Abstract
Long intergenic non-coding RNA (lincRNA) genes have diverse features that distinguish them from mRNA-encoding genes and exercise functions such as remodelling chromatin and genome architecture, RNA stabilization and transcription regulation, including enhancer-associated activity. Some genes currently annotated as encoding lincRNAs include small open reading frames (smORFs) and encode functional peptides and thus may be more properly classified as coding RNAs. lincRNAs may broadly serve to fine-tune the expression of neighbouring genes with remarkable tissue specificity through a diversity of mechanisms, highlighting our rapidly evolving understanding of the non-coding genome.
Long intergenic non-coding RNAs (lincRNAs) are defined as autonomously transcribed non-coding RNAs longer than 200 nucleotides that do not overlap annotated coding genes. lincRNAs share features with the other transcripts of the long non-coding RNA (lncRNA) family and constitute more than half of lncRNA transcripts in humans (TABLE 1). The existence of lincRNAs was first suggested by studies using tiling arrays across genomic sequences, which observed pervasive transcription1,2 from regions with no known coding genes3–6. Assessment of chromatin state signatures in murine cell types provided early support for the presence of active transcription units at the putative loci of these transcripts7. lincRNAs have been distinguished from the broader lncRNA class of transcripts, as many lncRNAs share sequence with coding loci. Many publications, however, do not distinguish between these two sets of transcripts and group them collectively as ‘lncRNAs’. The accelerating pace with which both intergenic and genic lncRNAs have been discovered and annotated has contributed to an evolving understanding of the functions of the non-coding RNA world4,8,9.
Table 1.
Definitions and classifications of lncRNA and lincRNA
| RNA | Definition | Number of transcripts in the human genome | Diagram |
|---|---|---|---|
| LncRNA | Autonomously transcribed RNAs longer than 200 nucleotides with minimal coding potential | Key: Blue = lncRNA Red = coding RNA Box = exon Line = intron |
|
| Genic lncRNA |
LncRNA overlapping a protein-coding transcript at one or more nucleotides | 7,169 (of which 1,317 are e-lncRNAs) |
|
| Nested | LncRNA genes contained entirely within protein-coding transcripts | 1,256 (S), 2,917 (AS) |
|
| Containing | Protein-coding transcripts contained entirely within lncRNAs | 38 (S), 138 (AS) |
|
| Overlapping | LncRNA genes neither ‘nested’ nor ‘containing’ | 206 (S), 1,867 (AS) |
|
| Multiple relationships | LncRNA with more than one of the above relationships to protein-coding transcripts | 747 |
|
| LincRNA | LncRNA not overlapping a protein-coding transcript | 8,598 (2,488 e-lincRNAs) | |
| Same strand | LincRNA within 50 kb of and transcribed from the same strand and in the same direction as the nearest protein-coding transcript | 2,059 |
|
| Convergent | LincRNA within 50 kb of and transcribed head-to-head with the nearest protein-coding transcript | 830 |
|
| Divergent | LincRNA within 50 kb of and transcribed tail-to-tail with the nearest protein-coding transcript | 1,710 |
|
| Isolated | LincRNA >50 kb from the nearest protein-coding gene | 3,999 |
|
We define long intergenic non-coding RNA (lincRNA) genes as those giving rise to exclusively intergenic transcripts; 663 GENCODE-annotated lincRNA genes give rise to at least one genic transcript and so should be categorized as genic lncRNA genes. We define a lincRNA as ‘isolated’ if it is >50 kb from the nearest protein-coding gene; a specific distance cut-off has not yet been described in the literature. This distance divides the lincRNA class in half between isolated and non-isolated transcripts, with non-isolated transcripts further subtyped by their positional relationship with respect to the nearest protein-coding gene. We intersected GENCODE transcripts with a comprehensive enhancer-derived transcript data set126,127 to quantify the lincRNA and lncRNA transcripts with enhancer-like functions (e-lincRNAs and e-lncRNAs, respectively). AS, antisense; S, sense.
By conservative estimates from GENCODE v25 annotations, 51.8% of the human genome is transcribed, but only 1.2% encodes proteins. The discovery of the role of the lincRNA X-inactive specific transcript (Xist) in X chromosome inactivation (XCI) and gene dosage compensation in the early 1990s ( REFS 10,11) was followed in the 2000s by that of HOTAIR, which represses the transcription of HOX family genes12. These studies stimulated interest in considering linc-RNA function in specific cellular contexts, cell types, developmental stages and disease13. Of the thousands of lincRNAs subsequently annotated by next-generation sequencing technologies, a small fraction have been functionally interrogated and assigned roles in diverse gene regulation processes, organisms and human disease14,15–26.
In addition to the lack of physical overlap between lincRNAs and protein-coding genes, the distinction between lincRNAs and genic lncRNAs was supported by gene expression analyses, evolutionary conservation patterns and targeted gene disruptions that did not alter adjacent protein-coding genes or genic RNAs. Whether lincRNAs and genic lncRNAs share features is an area of interest. The presence of lincRNAs within gene deserts may weaken the burden of evolutionary conservation, thereby enabling rapid functional diversification of lincRNA loci and resulting in transcripts with fewer mRNA characteristics compared with genic lncRNAs. Alternatively, the separation of lincRNAs from genic lncRNAs may reflect a semantic distinction that is biologically inconsequential14. Characterization of potential systematic differences between lincRNAs and genic lncRNAs is an area of active investigation, as is the relationship of these RNAs to gene regulation and enhancer function.
In this Review, we first provide an evolutionary perspective of lincRNAs and discuss its relevance to lincRNA annotation and to understanding their function. We then focus on the emerging unique features of lincRNAs compared with mRNAs and finally discuss the diverse functions of lincRNAs and propose a mechanistic framework for understanding them.
Evolutionary conservation of lincRNAs
GENCODE currently annotates 13,255 lincRNA transcripts arising from 8,598 genes. Despite a notable lack of primary sequence conservation, lincRNAs are not evolutionarily neutral, showing greater conservation than ancient repeat sequences but less than protein-coding genes. Their poor conservation has confounded efforts to predict lincRNA functions across species17,27,28 but has also helped to highlight focal areas of potential functional importance29,30.
Compared with protein-coding RNAs and other non-coding RNAs, mammalian lincRNAs have fewer invertebrate orthologues and have undergone rapid evolution14. Approximately 5% of mammalian lincRNAs are conserved in zebrafish, and conservation is typically restricted to short polynucleotide stretches31. Some mouse and human orthologues, however, have been shown to phenotypically rescue zebrafish lincRNA loss of function, as in the case of the lincRNA cyrano14, which is crucial for embryonic development, indicating that at least some lincRNAs are functionally conserved across species.
LincRNAs have functional roles across a spectrum of species. In the budding yeast Saccharomyces cerevisiae, lincRNAs participate in various stress responses, including nutrient starvation32. In Arabidopsis thaliana, lincRNAs are expressed at lower levels than protein-coding genes, but subsets of lincRNAs exhibit tissue-specific and stress-responsive expression patterns33,34. Caenorhabditis elegans lincRNAs share features with those of other invertebrates and vertebrates. C. elegans lincRNAs harbour short conserved regions not subject to the swift sequence evolution of adjacent gene regions. C. elegans-specific lincRNA roles are often reproductive and reminiscent of those in yeast32, including dauer and sperm formation, establishment of male identity and interacting with sperm-specific transcripts35. Thus, across invertebrate species, lincRNAs function in metabolic and tissue-specific processes.
The degree of similarity between mouse and human lincRNAs varies between annotation methods. Nearly half of the mouse non-coding transcripts from cDNA libraries map to the human genome. Only 14%, however, have evidence of expressed sequence tags or cDNA indicating the existence of a human orthologous transcript36, suggesting that the homologous subset is modest37,38. Although low conservation does not obviate similar function, it is unlikely that evolutionary clades would independently develop similar functions for such substantial genomic fractions. These findings called into question the conservation of lincRNA functions and motivated the development of alternative annotation methods14,38. Categorization of transcripts based on patterns of histone post-translational modifications at the loci that encode them enabled the identification of >1,000 previously unannotated mouse lincRNAs with substantial (>95%) cross-mammalian conservation7.
Mouse lincRNA knockout models have revealed lincRNAs with perinatal or postnatal lethal phenotypes, as well as growth and organ-restricted developmental defect phenotypes that cannot be analogously studied in humans39. The majority of mouse non-coding RNAs are expressed in neuronal tissue and distinct cortical regions40. Many have distant chicken and opossum orthologues41, which, with their expression patterns42, suggest they have roles in neurogenesis and the tissue-specific tuning of gene expression, although definitive assignment of lincRNA function based on a knockout phenotype is not straightforward because the phenotype could also result from disruption of a gene regulatory element within a lincRNA locus. Murine genetic studies are thus helpful to model human lincRNA biology, although they are limited by both evolutionary divergence and by the experimental challenges of excluding RNA-independent effects. In both murine models and human cell lines, it is generally difficult to determine whether a rescue phenotype observed by providing lincRNA-encoding DNA in trans is due to a transcript-dependent effect, the act of transcription itself, effects on other genes or transcripts, inter actions with other proteins, or lincRNA-derived peptides. Furthermore, transexpressed lincRNAs capable of rescue may not produce an accurate phenotype if expressed at non-physiological levels or if expressed at an incorrect subcellular location, highlighting the difficulties inherent to assigning mechanisms to lincRNAs.
Mapping of human lncRNA transcripts, including lincRNAs, across species ranging from mice to fish reveals that ~12% have non-human homologues43. The orthologous segments span 21–56% of transcript lengths and globally exhibit modest conservation. The fraction of identical bases between lncRNAs that are mapped to different genomic regions is lower than that between orthologous coding gene pairs, greater than random genomic regions and similar to syntenic blocks28. Greater conservation at lincRNA promoters than at their downstream regions44 suggests selective pressure for enhancer-like functions7,27,45, which has indeed been predicted for a substantial subset of lincRNAs46, termed e-lincRNAs (TABLE 1).
