;42(15):9717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Reynolds D, Hofmeister BT, Cliffe L, et al. Histone H3 variant regulates RNA polymerase II transcription termination and dual strand transcription of siRNA loci in Trypanosoma brucei. PLoS Genet 2016;12(1):e1005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Jackson AP, Otto TD, Aslett M, et al. Kinetoplastid phylogenomics reveals the evolutionary innovations associated with the origins of parasitism. Curr Biol 2016;26(2):161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Zysset-Burri DC, Müller N, Beuret C, et al. Genome-wide identification of pathogenicity factors of the free-living amoeba Naegleria fowleri. BMC Genomics 2014;15:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Fritz-Laylin LK, Prochnik SE, Ginger ML, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 2010;140(5):631–42. [DOI] [PubMed] [Google Scholar]
- 171. Lukes J, Leander BS, Keeling PJ.. Cascades of convergent evolution: the corresponding evolutionary histories of euglenozoans and dinoflagellates. Proc Natl Acad Sci USA 2009;106(Suppl 1):9963–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Song MJ, Kim M, Choi Y, et al. Epigenome mapping highlights chromatin-mediated gene regulation in the protozoan parasite Trichomonas vaginalis. Sci Rep 2017;7:45365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Balandina A, Kamashev D, Rouviere-Yaniv J.. The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J Biol Chem 2002;277(31):27622–8. [DOI] [PubMed] [Google Scholar]
- 174. Dillon SC, Dorman CJ.. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 2010;8(3):185–95. [DOI] [PubMed] [Google Scholar]
- 175. Triana O, Galanti N, Olea N, et al. Chromatin and histones from Giardia lamblia: a new puzzle in primitive eukaryotes. J Cell Biochem 2001;82(4):573–82. [DOI] [PubMed] [Google Scholar]
- 176. Teodorovic S, Walls CD, Elmendorf HG.. Bidirectional transcription is an inherent feature of Giardia lamblia promoters and contributes to an abundance of sterile antisense transcripts throughout the genome. Nucleic Acids Res 2007;35(8):2544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Ngan CY, Wong CH, Choi C, et al. Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. Nat Plants 2015;1(1):15107. [DOI] [PubMed] [Google Scholar]
- 178. Veluchamy A, Rastogi A, Lin X, et al. An integrative analysis of post-translational histone modifications in the marine diatom Phaeodactylum tricornutum. Genome Biol 2015;16:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Xiong J, Gao S, Dui W, et al. Dissecting relative contributions of cis- and trans-determinants to nucleosome distribution by comparing Tetrahymena macronuclear and micronuclear chromatin. Nucleic Acids Res 2016;44(21):10091–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Beh LY, Müller MM, Muir TW, et al. DNA-guided establishment of nucleosome patterns within coding regions of a eukaryotic genome. Genome Res 2015;25(11):1727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Allen SE, Hug I, Pabian S, et al. Circular concatemers of ultra-short DNA segments produce regulatory RNAs. Cell 2017;168(6):990–9.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Canzio D, Larson A, Narlikar GJ.. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol 2014;24(6):377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Nishibuchi G, Nakayama J.. Biochemical and structural properties of heterochromatin protein 1: understanding its role in chromatin assembly. J Biochem 2014;156(1):11–20. [DOI] [PubMed] [Google Scholar]
- 184. Kataoka K, Mochizuki K.. Phosphorylation of an HP1-like protein regulates heterochromatin body assembly for DNA elimination. Dev Cell 2015;35(6):775–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Suhren JH, Noto T, Kataoka K, et al. Negative regulators of an RNAi-heterochromatin positive feedback loop safeguard somatic genome integrity in Tetrahymena. Cell Rep 2017;18(10):2494–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Swart EC, Bracht JR, Magrini V, et al. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol 2013;11(1):e1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Chen X, Bracht JR, Goldman AD, et al. The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell 2014;158(5):1187–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Riley JL, Katz LA.. Widespread distribution of extensive chromosomal fragmentation in ciliates. Mol Biol Evol 2001;18(7):1372–7. [DOI] [PubMed] [Google Scholar]
