Abstract
Obesity has been linked to several types of cancer. Access to adequate health information activates people’s participation in managing their own health, which ultimately improves their health outcomes. Nevertheless, the existing online information about the relationship between obesity and cancer is heterogeneous and poorly organized. A formal knowledge representation can help better organize and deliver quality health information. Currently, there are several efforts in the biomedical domain to convert unstructured data to structured data and store them in Semantic Web knowledge bases (KB). In this demo paper, we present, OC-2-KB (Obesity and Cancer to Knowledge Base), a system that is tailored to guide the automatic KB construction for managing obesity and cancer knowledge from free-text scientific literature (i.e., PubMed abstracts) in a systematic way. OC-2-KB has two important modules which perform the acquisition of entities and the extraction then classification of relationships among these entities. We tested the OC-2-KB system on a data set with 23 manually annotated obesity and cancer PubMed abstracts and created a preliminary KB with 765 triples. We conducted a preliminary evaluation on this sample of triples and reported our evaluation results.
Keywords: Software, obesity and cancer, Semantic Web knowledge base, Resource Description Framework
I. Introduction
Obesity and overweight have been linked to several types of cancer, such as endometrium, breast, kidney, colorectal, pancreas, esophagus, ovaries, gallbladder, thyroid, and possibly prostate cancer [1], [2]. Interventions that reduce the excess weight can be used for cancer prevention and treatment. Access to adequate health information activates people’s participation in their care management such as joining weight loss programs, which ultimately leads to improved health outcomes. Nevertheless, the existing online information about the relationship between obesity and cancer is heterogeneous, poorly organized, not evidenced-based, and of poor quality. Further, typical consumers cannot translate the vast amounts of health information into usable knowledge. Thus, there is a need to organize the obesity and cancer information in a meaningful way that helps consumers make informed health decisions. A formal knowledge representation, using the Resource Description Framework (RDF) and the Web Ontology Language (OWL), can help better organize and deliver quality health information. In the biomedical domain, there has been several efforts making Semantic Web knowledge bases (KBs), such as BioNELL [3] and SemMedDB [4].
In this demo paper, we present, OC-2-KB (Obesity and Cancer to Knowledge Base), a system (available at: https://github.com/juanlossio/OC-2-KB) that automatically builds an obesity and cancer KB from text. Our main information sources are PubMed abstracts related to obesity and cancer literature. OC-2-KB has two important modules which perform the acquisition of entities and the extraction then classification of relationships among these entities. Our preliminary knowledge base consists of 765 triples extracted from 23 annotated abstracts (it contains 347 annotated triplets). We conducted an evaluation on this sample of triples and report results.
II. Related Work
In biomedicine and life sciences, there are a few studies in the construction of Semantic Web KBs such as BioNELL [3] that uses six biomedical ontologies to guide the entity and relation extraction processes. Another related study is KnowLife [5], which constructed a KB from life science publications and health-related social media content. Luo et al. [6] proposed an algorithm that translates free-text sentences from pathology reports into a graph representation, where the nodes of the graph represent the concepts (e.g., genes and proteins) and the edges indicate the syntactic dependency links between these concepts. Further, a number of researchers have attempted to integrate different information sources to form a large KB [7].
III. System overview
Extracting knowledge from free-text sources and then integrating these knowledge into a coherent knowledge base is a multi-step process related to many different research areas, including natural language processing (NLP), information extraction, information integration, semantic databases, and machine learning. In our previous study, we explored algorithms and methods to extract triple statements from free-text scientific articles [8]. Built upon our previous study, OC-2-KB is a complete software pipeline that can automatically build an obesity and cancer knowledge base from PubMed abstracts.
