Abstract
One of the central goals in molecular biology is to understand how cell-type-specific expression patterns arise through selective recruitment of RNA polymerase II (Pol II) to a subset of gene promoters. Pol II needs to be recruited to a precise genomic position at the proper time to produce messenger RNA from a DNA template. Ostensibly, transcription is a relatively simple cellular process; yet, experimentally measuring and then understanding the combinatorial possibilities of transcriptional regulators remain a daunting task. Since its introduction in 1985, chromatin immunoprecipitation (ChIP) has remained a key tool for investigating protein–DNA contacts in vivo. Over 30 years of intensive research using ChIP have provided numerous insights into mechanisms of gene regulation. As functional genomic technologies improve, they present new opportunities to address key biological questions. ChIP-exo is a refined version of ChIP-seq that significantly reduces background signal, while providing near base-pair mapping resolution for protein–DNA interactions. This review discusses the evolution of the ChIP assay over the years; the methodological differences between ChIP-seq, ChIP-exo and ChIP-nexus; and highlight new insights into epigenetic and transcriptional mechanisms that were uniquely enabled with the near base-pair resolution of ChIP-exo.
Keywords: ChIP-exo, ChIP-nexus, epigenetics, gene regulation, transcription, bioinformatics
Introduction
Over 200 distinct cell types in the human body are generated from the complex process of cellular differentiation, which remains poorly understood. Cell identity is established and propagated through tissue-specific expression of transcription factors (TFs) and epigenetic mechanisms, such as cell-type-specific enhancers [1–5]. Enhancers are cis-regulatory elements dispersed throughout the genome that serve to increase transcription of genes, often in response to extracellular stimuli or developmental signals. Enhancer elements operate from promoter distal regions of the genome independent of their orientation relative to genes [6, 7]. Tens of thousands of enhancers are scattered across mammalian genomes in a cell-type-specific manner and are dynamically shaped in response to environmental stimuli [2]. Collectively, TFs orchestrate cell-type-specific transcription programs by binding to their cognate sequence motifs within specific enhancers, and recruiting co-regulators and RNA polymerase II (Pol II) to promoters to initiate transcription [8, 9]. Enhancer segments are typically co-occupied by combinations of TFs that influence gene expression programs, often integrating signals from multiple pathways. Shaping the epigenome in a given cell type involves coordinate activities of TFs, together with nucleosome modifiers and remodelers [10].
Understanding mechanisms of gene regulation remains an important, fundamental goal of research efforts over the past 50 years. Chromatin immunoprecipitation (ChIP) is a powerful method to study mechanisms of gene regulation by selectively enriching for DNA fragments that interact with a given protein in living cells. Detection methods of ChIP-enriched DNA fragments have evolved as technology improves, from detection of a single locus [standard ChIP-polymerase chain reaction (PCR)] to hybridization on oligonucleotide microarrays (ChIP-chip) to high-throughput sequencing (ChIP-seq). More recently, clever improvements in the ChIP-seq assay have yielded near base-pair resolution mapping (e.g. ChIP-exo) of transcriptional regulators.
This review discusses the evolution of ChIP-based assays, the experimental differences between global mapping assays and focuses on unique biological insights enabled by the near base-pair resolution of ChIP-exo. Many excellent bioinformatics tools have been developed specifically for ChIP-exo data analysis [11–22], but are outside the scope of this review (please refer to a recent review [23] for a nice discussion of ChIP-exo analysis tools and considerations). Briefly, a number of peak finding algorithms have been developed specifically for ChIP-exo/nexus data analysis [11–13, 17, 20–22]. Additionally, ChIP-exo footprint and motif recognition tools have been reported [14–16, 18].
Historical perspective on protein–DNA interaction assays
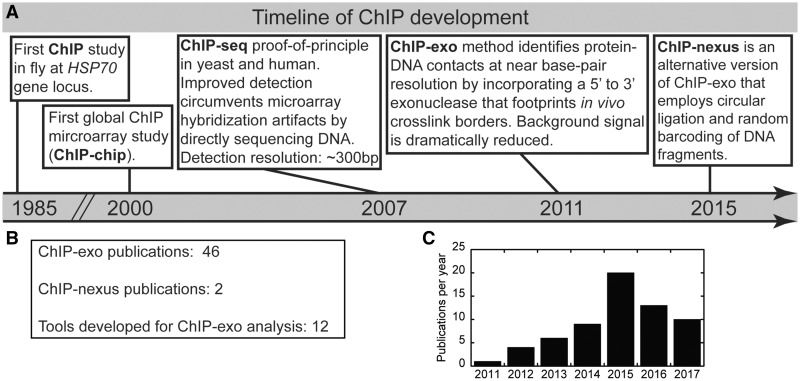
Tracking the genomic spatiotemporal patterns of transcriptional regulators is critical to understanding their function. Over 30 years ago, Drs Gilmour and Lis [24] described a method to examine the in vivo distribution of proteins on genomic DNA, which is now referred to as ChIP (Figure 1A). In this study, the authors tracked the density of Pol II over the HSP70 gene before and after heat shock treatment in fly tissue culture cells. The ChIP methodology represented a technological breakthrough at the time because such protein–DNA interactions were primarily defined in vitro using DNA-binding assays and purified transcription systems [30, 31]. Nevertheless, the ChIP assay is not without general conceptual problems. For example, successful ChIP enrichment of protein-DNA interactions critically depend on the efficiency of a cross-linking reagent to preserve in vivo contacts [32], which vary from protein to protein.
Figure 1.
Historical perspective on the development of the ChIP assay and detection methods. (A) Key technological advancements that ultimately led to ChIP-exo/nexus occurred in the following years 1985 [24], 2000 [25], 2007 [26, 27], 2011 [28] and 2015 [29]. (B) Current tallies of publications are shown using ChIP-exo, ChIP-nexus or developing bioinformatic tools specifically for ChIP-exo analysis. (C) Bar graph representation of publications with ‘ChIP-exo’ keyword by year as returned by PubMed.
