Abstract
The Integrated Stress Response (ISR) is a conserved translational and transcriptional program affecting metabolism, memory and immunity. The ISR is mediated by stress-induced phosphorylation of translation initiation factor 2α (eIF2α) that attenuates the guanine nucleotide exchange factor eIF2B. A chemical inhibitor of the ISR, ISRIB, reverses the attenuation of eIF2B by phosphorylated eIF2α, protecting mice from neurodegeneration and traumatic brain injury. We describe a 4.1 Å resolution cryo-electron microscopy structure of human eIF2B with an ISRIB molecule bound at the interface between the β and δ regulatory subunits. Mutagenesis of residues lining this pocket altered the hierarchical cellular response to ISRIB analogs in vivo and ISRIB-binding in vitro. Our findings point to a site in eIF2B that can be exploited by ISRIB to regulate translation.
The Integrated Stress Response (ISR) has homeostatic functions that increase fitness. However, in some pathological circumstances benefit arises from attenuated signaling in the ISR (1). A search for ISR inhibitors led to the discovery of ISRIB (2), a small molecule efficacious in mouse models of neurodegeneration (3) and traumatic brain injury (4).
ISRIB action converges on eIF2B, a protein complex with guanine nucleotide exchange (GEF) activity towards eIF2 (5) that is inhibited by phosphorylated eIF2 (6, 7). Addition of ISRIB accelerates eIF2B GEF activity in vitro and targeting eIF2B’s δ regulatory subunit can impart ISRIB resistance (8, 9). However, known ISRIB-resistant mutations in eIF2B cluster at a distance from both the regulatory site engaged by eIF2(αP) and the catalytic site engaged by eIF2γ (10). Thus, whilst the bulk of the evidence suggests that ISRIB binds eIF2B to regulate its activity, indirect modes of action are not excluded.
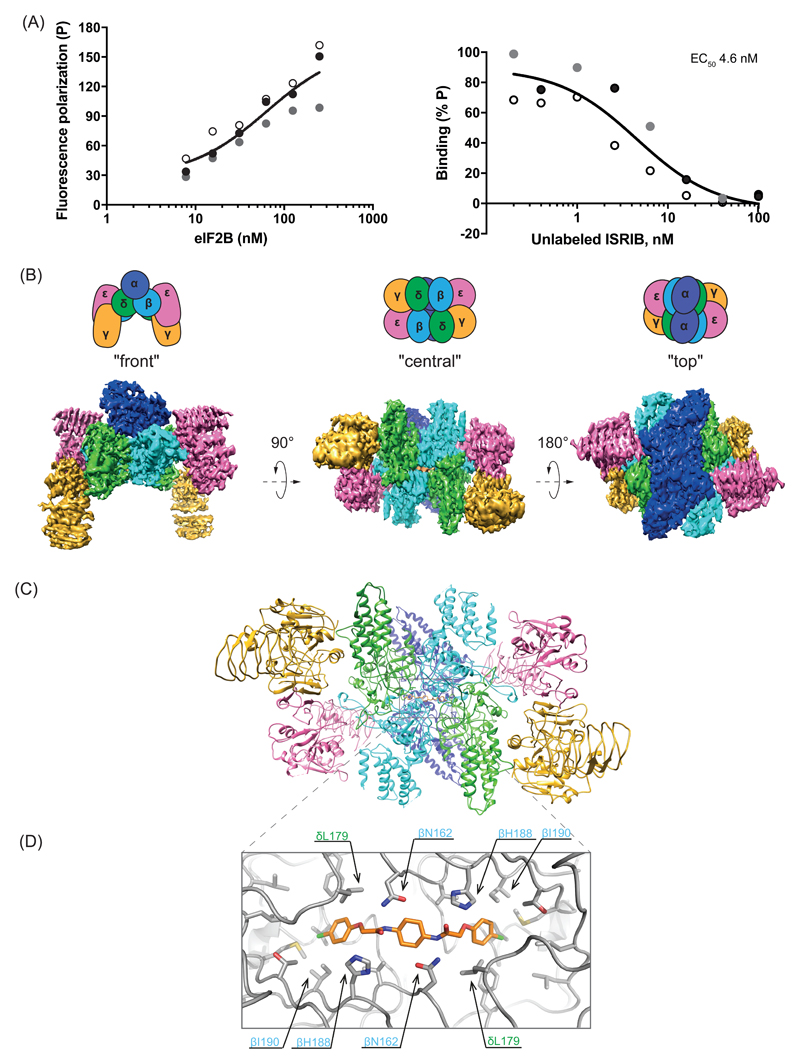
ISRIB stabilized the eIF2B complex from HeLa cells (Fig. S1), as expected (9). Thus, we added a fluorescently labeled derivative of ISRIB (AAA2-101) (Fig. S2A) to purified eIF2B and observed increased fluorescence polarization (FP) (Fig. 1A-left panel). Unlabeled ISRIB competed for eIF2B in the FP assay with an EC50 in the nanomolar range (Fig. 1A-right panel). As observed for ISRIB action in cells, less active analogs competed less successfully (Fig. S2B & S2C).