LincRNAs exhibit a range of conservation patterns. Variable conservation at the nucleic acid level may underlie greater conservation at the (secondary or tertiary) structural level. A minority of lincRNAs are highly conserved at both the sequence and the RNA secondary structure level. This subset includes several well-studied molecules, including MALAT1 ( REF. 47), which is under selection to preserve intron–exon organization as well as secondary structure; MALAT1 is stabilized at its 3′ end by a triple helix structure14. Other lincRNAs have conservation biased towards their 5′ ends and exhibit rapid changes at their 3′ ends across vertebrates. TINCR, for example, is conserved at its 5′ end across vertebrates. Its 3′ end is conserved in rhesus macaques but not in mice48; another well-studied lincRNA with 5′ conservation bias is the MYC proto-oncogene protein-associated PVT1 ( REF. 49). Such a pattern suggests sequence-specific functions may cluster towards the 5′ end and potentially that species-specific functions could derive from the more divergent 3′ end. Other lincRNAs exhibit focal conservation, which may reflect the fact that some genes currently annotated as lincRNAs are actually not non-coding RNAs but in fact contain small open reading frames (smORFs)29,40. A subset of lincRNAs harbour conserved smORFs that give rise to polypeptides with biological function50. For example, the 90-amino-acid polypeptide small regulatory polypeptide of amino acid response (SPAR) is encoded by a smORF, which is conserved between mice and humans in the lincRNA LINC00961 ( REF. 18). Alternatively, focal conservation could underlie sequence-dependent lincRNA functions such as interactions with other nucleic acids or proteins or translation-linked but peptide-independent activity. Finally, sequence divergence may suggest transcript dispensability, at least for some lincRNAs19,20, for example, at the mouse Blustr locus, where transcriptional activity largely independent of the produced transcript affects the transcription of a neighbouring gene19.
To summarize, lincRNAs have diverse conservation patterns and have undergone rapid evolution across species. Although they exhibit less cross-species conservation than do protein-coding genes, lincRNAs are not evolutionarily neutral and may have transcript-dependent and/or transcript-dispensable, species-specific functions.
Comparison of lincRNA and mRNA features
LincRNAs differ from mRNAs in their abundance, genomic localization and subcellular localization, as well as in their metabolic profiles, epigenetic regulation and tissue specificity. In this section, we distinguish, where possible, between lincRNAs and genic lncRNAs.
Abundance, size and genomic localization
The number of lincRNAs continues to expand, with GENCODE v25 annotating 8,598 genes that meet our criteria for lincRNAs (TABLE 1), compared with 19,950 protein-coding genes. Cap analysis of gene expression by the FANTOM5 consortium was recently used to identify 27,919 human lncRNA genes, of which 13,105 are lincRNA genes46. Specific lincRNA inclusion criteria included chromatin signatures7 and distance from adjacent coding genes (to account for unannotated, alternatively spliced transcripts)27, although a uniform distance cut-off is lacking. lincRNAs are located from several bases to >3 Mb away from the nearest protein-coding gene, at a median distance of 40 kb — 28% are within 10 kb ( REF. 28), and nearly half are >50 kb away, which we define as ‘isolated’ lincRNAs (TABLE 1). Relative to mRNAs, lincRNA transcripts have fewer exons (an average of 2.9 compared with 10.7), are shorter (average length of 1 kb compared with 2.9 kb)27,44 and are expressed at a tenfold lower level44. The low expression of lincRNAs in whole-organ tissues — that is, tissue samples derived from complex organs containing multiple tissue subtypes — may be driven by cell-type-specific expression, at least in complex tissues44,51, consistent with lincRNAs having tissue-defining roles.
Subcellular localization
mRNAs are chiefly trafficked to the cytoplasm, where they undergo translation. lincRNAs, by contrast, are more often located in the nucleus than in the cytoplasm, as demonstrated by fluorescent in situ hybridization48 and ribosome profiling52. lncRNAs in general have similar nuclear:cytoplasmic enrichment ratios across cell types27, although genome-wide subcellular (compartment) lncRNA quantification has not yet been published. The nuclear enrichment of lincRNAs may suggest increased stability and function in the nucleus, although evidence for greater cytoplasmic stability44 and for degradation by the nuclear exosome53 exist. The latter may account for relative lincRNA enrichment at the chromatin but depletion in the nucleoplasm compared with mRNAs53. Recent work proposed a lncRNA classification schema based on RNA metabolic profiles, under the hypothesis that RNA species that share such metabolic profiles may also share functional profiles54. In this work, which found lncRNAs to be synthesized less efficiently and degraded more efficiently and to undergo slower splicing than mRNAs, lincRNAs were found to be evenly distributed across broadly defined RNA functional classes, suggesting that neither genomic position nor metabolic profile globally correlate with RNA functional classification54.
Transcriptional regulation, biogenesis and splicing
RNA polymerase II (Pol II) transcribes approximately 150,000 pre-mRNAs in the human genome, which undergo 5′ capping, splicing and 3′ cleavage and poly-adenylation55. To varying degrees, lincRNAs share these processing features, but whereas mRNAs are more robustly co-transcriptionally spliced and polyadenylated, lincRNAs are more often co-transcriptionally cleaved and prematurely terminated (FIG. 1a). Recently, mammalian native elongating transcript sequencing (mNET-seq) was used to profile lincRNAs53. mNET-seq surveys genome-wide Pol II density by detecting the phosphorylation status of the Pol II C-terminal domain (CTD) at single-base resolution. Phosphorylation of the CTD on Thr4 is associated with transcription termination and is enriched in Pol II located downstream of mRNA polyadenylation sites, but in lincRNAs it is found across entire transcription units. This suggests that whereas Pol II pauses inefficiently at lincRNA promoters, it pauses throughout lincRNA transcription units, resulting in more frequent transcription termination than observed in protein-coding genes53. lincRNAs broadly lack the phospho-CTD features of protein-coding genes and therefore are not consistently associated with CTD phosphorylation forms that increase mRNA-like processing53.
Figure 1. Distinguishing features of long intergenic non-coding RNAs (lincRNAs) and mRNAs.
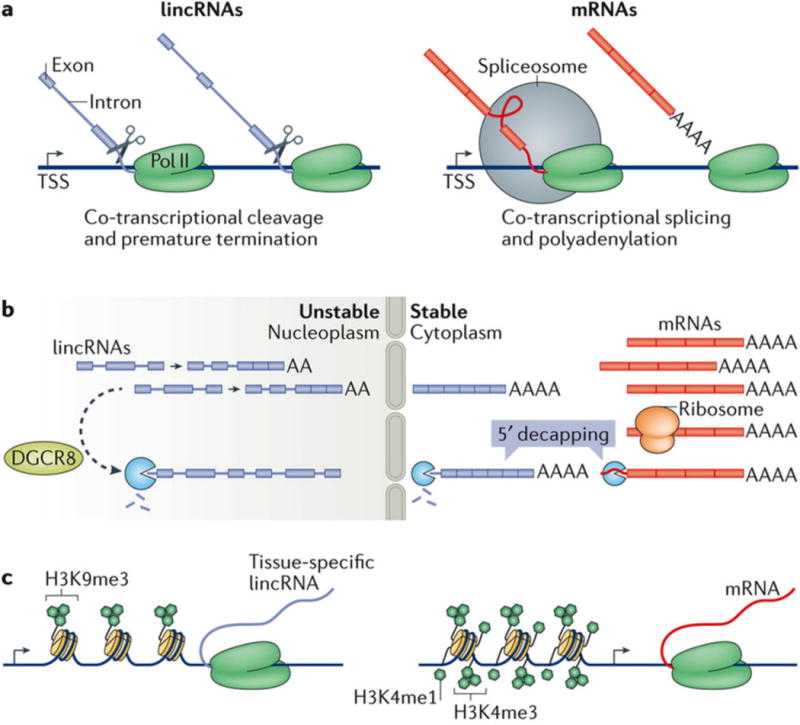
a | During transcription, some lincRNAs undergo cleavage and premature termination, whereas others are spliced and polyadenylated similarly to mRNAs53. b | LincRNAs are generally more abundant in the nucleus, whereas mRNAs are generally more abundant in the cytoplasm, where they associate with ribosomes. LincRNAs and mRNAs have similar occupancy (residence) at the chromatin, whereas lincRNAs are relatively depleted in the nucleoplasm compared with mRNAs, possibly as a result of degradation by the nuclear exosome53. Poorly processed lincRNAs may be targeted to the nuclear exosome for degradation at least partially by microprocessor complex subunit DGCR8, whereas those more similar to mRNAs may remain in the cytoplasm, where they are stable. Cytoplasmic lincRNAs and mRNAs can be degraded following 5ʹ decapping. c | LincRNA-coding and protein-coding genes have globally similar chromatin profiles7,53. LincRNAs are distinguished by enrichment in histone H3 Lys9 trimethylation (H3K9me3) at their promoters. This is a canonically repressive mark, but in lincRNAs, it is instead associated with greater tissue specificity; the mechanism has not yet been described. H3K4me1 is associated with enhancers and H3K4me3 is associated with promoters. Pol II, RNA polymerase II; TSS, transcription start site.
Pol II transcripts can be defined by their positional relationship to coding genes: promoter upstream transcripts in the antisense direction (PROMPTs)56 and enhancer RNAs (eRNAs)57 derive from coding gene promoters and enhancer elements, respectively. Most eukaryotic promoters are divergent (bidirectional) and can generate transcripts in both sense (mRNA) and anti-sense directions. Divergent transcription produces approximately 13% of the annotated lincRNAs located within 10 kb of coding gene promoters, of which 65% are within 1 kb of transcription start sites28. Divergent transcription is associated with histone H3 Lys56 (H3K56) acetylation and Pol II Tyr1 phosphorylation, is promoted by SWI/SNF chromatin remodellers and is repressed by the RNA deadenylase CAF1 ( REFS 17,58,59), features that may indicate distinct divergent RNA transcription and RNA processing mechanisms.