- 189. Ricard G, de Graaf RM, Dutilh BE, et al. Macronuclear genome structure of the ciliate Nyctotherus ovalis: single-gene chromosomes and tiny introns. BMC Genomics 2008;9:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Dodge JD. Chromosome structure in the dinoflagellates and the problem of the mesocaryotic cell. Prog Protozool 1965;2:264. [Google Scholar]
- 191. Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R.. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta 2014;1839(3):155–68. [DOI] [PubMed] [Google Scholar]
- 192. Hackett JD, Anderson DM, Erdner DL, Bhattacharya D.. Dinoflagellates: a remarkable evolutionary experiment. Am J Bot 2004;91(10):1523–34. [DOI] [PubMed] [Google Scholar]
- 193. Rizzo PJ. Those amazing dinoflagellate chromosomes. Cell Res 2003;13(4):215–17. [DOI] [PubMed] [Google Scholar]
- 194. Lin S, Cheng S, Song B, et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 2015;350(6261):691–4. [DOI] [PubMed] [Google Scholar]
- 195. Shoguchi E, Shinzato C, Kawashima T, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol 2013;23(15):1399–408. [DOI] [PubMed] [Google Scholar]
- 196. Archibald JM. Nucleomorph genomes: structure, function, origin and evolution. Bioessays 2007;29(4):392–402. [DOI] [PubMed] [Google Scholar]
- 197. Marinov GK, Lynch M.. Conservation and divergence of the histone code in nucleomorphs. Biol Direct 2016;11(1):18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Cerutti H, Casas-Mollano JA.. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet 2006;50(2):81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Volpe T, Martienssen RA.. RNA interference and heterochromatin assembly. Cold Spring Harb Perspect Biol 2011;3(9):a003731.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Tóth KF, Pezic D, Stuwe E, Webster A.. The piRNA pathway guards the germline genome against transposable elements. Adv Exp Med Biol 2016;886:51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Ryan JF, Pang K, Schnitzler CE, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 2013;342(6164):1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Moroz LL, Kocot KM, Citarella MR, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature 2014;510(7503):109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Whelan NV, Kocot KM, Moroz LL, Halanych KM.. Error, signal, and the placement of Ctenophora sister to all other animals. Proc Natl Acad Sci USA 2015;112(18):5773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Pisani D, Pett W, Dohrmann M, et al. Genomic data do not support comb jellies as the sister group to all other animals. Proc Natl Acad Sci USA 2015;112(50):15402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Simion P, Philippe H, Baurain D, et al. A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr Biol 2017;27(7):958–67. [DOI] [PubMed] [Google Scholar]
- 206. Sperling EA, Peterson KJ, Pisani D.. Phylogenetic-signal dissection of nuclear housekeeping genes supports the paraphyly of sponges and the monophyly of Eumetazoa. Mol Biol Evol 2009;26(10):2261–74. [DOI] [PubMed] [Google Scholar]
- 207. Ball EE, Miller DJ.. Phylogeny: the continuing classificatory conundrum of chaetognaths. Curr Biol 2006;16(15):R593–6. [DOI] [PubMed] [Google Scholar]
- 208. Egger B, Steinke D, Tarui H, et al. To be or not to be a flatworm: the acoel controversy. PLoS One 2009;4(5):e5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Cannon JT, Vellutini BC, Smith J 3rd, et al. Xenacoelomorpha is the sister group to Nephrozoa. Nature 2016;530(7588):89–93. [DOI] [PubMed] [Google Scholar]
- 210. Rouse GW, Wilson NG, Carvajal JI, Vrijenhoek RC.. New deep-sea species of Xenoturbella and the position of Xenacoelomorpha. Nature 2016;530(7588):94–7. [DOI] [PubMed] [Google Scholar]
- 211. Lu TM, Kanda M, Satoh N, Furuya H.. The phylogenetic position of dicyemid mesozoans offers insights into spiralian evolution. Zoological Lett 2017;3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Dujardin F. Histoire Naturelle Des Zoophytes. Infusoires, Comprenant La Physiologie Et La Classification De Ces Animaux, Et La Manière De Les Étudier á L’aide Du Microscope. Paris: Roret, 1841. [Google Scholar]