Figure 1. Biophysical and structural analysis of ISRIB binding to human eIF2B.
(A) Fluorescence polarization (FP) assays showing binding of ISRIB to human eIF2B. Left panel: a plot of the FP signal arising from fluorescein-labeled ISRIB analog (AAA2-101) (2.5 nM) as a function of the concentration of eIF2B in the sample. Right panel: a plot of the relative FP signal arising from samples with fluorescein-labeled AAA2-101 (2.5 nM) bound to purified human eIF2B (30 nM) in the presence of the indicated concentration of unlabeled trans-ISRIB introduced as a competitor. Concentrations of eIF2B and ISRIB on respective plots are represented on a log10 scale. Curve fitting and EC50 was generated using agonist vs. response function on GraphPad Prism, shown are values of three independently-acquired measurements.
(B) Representative views of the cryo-EM map of the ISRIB-bound decameric human eIF2B complex. Density is colored according to the subunit architecture indicated in the cartoons: α – blue, β – cyan, δ – green, γ – gold, ε – pink, ISRIB - orange.
(C) Ribbon representation of ISRIB-bound human eIF2B “central” view of the (βδ)2 dimer interface with a single molecule of ISRIB.
(D) Close-up of the “central” view showing the ISRIB binding site. An ISRIB molecule is docked into the cavity at the (βδ)2 dimer interface. Residues contacting ISRIB in the central part of the pocket from the β (blue) and δ (green) subunits are indicated. ISRIB is represented in orange sticks.
We purified endogenous eIF2B from HeLa cell lysates in the presence of ISRIB and determined the structure of the complex by single particle cryo-electron microscopy at an overall resolution of 4.1 Å (Fig. 1B, S1A, S3 and table S6). Within the β and δ regulatory core, protein side chains were clearly resolved, resulting in a near complete atomic model of this region (Fig. 1C, S4A & S4B). The resolution of the γ and ε human catalytic subcomplex was lower compared with the regulatory core (Fig S4A) and the catalytically important C-terminal HEAT domain of the ε subunit remained unresolved in the cryo-EM map.
Owing to a lack of sufficient high-quality cryo-EM images of an apo-eIF2B complex, we were unable to calculate a difference map of eIF2B with and without ISRIB. However, a nearly continuous density with a shape and size of a single ISRIB molecule was conspicuously present at the interface of the β and δ regulatory subunits (Fig. 1B, “central view” & S4C). The ISRIB binding pocket was located at the plane of symmetry between the β and δ subunits. In the central part of the pocket, the side chain of βH188 was positioned in the vicinity of the essential carbonyl moiety of ISRIB (11) and βN162 was poised to stabilize the diaminocyclohexane moiety of ISRIB through hydrogen bonding interactions (Fig. 1D). More distally the side chains of δL179, δF452, δL485, δL487, βV164, βI190, βT215, and βM217 formed the hydrophobic end of the symmetrical pocket that accommodated the aryl groups of ISRIB, with βI190 and δL179 located within Van der Waals interaction distance to the aryl group (Fig. S4D). A hamster Eif2b4L180F mutation (δL179 in the human) disrupts ISRIB action in cells (8), which is consistent with a potential loss of these interactions as well as a clash between the bulkier side chain of phenylalanine and the bound ISRIB molecule (Fig. 1D).