The binding of nascent transcripts by the spliceosomal U1 small nuclear RNA (snRNA) inhibits the use of alternative polyadenylation signals, thus preventing premature transcript degradation. Bidirectional promoters give rise to asymmetric enrichment in polyadenylation sites (PASs) in the antisense lincRNA transcript and to enrichment in U1 binding sites in the sense mRNA transcript, an asymmetry that would favour premature transcription termination and polyadenylation of antisense lincRNAs, but efficient elongation and splicing of mRNAs60. Contradicting the initial descriptions of fewer U1 sites in divergently transcribed lncRNAs than in their promoter-paired mRNAs60, a more recent study of lincRNAs specifically found comparable U1 binding motif enrichment and polyadenylation motif depletion downstream of the transcription start site — the so-called U1–PAS axis — in lincRNAs and mRNAs44. Despite this evidence, lincRNAs are processed less efficiently: they undergo less co-transcriptional splicing and 3′ end cleavage and polyadenylation53. They do, however, undergo alternative splicing, with an average of 2.3 isoforms produced per locus28. Decreased splicing in lincRNAs compared with mRNAs may be the consequence of weaker internal 3′ splice site signals and less binding by the splicing factor U2AF65 ( REF. 44). The subset of lincRNAs processed most similarly to mRNAs may be more stable than other lincRNAs and have transcript-dependent roles. For example, Xist and Firre, which have well-characterized transcript functions, have greater splice site conservation and are spliced more efficiently than lincRNAs in general44. However, on lincRNAs, splicing efficiency does not correlate with ribosomal association52, suggesting that, unlike mRNAs, efficiently spliced lincRNAs are not better recognized by the translation machinery. The majority of divergently transcribed lincRNAs are spliced in their tissue of maximal expression, but their expression is not highly correlated with that of neighbouring coding genes, as half of the tissue-specific divergent lincRNAs are paired with ubiquitously expressed neighbouring coding genes; this finding implies that lincRNA processing does not invariably influence or reflect nearby gene expression28.
In addition to possessing unique splicing features, lincRNAs also influence the splicing of other RNAs by interacting with splicing factors or masking splicing signals61. For example, the lincRNAs MALAT1 ( REF. 62) and GAPLINC63 bind the splicing-associated factor PSF, suggesting that lincRNA-mediated splicing of other genes affects tumour invasion and metastatic capability by regulating oncogene or tumour suppressor activity. An intriguing class of lncRNAs termed 5′ small nucleolar RNA-capped and 3′ polyadenylated (SPAs) were recently implicated in the pathogenesis of Prader–Willi syndrome. SPAs affect the binding of several RNA-binding proteins, including TDP43 and RBFOX2, to their RNA interactors, thereby supporting alternative splicing patterns that are consistent with the disease state64. Additionally, principal component analysis of differential lncRNA gene expression and differential splicing in autism has revealed a high correlation between altered expression and splicing activity, again suggesting a role for lncRNAs in modulating splicing activity with functional impact65.
Stability and degradation
Whereas cytoplasmic mRNAs are often stable, the degree of stability of lincRNAs is less clear. Early work found relative lincRNA instability compared with mRNAs66, but more recent analyses of lincRNAs paired to mRNAs with similar expression levels identified similar levels of stability for lincRNAs and mRNAs44, suggesting that mRNAs expressed at relatively low levels that match lincRNA expression levels may be less stable than mRNAs expressed at relatively high levels. Somewhat surprisingly, ribosome footprinting analyses have identified robust cytoplasmic lncRNA–ribosomal association67. Over half of lncRNAs are found in the cytoplasm, and 70% of these lncRNAs have over half of their cytoplasmic transcripts associated with ribosomes52. Ribosome-associated lncRNAs have a long pseudo-5′-UTR cap structure and lack repetitive sequences. Ribosome engagement stabilizes some lncRNAs, as ribosome stalling following translation inhibition increases the stability of some ribosome-bound lncRNAs52. This engagement may additionally reflect the translation of smORFs into functional micropeptides68. Overall, cytoplasmic mRNAs are degraded by three main mechanisms: 3′ deadenylation, 5′ decapping followed by 5′-to-3′ exonuclease-mediated decay, and endoribonuclease cleavage. lincRNAs are degraded by these mechanisms but also by independent mechanisms, such as being targeted to the nuclear exosome17,53.
As they globally lack canonical ORFs and the ORFs that they do have contain premature stop codons, many translated lincRNAs undergo early translation termination and, like mis-translated mRNAs, may undergo nonsense-mediated decay69. lincRNAs can also undergo translation-independent destabilization, for example, by microRNAs (miRNAs; the miRNA let-7 destabilizes HOTAIR70), RNA-binding proteins (HuR promotes lincRNA decay71) and 3′-end processing72. It was recently proposed that lincRNAs that undergo sporadic transcription termination are targeted by the pre-miRNA-processing factor DGCR8 for degradation by the nuclear exosome (FIG. 1b), based on evidence that the inhibition of the nuclear exosome stabilizes nucleoplasmic lincRNAs53. An orthogonal study found that, following transcription inhibition, lincRNAs and mRNAs have indistinguishable half-lives44. Regarding transcript stabilization by HuR, mRNAs are preferentially bound by HuR at their 3′ ends, but lincRNAs are bound equivalently along their transcripts44, suggesting that differential HuR binding patterns do not influence transcript stability. Reconciling the apparent contradiction in lincRNA stability between the different studies may depend on analysing specific lincRNA subsets in greater detail rather than making global class assertions. It is possible that partially transcribed and poorly spliced lincRNAs are degraded by the nuclear exosome, whereas those processed more like mRNAs escape this fate, thus remaining cytoplasmic and stable.
Epigenetic regulation
LincRNAs and mRNAs can positively or negatively regulate the expression of their own genes, or target other genes, by interacting with chromatin-modifying complexes to modulate the epigenetic landscape of chromatin. lincRNA-coding genes and protein-coding genes are similarly globally enriched at their promoters in transcription-activating histone modifications such as H3K27ac, H3K4me3 and H3K9ac17,27, and many lincRNA genes are characterized by H3K4me3 at the transcription start site and H3K36me3 along the gene body. Conversely, a number of lincRNAs bind EZH2, the catalytic component of polycomb recessive complex 2 (PRC2), which deposits the repressive H3K27me3 modification73, although the specificity74 and requirement75,76 of lincRNA–PRC2 interactions for lincRNA function have recently been called into question. A subset of active lincRNA promoters are enriched in the repressive H3K9me3 modification, here associated with greater tissue specificity rather than differential expression; neither the importance nor mechanism of this modification are currently known44 (FIG. 1c). In mice, knockout of Dicer1, which encodes the miRNA-processing RNase Dicer, decreased lncRNA expression, particularly that of divergent transcripts. This effect is at least partially mediated by the activation by Dicer of oncogenic MYC, implicating miRNA and MYC circuitry in the maintenance of lncRNA expression, independently of mRNA regulation77. Reduced lncRNA expression in Dicer1-knockout mice was associated with decreased H3K4me3 and H3K36me levels at the downregulated lncRNA loci, suggesting that Dicer has a chromatin-modifying function that maintains lncRNA expression77. In yeast, four ATP-dependent chromatin remodelling factors — Isw2, Swr1, Ino80 and Rsc — repress the transcription of a set of antisense lncRNAs to regulate the expression of overlapping mRNAs78.
Tissue specificity and developmental patterning
LincRNAs often exhibit remarkable tissue specificity and may function to fine-tune the expression of their target genes in a tissue-specific manner. One study performed unsupervised clustering analyses to calculate the tissue specificity expression scores of individual transcripts. By this metric, 78% of lincRNAs were tissue-specific compared with 19% of mRNAs. This result was independent of expression differences between transcripts, as higher scores were assigned to more highly expressed lincRNAs28. Using RNA-sequencing (RNA-seq) data of 30 tissues from the most recent data release of the genotype-tissue expression (GTEx) consortium, a median tau score of tissue specificity of 0.90 was calculated for lincRNAs, compared with 0.77 for mRNAs (FIG. 2a), with 60.8% of lincRNAs and 29.4% of mRNAs found to be tissue-specific (FIG. 2b). The absolute number of lincRNAs with a distinct expression signature in a single tissue, defined by their tissue enrichment or depletion patterns, revealed an over-representation of tissue-specific lincRNAs in the brain and testis (FIG. 2c). Other studies have similarly found that one-third of lincRNAs have testis-specific expression28; these lincRNAs may tightly regulate reproductive functions, as do lincRNAs in yeast32 and C. elegans35. Recent work raised the possibility that this tissue specificity may reflect the function of nearly half of lincRNAs as e-lincRNAs, thus representing enhancer-specific cell type expression46.
Figure 2. Tissue specificity of long intergenic non-coding RNA (lincRNA) expression.