With the development of microarray printing of oligonucleotides onto a glass slide in the late 1990s, the ChIP assay was applied genome wide for the first time, which targeted the yeast TFs Gal4 and Ste12 [25]. Applying the ChIP assay to microarray chip detection was termed ChIP-chip. The ability to systematically interrogate an entire genome was fully realized with the advent of next-generation sequencing technologies. Coupling ChIP to massively parallel sequencing detection (ChIP-seq) enabled much larger mammalian genomes to be probed for protein–DNA interactions. In 2007, ChIP-seq was first used to determine the genome-wide locations for the histone variant H2A.Z in yeast, and to identify global Stat1-binding locations in human tissue culture cells upon interferon gamma stimulation [26, 33]. Although numerous ChIP-seq studies have advanced our understanding for how transcription is controlled at the molecular level in living cells, the ChIP-seq method remains hampered by relatively low mapping resolution of several hundred base pairs and high background signal [34].
ChIP-exo maps genomic locations of proteins at near base-pair resolution
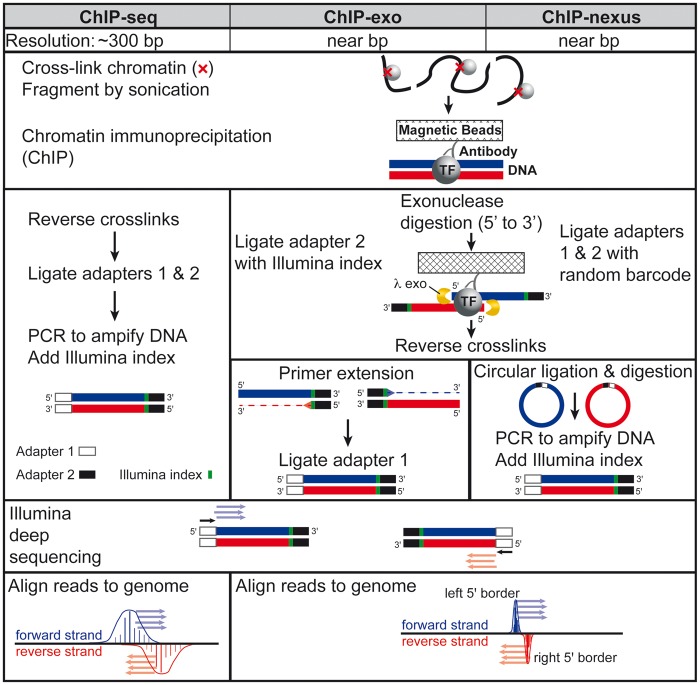
The ChIP-exo method was developed by Dr Pugh in 2011 [28], and represents a refined version of ChIP-seq that substantially improves on both resolution and noise. Several excellent versions of the ChIP-exo protocol are available [35–38], including one in video form [39]. Although ChIP-exo is more technically challenging to master than ChIP-seq, it is now widely adopted as studies aim to gain unique ultra high-resolution insights using diverse biological systems (Figure 1B and C). The key distinction of the ChIP-exo methodology is the incorporation of lambda exonuclease digestion in the library preparation workflow to effectively footprint the left and right 5' DNA borders of the protein–DNA cross-link site (Figure 2). The ChIP-exo libraries are then subjected to high-throughput sequencing. The resulting data can be leveraged to provide unique and ultra high-resolution insights into the functional organization of the genome. Although the methodology requires more steps, transitions between steps are achieved by simple bead washing. In contrast, ChIP-seq DNA is often purified between steps, resulting in sample loss and experimental variability. However, several modifications to the ChIP-seq methodology have recently been developed to mitigate these issues [40, 41].
Figure 2.
Comparative workflow of ChIP-seq, ChIP-exo and ChIP-nexus technologies.
Recently, ChIP-nexus was developed as a variation on the ChIP-exo method, which sought to improve ChIP-exo library complexity by replacing traditional double-stranded DNA linear ligation with a circular ligation step [29]. As circular ligation is more thermodynamically favorable than linear ligation, a higher yield of ChIP-enriched DNA fragments would be expected. This in turn should translate into higher library complexity. It is important to note that spatial resolution is not improved with ChIP-nexus, as it relies on the same lambda exonuclease step as ChIP-exo. Another key innovation of ChIP-nexus was the inclusion of unique molecular identifiers that enable bioinformatic identification of PCR amplification artifacts, which can be problematic in some high-resolution mapping libraries [42, 43].
Despite the increasing adoption of the ChIP-exo/nexus methods, the limitations of these technologies should be noted. First, ChIP-exo and ChIP-nexus are experimentally more complex than ChIP-seq, which increases the cost and time for their experimental workflows. For instance, ChIP-nexus uses a circular ligation enzyme (CircLigase) that is currently 5-6x more expensive than T4 DNA ligase (Epicentre CL4115K and NEB M0202L, when normalized to units per library prep). In addition, one study reported circLigase sequence specificity [44], which could lead to biased libraries. ChIP-exo protocols typically contain ∼10 sequentially dependent enzymatic reactions, which are exchanged by simple bead washing. Additionally, on resin reactions containing protein-DNA cross-linked complexes are performed at elevated temperatures, which can reverse crosslinks at a rate of 3–4% per hour at 37 °C [45]. In most ChIP-seq workflows, these elevated temperature incubations of protein-DNA cross-links are completely avoided. Taken together, the issues specific to ChIP-exo/nexus methodologies can contribute to lower library complexity, which can lead to PCR artifacts [42]. For ChIP-exo, low library complexity can be mitigated by using more nuclear extract in the ChIP step, but this may not be feasible for experiments using cells derived from primary tissues. To assist in artifact removal in ChIP-exo data, a technique-specific input control strategy has been proposed [43]. For ChIP-nexus, PCR artifacts can simply be computationally filtered out based on a DNA barcoding strategy. Given these limitations, it is important to note that not all proteins that can be assayed by other ChIP-based approaches will necessarily generate successful ChIP-exo/nexus libraries.