Overall the human structure is highly similar to the published S. pombe eIF2B structure (10) (r.m.s.d. of 2.57 Å over 3049 alpha carbons, Fig. S4E). However, there is no density in the corresponding region in the S. pombe eIF2B map, indicating that the density found in the human structure was the bound ISRIB.
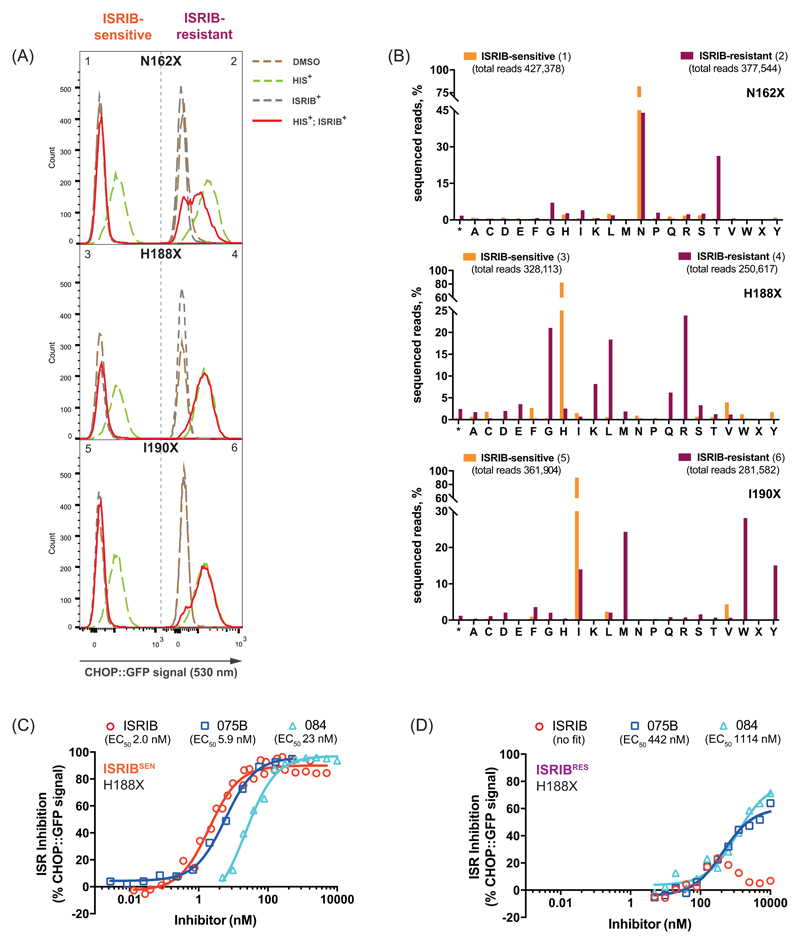
To test these features of ISRIB binding we used CRISPR/Cas9 to randomize residues lining the ISRIB-binding pocket (Eif2b2N162, Eif2b2H188 or Eif2b2I190) and correlated amino acid substitutions to ISRIB activity in the mutagenized cells. Histidinol, an agent that activates the eIF2α kinase GCN2 and induces the ISR, normally activates a CHOP::GFP reporter gene, whilst ISRIB represses the reporter (8). Fluorescence activated cell sorting (FACS) of histidinol-treated, mutagenized cells, segregated them into ISRIB sensitive, ISRIBSEN (CHOP::GFP inhibited), and ISRIB resistant, ISRIBRES (CHOP::GFP activated) classes (Fig. 2A – left and right panels, respectively).
Figure 2. Structure-directed chemogenetic analysis of ISRIB and its analogs’ binding to eIF2B.