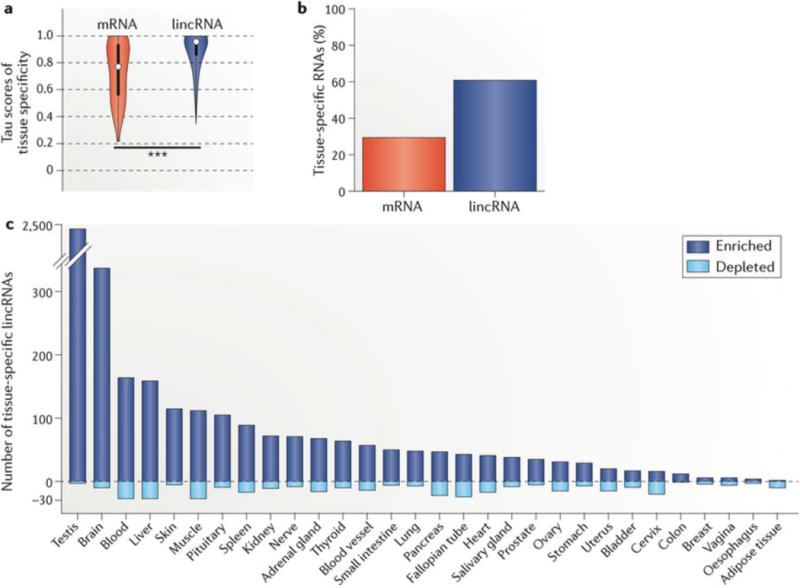
a | We calculated the individual tau (tissue specificity) scores, which range from 0 to 1 (0 for uniform expression; 1 for single-tissue expression), of 7,842 lincRNAs (blue) and 22,285 mRNAs (red) across 30 human tissues from the GTEx Analysis v6 data set (dbGaP Accession phs000424.v6.p1 based on GENCODE v19 (hg19; July 2013)). We additionally calculated tau scores after permutation of each RNA to its assigned tissue to estimate the false discovery rate (FDR). The results are depicted as a violin plot of tau score distributions, demonstrating greater tissue specificity for lincRNAs than mRNAs (median 0.90 for lincRNA; 0.77 for mRNA; 0.47 for permutation; one-sided Wilcoxon test, ***P < 0.001). The white dots represent the median; thick and thin bars represent one or two standard deviations from the median, respectively. b | Proportion of lincRNAs and mRNAs that are tissue-specific by tau score. c | For each lincRNA with significant tissue specificity (tau >0.88; FDR <0.05 by permutation distribution), we re-calculated its tau score 30 additional times, each time excluding one tissue to estimate the individual contribution of the tissue. If tissue specificity no longer reached significance with exclusion of one tissue, we designated the lincRNA as specific to that tissue. Given that such a lincRNA may have tissue specificity through enrichment or depletion, we also calculated the direction of its contribution (one-sided Student’s t-test, P < 0.05). Shown is the number of lincRNAs with a distinct expression signature in a single tissue owing to enrichment (positive y axis) or depletion (negative y axis), which highlights the abundance of tissue-specific lincRNAs in the testis and brain.
Protein-coding genes with a lincRNA located within 10 kb are enriched for transcription regulation and developmental patterning functions28. Although some lincRNAs demonstrate regulatory relationships with their neighbouring loci19, the overall expression of lincRNAs in proximity to coding genes is not invariably more correlated with that of their neighbours than is expression of adjacent coding loci28. The attempted characterization of lincRNA functions by clustering with expression-matched rather than proximal coding genes has specified lincRNAs with putative tissue-specific functions28. Furthermore, trait-associated lincRNAs are preferentially located at the boundaries of topologically associated domains and are frequently derived from evolutionarily conserved enhancer regions. Their expression correlates with protein-coding genes associated with the same traits, suggesting the presence of a shared cis regulatory mechanism that involves the modulation of chromatin architecture79 and that shared regulation and tissue expression may be related to shared function.
In summary, the tissue specificity of lincRNAs suggests that they fine-tune expression of other genes through physical proximity, similar expression patterns or shared phenotypic contributions28. Interestingly, the repressive H3K9me3 modification enriched at lincRNA gene promoters correlates with greater tissue specificity, not with lower expression as is the case for mRNAs44. Taken together, these data indicate that the specificity of lincRNA expression may promote tissue establishment and maintenance and that lincRNA functions may be intimately linked to those of mRNAs or other non-coding RNAs expressed in the same tissue.
The functions of lincRNAs
As of its latest data release (January 2015), the database lncRNAdb catalogued 156 human lncRNAs with a putative function. This represents a small minority of the thousands of annotated lncRNAs. In this section, we discuss the range of lincRNA functions described to date, which include regulating chromatin topology by both cis and trans mechanisms, scaffolding of proteins and other RNAs, acting as protein and RNA decoys, regulating neighbouring genes and producing micropeptides. lincRNAs function broadly to tune gene expression by directly affecting nuclear architecture and by sequestering intracellular molecules or promoting their function, as well as more indirectly via the effects of their transcription or translation. The emerging evidence for the existence of smORFs in a subset of annotated lincRNAs highlights the need to revise categorizations of some lincRNAs to coding RNAs. Notable examples of lincRNAs directly implicated in human disease pathogenesis and their potential as biomarkers and therapeutic targets have been reviewed elsewhere16,80 but are summarized in TABLE 2. Computational and methodological advances facilitating lincRNA annotation and functional characterization are summarized in TABLE 3.
Table 2.
LincRNAs in human disease and development
| LincRNA | Role | Refs |
|---|---|---|
| In disease | ||
| MALAT1 |
|
25,128 |
| PVT1 |
|
49,129 |
| LINK-A (LINC01139) |
|
26 |
| In development | ||
| Linc-RoR | Establishment and maintenance of pluripotency | 130,131 |
| TINCR | Promotion of epidermal differentiation | 48 |
| ANCR | Maintenance of epidermal progenitor state | 132 |
| BANCR | Melanoma cell migration | 133 |
| LincR-Ccr2-5′AS | T cell development | 134 |
| ALIEN | Cardiac lineage specification | 135,136 |
| PNKY | Neurogenesis | 42,137 |
| LincRNA-EPS | Immunomodulation | 138 |
| LINC00948; LINC00961 |
Micropeptide (myoregulin and SPAR) regulation of muscle activity and regeneration | 18,121 |
| As biomarkers | ||
| PCA3 (urine PCR) | Greater specificity and positive predictive value than prostate-specific antigen for prostate cancer | 139 |
| HULC (blood PCR) |
|
140,141 |
| HOTAIR (tumour PCR) |
|
142,143 |
| MALAT1 (tumour PCR) | Expression correlates with metastasis in early-stage non-small-cell lung cancer | 144 |
| As therapeutics (mechanisms and examples) | ||
| Post-transcriptional degradation or silencing by antisense pairing |
|
145–147 |
| Harnessing expression specificity for directed drug delivery | Delivery of diphtheria toxin to ovarian cancer cells under the control of the H19 promoter | 148 |
| Modulating action on target |
|
149,150 |
Long intergenic non-coding RNAs (lincRNAs) directly participate in ordered and disordered cellular and developmental processes. In cancer, lncRNA functions derive from copy number alteration, somatic mutation or differential regulation and promote tumorigenesis, metastasis and invasion16,80,135. LncRNA cancer16,80, cardiovascular and renal functions135 have been comprehensively reviewed; recent work has explored non-coding transcriptome alterations in autism65. We highlight several lincRNAs here that illustrate the spectrum of involvement in disease and development. There is emerging evidence for lincRNA variants implicated in depression and cancer, which may affect disease-implicated gene expression and disease subpathways151–153. Of lncRNAs, 18% have breast cancer subtype specificity, compared with 10% of protein-coding genes154, suggesting that their profiling would augment unknown primary tumour assignments and contribute to cancer subtype expression. We additionally provide an overview of lincRNAs as biomarkers and their therapeutic horizons. We direct readers elsewhere for a broader perspective on RNA therapies155,156. LNA, locked nucleic acid; PIP3, phosphatidylinositol-3,4,5-trisphosphate; SPAR, small regulatory polypeptide of amino acid response; Xi, inactive X chromosome.
Table 3.
Profiling lincRNA biogenesis, coding potential and interactomes
| Method | Description | Discovery |
|---|---|---|
| Protein-centric | ||
| nRIP, nRIP-seq |
|
|
| CLIP CLIP-seq, irCLIP | Firre interacts with hnRNPU to affect nuclear architecture81 | |
| RNA-centric | ||
| RNA pull-down | An RNA probe is used to isolate RNA with associated proteins | |
| ChIRP. dChIRP |
|
|
| CHART | Probes identified using RNase H are used to empirically determine hybridization region | Definition of MALAT1 genome occupancy; enrichment at active genes and for nuclear paraspeckle components162 |
| RAP | Oligonucleotide hybridization using long probes (>60 nucleotides); more stable isolation | |
With increasing evidence for long intergenic non-coding RNA (lincRNA)-encoded micropeptides, attention has turned towards estimations of small open reading frame coding potential within lincRNAs by sequence conservation164 or ribosomal profiling124,165. Orthogonal approaches to predict effects of non-coding variants using deep convolutional neural networks have been proposed166. Multiple methods to characterize lincRNA interactomes have been described; several are highlighted here. CHART, capture hybridization of RNA targets; ChIRP, chromatin isolation by RNA purification; CLIP, crosslinking immunoprecipitation; dChIRP, domain-specific ChIRP; HDAC3, histone deacetylase 3; irCLIP, infrared CLIP; lncRNA, long non-coding RNA; nRIP, native RNA precipitation; nRIP-seq, nRIP followed by sequencing; Pol II, RNA polymerase II; RAP, RNA antisense purification; SHARP, enhancer-of-split and hairy-related protein (also known as BHLHE41); Xi, inactive X chromosome.
Chromatin topology
LincRNAs can enforce both stable and repressive chromatin states — that is, those that increase or repress transcriptional activation. They can locally regulate chromatin structure in cis as well as inter-chromosomal nuclear architecture in trans16,21,80,81. Cis lincRNA-mediated chromatin interactions include chromatin looping82 and transcription activation83 or repression84 of target genes. Trans lincRNA-mediated chromatin interactions are broad and include regulation of co-expressed coding genes by chromosomal looping82 or by directly binding chromatin-modifying complexes74 and transcription factors85.