Overview of biological insights enabled by ChIP-exo
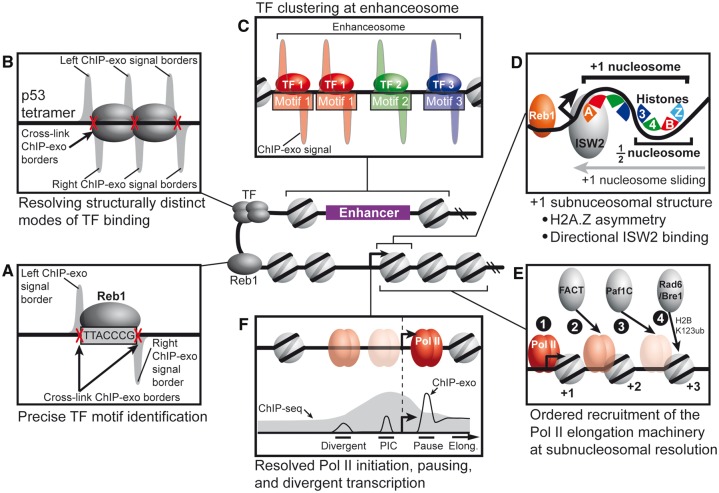
As functional genomic technologies improve, they provide unique opportunities to address key biological questions. Since its development, the ChIP-exo technique has provided a deeper understanding into three key areas of gene regulation (Figure 3): (1) TF-binding mechanics, (2) chromatin structure and regulation and (3) Pol II transcription cycle. Discussed below are the unique biological insights revealed by leveraging the near base-pair resolution of ChIP-exo.
Figure 3.
Models for how unique biological insights are revealed with the near base-pair resolution of ChIP-exo. (A) ChIP-exo signals delineate left and right protein–DNA cross-link borders, which enables precise TF motif identification. (B) ChIP-exo enables determination of structurally distinct modes of TF binding, such as the p53 tetrameric complex. (C) ChIP-exo distinguishes clustered TFs, such as the assembly of the enhanceosome. (D) Subnucleosomal structure of histone subunit–DNA contacts can be resolved by ChIP-exo. For example, the H2A.Z histone variant is asymmetrically incorporated into the +1 nucleosome. In addition, the ISW2 chromatin remodeler engages the +1 nucleosome on the promoter proximal side. (E) Ordered recruitment of Pol II elongation machinery at subnucleosomal resolution. (F) ChIP-exo resolves Pol II engaged at the PIC site, promotor proximal pause site and divergently oriented PIC.
Precise identification of TF motifs
TFs are the interpreters of the cellular genome. It is estimated that the human genome encodes over 2000 TFs [46]. Their DNA binding activity is often controlled by upstream signaling pathways that integrate responses to stimuli or other environmental cues that ultimately lead to changes in gene expression patterns. For example, the hormone erythropoietin activates the phosphatidylinositol-3-kinase (PI3K) signaling pathway that phosphorylates the TF Gata1 [47]. Gata1, as a master regulator of erythropoiesis, orchestrates the dynamic expression patterns during terminal erythroid differentiation [48]. Thus, to understand how TFs establish dynamic transcription programs in different developmental contexts, it is necessary to understand the binding patterns of TFs.
Short, often degenerate DNA sequences are recognized by TFs using a variety of oligomeric states (e.g. monomer, homodimer and heterodimer, tetramers and so on) [49–51]. In some cases, TFs can bind in a clustered manner with itself or other TFs, or bind to DNA indirectly through interactions with another TF [52]. Further confounding is the observation that TFs in the nuclear hormone receptor family can recognize distinct motifs in a ligand-dependent manner [53, 54]. While ChIP-seq can find TF-bound regions of the genome, its resolution is inadequate to unambiguously identify the precise sequence motif occupied by a given TF, discern clustered binding or assess the oligomeric state of DNA-bound TFs. These problems are further compounded for TFs whose motif is unknown or highly degenerate. Thus, the near base-pair mapping resolution of ChIP-exo is necessary for higher-confidence motif identification and for distinguishing TF-binding modalities.
Proof-of-principle for the ChIP-exo method focused on the precise identification of the yeast Reb1-binding locations, which were validated using the known Reb1 motif (Figure 3A) [28]. Remarkably, ChIP-exo peak calls were within 5 bp on average of the Reb1 motif, and nearly every peak occupied a Reb1 motif. Together, this analysis made a compelling case for the benefits of near base-pair mapping and with the added benefit of >20× lower background than ChIP-seq [28]. Other TFs examined in this study revealed in vivo mechanics of TF binding, such as compound motif usage and adjacent binding of the same TF. Since the development of ChIP-exo, numerous studies have sought to shed light on in vivo TF-binding mechanics using this technique [55–62].
Resolving structurally distinct modes of TF binding
TFs often bind to DNA in oligomeric states. For example, p53 is a well-known tumor suppressor that binds to DNA as a tetramer to paired half sites [63]. When ChIP-exo was applied to p53 in human U2OS cells under a variety of stresses [64], it became clear from the structure of the peak-pairs that the high-resolution mapping was able to resolve the individual tetrameric subunits (Figure 3B). Importantly, the resolution of ChIP-exo uniquely revealed the spatial relationship in vivo between p53 subunits and paired half sites. In particular, the left/right ChIP-exo signal borders were consistent with cross-links occurring at the tetramer edges and internally between dimers (Figure 3B).
The glucocorticoid receptor (GR) has been the focus of extensive study for decades [65]. GR behaves as a DNA-binding TF in response to hormonal stimulation; yet, how it activates some genes while repressing others remains an open question. The oligomeric status of GR has been proposed to determine whether it activates (GR dimer) or represses (GR monomer) transcription of target genes [66]. A recent report turned to ChIP-exo analysis of mouse livers exposed to corticosterone to test this model [67]. This study found that GR more commonly binds as a monomer to its half-site motif, and was not always associated with transcriptional repression as previously thought. This finding would not have been possible with ChIP-seq, as it would not have been able to resolve DNA-bound GR monomers from dimers.
