Abstract
At least 15% of the disease-causing mutations affect mRNA splicing. Many splicing mutations are missed in a clinical setting due to limitations of in silico prediction algorithms or their location in noncoding regions. Whole-transcriptome sequencing is a promising new tool to identify these mutations; however, it will be a challenge to obtain disease-relevant tissue for RNA. Here, we describe an individual with a sporadic atypical spinal muscular atrophy, in whom clinical DNA sequencing reported one pathogenic ASAH1 mutation (c.458A>G;p.Tyr153Cys). Transcriptome sequencing on patient leukocytes identified a highly significant and atypical ASAH1 isoform not explained by c.458A>G(p<10−16). Subsequent Sanger-sequencing identified the splice mutation responsible for the isoform (c.504A>C;p.Lys168Asn) and provided a molecular diagnosis of autosomal-recessive spinal muscular atrophy with progressive myoclonic epilepsy. Our findings demonstrate the utility of RNA sequencing from blood to identify splice-impacting disease mutations for nonhematological conditions, providing a diagnosis for these otherwise unsolved patients.
Keywords: ASAH1, next-generation sequencing, spinal muscular atrophy with progressive myoclonic epilepsy (SMA-PME), transcriptome sequencing
Mutations that affect mRNA splicing are estimated to account for between 15% and 60% of genetic diseases (Krawczak, Reiss, & Cooper, 1992; Lopez-Bigas, Audit, Ouzounis, Parra, & Guigo, 2005). These variants can trigger a variety of effects, including exon skipping, intron retention, activation of cryptic splice sites, or creation of new splice sites. Resultant defective transcripts may produce a partially or even nonfunctional protein, leading to disease.
It is now well established that next-generation sequencing (NGS) of DNA, by either gene panel or whole-exome sequencing, is an effective method to identify disease-causing mutations (Sawyer et al., 2016; Sun et al., 2015). However, these assays are usually limited to the coding sequence and minimal flanking DNA (±10 bp), and rely on computational algorithms to predict the effects of mutations at the mRNA and protein level. Many splice-impact prediction programs have been developed (e.g., MaxEntScan [Yeo & Burge, 2004], NNSplice [Reese, Eeckman, Kulp, & Haussler, 1997], Human Splicing Finder [Desmet et al., 2009], GeneSplicer [Pertea, Lin, & Salzberg, 2001], GENSCAN [Burge & Karlin, 1997], VEP [McLaren et al., 2016], SPANR [Xiong et al., 2015], and SplicePredictor [Brendel, Xing, & Zhu, 2004]), and while each program has strengths and limitations, there is no standard approach to assess potential splicing impact. Furthermore, while evaluating the reference genome, these programs incorrectly predict between 2.6% and 76.1% of splicing events (Jian, Boerwinkle, & Liu, 2014). Therefore, it is highly likely that many splice-impacting mutations are not appreciated due to their location in noncoding regions or limitations of current splice-impact prediction.
A patient with a sporadic and atypical form of spinal muscular atrophy presented to the regional Neurogenetics Clinic for evaluation. His history included weakness with onset at the age of 3 years, followed by a slow progressive deterioration in muscle strength in childhood and adolescence. During his late teen years, he had pronounced progressive weakness in the proximal muscles with marked atrophy of the periscapular, pectoralis, and triceps muscles. He also developed a prominent facial tremor, myoclonic jerks, and sensorineural hearing loss. There was no sensory or autonomic dysfunction, and brain MRI was normal. EEG demonstrated frequent bursts of paroxysmal polyspikes and slow-wave activity. Nerve conduction studies were consistent with a diffuse motor neuropathy/neuronopathy. Left deltoid muscle biopsy primarily demonstrated neurogenic changes. Cognition was normal and family history unremarkable. DNA was sent for SMN1 deletion testing, chromosomal microarray, and SMA gene panel testing (SMA panel at a CLIA-certified USA-based clinical laboratory that assesses 15 motor neuron-related disease genes). A single variant in ASAH1 (NM_004315.4: c.458A>G; p.Tyr153Cys) was reported and classified as a variant of unknown significance, but was highly suspicious based on its rarity in control cohorts (seen in one of 70,372 alleles in ExAC Browser), and it is predicted to be deleterious to protein function (PolyPhen-2 [1.0], SIFT [0.0]). Subsequent ASAH1 deletion and duplication testing to assess for a second mutation was negative.