HOTTIP is a cis-acting lincRNA that promotes the expression of the gene HOXA. Transcribed from the 5′ tip of the HOXA locus, HOTTIP directly interacts at the HOTTIP locus with the adaptor protein WD repeat-containing protein 5, which is a component of the myeloid/lymphoid or mixed-lineage leukaemia protein 1 (MLL1; also known as KMT2A) histone lysine methyltransferase complex and, through chromatin looping, targets MLL1 to the HOXA locus — which is up to 40 kb away — thereby inducing H3K4 trimethylation and HOXA transcription82 (FIG. 3a). Further demonstrating its cis activity, HOTTIP depletion decreases HOXA expression, but not the expression of the highly homologous gene HOXD82.
Figure 3. The diverse functions of long intergenic non-coding RNAs (lincRNAs).
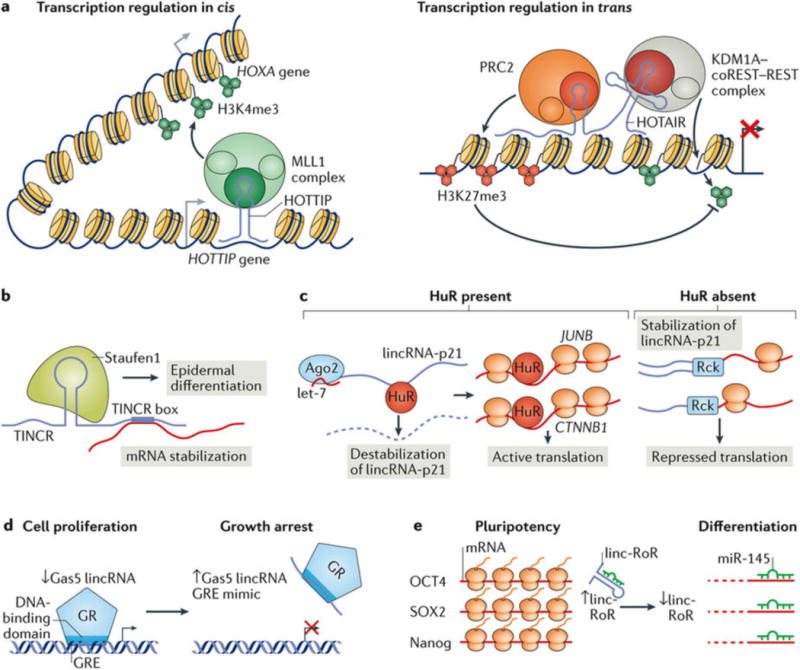
a | Regulation of chromatin structure and function in cis and in trans. HOTTIP associates with the myeloid/lymphoid or mixed-lineage leukaemia protein 1 (MLL1) complex, which catalyses histone H3 Lys4 trimethylation (H3K4me3) to activate HOTTIP transcription in cis (left). HOTAIR interacts in trans with the polycomb recessive complex 2 (PRC2) to mediate its deposition of the repressive H3K27me3 modification and with the KDM1A–CoREST–REST complex to mediate H3K4 demethylation, to coordinate transcription repression at target loci (right)125. b,c | LincRNAs scaffold proteins and RNAs in the nucleus and cytoplasm. b | TINCR binds Staufen1 in the cytoplasm, and binds and stabilizes mRNAs through its TINCR box motif to promote epidermal differentiation48. c | In the presence of HuR, lincRNA-p21 is destabilized by recruitment of the microRNA (miRNA) let-7 in complex with Argonaute 2 (Ago2). HuR association with the lincRNA-p21 target mRNAs JUNB and CTNNB1 results in their translation. In the absence of HuR, lincRNA-p21 remains stable, accumulates and associates with the JUNB and CTNNB1 transcripts in a mechanism that is at least partially mediated by co-association with the RNA-binding protein Rck and represses their translation by decreasing their ribosome association71. d,e | LincRNAs act as protein and RNA decoys. d | The expression of the lincRNA Gas5 is induced by growth arrest. Gas5 mimics the glucocorticoid response element (GRE) and binds the DNA-binding domain of the glucocorticoid receptor (GR), which sequesters the glucocorticoid receptor from its target genes103. e | Linc-RoR is abundant in pluripotent stem cells, where it acts as a decoy of the miRNA miR-145, thereby inhibiting the targeting and downregulation of the mRNAs of the pluripotency factors octamer-binding protein 4 (OCT4), the transcription factor SOX2 and homeobox protein Nanog. As linc-RoR levels decrease during differentiation, miR-145 is released and mediates the degradation of its targets to promote differentiation106. Part a is from REF. 125, Macmillan Publishers Limited.
LincRNAs can modulate developmentally regulated genes in trans45. For example, HOTAIR12, which is transcribed from the HOXC locus, silences HOXD as well as genes on other chromosomes86. HOTAIR scaffolds PRC2 with the KDM1A–coREST–REST complex, thereby inducing H3K27 trimethylation and H3K4 demethylation87, respectively, to coordinately repress transcription (FIG. 3a). More recently, the PRC2 dependency of HOTAIR-mediated transcriptional repression has been challenged by evidence that the HOTAIR–PRC2 interaction is dispensable for HOTAIR function76. Such findings suggest there is a need to broadly interrogate and revisit prior evidence for the specificity and functional relevance of lincRNA–PRC2 interactions75.
LincRNAs are themselves regulated by cis and trans mechanisms. For example, Xist orchestrates XCI in cis10. It is a 17 kb transcript88 expressed from the X inactivation centre89, which is a 500 kb region that also contains the Xist upstream regulatory non-coding RNAs Jpx and Ftx90. In female mammals, Xist is required for the random silencing of one of two X chromosomes, by spreading along and recruiting PRC proteins to the future inactive X chromosome (Xi)91. The Jpx transcript transactivates Xist on the Xi92 by titrating CTCF away93; conversely, on the active X, Xist is antagonized by its antisense non-coding RNA Tsix. Tsix induces chromatin asymmetry between the X chromosomes, establishing a binary Xist transcriptional state94,95. Xist nucleation on the Xi requires binding of the autosomal YY1 transcription factor to YY1-binding motifs at the repeat C domain of Xist85. Although Xist is stable and can diffuse in the nucleoplasm when not chromatin-bound, it is not promiscuous in its binding of chromatin, as is evident by the lack of Xist binding to YY1-binding motifs present at autosomes. This suggests that factors other than YYI contribute to the specificity of Xist tethering in cis85. Xist was recently demonstrated to affect nuclear architecture by directly binding the Lamin B receptor, which tethers Xist to the nuclear lamina, thereby limiting the mobility of Xi to sequester Xist-coated regions from the active X and maintain a repressive gene state96. The Xi is partitioned into two repressive chromosomal megadomains by the DXZ4 boundary element, which is required for the establishment of the Xi chromosomal conformation, but not its silencing97,98.
Recently, lincRNAs that utilize both cis and trans mechanisms of action were shown to have roles in the three-dimensional organization of the nucleus. The lincRNA Firre, which is required for adipogenesis, is localized to a 5 kb region around its transcription start site and is required for mediating inter-chromosomal binding in trans of at least five loci, thereby establishing nuclear compartments that may support its role in adipogenesis81. The nuclear matrix protein HNRNPU binds a 156-nucleotide repeat sequence in Firre and is required for the establishment of the multi-chromosome interactions, possibly by anchoring Firre to chromatin to initiate the formation of the nuclear compartment81.
Scaffolding and modulating the activity of proteins and RNA
LincRNAs may interact with nucleic acids through sequence-complementarity and with proteins through RNA structural elements. Protein scaffolding and modulation by lincRNAs often defines lincRNA function. In the nucleus, lincRNAs regularly scaffold PRC proteins, thereby affecting chromatin accessibility, nuclear architecture and gene expression. The non-coding RNA RepA, located in the Xist locus, directly binds EZH2, which is the catalytic subunit of PRC2; this scaffolding is required for initiating XCI and Xist spreading99. The Xist-bound proteome was recently described by mass spectrometry and implies the existence of a dynamic lincRNA–protein interaction landscape that changes during cellular differentiation100. In addition to the scaffolding by HOTAIR of PRC2 and chromatin at the HOXD locus12, more than 20% of lincRNA transcripts (and <2% of mRNAs) interact with PRC2 ( REF. 74), suggesting the existence of nonspecific PRC2 binding of RNA that warrants further investigation75.
Given their low abundance, particularly in the cytoplasm44,53, lincRNAs may efficiently and temporarily scaffold multiple proteins or participate in the formation of stable, low-abundance complexes. Cytoplasmic lincRNAs alter the stability48, degradation101 and translation status71 of target mRNAs. For example, TINCR scaffolds the RNA-binding protein staufen1 with epidermal differentiation-promoting mRNAs that bind the TINCR box motif, thereby facilitating their post-transcriptional stabilization and accumulation48 (FIG. 3b). Whereas in myogenesis staufen1 promotes both mRNA stabilization and decay, in epidermal differentiation its interaction with differentiation-induced RNAs promotes only mRNA stabilization by an unknown mechanism102. lincRNA-p21 binds the JUNB and CTNNB1 transcripts and represses their translation; this effect is countered by the binding of lincRNA-p21 by HuR, which recruits the RNA-induced silencing complex (RISC) to destabilize lincRNA-p21 and promote the translation of JUNB and CTNNB1 ( REF. 71) (FIG. 3c).
Protein and RNA decoys
LincRNAs can inhibit protein, mRNA and miRNA activity by their sequestration. The lincRNA Gas5 mimics the glucocorticoid response element (GRE) by forming a double-stranded structure that binds the DNA-binding domain of the glucocorticoid receptor. This interaction prevents the glucocorticoid receptor from binding its target genes and activating their transcription (FIG. 3d). Gas5 levels are low in proliferating cells and increase upon growth arrest, during which Gas5 represses GRE-mediated expression of enzymes involved in the rate-limiting steps of gluconeogenesis and glycogenolysis103. As part of the stress response to proteasome inhibition, upregulation of the lincRNA NEAT1 promotes the assembly of nuclear paraspeckles and the sequestration of transcription factors to control gene expression104. MALAT1, which is required for proper splicing, can similarly sequester splicing factors within nuclear paraspeckles to modulate splicing efficiency47.