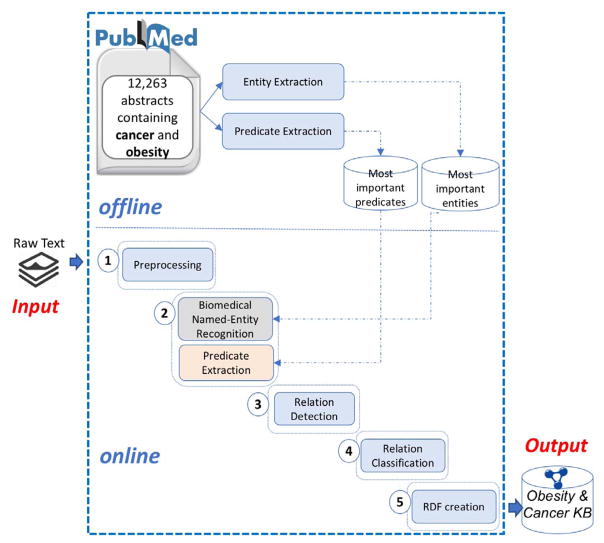
Fig. 1 shows the overview of the OC-2-KB system. Currently, OC-2-KB has two independent processes: (1) an offline process that creates dictionaries of candidate entities and predicates; and (2) an online process that extract facts from biomedical literature.
Fig. 1.
An overview of the OC-2-KB system.
A. The Offline Process: Create Dictionaries of Domain-Relevant Entities and Predicates
There are two main challenges associated with domain-specific KB constructions: (1) building a dictionary of the most representative entities, and (2) building a dictionary of the most representative predicates of the specific domain mined from domain relevant literature. Since our demo data set is a small sample (i.e., 23 abstracts), we used an offline process to create the dictionaries of entities and predicates over all PubMed abstracts related to obesity and cancer to improve their coverages. The offline process is only done once and considered as part of the setup process of the OC-2-KB system. This process is composed of the following parts.
The PubMed corpus
To create an evidence-based obesity and cancer KB, we used PubMed articles titles and abstracts. Our corpus consists of all articles containing the keywords “obesity” and “cancer” in the titles and abstracts. A total of 12,263 articles were extracted.
Entity extraction
The objective of this step is to extract and construct a dictionary of the most representative entities of our PubMed data set. We used the LIDF-value [9] measure implemented in BioTex [10]. This baseline measure can recognize both biomedical entities that already exist in standard terminology (i.e., Unified Medical Language System, UMLS) and potential new biomedical entities, which are also important in KB construction such that new biomedical knowledge can be incorporated into the KB as the field evolves. For instance, LIDF-value extracted “dose-response meta-analysis” as a representative entity which does not exist in UMLS. We extracted approximately 34,500 entities relevant to obesity and cancer.
Predicate extraction
The aim of this step is to extract the most representative predicates in the obesity and cancer literature. In a similar way, we used the LIDF-value and extracted approximately 8,200 predicates.
Dictionaries of domain-relevant entities and predicates
The last two steps created two dictionaries which will be taken as inputs to the online process described below.
B. The Online Process: Extracting Facts from Scientific Literature
The online process is used to extract facts from scientific literature related to obesity and cancer and to construct a Semantic Web KB as output. The components of this process are described below.
Input
The input of this process is the PubMed titles and abstracts relevant to obesity and cancer.
Step 1: Preprocessing
In the preprocessing step, each abstract is split into sentences, and the facts are extracted from each sentence. We incorporated the Stanford Tokenizer tool[11] into our software for sentence segmentation. A sentence is detected when a sentence-ending character (i.e., ., !, or ?) is found and it is not grouped with other characters such as in an abbreviation or a number.
Step 2: Biomedical Named-Entity Recognition and Predicate Extraction
This step extracts the entities and predicates from each sentence. As discussed in our previous work [8], our methods for biomedical named-entity recognition and predicate extraction are based on both linguistic and statistic features. The input of this module is a list of sentences generated in Step 1 and the output is the list of entities and predicates extracted from each sentence, as shown in Fig. 2. Our experiments indicated that the performance of these extractions is limited without the a prior knowledge source (i.e., the entity and predicate dictionaries created during the offline process).