TF clustering at enhanceosomes
Enhancers are critical determinants of cell identity that together with tissue-specific TFs serve to maintain gene expression patterns for a given cell type. Enhanceosome is a term used to describe a higher-order complex of multiple TFs that bind DNA in close proximity to one another to regulate gene expression (Figure 3C) [68]. As TFs within the enhanceosome tend to be in close proximity to one another, conventional protein–DNA mapping methods lack the resolution to discern the spatial arrangement of TFs in vivo. Thus, the near base-pair resolution of ChIP-exo is essential to understand the molecular mechanisms of adjacent TF-binding.
Although the compact yeast genome lacks conventional enhancers, upstream activating sequences serve a similar role as a platform for TFs to assemble to regulate gene expression. To mechanistically understand how yeast ribosomal protein genes (RPGs) are coordinately regulated by an assemblage of TFs at an enhanceosome-like structure, ChIP-exo was applied to five TFs known to influence expression of yeast RPGs [69]. The report found that the TFs displayed a well-defined spatial organization, with multiple molecules of some TFs, such as Hmo1, occupying a subset of RP genes. The authors suggested that the spatial arrangement of these clustered TFs in part influence the local nucleosome positions by serving as barriers against which chromatin remodelers use to position adjacent promoter nucleosomes.
Another report focused on two master TF regulators (Gata1 and Tal1) of erythropoiesis that are known to co-occupy many genomic locations [70] and form higher-order complexes [71]. How Gata1 and Tal1 are coordinately and spatially organized across the genome to recognize their cognate DNA-binding sites remains poorly understood. Using a mouse cell system that synchronously undergoes erythropoiesis upon Gata1 induction, ChIP-exo analysis of Gata1 and Tal1 showed that they co-occupy ∼3000 locations throughout the mouse genome in a positionally constrained manner in relation to the Gata and partial E-box motifs [72]. Furthermore, the study revealed that homotypic clustering of Gata1 and Tal1 binding are common elsewhere in the mouse genome, suggesting that these TFs display distinct combinatorial binding modes throughout the genome.
Other studies are using ChIP-exo to identify candidate enhancer regions in the mammalian genome, and then address spatiotemporal questions about how TFs assemble within enhancer regions to influence gene expression [73, 74]. Taken together, the ultra high-resolution afforded by ChIP-exo is revealing new concepts into how TFs mechanistically interpret the cellular genome in living cells.
Subnucleosomal structure and chromatin regulator interactions
Genomic DNA is a nucleic acid polymer that is packaged in the form of nucleosomes. Each nucleosome contains 146 bp of DNA wrapped around a histone octamer, which includes two copies of H2A, H2B, H3 and H4 [75]. MNase-seq mapping of nucleosome positions genome-wide provided the foundation for understanding how the transcription machinery operates in the context of chromatin [76–78]. Similarly, near base-pair mapping of individual histone subunits would provide the framework for understanding how chromatin regulators interact with the individual histone subunits within the subnuceosomal structure. The histone variant H2A.Z marks promoter regions through nucleosomal exchange with its canonical counterpart, histone H2A [79]. H2A.Z is preferentially incorporated into the +1 nucleosome that resides just downstream of the transcriptional start site (TSS). Interestingly, deposition and removal of H2A.Z by the SWR-C and INO80 complexes, respectively, were shown to occur through asymmetrical interactions with the +1 nucleosome [80]. This finding raised the question of whether both copies of H2A in the +1 nucleosome are exchanged for H2A.Z, or the H2A.Z variant is incorporated in an asymmetrical manner.
A recent ChIP-exo study in yeast focused on the four canonical histone subunits and the H2A.Z variant [81]. Remarkably, ChIP-exo effectively resolved the protein–DNA cross-link sites for both copies of each of the four canonical histones. Consistent with the crystal structure for the nucleosome, H3/H4 subunits cross-linked predominantly to the nucleosomal DNA dyad, whereas each copy of H2A or H2B histones cross-linked to opposing edges of the nucleosomal DNA (Figure 3D). Interestingly, H2A.Z was predominantly incorporated into the promoter distal copy of H2A within the +1 nucleosome, suggesting that the direction of transcription may influence which copy of H2A is exchanged for H2A.Z.
The manner by which chromatin remodelers cooperate to organize nucleosomes around the TSS in living cells remains unclear. The directional incorporation of H2A.Z suggests that chromatin regulators may also interface with the +1 nucleosome in a directional manner. Indeed, ChIP analysis showed that the multiple subunits of the chromatin remodeler complex ISW2 (Imitation Switch 2) cross-linked to the promoter proximal side of the +1 nucleosome [82]. The study further showed that Reb1 co-occupied many of these promoters, suggesting that Reb1 and ISW2 may cooperate to properly position and space promoter-flanking nucleosomes (Figure 3D).
Ordered recruitment of Pol II elongation machinery at subnucleosomal resolution
Post-translational modifications to the nucleosome by chromatin modifier complexes often occur in a co-transcriptional manner through association with Pol II [83, 84]. H2B123-ubiquitylation is one such modification that is stimulated by the Paf1C complex in association with the ubiquitin ligase Rad6/Bre1 complex [84]. In association with Pol II, FACT (facilitates chromatin transcription) assists transcription through nucleosomes by dismantling H2A/H2B dimers [84]. How epigenetic changes are mechanistically coupled with transcriptional elongation remains an open question. A recent study sought to address this question in yeast by tracking the subnucleosomal distributions of Paf1C and Rad6/Bre1 subunits using ChIP-exo [85]. The high-resolution mapping from this study supported an ordered recruitment model for early elongation, wherein FACT is first recruited to the +2 nucleosome via Pol II, followed by Rad/Bre1 recruitment to the +2/+3 nucleosomes via Paf1C mediated tethering to Pol II (Figure 3E). In summary, this study provided new mechanistic insights for how the H2BK123ub mark regulated and associated with early elongation events.