Competing endogenous RNAs (ceRNAs) sequester mi RNAs from their mRNA targets, thereby increasing the expression of target mRNAs. The proposed ceRNA mechanism of function holds that some lincRNAs, pseudogene transcripts and circular RNAs accomplish this goal by containing multiple miRNA target sites that sequester miRNAs from binding RISC complexes and thus prevent the targeting and degradation of mRNAs105. This mechanism is well illustrated by linc-RoR, which maintains stem cell pluripotency. In pluripotent stem cells, linc-RoR sequesters miR-145, thereby promoting the accumulation of the pluripotency factors octamer-binding protein 4 (OCT4; also known as POU5F1), the transcription factor SOX2 and the homeobox protein Nanog, which are miR-145 targets (FIG. 3e). The levels of linc-RoR decrease during differentiation, and miR-145 is released and promotes the degradation of its pluripotency-promoting targets106. Similarly, the lincRNA TUG1, which associates with PRC2 in the nucleus, acts as a sponge of PTEN-targeting mi RNAs in the cytoplasm22, thereby adopting compartment-specific roles.
Regulators of neighbouring transcription
Less than 1% of lincRNAs have been functionally characterized, raising the possibility that many lincRNA transcripts have no intrinsic function. Although large-scale screening for ORFs through CRISPR–Cas9-mediated disruption is efficient, it does not naturally translate to lincRNA screening because most lincRNAs do not function via ORF-derived activity. Alternatively, CRISPR interference (CRISPRi), which employs a nuclease-dead, catalytically inactive Cas9 fused to a protein repressor domain to repress transcription, was recently used to screen 17,000 lncRNA loci across seven human cell lines for functions in cell growth. Impairing the transcription at 449 lincRNA loci affected cell growth; suppression of 89% of these lincRNAs produced phenotypes only in one cell type, in contrast to protein-coding genes, which generally share growth requirements across cell types. Disruption of 14 of these lincRNAs resulted in local transcriptional changes within 20 gene windows surrounding each lncRNA, suggesting that a small fraction of lncRNAs affect transcription locally20. This study underscores the importance of broadly interrogating both transcript-dependent and transcript-independent locus functions, as well as the importance of cellular context.
Another recent study explored sequence-dispensable lincRNA roles. Knockout of 5 of 12 loci in mouse cells affected gene expression in cis, largely owing to loss of enhancer-like activity from DNA elements in lincRNA promoters. A transcript-sequence-dependent function was dispensable at all loci. At one such lincRNA locus, Blustr, transcript length and the level of transcriptional activity correlated with the expression of the upstream Sfmbt2 locus, but Sfmbt2 activation did not require Blustr-specific sequences. Furthermore, the transcription of both Blustr and Sfmbt2 requires splicing only of the 5′ end of Blustr, implying that the spliceosome at the nascent Blustr transcript participates in Sfmbt2 regulation (FIG. 4a). Aside from 5′ splice site designation, this is a sequence-independent function associated with general Blustr transcriptional activity, which may include cofactor recruitment, altering of chromatin structure and transcription-promoting protein accumulation19.
Figure 4. Long intergenic non-coding RNAs (lincRNAs) that function through their transcriptional and translational activity.
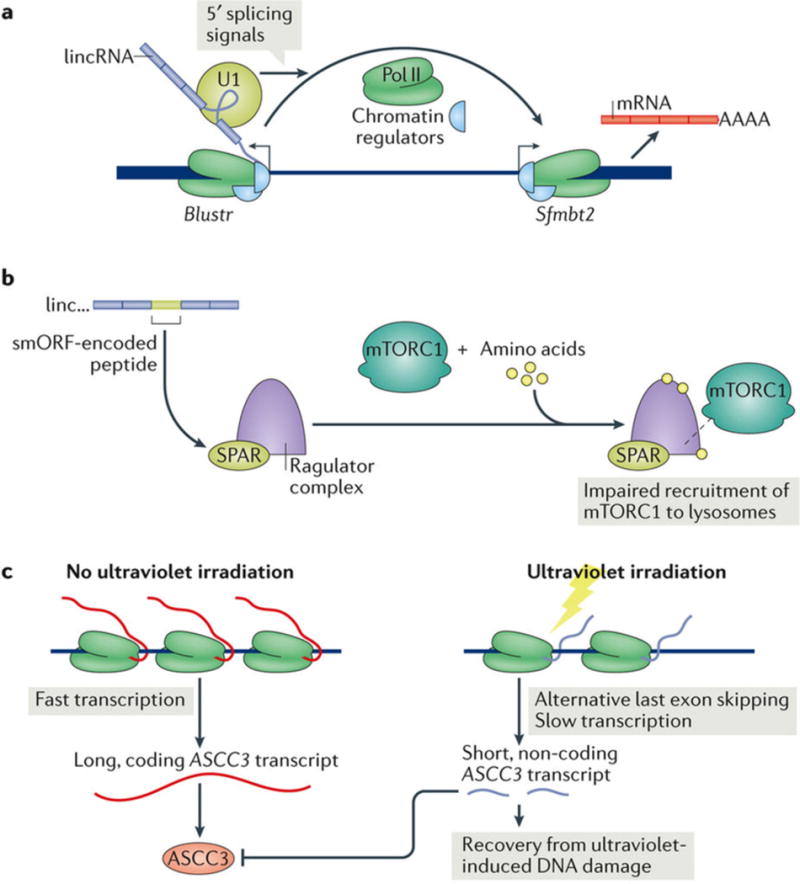
a | LincRNAs can regulate neighbouring genes by sequence-independent regulatory functions. In mice, splicing of the 5′ end of the lincRNA Blustr as well as general transcriptional activity at the Blustr locus activate the transcription of the upstream locus Sfmbt2, possibly by recruiting cofactors, accumulation of pro-transcription proteins and chromatin alteration19. b | Some lincRNAs encode micropeptides. A conserved, small open reading frame (smORF) in the lincRNA LINC00961 produces the peptide small regulatory polypeptide of amino acid response (SPAR). In the presence of amino acids, SPAR interacting with the Ragulator complex impairs the recruitment of mTORC1 to the lysosome and thus inhibits mTORC1 activation18. c | In the absence of ultraviolet irradiation, ASCC3 is efficiently transcribed to produce a long, coding transcript that produces the ASCC3 protein, which is a component of the ASCC complex that represses ultraviolet-induced DNA damage repair. Ultraviolet irradiation leads to slower transcription at the ASCC3 locus and induces the use of an alternative, proximal last exon and the expression of a short, non-coding RNA. This non-coding RNA antagonizes the function of the ASCC complex, thereby promoting recovery from ultraviolet-induced DNA damage24. Pol II, RNA polymerase II; U1, U1 small nuclear RNA.
Recently, the act of transcription was proposed to be the cause and not the consequence of nuclear conformation changes that affect gene expression107. Supporting this model, transcription from the active X chromosome is sufficient to maintain an open chromatin state108. lincRNA transcription may alter the epigenetic state at neighbouring coding loci, promoting interactions with nuclear-organizing protein complexes and obviating the requirement for lincRNA accumulation or stability, that is, even low levels of lincRNA transcription could promote the formation of multiple macromolecular complexes — consisting of proteins, nucleic acids, carbohydrates or lipids — without the lincRNAs themselves being in high abundance or comparatively stable. By contrast, macromolecular complex stabilization by lincRNAs would require high lincRNA abundance or stability. These findings underscore the need to perform genetic rescue experiments for any lincRNA with putative functions that are intrinsic to its RNA transcript.
The potential widespread function of lincRNAs as eRNAs has received increasing attention. Because they often do not overlap introns or exons of coding genes, many eRNAs are considered intergenic109. eRNAs can act by orchestrating enhancer-directed chromatin looping through mediator complex assembly110,111. Both PROMPTs and eRNAs may be the consequence of Pol II accumulation at the sites of transcription initiation of coding genes and may therefore not represent classical, independent transcriptional units53. In agreement with this, recent 5′ end lincRNA mapping indicated that 45.8% of the analysed lincRNAs originated from enhancers at open chromatin and may thus repre sent an important subclass of eRNAs46 (e-lincRNAs; TABLE 1). In this regard, lncRNAs encompassing disease-associated expression quantitative trait loci (eQTL) are expressed in disease-relevant cell types and co-expressed with their overlapping, eQTL-containing mRNAs; such e -lncRNAs are implicated in regulating the expression of their corresponding coding genes46.
Proteins such as the transcription factors OCT4, SOX2 and Nanog112 can regulate their own expression by interacting with cis-regulatory elements of their own loci. Because proteins need to be imported to the nucleus from the cytoplasmic sites of protein synthesis in order to directly regulate transcription, the mechanism of protein auto-regulation of transcription is not strictly in cis. lincRNAs, by comparison, are produced and can act in the nucleus and are therefore poised to more intimately regulate their own expression or the expression of nearby loci purely in cis17. A range of lncRNA cis-regulatory mechanisms of gene expression not yet described for the intergenic subset were recently reviewed17. They include regulation by eRNAs111, lncRNAs transcribed from imprinted loci113, autoregulatory lncRNAs and antisense lncRNAs17. Aside from antisense lncRNAs, which by definition overlap coding loci, these mechanisms may also be shared by lincRNAs.
Encoding functional micropeptides
Ribosome profiling with deep sequencing (Ribo–seq)67,114 and proteomic approaches115 have enabled the identification of functional smORF-encoded micropeptides of <100 codons in length derived from non-coding loci in flies, mice and humans115,116. Non-coding ORF-harbouring loci may constitute up to 10% of the genome, and hundreds to thousands of putative micropeptides originate from genes currently annotated as non-coding115,117.