Fig. 2.
An example of the entities and predicates detected from each sentence (highlighted in blue and purple).
Step 3: Relation Detection
With a list of entities and predicates, this step determines whether a pair of two biomedical entities and a predicate can form a valid relation (i.e., that can lead to a valid assertion as a subject-predicate-object statement). Our method for relation detection is based on a supervised machine learning algorithm. In our previous work [8], our evaluation has shown that our relation detection method can achieve promising results. All possible combinations of the entities and predicates (as shown in Fig. 3) are evaluated by the relation detection classifier. The result is a list of combinations predicted to be correct (i.e., that form valid relations).
Fig. 3.
An example of potential triples to be evaluated.
Step 4: Relation Classification
After relation detection, this step normalizes the extracted predicate to one of the twelve specific relation classes we adapted from the Relation Ontology (RO) [12] shown in Table I. RO contains a collection of relations intended primarily for standardization across ontologies in the OBO Foundry [13]. We chose these twelve relations based on an evaluation of the predicates often used in the obesity and cancer related literature. Fig. 4 shows an example where the extracted predicate “can lead” was normalized to “causes or contributes to condition”. This step is also based on a supervised machine learning algorithm.
TABLE I.
The twelve most frequently used relation classes in the obesity and cancer literature normalized to the Relation Ontology.
| 1 | associated, linked |
| 2 | is a |
| 3 | causally upstream of, negative effect |
| 4 | contributes to |
| 5 | benefits |
| 6 | causally upstream of, positive effect |
| 7 | treats |
| 8 | correlated with |
| 9 | has |
| 10 | causes or contributes to condition |
| 11 | alters |
| 12 | positively regulates |
Fig. 4.
An example of relation classification.
Step 5: RDF Creation
Finally we create an RDF file of the triples extracted in the previous steps. We used the Jena RDF API [14] to extract data from and write to RDF graphs, and to interact with the underline graph database.
Output
The output of the online process is the knowledge base, i.e. the RDF graphs, created from the textual data received as input. In our study, we used GraphDB [15], a semantic graph database compliant with W3C standards, as our underline data store of the KB.
IV. A demo of the OC-2-KB system
Our initial testing of the OC-2-KB system is done with a small collection of obesity and cancer PubMed abstracts to show the feasibility of the proposed KB construction process (see Fig. 1).
System Input
Data collection: we randomly collected 23 PubMed abstracts based on the search keywords “obesity” and “cancer”. From these 23 abstracts, our system extracted 214 sentences. This data set is available freely online1.
System Output
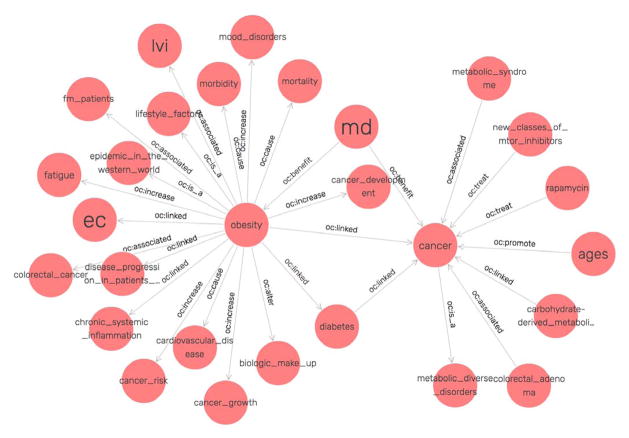
Our system found 765 facts (i.e., triples) from the 23 PubMed abstracts, where only 259 of them could be mapped to the twelve RO classes. Fig. 5 shows parts of the initial obesity and cancer KB using the visualization tool built in GraphDB.
Fig. 5.
A visualization of the initial obesity and cancer KB.