Given the highly suspicious ASAH1 variant and similarity between our patient phenotype and spinal muscular atrophy with progressive myoclonic epilepsy (SMA-PME) (OMIM 159950), a recessive condition caused by mutations in ASAH1 (Zhou et al., 2012), we hypothesized the existence of a noncoding variant that was missed by the clinical panel sequencing. We chose to perform whole-transcriptome sequencing on a research basis. The patient was enrolled in the Care4Rare Canada research project, which was approved by the institutional research ethics board (Children’s Hospital of Eastern Ontario); free and informed consent was obtained from the patient.
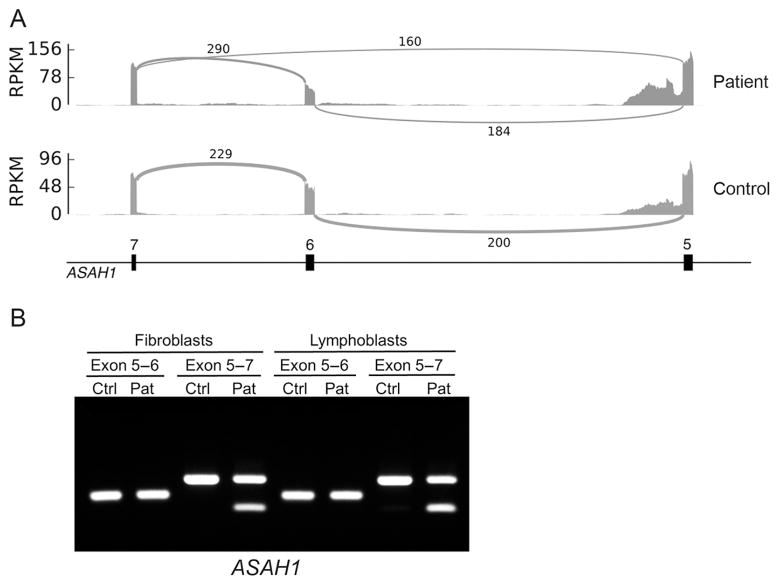
An important consideration in our approach to transcriptome sequencing was to utilize the most readily available and least invasive cell type for evaluation. Therefore, we chose to conduct RNA sequencing from blood. Importantly, we have a control dataset of 909 control samples with which to compare patient data (Battle et al., 2014). Leukocytes were selected, transcriptome libraries were prepared (including Globin depletion and polyA capture), and the RNA was sequenced to a read depth of 63 million reads on an Illumina NextSeq500, using a combination of 150 and 75 bp paired-end technology. RNA reads were aligned to the genome, PCR duplicates were removed, and splice junctions were assessed (see Supp. Information for detailed RNA sequencing and analysis methods). We then tested for differential exon usage between the patient and all other individuals in order to identify aberrant splicing events unique to the patient (allowing a junction to be considered if it is observed in at least one sample with a minimum of 20 supporting reads). The most significant splicing difference between the patient and the 909 unaffected individuals was a skipping of the sixth exon in the ASAH1 gene (Fig. 1A). This event was remarkable because it was not observed in any of the control samples, and is orders of magnitude more significant than any other differential splicing event in the patient (P value < 1e-16 after Benjamini–Hochberg correction). Haplotype analysis revealed that while there was a proportion of the normal allele from both haplotypes, 90% of the reads from one haplotype were of the alternative transcript. RT-PCR analysis on RNA extracted from fibroblast and lymphoblast cell lines derived from the patient was conducted to confirm this effect (Fig. 1B).
FIGURE 1.
ASAH1 differential splicing. A: Sashimi plot of ASAH1 exons 5–7 in control and patient transcriptomes demonstrates the novel splice isoform observed in our patient lacking exon 6. Data are displayed as RPKM (1e3 * [depth/read_length]/[n_mapped_reads/1e6]), and the number of reads for each junction is noted on each junction arc. B: The novel isoform lacking exon 6 was confirmed by RT-PCR on RNA from patient-derived fibroblasts and lymphoblasts
The initial ASAH1 c.458A>G variant identified by clinical testing was not located near an exon 6 boundary and was therefore unlikely to have caused this splicing effect. We conducted Sanger sequencing in the Care4Rare research laboratory to assess for a second ASAH1 variant. A c.504A>C, p.Lys168Asn (NM_004315.4) variant located at the 3′ end of exon 6 (−2 bp from the splice junction) was identified. While informatics scores do not predict this variant to be damaging for protein function (PolyPhen-2 [0], SIFT [0.53]), this variant has been previously reported as a mutation in SMA-PME that effects splicing (Dyment et al., 2014). It is unclear why the second mutation was missed by clinical DNA testing and an explanation was not provided by the clinical laboratory. We conclude that the differential splicing effect identified by trancriptome sequencing is likely a consequence of the c.504A>C ASAH1 variant.