The primary sequences of smORFs are less conserved than protein-coding genes but more conserved than introns50, non-ORF lincRNA transcript regions and lincRNA transcripts as a class. smORF regions are depleted of non-synonymous mutations and lack insertions and deletions, suggesting conservation at the peptide level118,119. Putative micropeptide-harbouring lincRNAs are expressed at higher levels and in a less restricted pattern than lincRNAs lacking smORFs119, implying that their translation or their peptide products may function more broadly than the transcription or transcripts of lincRNAs lacking smORFs. The most comprehensive smORF catalogue was derived from cross-species conservation-based in silico smORF predictions119. Other studies have reported varying putative lincRNA-derived smORF numbers without applying conservation criteria50. As the functional characterization of smORFs is in the early stages of experimentation, their quantification and definitions will be an exciting area of study.
Two polypeptides derived from annotated lincRNAs regulate the function of the SERCA membrane pump to control muscle relaxation: myoregulin and DWORF. Myoregulin is a conserved, transmembrane α-helix micropeptide encoded by LINC00948 and expressed exclusively in skeletal muscle. It directly interacts with SERCA to inhibit calcium uptake in the sarcoplasmic reticulum. DWORF is a human LOC100507537-encoded micropeptide that displaces SERCA-inhibitory peptides to increase sarcoplasmic reticulum calcium uptake and promote myocyte contractility120. The NoBody peptide is translated from the LINC01420 lincRNA and interacts with mRNA decapping proteins to control the number of P-bodies118. SPAR is a peptide conserved between humans and mice and encoded by LINC00961, which inhibits mTORC1 activation and muscle regeneration18 (FIG. 4b).
Importantly, only a small fraction of conserved lncORFs have peptidomic evidence in mass spectrometry data sets, probably underestimated by the origin of these data sets from only a few cell types119. We identified 1,179 lincRNAs that contain 1,432 conserved smORFs, which to date include six lincRNAs that produce six experimentally verified micropeptides. These lincRNAs include RP1-302G2.5 (84 aa product, unnamed); RP11-672F9.1 (40 aa product, unnamed); RP11-451G4.2 (34 aa product, DWORF); LINC00948 (also known as MRLN; 46 aa product, myoregulin); SPAAR (also known as LINC00961; 75 aa product, SPAR); and NBDY (also known as LINC01420; 68 aa product, NoBody). The lincRNAs encoding these smORFs should be more properly categorized as coding rather than non-coding genes18,68,81,118–121. Given that more than 80% of the transcript regions of these parental lincRNAs are non-coding — that is, lack translational activity — it is intriguing to speculate that some loci could support both coding and non-coding functions.
The majority of predicted smORF products lack homology with known peptides, which highlights the fact that conservation criteria may not appropriately define smORFs. Furthermore, little information exists on the functions of peptides with homologues, thereby necessitating de novo peptide characterization, which is not straightforward in the absence of a reliable screen119. Chemical biology expertise could greatly contribute to the identification of more micropeptides, the elucidation of their functions and the exploration of micropeptides as drug targets67.
By chance, any long-enough sequence will contain an ORF, although lincRNAs are defined by their lack of canonical ORFs. Can a lincRNA with coding capacity also have a non-coding function? A fascinating recent example is the switch induced by ultraviolet irradiation from the expression of long, protein-coding ASCC3 transcripts to the expression of a short non-coding ASCC3 isoform. The non-coding isoform antagonizes the ASCC3 protein to facilitate transcriptional recovery following ultraviolet-induced damage24 (FIG. 4c).
Conclusions and future perspectives
How can coding functions evolutionarily replace or exist alongside non-coding functions? Protein-coding genes may be under stringent selective pressure to retain coding capacity. During evolution, some loci may lose coding functions and gain non-coding functions, possibly subsequently to gene duplication. These non-coding loci, including lincRNAs, may regulate three-dimensional nuclear organization107 and affect gene expression solely by their transcriptional activity and may modulate protein and nucleic acid interactions directly by their transcripts. Some lincRNA loci may undergo additional changes to produce smaller RNAs or regain coding capacity and produce micropeptides122 that have peptide-dependent or translation-linked functions. In this fashion, a single locus may evolve both non-coding and coding regulatory capacities123. The act of lincRNA transcription and translation, as well as the resulting products, may thus evolve to regulate activity at other coding loci124.
The lincRNA landscape continues to expand and exhibit the diversity of identity and function of lincRNAs. Recent work has demonstrated the dispensability of some lincRNA transcripts for local gene regulation, their role in encoding micropeptides and their implication as enhancer RNAs. Modelling lincRNAs as genetic repositories with the capacity to develop multiple functions, freed from the evolutionary constraints of mRNAs, offers a framework for integrating the diverse functions of the non-coding transcriptome.
Acknowledgments
This work was supported by the US Department of Veterans Affairs (USVA) Office of Research and Development and by the US National Institutes of Health (NIH) AR049737 to P.A.K. and by a Howard Hughes Medical Institute (HHMI) Medical Research Fellowship award to J.D.R. We thank H. Y. Chang and members of the Khavari laboratory for helpful discussions.
Glossary
- Tiling arrays
A method used to probe the transcriptome without prior knowledge of the transcribed loci by hybridizing it to DNA or RNA probes.
- Dauer
An alternative developmental stage in nematodes induced by nutrient starvation and characterized by distinct metabolic and morphological features.
- Prader–Willi syndrome
A multisystem, genetic disorder secondary to maternal uniparental disomy of the long arm of chromosome 15, or disruption or deletion of the paternal long arm of chromosome 15; characterized by decreased fetal activity, hyperphagia, short stature, mental retardation and hypogonadotropic hypogonadism.
- Pseudo-5′-UTR
The region immediately upstream of the first AUG sequence in a long intergenic non-coding RNA.
- Tau score
A rank correlation coefficient used to conduct a non-parametric hypothesis test for the statistical correlation between data variables.
- Nuclear paraspeckles
Mammalian nuclear organelles, the formation of which is dependent on non-coding RNA species that may have a role in nuclear retention of translatable RNA.
- Circular RNAs
Conserved RNAs formed by pre-mRNA back splicing, the function of which may be linked to that of their host genes.
- Mediator complex
A conserved protein complex involved in transcription regulation and with multiple roles in gene regulation.
- Expression quantitative trait loci
(eQTL). Genetic sequence variants associated with expression changes in one or more local or distant genes; that is, genetic variants that may account for variable gene expression levels.
- P-bodies
Cytoplasmic aggregates of messenger ribonucleoproteins associated with mRNA decay machinery and translation inhibition.
Footnotes
Author contributions
J.D.R., Y.W. and P.A.K. researched data for the article. J.D.R. and P.A.K. made substantial contributions to discussions of the content and wrote and edited the manuscript before submission.
Competing interests statement
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venter CJ, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 4.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 5.Rinn JL, et al. The transcriptional activity of human chromosome 22. Genes Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda N, et al. Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS Genet. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salditt-Georgieff M, Darnell JE. Jr Further evidence that the majority of primary nuclear RNA transcripts in mammalian cells do not contribute to mRNA. Mol Cell Biol. 1982;2:701–707. doi: 10.1128/mcb.2.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 10.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 11.Brockdorff N, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 12.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. This work uses tiling microarrays to discover hundreds of non-coding RNAs transcribed from HOX gene clusters, focusing on HOTAIR, which regulates HOX gene expression in trans on distant chromosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulitsky I, Bartel DP. LincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–1465. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto A, et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–232. doi: 10.1038/nature21034. This paper describes a lincRNA-encoded micropeptide, termed SPAR, which derives from a highly conserved region of its parental lincRNA and functions in muscle regeneration distinctly from its lincRNA transcript, implying a coding function from a previously annotated non-coding transcript. [DOI] [PubMed] [Google Scholar]
- 19.Engreitz JM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. This paper dissects 12 lincRNA loci, identifying five that regulate the activity of neighbouring genes via their transcriptional activity, in largely transcript-independent roles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu SJ, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:aah7111. doi: 10.1126/science.aah7111. CRISPRi employs a nuclease-dead Cas9 fused to a repressor domain to create a transcriptional roadblock around the transcription start site of a target region. It is used here to interrogate and screen for cis-acting and trans-acting lncRNA products, function secondary to transcriptional activity, and enhancer-like lncRNA function; the authors identified 449 lncRNA loci that affect cancer cell line growth by one or several of these mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18:499–508. doi: 10.1038/ni.3712. [DOI] [PubMed] [Google Scholar]
- 22.Du Z, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun. 2016;7:10982. doi: 10.1038/ncomms10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson KM, et al. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williamson L, et al. UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell. 2017;168:843–855.e13. doi: 10.1016/j.cell.2017.01.019. This paper describes dual coding and non-coding functions of the ASCC3 locus, with a switch from long, coding transcript production induced by ultraviolet irradiation to produce the short, non-coding RNA product. The non-coding RNA promotes recovery from ultraviolet-induced DNA damage, antagonizing the function of the protein product of the coding transcript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khurana E, et al. Role of non-coding sequence variants in cancer. Nat Rev Genet. 2016;17:93–108. doi: 10.1038/nrg.2015.17. [DOI] [PubMed] [Google Scholar]
- 26.Lin A, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol. 2017;19:238–251. doi: 10.1038/ncb3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014;30:121–123. doi: 10.1016/j.tig.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Hezroni H, et al. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11:1110–1122. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita A, Shichino Y, Yamamoto M. The long non-coding RNA world in yeasts. Biochim Biophys Acta. 2016;1859:147–154. doi: 10.1016/j.bbagrm.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, et al. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–4345. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Yamada M, Han X, Ohler U, Benfey PN. High-resolution expression map of the arabidopsis root reveals alternative splicing and lincRNA regulation. Dev Cell. 2016;39:508–522. doi: 10.1016/j.devcel.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nam JW, Bartel DP. Long noncoding RNAs in C. elegans. Genome Res. 2012;22:2529–2540. doi: 10.1101/gr.140475.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 37.Church DM, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauvageau M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 41.Chodroff RA, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goff LA, et al. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2015;112:6855–6862. doi: 10.1073/pnas.1411263112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melé M, et al. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017;27:27–37. doi: 10.1101/gr.214205.116. This detailed study of lincRNAs explores their pre-transcriptional and post-transcriptional regulation across human cell lines, identifying H3K9me3 as enriched at lincRNA promoters and associated with tissue specificity, as well as equivalent U1 binding enrichment at lincRNAs and mRNAs, suggesting that factors other than U1 mediate lincRNA splicing inefficiency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hon CC, et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. This work describes the first lincRNA involved in terminal epithelial differentiation. It functions by scaffolding staufen1 and stabilizing differentiation-promoting mRNAs by their interaction with its 25-nucleotide ‘TINCR box’ motifs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tseng YY, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slavoff SA, et al. Peptidomic discovery of short open reading frame–encoded peptides in human cells. Nat Chem Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu SJ, et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:67. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlevaro-Fita J, Rahim A, Guigo R, Vardy LA, Johnson R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA. 2016;22:867–882. doi: 10.1261/rna.053561.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlackow M, et al. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol Cell. 2017;65:25–38. doi: 10.1016/j.molcel.2016.11.029. This study applies mNET-seq to analyse 285 highly expressed lincRNAs, identifying weak co-transcriptional lincRNA splicing and nuclear exosomal depletion, suggesting that stable lincRNAs escape these fates and are instead more mRNA-like. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukherjee N, et al. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat Struct Mol Biol. 2017;24:86–96. doi: 10.1038/nsmb.3325. [DOI] [PubMed] [Google Scholar]