(A) Histograms of the ISR-responsive CHOP::GFP fluorescent reporter activity, induced by histidinol (HIS+, 0.5 mM) in ISRIB-sensitive (ISRIBSEN) (left panels) and ISRIB-resistant (ISRIBRES) (right panels) pools of CHO-K1 cells, selected for their responsiveness to ISRIB (200 nM), following CRISPR/Cas9-induced random mutagenesis of the indicated codon of Eif2b2.
(B) Bar graph of the distribution of residues identified at the indicated positions of mutagenized Eif2b2, analyzed by the next generation sequencing (NGS). Shown is the number of sequenced reads in ISRIBSEN pools (orange bars) or ISRIBRES pools (plum bars) encoding each amino acid (* - stop codon, X – ambiguous sequence).
(C) Graphs showing inhibition of the ISR-activated CHOP::GFP reporter (induced as in panel “A”) by ISRIB or two related analogs, compound AAA1-075B (075B) and compound AAA1-084 (084), in ISRIBSEN (left) and ISRIBRES (right) mutant pools of EIf2b2H188X. Shown is a representative from three independent experiments for each of the compounds. Concentration of inhibitor is represented on a log10 scale. Curve fitting and EC50 was generated using agonist vs. response function on GraphPad Prism.
To determine if the phenotypically-distinguished pools of mutagenized cells (Fig. 2A) were enriched in different mutations, we subjected genomic DNA derived from each population to deep sequencing analysis (Fig. 2B & table S4). The ISRIBRES pool targeted at Eif2b2H188 diverged dramatically from the parental sequence (Fig. 2B, middle panel). Of a total of 250,617 sequencing reads, histidine was present in only 6,443 (2.6%), with arginine, glycine, leucine, lysine and glutamine dominating (24%, 21%, 18%, 8.2% and 6.2%, respectively). Histidine was preserved in the ISRIBSEN pool (269,253 of 328,113 reads, 82%). The ISRIBRES pool of cells targeted at Eif2b2I190 was dominated by tryptophan, methionine and tyrosine (28%, 24% and 15%, respectively) consistent with a role for these bulky side chains in occluding the ISRIB binding pocket (Fig. 2B – bottom panel). Mutagenesis of Eif2b2N162 was less successful in generating a pool of strongly ISRIB-resistant cells; nonetheless, threonine was enriched in the ISRIBRES pool (26%) (Fig. 2B, top panel). Thus, residues lining the ISRIB binding pocket play a role in ISRIB action.
Despite considerable allele diversity, the Eif2b2H188X ISRIBRES pool exhibited selective loss of sensitivity to ISRIB, while retaining a measure of responsiveness to certain ISRIB analogs (Fig. 2D). The residual response to analogs AAA1-075B (075B) and AAA1-084 (084) (albeit with much reduced affinity compared to ISRIBSEN population (Fig. 2C), pointed to a shift in the binding properties of the ISRIB pocket, induced by the mutations at β188.