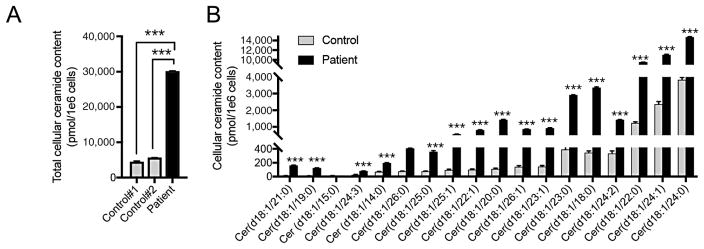
Finally, given the variant of unknown significance classification of the c.458A>G change and the patient’s atypical SMA-PME clinical presentation, we wanted to assess ASAH1 function to conclusively provide a SMA-PME diagnosis for this patient. ASAH1 encodes a member of the acid ceramidase family, which catalyzes the degradation of ceramide into sphingosine and free fatty acids. Cellular ceramide content was quantified by high-performance liquid chromatography electrospray ionization tandem mass spectrometry in extracts from patient and control fibroblast cells. Eighteen distinct ceramide species with a d18:1 sphingosine backbone were detected; a significant increase was observed in 17 of the 18 ceramides (Fig. 2A and B), confirming the loss of ASAH1 function, and thus the pathogenicity of the ASAH1 variants. We conclude that our patient has SMA-PME caused by biallelic ASAH1 mutations, c.504A>C (p.Lys168Asn) and c.458A>G (p.Tyr153Cys).
FIGURE 2.
Defective ASAH1 function confirms the SMA-PME diagnosis. Total ceramide levels (A) and detailed ceramide profile (B) demonstrate a loss of ASAH1 function resulting in increased ceramides in patient-derived fibroblast cells compared with controls
Genes highly implicated in disease have an increased number of splice junctions and transcripts per gene, when compared with nondisease genes (Jian et al., 2014), speaking to the importance of splicing for human health. At present, approximately 10% of disease mutations cataloged in the Human Gene Mutation Database (HGMD) are annotated as splice impacting (Stenson et al., 2003). However, splice-impacting mutations may be missed by DNA NGS due to (1) failed detection of the DNA variant (due to location in a region not sequenced or in a GC-rich, AT-rich, or repetitive region difficult to capture and analyze), (2) failed reporting (due to sequence quality, population variant frequency, or human error), or (3) failed annotation of splice impact (due to limitations in prediction algorithms). Given these limitations, it is likely that the true proportion of splice-impacting mutations in rare disease is much higher than 10%. There is a clear need for improved methods to identify splice-impacting mutations.
Whole-transcriptome sequencing is a promising new tool to assess mRNA splicing. This may be particularly effective in recessive conditions when one pathogenic or likely pathogenic mutation is identified by DNA NGS and the second remains elusive. Additionally, the use of transcriptome sequencing could play a key role in interpreting variants of unknown significance by establishing any splicing effects in a gene of interest (splicing effects is an important ACMG pathogenicity classification criteria [Richards et al., 2015]). Whole-transcriptome sequencing is an ideal assay as this can be used for all disorders, as opposed to targeted approaches that must be individually designed and validated. The use of transcriptome sequencing in blood in the patient reported here with a neurological disease demonstrates the power of transcriptome sequencing to aid in the identification of splice-impacting DNA mutations. Interestingly, of the genes on the motor neuron panel, 71% are expressed in at least half of our control samples, and of the 78 genes in OMIM linked to myoclonic seizures, 40% are expressed in at least half of the control samples (a gene was considered expressed if at least 50% of the control cohort samples showed FPKM ≥ 1). This indicates that blood transcriptome sequencing would be a viable option for many of these disorders.