- 213. King N, Westbrook MJ, Young SL, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 2008;451(7180):783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. de Mendoza A, Sebé-Pedrós A, Šestak MS, et al. Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc Natl Acad Sci USA 2013;110(50):E4858–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Sebé-Pedrós A, Ballaré C, Parra-Acero H, et al. The dynamic regulatory genome of Capsaspora and the origin of animal multicellularity. Cell 2016;165(5):1224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Gaiti F, Jindrich K, Fernandez-Valverde SL, et al. Landscape of histone modifications in a sponge reveals the origin of animal cis-regulatory complexity. Elife 2017;6:e22194. pii: e22194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Schwaiger M, Schönauer A, Rendeiro AF, et al. Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res 2014;24(4):639–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Heger P, Marin B, Bartkuhn M, et al. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci USA 2012;109(43):17507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Gaiti F, Calcino AD, Tanurdžić M, Degnan BM.. Origin and evolution of the metazoan non-coding regulatory genome. Dev Biol 2017;427:193–202. [DOI] [PubMed] [Google Scholar]
- 220. Daugherty AC, Yeo R, Buenrostro JD, et al. Chromatin accessibility dynamics reveal novel functional enhancers in C. elegans. Genome Res 2017;27(12):2096–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Heger P, George R, Wiehe T.. Successive gain of insulator proteins in arthropod evolution. Evolution 2013;67(10):2945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Roquis D, Lepesant JM, Picard MA, et al. The epigenome of Schistosoma mansoni provides insight about how cercariae poise transcription until infection. PLoS Negl Trop Dis 2015;9(8):e0003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Kao D, Mihaylova Y, Hughes S, et al. Epigenetic analyses of the planarian genome reveals conservation of bivalent promoters in animal stem cells. bioRxiv 2017, https://doi.org/10.1101/122135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006;125(2):315–26. [DOI] [PubMed] [Google Scholar]
- 225. Navratilova P, Danks GB, Long A, et al. Sex-specific chromatin landscapes in an ultra-compact chordate genome. Epigenetics Chromatin 2017;10(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Ho JW, Jung YL, Liu T, et al. Comparative analysis of metazoan chromatin organization. Nature 2014;512(7515):449–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Bártfai R, Hoeijmakers WA, Salcedo-Amaya AM, et al. H2A.Z demarcates intergenic regions of the Plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog 2010;6(12):e1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Shankaranarayanan P, Mendoza-Parra MA, Walia M, et al. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nat Methods 2011;8(7):565–7. [DOI] [PubMed] [Google Scholar]
- 229. Hoeijmakers WA, Bártfai R, Françoijs KJ, Stunnenberg HG.. Linear amplification for deep sequencing. Nat Protoc 2011;6(7):1026–36. [DOI] [PubMed] [Google Scholar]
- 230. Sos BC, Fung HL, Gao DR, et al. Characterization of chromatin accessibility with a transposome hypersensitive sites sequencing (THS-seq) assay. Genome Biol 2016;17:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Teytelman L, Ozaydin B, Zill O, et al. Impact of chromatin structures on DNA processing for genomic analyses. PLoS One 2009;4(8):e6700.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Auerbach RK, Euskirchen G, Rozowsky J, et al. Mapping accessible chromatin regions using Sono-Seq. Proc Natl Acad Sci USA 2009;106(35):14926–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Park D, Lee Y, Bhupindersingh G, Iyer VR.. Widespread misinterpretable ChIP-seq bias in yeast. PLoS One 2013;8(12):e83506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Marinov GK, Kundaje A, Park PJ, Wold BJ.. Large-scale quality analysis of published ChIP-seq data. G3 2014;4(2):209–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Carroll TS, Liang Z, Salama R, et al. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front Genet 2014;5:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Marinov GK, Wang YE, Chan D, Wold BJ.. Evidence for site-specific occupancy of the mitochondrial genome by nuclear transcription factors. PLoS One 2014;9(1):e84713.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Shema E, Jones D, Shoresh N, et al. Single-molecule decoding of combinatorially modified nucleosomes. Science 2016;352(6286):717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Savic D, Partridge EC, Newberry KM, et al. CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins. Genome Res 2015;25(10):1581–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Egelhofer TA, Minoda A, Klugman S, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol 2011;18(1):91–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Busby M, Xue C, Li C, et al. Systematic comparison of monoclonal versus polyclonal antibodies for mapping histone modifications by ChIP-seq. Epigenetics Chromatin 2016;9:49. [DOI] [PMC free article] [PubMed] [Google Scholar]