Pol II initiation, pausing and divergently oriented preinitiation complex resolved by ChIP-exo
Over three decades ago, parallel work from Drs Lis and Groudine showed that Pol II had initiated transcription but paused elongation at the promoter proximal region, ∼30–50 bp downstream of the TSS [86, 87]. Accumulating evidence points to the process of transcriptional elongation as a critical regulatory point in the control of gene expression, particularly during development [88]. The prevailing models for the rate-limiting steps in the eukaryotic transcription cycle are the formation of the preinitiation complex (PIC) in yeast and promoter-proximal Pol II pausing in metazoans [89–91]. Indeed, there is no evidence that PIC release is a rate-limiting step in metazoans, nor is there evidence for Pol II pausing yeast [92]. However, understanding the spatiotemporal relationships between PIC and pausing in vivo has been hampered by limitations in the resolving power of ChIP-seq. To address this issue, several recent studies in yeast, fly and human have investigated the spatial organization of Pol II at promoters in high resolution using ChIP-exo/nexus [93–95]. In principle, the near base-pair resolution of ChIP-exo/nexus should discern Pol II binding at the PIC assembly site, pause site and divergently initiating sites (Figure 3F).
Previously, we reported the genomic organization of human initiation complexes using ChIP-exo for Pol II and subunits of the PIC [94]. We found that divergent transcription was driven by separate initiation complexes in opposing orientations. This also seems to be the case in yeast for antisense transcription, wherein it was reported that divergent transcription from yeast promoters was accompanied by separate, resolvable Pol II PICs [95]. In our study of human initiation complexes, we detected little Pol II occupancy at the PIC site in our meta-genomic analyses, consistent with the notion that PIC-associated Pol II is a transient step in the transcription cycle of metazoans.
A ChIP-nexus study in fly for Pol II and several GTFs (general TFs) reported that Pol II PIC assembly and pausing were mutually exclusive events at some promoters [93]. By using the near base-pair resolution of ChIP-nexus coupled to pharmacological inhibition of transcription, this study spatially resolved Pol II-engaged PICs from Pol II pause sites, and suggested that Pol II pausing inhibits new initiation. However, this conclusion runs counter to a previous report in human tissue culture cells using another high-resolution method called PIP-seq (permanganate assay followed by ChIP and sequencing). Contrary to prevailing models, PIP-seq for TFIIB and Pol II in human cells showed that Pol II initiation and pausing co-occurred within the same promoter region [96]. It remains an open question whether these differences are reconciled by methodological considerations or whether it reflects evolutionary divergence from fly to human in relation to Pol II initiation and pausing.
Discussion
The development of new, more sensitive functional genomic technologies provides the opportunity to address key biological questions with increasing molecular detail. In this review, we have highlighted unique insights that were enabled by the near base-pair resolving power of protein–DNA interactions in a diversity of organisms by ChIP-exo/nexus. As with any method, ChIP-exo as a functional genomics tool is not without its limitations, which were discussed above. Yet another complicating issue that broadly affects the ChIP assay is the limited availability of ChIP-grade antibodies. While a detailed discussion on this issue is outside the scope of this review, this issue poses a major challenge to the production of high-quality ChIP libraries, especially for TFs that have not previously been ChIP’d. The ENCODE (Encyclopedia of DNA Elements) consortium has sought to standardize validation methods [97], but this does not solve the broader challenge of sparse antibody availability directed against the ∼2000 TFs encoded in mammalian genomes. Presumably, systematic mapping of all human TFs will be a long-term effort, and several alternative approaches to facilitate this goal are already in development. For example, one group is using the high-throughput genome editing with Clustered Regularly Interspaced Short Palindromic Repeats - CRISPR associated protein 9 (CRISPR-Cas9) to systematically epitope tag nearly every TF [98]. However, this approach is largely limited to cell systems in which CRISPR can currently be applied (e.g. tissue culture cell lines), which currently excludes nearly all primary tissue sources.
New biological insights often beget new questions. The uniquely high-resolution of ChIP-exo opens the door to more detailed analyses that integrate complementary approaches to enhance biological discovery. Potential integrative approaches with ChIP-exo mapping include (1) single-cell imaging, (2) mathematical modeling, (3) human genetics and (4) protein structure data. For example, by combining Sox2 ChIP-exo mapping with single-molecule imaging, an integrative analysis enabled a deeper understanding of the kinetics for TF assembly in the context of the enhanceosome [99, 100]. Published ChIP-exo data can also provide a rich set of data for mathematical modeling, such as modeling the influence that DNA triplets exert on PIC positioning in human cells [101].
Single-nucleotide polymorphisms (SNPs) underlie genetic human genetic diversity, and, in some cases, are disease-associated [102, 103]. The majority of disease-associated SNPs reside in noncoding regions of the genome that include many enhancers and TF-binding sites [104]. Yet the extent to which SNPs alter TF-binding and regulatory potential remain poorly understood. ChIP-exo is well suited to pinpoint whether TF occupancy coincides with a given SNP location. Several recent studies have sought to clarify the mechanistic basis for complex disease using ChIP-exo mapping of TFs [105, 106]. In these studies, the near base-pair resolution was critical to link altered TF binding to a given SNP. For example, a recent report investigated the extent to which ligand-activated vitamin D receptor (VDR) TF binding was altered by SNP variants [105]. Remarkably, this study found that over 40 000 genetic variants altered VDR-binding across 27 human lymphoblastoid cell lines. This finding underscores the potential for future ChIP-exo studies that provide new insights into the molecular mechanisms underlying complex human disease. In mouse TH2 cells, ChIP-exo mapping of the Batf-Irf4 TF heterodimer enabled the discovery of a previously unknown recognition motif that encompassed a human SNP associated with autoimmunity resistance in humans [106].