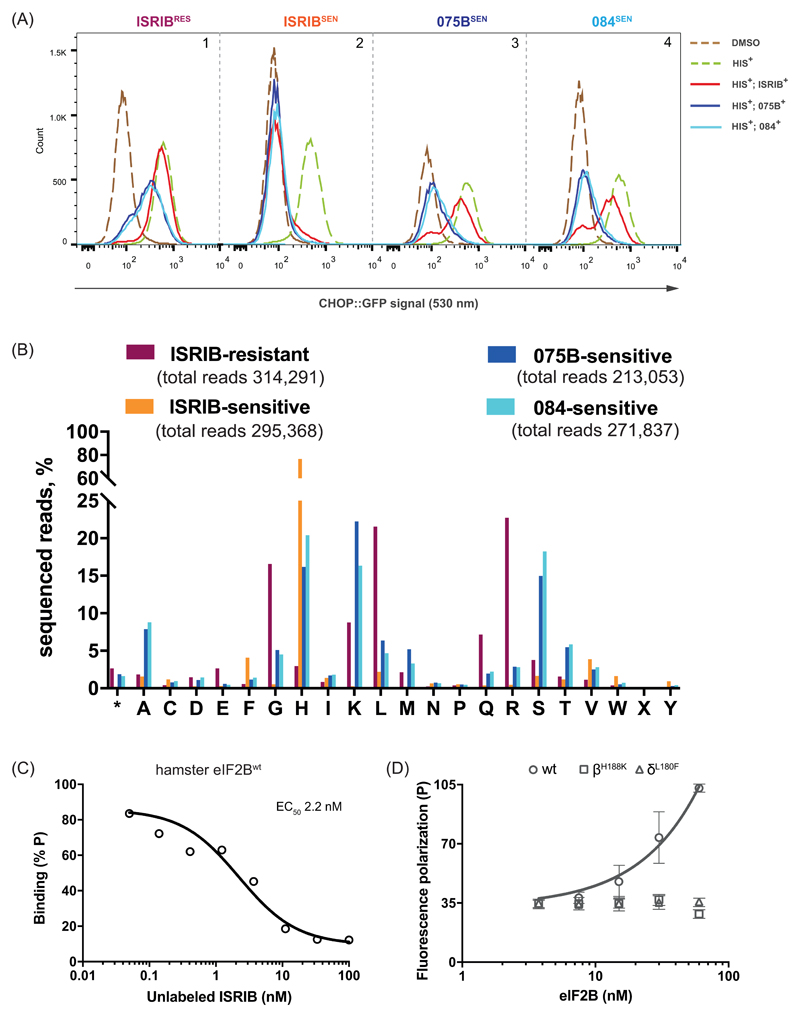
Next, we exploited the diversity of ISRIBRES mutations in the Eif2b2H188X population to select for sub-pools that either acquired sensitivity to compounds AAA1-075B or AAA1-084, regained sensitivity to ISRIB, or retained ISRIB resistance (Fig. 3A) and sequenced their Eif2b2 alleles (Fig. 3B & table S5). As expected, sorting for ISRIB sensitivity enriched, by over 20-fold, rare wildtype H188 alleles that persisted the ISRIBRES Eif2b2H188X pool (Fig. 3B, compare plum and orange bars). H188 was also somewhat enriched in the pools of 075BSEN or 084SEN cells, but unlike the ISRIBSEN these pools were also enriched for residues other than histidine (Fig. 3B & table S5). Importantly, selecting for sensitivity to these ISRIB analogs enriched for different residues than those found in the original ISRIBRES pool: arginine, glycine and leucine were depleted and replaced by lysine, serine, alanine and threonine (Fig. 3B, compare plum to blue and cyan bars). The correlation between mutations in residues lining the ISRIB binding pocket and selective responsiveness to ISRIB analogs, observed in the pools, was confirmed in individual mutant clones (Fig. S5).
Figure 3. Sensitivity to ISRIB analogs selects for a divergent palette of mutations in codon 188 of Eif2b2.
(A) Histograms of the ISR-responsive CHOP::GFP fluorescent reporter activity, induced by histidinol (HIS+, 0.5 mM) in ISRIBRES, ISRIBSEN, compound 075BSEN and compound 084SEN sub-pools, selected for their responsiveness to ISRIB or its analogs (2.5 µM) from a population of originally ISRIBRES Eif2b2H188X mutant cells.
(B) Bar graph of the distribution of residues identified at Eif2b2 codon 188 in phenotypically divergent pools of CHO-K1 cells. The number of sequenced reads in ISRIBRES (plum), ISRIBSEN (orange), compound 075BSEN (blue) and compound 084SEN (cyan) pools encoding each amino acid (* - stop codon, X – ambiguous) is plotted.
(C) Plot of the relative FP signal arising from samples with fluorescein-labeled AAA2-101 (2.5 nM) bound to purified hamster eIF2B (30 nM) in the presence of the indicated concentration of unlabeled ISRIB introduced as a competitor (represented on a log10 scale). Shown is a representative of two independent experiments. The fitting curve and EC50 was generated using “agonist vs. response” function on GraphPad Prism.