A recent study used transcriptome sequencing of muscle-derived RNA to identify a splice-impacting DMD mutation in a muscular dystrophy patient (Gonorazky et al., 2016). To our knowledge, our study is the first to identify a hereditary Mendelian disease mutation via transcriptome sequencing in blood. Importantly, the approach was successful in identifying a splice-impacting mutation for SMA-PME, a non-hematological condition. Relevant disease tissues are often difficult to obtain, whereas blood is a more readily available and less invasive sample. It is noted that some causative mutations may exist in transcripts that are not expressed in blood (e.g., some muscle or neuron-specific); conversely, many tissue-specific transcripts are expressed at low levels elsewhere, including blood. Given the limitations of sample acquisition from living patients, transcriptome sequencing from blood likely offers a feasible approach to identify splice-impacting DNA mutations for many hereditary Mendelian diseases.
In conclusion, we used whole-transcriptome sequencing from blood to identify a splice-impacting disease mutation and provide a diagnosis of SMA-PME in a patient where only a single variant of uncertain significance had been identified by the clinical laboratory. While DNA-based NGS applications are employed in many clinical diagnostic laboratories, RNA-based NGS applications have yet to be leveraged in the same fashion. Our results highlight the potential of combined multiomic approaches, including transcriptome sequencing, to aid the diagnosis of rare disease patients.
Supplementary Material
Acknowledgments
Contract grant sponsors: Genome Canada; Canadian Institutes of Health Research; Ontario Genomics Institute; Ontario Research Fund; Genome Quebec; Children’s Hospital of Eastern Ontario Foundation (OGI-064); NIH (R01HG008150, R01MH101814, U01HG007436, T32HG000044, and U01HG009080); National Science Foundation GRFP (DGE-114747); Stanford Center for Computational, Evolutionary, and Human Genomics (CEHG).
The authors would first and foremost like to thank the patient and their family for their active participation in this study. This work was performed under the Care4Rare Canada Consortium (Enhanced Care for Rare Genetic Diseases in Canada).
Footnotes
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- Battle A, Mostafavi S, Zhu X, Potash JB, Weissman MM, McCormick C, … Koller D. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Research. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V, Xing L, Zhu W. Gene structure prediction from consensus spliced alignment of multiple ESTs matching the same genomic locus. Bioinformatics. 2004;20:1157–1169. doi: 10.1093/bioinformatics/bth058. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. Journal of Molecular Biology. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Research. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyment DA, Sell E, Vanstone MR, Smith AC, Garandeau D, Garcia V, … Boycott KM. Evidence for clinical, genetic and biochemical variability in spinal muscular atrophy with progressive myoclonic epilepsy. Clinical Genetics. 2014;86:558–563. doi: 10.1111/cge.12307. [DOI] [PubMed] [Google Scholar]
- Gonorazky H, Liang M, Cummings B, Lek M, Micallef J, Hawkins C, … Dowling JJ. RNAseq analysis for the diagnosis of muscular dystrophy. Annals of Clinical and Translational Neurology. 2016;3:55–60. doi: 10.1002/acn3.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Research. 2014;42:13534–13544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Human Genetics. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Lopez-Bigas N, Audit B, Ouzounis C, Parra G, Guigo R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Letters. 2005;579:1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, … Cunningham F. The ensembl variant effect predictor. Genome Biology. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Lin X, Salzberg SL. GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Research. 2001;29:1185–1190. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. Journal of Computational Biology. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J … ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Hartley T, Dyment DA, Beaulieu CL, Schwartzentruber J, Smith A, … Boycott KM. Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: Time to address gaps in care. Clinical Genetics. 2016;89:275–284. doi: 10.1111/cge.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Human Mutation. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Sun Y, Ruivenkamp CA, Hoffer MJ, Vrijenhoek T, Kriek M, van Asperen CJ, Santen GW. Next-generation diagnostics: Gene panel, exome, or whole genome? Human Mutation. 2015;36:648–655. doi: 10.1002/humu.22783. [DOI] [PubMed] [Google Scholar]
- Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RK, Frey BJ. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. Journal of Computational Biology. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Zhou J, Tawk M, Tiziano FD, Veillet J, Bayes M, Nolent F, … Melki J. Spinal muscular atrophy associated with progressive myoclonic epilepsy is caused by mutations in ASAH1. American Journal of Human Genetics. 2012;91:5–14. doi: 10.1016/j.ajhg.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.