Given that ChIP-exo cross-linking patterns occur within the structural constraints of a given protein–DNA interaction, a number of studies have used published crystallographic structure to provide biochemical evidence for the observed in vivo protein–DNA cross-linking patterns [72, 80–82, 95]. Furthermore, a few reports have coupled new crystallographic structures with in vivo ChIP-exo mapping to validate in vitro biochemical and structural data [85, 107, 108]. In particular, a recently published crystal structure for the histone chaperone Nap1 in complex with an H2A-H2B dimer revealed new concepts underlying how histones are chaperoned to the nucleus and packaged into nucleosomes [108]. ChIP-exo analysis for wild-type and mutant Nap1 complemented structural studies by showing that Nap1 is globally required for subnucleosomal assembly of H2A-H2B dimers into the nucleosome.
Although many fundamental gene regulatory mechanisms are conserved across eukaryotes, notable differences exist between yeast, fly and mammals. For example, the nucleosome organization with respect to the TSS is distinct [74–78], and mechanisms of Pol II initiation/pausing fundamentally differ across eukaryotes [89]. Therefore, moving forward it will be critical to examine the extent to which new gene regulatory concepts from yeast and fly are evolutionarily conserved in mouse and human. Finally, we look forward to new technological and methodological advances that should enable new insights into gene regulatory mechanisms.
Key Points
ChIP-exo identifies in vivo protein–DNA interactions at near base-pair resolution with low background.
Ultra high-resolution mapping enables new insights into gene regulation from diverse model organisms.
Emerging integrative approaches with ChIP-exo provides deeper understanding of molecular mechanisms.
Acknowledgements
Special thanks to Andrea Perreault for helpful discussions and critical reading of the manuscript.
Funding
This work was supported by the Vanderbilt Molecular Endocrinology Training Program grant 5T32 DK07563 from the National Institutes of Health.
Bryan J. Venters is an Assistant Professor at Vanderbilt University. His laboratory is focused on understanding how Epo signaling regulates enhancer activity and transcription during erythropoiesis, using functional genomic approaches.
References
- 1. Calo E, Wysocka J.. Modification of enhancer chromatin: what, how, and why? Mol Cell 2013;49(5):825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rivera CM, Ren B.. Mapping human epigenomes. Cell 2013;155(1):39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Plank JL, Dean A.. Enhancer function: mechanistic and genome-wide insights come together. Mol Cell 2014;55(1):5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hnisz D, Day DS, Young RA.. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell 2016;167(5):1188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li W, Notani D, Rosenfeld MG.. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 2016;17(4):207–23. [DOI] [PubMed] [Google Scholar]
- 6. Bulger M, Groudine M.. Functional and mechanistic diversity of distal transcription enhancers. Cell 2011;144(3):327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartman CR, Blobel GA.. Perturbing chromatin structure to understand mechanisms of gene expression. Cold Spring Harb Symp Quant Biol 2015;80:207–12. [DOI] [PubMed] [Google Scholar]
- 8. Venters BJ, Pugh BF.. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol 2009;44(2–3):117–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee TI, Young RA.. Transcriptional regulation and its misregulation in disease. Cell 2013;152(6):1237–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang C, Pugh BF.. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 2009;10(3):161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Albert I, Wachi S, Jiang C, et al. GeneTrack–a genomic data processing and visualization framework. Bioinformatics 2008;24(10):1305–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo Y, Mahony S, Gifford DK, Aerts S.. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol 2012;8(8):e1002638.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bardet AF, Steinmann J, Bafna S, et al. Identification of transcription factor binding sites from ChIP-seq data at high resolution. Bioinformatics 2013;29(21):2705–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grau J, Posch S, Grosse I, et al. A general approach for discriminative de novo motif discovery from high-throughput data. Nucleic Acids Res 2013;41(21):e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun W, Hu X, Lim MH, et al. TherMos: estimating protein-DNA binding energies from in vivo binding profiles. Nucleic Acids Res 2013;41(11):5555–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jia C, Carson MB, Wang Y, et al. A new exhaustive method and strategy for finding motifs in ChIP-enriched regions. PLoS One 2014;9(1):e86044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang L, Chen J, Wang C, et al. MACE: model based analysis of ChIP-exo. Nucleic Acids Res 2014;42(20):e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Starick SR, Ibn-Salem J, Jurk M, et al. ChIP-exo signal associated with DNA-binding motifs provides insight into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res 2015;25(6):825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tebaldi T, Zaccara S, Alessandrini F, et al. Whole-genome cartography of p53 response elements ranked on transactivation potential. BMC Genomics 2015;16(1):464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hansen P, Hecht J, Ibn-Salem J, et al. Q-nexus: a comprehensive and efficient analysis pipeline designed for ChIP-nexus. BMC Genomics 2016;17(1):873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hartonen T, Sahu B, Dave K, et al. PeakXus: comprehensive transcription factor binding site discovery from ChIP-Nexus and ChIP-Exo experiments. Bioinformatics 2016;32(17):i629–38. [DOI] [PubMed] [Google Scholar]
- 22. Tang X, Srivastava A, Liu H, et al. annoPeak: a web application to annotate and visualize peaks from ChIP-seq/ChIP-exo-seq. Bioinformatics 2017;33(10):1570–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mahony S, Pugh BF.. Protein-DNA binding in high-resolution. Crit Rev Biochem Mol Biol 2015;50(4):269–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gilmour DS, Lis JT.. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol Cell Biol 1985;5(8):2009–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ren B, Robert F, Wyrick JJ, et al. Genome-wide location and function of DNA binding proteins. Science 2000;290(5500):2306–9. [DOI] [PubMed] [Google Scholar]
- 26. Albert I, Mavrich TN, Tomsho LP, et al. Translational and rotational settings of H2A.Z nucleosomes across the saccharomyces cerevisiae genome. Nature 2007;446(7135):572–6. [DOI] [PubMed] [Google Scholar]
- 27. Robertson G, Hirst M, Bainbridge M, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 2007;4:651–7. [DOI] [PubMed] [Google Scholar]