(D) A plot of the FP signal arising from fluorescein-labeled AAA2-101 (2.5 nM) as a function of the concentration of wildtype (wt) or mutant eIF2B (δL180F or βH188K) in the sample. Shown are mean ± SD (n=3). Concentrations of eIF2B are represented on a log10 scale.
To directly address the effect of ISRIB-resistant mutations on ISRIB binding, we purified eIF2B from wildtype, Eif2b4L180F and Eif2b2H188K CHO cells (Fig. S6). The wildtype eIF2B gave rise to a concentration-dependent FP signal in the presence of a fluorescein-labeled AAA2-101 that was readily competed with unlabeled ISRIB (Fig. 3C). However, eIF2B purified from the mutant cells failed to give rise to an FP signal (Fig. 3D), thereby establishing a correlation between ISRIB resistance in cells and defective ISRIB binding in vitro.
The ISRIB binding pocket, defined structurally and validated chemogenetically, straddles the two-fold axis of symmetry of the core regulatory subcomplex, and a single molecule of ISRIB appears to engage the same residues from opposing protomers of the (βδ)2 dimer. These features fit with ISRIB’s own symmetry and could explain the ability of ISRIB to stabilize the eIF2B decamer, possibly increasing its abundance in ISRIB-treated cells. Our findings are also consistent with ISRIB’s ability to stabilize a rate-limiting assembly intermediate of the active decamer, as demonstrated biochemically in an accompanying manuscript (12). Indeed, comparison of the S. pombe (10) and ISRIB-bound human eIF2B speaks against large domain movements associated with ISRIB binding. However, an important allosteric effect of ISRIB binding might easily have been missed, as the critical catalytic domain of the ε subunit is resolved in neither structure. Similar considerations apply to the potential effect of ISRIB on the inhibitory interaction between eIF2B with eIF2(αP). These might arise from subtle ISRIB induced conformational changes propagated through the regulatory core to the eIF2(αP) binding cavity formed by the convergence of the tips of the α, β and δ subunits (10, 13–15). The lower resolution of the cryo-EM density in that region might have masked important allosteric changes. Whilst the relative contribution of accelerated assembly, enhanced stability or allostery to ISRIB action remain to be resolved, it is intriguing to consider that endogenous ligands might engage the ISRIB binding site to regulate eIF2B in yet to be determined physiological states.
Supplementary Material
One Sentence Summary.
Cryo-electron microscopy and chemogenetic selection define an ISRIB binding pocket in the core of the eIF2B decamer.
Acknowledgements
We thank D. Barrett and C. Ortori (University of Nottingham) for measuring the ISRIB content of eIF2B, P. Sterk (University of Cambridge) for NGS analysis, S. Chen, C. Savva, G. McMullan, T. Darling and J. Grimmett (MRC LMB) for technical support with cryo-EM, movie data acquisition and help with computing, S. Preissler (University of Cambridge) for editorial comments and Diamond for access and support of the Cryo-EM facilities at the UK national electron bio-imaging centre (eBIC), (proposals EM-14606 and EM-17057), funded by the Wellcome Trust, MRC and BBSRC.
Funding: A Wellcome Trust Principal Research Fellowship (Wellcome 200848/Z/16/Z) to D.R. A Specialist Programme from Bloodwise (12048), the UK Medical Research Council (MC_U105161083) and core support from the Wellcome Trust Medical Research Council Cambridge Stem Cell Institute to A.J.W. and a Wellcome Trust Strategic Award to the Cambridge Institute for Medical Research (Wellcome 100140). A Higher Committee for Education Development, Iraq, Scholarship (4241047) to A.A.A.
Footnotes
Author contributions: A.Z. and D.R. designed and implemented the study and wrote the manuscript. F.W., A.F. and A.J.W. obtained and interpreted the structural data and edited the manuscript. A.A.A., C.F. and P.F. designed and synthesized compounds, interpreted the chemogenetic data and edited manuscript. A.C.C., Y.S. and H.P.H. designed and implemented the somatic cell genetics, cell phenotyping and deep sequencing and edited the manuscript. F.A. and L.P. analyzed NGS sequencing data and edited manuscript.