Initial Evaluation of the Knowledge Base
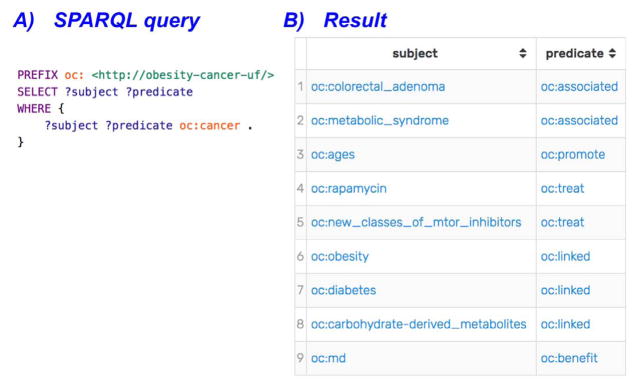
In this section, we present a few examples of possible semantic (i.e., SPARQL, a recursive acronym for SPARQL Protocol and RDF Query Language) queries to interact with the KB. Fig. 6 shows an example SPARQL query to extract all the subjects and predicates related to the class “cancer”, which forms assertions in the following triple statement format: <?subject-?predicate-oc:cancer>.
Fig. 6.
A SPARQL query and its results for extracting the subjects and predicates related to the “cancer” class.
The obesity and cancer KB also contains invalid triples because the accuracy of our automated relation extraction method, although comparable to state-of-the-art algorithms, is suboptimal (i.e., precision 25.5%, recall 56.2%, F-measure 35.1%). Fig. 7 shows an example of an invalid triple related to “measures of insulin resistance’. From the original sentence (“Conclusions: Our findings show IHF is associated with measures of insulin resistance, but not measures of visceral adiposity.”), OC-2-KB extracted <“conclusions” oc:associated “measures of insulin resistance”>, which is obviously incorrect.
Fig. 7.
An example of an invalid triple associated with “measures of insulin resistance”.
V. Discussion and Conclusion
We presented OC-2-KB, a system that can automatically build an evidence-based obesity and cancer KB from scientific literature (i.e., PubMed titles and abstracts). As shown in this paper, OC-2-KB can extract useful facts of obesity and cancer with promising results. There are a large number of hypotheses about how obesity affects cancer, and claims with varying degrees of clinical evidence. Given the frequency with which consumers turn to online resources for health information, it is important to organize the increasing amount of information on obesity and cancer in a way that helps consumers of the information. A Semantic Web KB with formal knowledge representations as we have built with OC-2-KB is such a tool that renders the evidence data collected from scientific literature in a well-organized and computable manner. Further, a well-designed interactive visualization system built upon the visualizations that we have presented in this paper can facilitate the thinking process and enhance consumers understanding of the knowledge. The ultimate goal of this project is a paradigm shift in how the general public access, read, digest, and use online health information. Rather than requiring the laypeople find and read static documents on the Internet via regular searches, we propose a dynamic knowledge acquisition model, where the content is routinely mined from the scientific literature, users interact with the KB via semantic queries, and consumers navigate the network of knowledge through interactive visualizations.
Our current study is still limited. First, the performance of our relation extraction tool is suboptimal (i.e., precision 25.5%, recall 56.2%, F-measure 35.1%). As shown in Fig. 7, many of these issues arose from the noisy nature of free-text data (e.g., inconsistent formatting, alternative spellings, and misspellings). Our NLP methods need to be further tailored to address these different scenarios. One simple solution to the issue presented in Fig. 7 is to filter out “Conclusion: in the preprocessing stage. Nevertheless, automatic information extraction (IE) is error prone with even the best machine learning models. An ideal IE system should leverage the advantages of both human and machine computation in a cohesive unit. Thus, in future work, we aim to use crowdsourcing to validate extractions that the machine identifies as likely candidates. Second, we built our initial KB with a small data set to test its feasibility. In future work, we plan to create a KB using all cancer and obesity related abstracts in PubMed.