- 28. Rhee HS, Pugh BF.. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 2011;147(6):1408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He Q, Johnston J, Zeitlinger J.. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol 2015;33(4):395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Payvar F, Firestone GL, Ross SR, et al. Multiple specific binding sites for purified glucocorticoid receptors on mammary tumor virus DNA. J Cell Biochem 1982;19(3):241–7. [DOI] [PubMed] [Google Scholar]
- 31. Dynan WS, Tjian R.. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell 1983;32(3):669–80. [DOI] [PubMed] [Google Scholar]
- 32. Lu K, Ye W, Zhou L, et al. Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J Am Chem Soc 2010;132(10):3388–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Euskirchen GM, Rozowsky JS, Wei CL, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res 2007;17(6):898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet 2009;10(10):669–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rhee HS, Pugh BF.. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr Protoc Mol Biol 2012;Chapter 21:Unit 21.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Serandour AA, Brown GD, Cohen JD, et al. Development of an Illumina-based ChIP-exonuclease method provides insight into FoxA1-DNA binding properties. Genome Biol 2013;14(12):R147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matteau D, Rodrigue S.. Precise identification of DNA-binding proteins genomic location by exonuclease coupled Chromatin Immunoprecipitation (ChIP-exo). Methods Mol Biol 2015;1334:173–93. [DOI] [PubMed] [Google Scholar]
- 38. Barfeld SJ, Mills IG.. Mapping protein-DNA interactions using ChIP-exo and illumina-based sequencing. Methods Mol Biol 2016;1443:119–37. [DOI] [PubMed] [Google Scholar]
- 39. Perreault AA, Venters BJ.. The ChIP-exo method: identifying protein-DNA interactions with near base pair precision. J Vis Exp 2016;118. doi:10.3791/55016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmidl C, Rendeiro AF, Sheffield NC, et al. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods 2015;12(10):963–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wallerman O, Nord H, Bysani M, et al. lobChIP: from cells to sequencing ready ChIP libraries in a single day. Epigenetics Chromatin 2015;8(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carroll TS, Liang Z, Salama R, et al. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front Genet 2014;5:75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Terooatea TW, Pozner A, Buck-Koehntop BA.. PAtCh-Cap: input strategy for improving analysis of ChIP-exo data sets and beyond. Nucleic Acids Res 2016;44(21):e159.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kwok CK, Ding Y, Sherlock ME, et al. A hybridization-based approach for quantitative and low-bias single-stranded DNA ligation. Anal Biochem 2013;435(2):181–6. [DOI] [PubMed] [Google Scholar]
- 45. Kennedy-Darling J, Smith LM.. Measuring the formaldehyde Protein-DNA cross-link reversal rate. Anal Chem 2014;86(12):5678–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Babu MM, Luscombe NM, Aravind L, et al. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol 2004;14(3):283–91. [DOI] [PubMed] [Google Scholar]
- 47. Zhao W, Kitidis C, Fleming MD, et al. Erythropoietin stimulates phosphorylation and activation of GATA-1 via the PI3-kinase/AKT signaling pathway. Blood 2006;107(3):907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng Y, Wu W, Kumar SA, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res 2009;19(12):2172–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Amoutzias GD, Robertson DL, Van de Peer Y, et al. Choose your partners: dimerization in eukaryotic transcription factors. Trends Biochem Sci 2008;33(5):220–9. [DOI] [PubMed] [Google Scholar]
- 50. Vaquerizas JM, Kummerfeld SK, Teichmann SA, et al. A census of human transcription factors: function, expression and evolution. Nat Rev Genet 2009;10(4):252–63. [DOI] [PubMed] [Google Scholar]
- 51. Andrilenas KK, Penvose A, Siggers T.. Using protein-binding microarrays to study transcription factor specificity: homologs, isoforms and complexes. Brief Funct Genomics 2015;14(1):17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet 2009;10(9):605–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kininis M, Kraus WL.. A global view of transcriptional regulation by nuclear receptors: gene expression, factor localization, and DNA sequence analysis. Nucl Recept Signal 2008;6:e005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen Z, Lan X, Thomas-Ahner JM, et al. Agonist and antagonist switch DNA motifs recognized by human androgen receptor in prostate cancer. Embo J 2015;34(4):502–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carraro N, Matteau D, Luo P, et al. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet 2014;10(10):e1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fang M, Bauer CE, Harwood CS.. The Vitamin B12-dependent photoreceptor AerR relieves photosystem gene repression by extending the interaction of CrtJ with photosystem promoters. MBio 2017;8(2):e00261-17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cho S, Cho YB, Kang TJ, et al. The architecture of ArgR-DNA complexes at the genome-scale in Escherichia coli. Nucleic Acids Res 2015;43(6):3079–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou X, Yan Q, Wang N.. Deciphering the regulon of a GntR family regulator via transcriptome and ChIP-exo analyses and its contribution to virulence in Xanthomonas citri. Mol Plant Pathol 2017;18(2):249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Poulin-Laprade D, Matteau D, Jacques PE, et al. Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon. Nucleic Acids Res 2015;43(4):2045–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zere TR, Vakulskas CA, Leng Y, et al. Genomic targets and features of BarA-UvrY (-SirA) signal transduction systems. PLoS One 2015;10(12):e0145035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Barfeld SJ, Urbanucci A, Itkonen HM, et al. c-Myc antagonises the transcriptional activity of the androgen receptor in prostate cancer affecting key gene networks. EBioMedicine 2017;18:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wales S, Hashemi S, Blais A, et al. Global MEF2 target gene analysis in cardiac and skeletal muscle reveals novel regulation of DUSP6 by p38MAPK-MEF2 signaling. Nucleic Acids Res 2014;42(18):11349–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kitayner M, Rozenberg H, Kessler N, et al. Structural basis of DNA recognition by p53 tetramers. Mol Cell 2006;22(6):741–53. [DOI] [PubMed] [Google Scholar]
- 64. Chang GS, Chen XA, Park B, et al. A comprehensive and high-resolution genome-wide response of p53 to stress. Cell Rep 2014;8(2):514–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Weikum ER, Knuesel MT, Ortlund EA, et al. Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat Rev Mol Cell Biol 2017;18(3):159–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Glass CK, Saijo K.. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol 2010;10(5):365–76. [DOI] [PubMed] [Google Scholar]