Competing interests: none
Data and materials availability: Cryo-EM density maps are deposited in the Electron Microscopy Data Bank (EMD-4162), and atomic coordinates are deposited in the Protein Data Bank (PDB) (6EZO).
References
- 1.Pakos-Zebruck K, et al. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sidrauski C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife. 2013;2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halliday M, et al. Partial restoration of protein synthesis rates by the small molecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis. 2015;6:e1672. doi: 10.1038/cddis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou A, et al. Inhibition of the integrated stress response reverses cognitive deficits after traumatic brain injury. Proc Natl Acad Sci U S A. 2017;114:E6420–E6426. doi: 10.1073/pnas.1707661114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panniers R, Henshaw EC. A GDP/GTP exchange factor essential for eukaryotic initiation factor 2 cycling in Ehrlich ascites tumor cells and its regulation by eukaryotic initiation factor 2 phosphorylation. J Biol Chem. 1983;258:7928–7934. [PubMed] [Google Scholar]
- 6.Siekierka J, Mitsui KI, Ochoa S. Mode of action of the heme-controlled translational inhibitor: relationship of eukaryotic initiation factor 2-stimulating protein to translation restoring factor. Proc Natl Acad Sci U S A. 1981;78:220–223. doi: 10.1073/pnas.78.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemens MJ, Pain VM, Wong ST, Henshaw EC. Phosphorylation inhibits guanine nucleotide exchange on eukaryotic initiation factor 2. Nature. 1982;296:93–95. doi: 10.1038/296093a0. [DOI] [PubMed] [Google Scholar]
- 8.Sekine Y, et al. Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science. 2015;348:1027–1030. doi: 10.1126/science.aaa6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sidrauski C, et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. Elife. 2015;4:e07314. doi: 10.7554/eLife.07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kashiwagi K, et al. Crystal structure of eukaryotic translation initiation factor 2B. Nature. 2016;531:122–125. doi: 10.1038/nature16991. [DOI] [PubMed] [Google Scholar]
- 11.Hearn BR, et al. Structure-Activity Studies of Bis-O-Arylglycolamides: Inhibitors of the Integrated Stress Response. ChemMedChem. 2016 doi: 10.1002/cmdc.201500483. [DOI] [PubMed] [Google Scholar]
- 12.Tsai J, et al. Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule. Science. 2018 doi: 10.1126/science.aaq0939. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vazquez de Aldana CR, Hinnebusch AG. Mutations in the GCD7 subunit of yeast guanine nucleotide exchange factor eIF-2B overcome the inhibitory effects of phosphorylated eIF-2 on translation initiation. Mol Cell Biol. 1994;14:3208–3222. doi: 10.1128/mcb.14.5.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavitt GD, Yang W, Hinnebusch AG. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol Cell Biol. 1997;17:1298–1313. doi: 10.1128/mcb.17.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogorad AM, Lin KY, Marintchev A. Novel mechanisms of eIF2B action and regulation by eIF2alpha phosphorylation. Nucleic Acids Res. 2017;45:11962–11979. doi: 10.1093/nar/gkx845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekine Y, et al. Paradoxical Sensitivity to an Integrated Stress Response Blocking Mutation in Vanishing White Matter Cells. PLoS One. 2016;11:e0166278. doi: 10.1371/journal.pone.0166278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Scheres SH. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. Elife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 27.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burnley T, Palmer CM, Winn M. Recent developments in the CCP-EM software suite. Acta Crystallogr D Struct Biol. 2017;73:469–477. doi: 10.1107/S2059798317007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dolle REC, Chu G-H. Preparation of pyrrolidinylphenethyl benzoxepine-, tetrahydronaphthalene-, chroman-, and benzofurancarboxamides as κ-opioid agonists. US 2005/54630 A1. 2005 [Google Scholar]
- 33.Carr JB. Lipogenesis control by esters of benzoxazinecarboxylic acids. U.S. Patent No. 4180572. 1979
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.