Acknowledgments
This work was supported in part by NIH grants UL1TR001427. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
References
- 1.Keum N, Greenwood DC, Lee DH, Kim R, Aune D, Ju W, Hu FB, Giovannucci EL. Adult weight gain and adiposity-related cancers: a dose-response meta-analysis of prospective observational studies. Journal of the National Cancer Institute. 2015;107(2):djv088. doi: 10.1093/jnci/djv088. [DOI] [PubMed] [Google Scholar]
- 2.Ligibel JA, Alfano CM, Hershman D, Ballard RM, Bruinooge SS, Courneya KS, Daniels EC, Demark-Wahnefried W, Frank ES, Goodwin PJ, et al. Recommendations for obesity clinical trials in cancer survivors: American society of clinical oncology statement. Journal of Clinical Oncology. 2015;33(33):3961–3967. doi: 10.1200/JCO.2015.63.1440. [DOI] [PubMed] [Google Scholar]
- 3.Movshovitz-Attias D, Cohen WW. Bootstrapping biomedical ontologies for scientific text using nell. Proceedings of the 2012 Workshop on Biomedical Natural Language Processing, ser. BioNLP’12; Stroudsburg, PA, USA: Association for Computational Linguistics; 2012. pp. 11–19. [Google Scholar]
- 4.Kilicoglu H, Shin D, Fiszman M, Rosemblat G, Rindflesch TC. Semmeddb: a pubmed-scale repository of biomedical semantic predications. Bioinformatics. 2012;28(23):3158–3160. doi: 10.1093/bioinformatics/bts591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst P, Meng C, Siu A, Weikum G. Knowlife: a knowledge graph for health and life sciences. Data Engineering (ICDE), 2014 IEEE 30th International Conference on; IEEE; 2014. pp. 1254–1257. [Google Scholar]
- 6.Luo Y, Sohani AR, Hochberg EP, Szolovits P. Automatic lymphoma classification with sentence subgraph mining from pathology reports. Journal of the American Medical Informatics Association. 2014;21(5):824–832. doi: 10.1136/amiajnl-2013-002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst P, Siu A, Milchevski D, Hoffart J, Weikum G. Deeplife: An entity-aware search, analytics and exploration platform for health and life sciences. Proceedings of ACL-2016 System Demonstrations; Berlin, Germany: Association for Computational Linguistics; Aug, 2016. pp. 19–24. [Online]. Available: http://anthology.aclweb.org/P16-4004. [Google Scholar]
- 8.Lossio-Ventura JA, Hogan W, Modave F, Hicks A, Hanna J, Guo Y, He Z, Bian J. Towards an obesity-cancer knowledge base: Biomedical entity identification and relation detection. 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); IEEE. Los Alamitos, CA, USA: IEEE Computer Society; 2016. pp. 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lossio-Ventura JA, Jonquet C, Roche M, Teisseire M. Biomedical term extraction: overview and a new methodology. Information Retrieval Journal. 2016;19(1–2):59–99. [Google Scholar]
- 10.Lossio-Ventura JA, Jonquet C, Roche M, Teisseire M. BioTex: A system for biomedical terminology extraction, ranking, and validation. Proceedings of the 13th International Semantic Web Conference, Posters & Demonstrations Track, ser. ISWC’14; 2014. pp. 157–160. [Google Scholar]
- 11. [accessed: April 15, 2017];Stanford Tokenizer. https://nlp.stanford.edu/software/tokenizer.shtml.
- 12. [accessed: December 1, 2016];The new OBO relations ontology. http://obofoundry.org/ontology/ro.html.
- 13.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The obo foundry: coordinated evolution of ontologies to support biomedical data integration. Nature biotechnology. 2007;25(11):1251. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. [accessed: September 25, 2017];An Introduction to RDF and the Jena RDF API. https://jena.apache.org/tutorials/rdfapi.html.
- 15. [accessed: May 20, 2017];Ontotext GraphDB. https://ontotext.com/products/graphdb/