- 67. Lim HW, Uhlenhaut NH, Rauch A, et al. Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res 2015;25(6):836–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Panne D. The enhanceosome. Curr Opin Struct Biol 2008;18(2):236–42. [DOI] [PubMed] [Google Scholar]
- 69. Reja R, Vinayachandran V, Ghosh S, et al. Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev 2015;29(18):1942–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ulirsch JC, Lacy JN, An X, et al. Altered chromatin occupancy of master regulators underlies evolutionary divergence in the transcriptional landscape of erythroid differentiation. PLoS Genet 2014;10(12):e1004890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Love PE, Warzecha C, Li L.. Ldb1 complexes: the new master regulators of erythroid gene transcription. Trends Genet 2014;30(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Han GC, Vinayachandran V, Bataille AR, et al. Genome-wide organization of GATA1 and TAL1 determined at high resolution. Mol Cell Biol 2016;36(1):157–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rhee HS, Closser M, Guo Y, et al. Expression of terminal effector genes in mammalian neurons is maintained by a dynamic relay of transient enhancers. Neuron 2016;92(6):1252–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Perreault AA, Benton ML, Koury MJ, et al. Epo reprograms the epigenome of erythroid cells. Exp Hematol 2017;51:47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Luger K, Mader AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997;389(6648):251–60. [DOI] [PubMed] [Google Scholar]
- 76. Mavrich TN, Ioshikhes IP, Venters BJ, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 2008;18(7):1073–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mavrich TN, Jiang C, Ioshikhes IP, et al. Nucleosome organization in the Drosophila genome. Nature 2008;453(7193):358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schones DE, Cui K, Cuddapah S, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 2008;132(5):887–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Subramanian V, Fields PA, Boyer LA.. H2A.Z: a molecular rheostat for transcriptional control. F1000Prime Rep 2015;7:01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yen K, Vinayachandran V, Pugh BF.. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 2013;154(6):1246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rhee HS, Bataille AR, Zhang L, et al. Subnucleosomal structures and nucleosome asymmetry across a genome. Cell 2014;159(6):1377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yen K, Vinayachandran V, Batta K, et al. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 2012;149(7):1461–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Allis CD, Jenuwein T.. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016;17(8):487–500. [DOI] [PubMed] [Google Scholar]
- 84. Fuchs SM, Laribee RN, Strahl BD.. Protein modifications in transcription elongation. Biochim Biophys Acta 2009;1789(1):26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Van Oss SB, Shirra MK, Bataille AR, et al. The histone modification domain of Paf1 complex subunit Rtf1 directly stimulates H2B ubiquitylation through an interaction with Rad6. Mol Cell 2016;64(4):815–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Giardina C, Perez-Riba M, Lis JT.. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev 1992;6(11):2190–200. [DOI] [PubMed] [Google Scholar]
- 87. Krumm A, Meulia T, Brunvand M, et al. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev 1992;6(11):2201–13. [DOI] [PubMed] [Google Scholar]
- 88. Nechaev S, Adelman K.. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle 2008;7(11):1539–44. [DOI] [PubMed] [Google Scholar]
- 89. Adelman K, Lis JT.. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 2012;13(10):720–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Core LJ, Waterfall JJ, Gilchrist DA, et al. Defining the status of RNA polymerase at promoters. Cell Rep 2012;2(4):1025–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sikorski TW, Buratowski S.. The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol 2009;21(3):344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kwak H, Lis JT.. Control of transcriptional elongation. Annu Rev Genet 2013;47:483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shao W, Zeitlinger J.. Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet 2017;49(7):1045–51. [DOI] [PubMed] [Google Scholar]
- 94. Pugh BF, Venters BJ.. Genomic organization of human transcription initiation complexes. PLoS One 2016;11(2):e0149339.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Rhee HS, Pugh BF.. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 2012;483(7389):295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lai WK, Pugh BF.. Genome-wide uniformity of human ‘open’ pre-initiation complexes. Genome Res 2017;27(1):15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Landt SG, Marinov GK, Kundaje A, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 2012;22:1813–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Savic D, Partridge EC, Newberry KM, et al. CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins. Genome Res 2015;25(10):1581–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chen J, Zhang Z, Li L, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 2014;156:1274–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Liu Z, Legant WR, Chen BC, et al. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. Elife 2014;3:e04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Goldshtein M, Lukatsky DB.. Specificity-determining DNA triplet code for positioning of human preinitiation complex. Biophys J 2017;112(10):2047–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009;461(7265):747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Motsinger-Reif AA, Jorgenson E, Relling MV, et al. Genome-wide association studies in pharmacogenomics: successes and lessons. Pharmacogenet Genomics 2013;23(8):383–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Nishizaki SS, Boyle AP.. Mining the unknown: assigning function to noncoding single nucleotide polymorphisms. Trends Genet 2017;33(1):34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Gallone G, Haerty W, Disanto G, et al. Identification of genetic variants affecting vitamin D receptor binding and associations with autoimmune disease. Hum Mol Genet 2017;26(11):2164–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Iwata A, Durai V, Tussiwand R, et al. Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF-IRF4 transcription factor complex. Nat Immunol 2017;18(5):563–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Murphy MW, Lee JK, Rojo S, et al. An ancient protein-DNA interaction underlying metazoan sex determination. Nat Struct Mol Biol 2015;22(6):442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Aguilar-Gurrieri C, Larabi A, Vinayachandran V, et al. Structural evidence for Nap1-dependent H2A-H2B deposition and nucleosome assembly. Embo J 2016;35(13):1465–82. [DOI] [PMC free article] [PubMed] [Google Scholar]