- 55.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 56.Jensen TH, Jacquier A, Libri D. Dealing with pervasive transcription. Mol Cell. 2013;52:473–484. doi: 10.1016/j.molcel.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 57.Kowalczyk MS, et al. Intragenic enhancers act as alternative promoters. Mol Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 58.Descostes N, et al. Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalian cells. eLife. 2014;3:e02105. doi: 10.7554/eLife.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marquardt S, et al. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell. 2014;157:1712–1723. doi: 10.1016/j.cell.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szczesniak MW, Makałowska I. LncRNA-RNA interactions across the human transcriptome. PLoS ONE. 2016;11:e0150353. doi: 10.1371/journal.pone.0150353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji Q, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang P, et al. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget. 2016;7:42183–42194. doi: 10.18632/oncotarget.9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu H, et al. Unusual processing generates SPA lncRNAs that sequester multiple RNA binding proteins. Mol Cell. 2016;64:534–548. doi: 10.1016/j.molcel.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 65.Parikshak NN, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540:423–427. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clark MB, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Couso JP, Patraquim P. Classification and function of small open reading frames. Nat Rev Mol Cell Biol. 2017;18:575–589. doi: 10.1038/nrm.2017.58. This Analysis-type Review characterizes lncORFs as lncRNAs (inclusive of lincRNAs) that give rise to smORFs with some coding potential. A small but growing number of these ORFs give rise to peptides with biological functions, suggesting that the lincRNAs from which they are derived are more properly characterized as coding than non-coding; we term the peptide-producing ORFs described herein as lincORFs. [DOI] [PubMed] [Google Scholar]
- 69.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 70.Yoon JH, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoon JH, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. tRNAs marked with CCACCA are targeted for degradation. Science. 2011;334:817–821. doi: 10.1126/science.1213671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blanco MR, Guttman M. Re-evaluating the foundations of lncRNA-Polycomb function. EMBO J. 2017;36:964. doi: 10.15252/embj.201796796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Portoso M, et al. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017;36:981–994. doi: 10.15252/embj.201695335. This paper challenges evidence that HOTAIR functions through a PRC2-dependent mechanism to repress target gene transcription. It calls into question more broadly the specificity and dependence of lincRNA interactions, specifically lncRNA–PRC2 interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng GX, Do BT, Webster DE, Khavari PA, Chang HY. Dicer-microRNA-Myc circuit promotes transcription of hundreds of long noncoding RNAs. Nat Struct Mol Biol. 2014;21:585–590. doi: 10.1038/nsmb.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alcid EA, Tsukiyama T. ATP-dependent chromatin remodeling shapes the long noncoding RNA landscape. Genes Dev. 2014;28:2348–2360. doi: 10.1101/gad.250902.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan JY, et al. Cis-acting complex-trait-associated lincRNA expression correlates with modulation of chromosomal architecture. Cell Rep. 2017;18:2280–2288. doi: 10.1016/j.celrep.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 80.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 81.Hacisuleyman E, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. This paper describes how the lincRNA Firre binds hnRNPU through a 156-nucleotide repeat domain to affect nuclear chromosomal architecture, which indicated that lincRNAs are involved in higher-order nuclear organization via RNA–protein and RNA–DNA interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dimitrova N, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maamar H, Cabili MN, Rinn J, Raj A. Linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 2013;27:1260–1271. doi: 10.1101/gad.217018.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brown CJ, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 89.Lee JT, Strauss WM, Dausman JA, Jaenisch RA. 450 kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- 90.Barakat TS, et al. The trans-activator RNF12 and cis-acting elements effectuate X chromosome inactivation independent of X-pairing. Mol Cell. 2014;53:965–978. doi: 10.1016/j.molcel.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 91.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 92.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun S, et al. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stavropoulos N, Lu N, Lee JT. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci USA. 2001;98:10232–10237. doi: 10.1073/pnas.171243598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 96.Chen CK, et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354:468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- 97.Giorgetti L, et al. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535:575–579. doi: 10.1038/nature18589. References 96 and 97 describe how the lincRNA Xist is divided into two repressive megadomains by the DXZ4 microsatellite region, which is required for maintenance of chromosomal architecture but, interestingly, not for Xist-mediated silencing activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Darrow EM, et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci USA. 2016;113:E4504–E4512. doi: 10.1073/pnas.1609643113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chu C, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gong C, Maquat LE. LncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kretz M. TINCR, staufen1, and cellular differentiation. RNA Biol. 2013;10:1597–1601. doi: 10.4161/rna.26249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kino T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hirose T, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 106.Wang Y, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 107.Melé M, Rinn JL. “Cat’s cradling” the 3D genome by the act of LncRNA transcription. Mol Cell. 2016;62:657–664. doi: 10.1016/j.molcel.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 108.Naughton C, Sproul D, Hamilton C, Gilbert N. Analysis of active and inactive X chromosome architecture reveals the independent organization of 30 nm and large-scale chromatin structures. Mol Cell. 2010;40:397–409. doi: 10.1016/j.molcel.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 110.Kung JT, Lee JT. RNA in the loop. Dev Cell. 2013;24:565–567. doi: 10.1016/j.devcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang X, et al. Maternally expressed gene 3, an imprinted non-coding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70:2350–2358. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saghatelian A, Couso JP. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat Chem Biol. 2015;11:909–916. doi: 10.1038/nchembio.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kondo T, et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 117.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 118.D’Lima NG, et al. A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol. 2017;13:174–180. doi: 10.1038/nchembio.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mackowiak SD, et al. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol. 2015;16:179. doi: 10.1186/s13059-015-0742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nelson BR, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Anderson DM, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu X, Sharp PA. Divergent transcription: a driving force for new gene origination? Cell. 2013;155:990–996. doi: 10.1016/j.cell.2013.10.048. This perspective piece puts forward the hypothesis that, because lncRNAs are under less stringent evolutionary pressure than are coding genes, they may function as ‘genetic repositories’ for continued biological adaptation and for novel gene derivation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dinger ME, Gascoigne DK, Mattick JS. The evolution of RNAs with multiple functions. Biochimie. 2011;93:2013–2018. doi: 10.1016/j.biochi.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 124.Fields AP, et al. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol Cell. 2015;60:816–827. doi: 10.1016/j.molcel.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 126.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.FANTOM Consortium and the RIKEN PMI and CLST (DGT) et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Millis MP, Bowen D, Kingsley C, Watanabe RM, Wolford JK. Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes. 2007;56:3027–3032. doi: 10.2337/db07-0675. [DOI] [PubMed] [Google Scholar]
- 130.Loewer S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guttman M, et al. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kretz M, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Flockhart RJ, et al. BRAF(V600E) remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22:1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hu G, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14:1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol. 2016;12:360–373. doi: 10.1038/nrneph.2016.51. [DOI] [PubMed] [Google Scholar]
- 136.Kurian L, et al. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 2015;131:1278–1290. doi: 10.1161/CIRCULATIONAHA.114.013303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ramos AD, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Atianand MK, et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165:1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hessels D, Schalken JA. The use of PCA3 in the diagnosis of prostate cancer. Nat Rev Urol. 2009;6:255–261. doi: 10.1038/nrurol.2009.40. [DOI] [PubMed] [Google Scholar]
- 140.Panzitt K, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 141.Yu X, Zheng H, Chan MTV, Wu WK. HULC: an oncogenic long non-coding RNA in human cancer. J Cell Mol Med. 2017;21:410–417. doi: 10.1111/jcmm.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Serghiou S, Kyriakopoulou A, Ioannidis JP. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol Cancer. 2016;15:50. doi: 10.1186/s12943-016-0535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Teschendorff AE, et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med. 2015;7:108. doi: 10.1186/s13073-015-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 145.Roux BT, Lindsay MA, Heward JA. Knockdown of nuclear-located enhancer RNAs and long ncRNAs using locked nucleic acid gapmeRs. Methods Mol Biol. 2017;1468:11–18. doi: 10.1007/978-1-4939-4035-6_2. [DOI] [PubMed] [Google Scholar]
- 146.Modarresi F, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Meng L, et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mizrahi A, et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med. 2009;7:69. doi: 10.1186/1479-5876-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci USA. 2010;107:22196–22201. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jiang J, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ye N, et al. Intergenic variants may predispose to major depression disorder through regulation of long non-coding RNA expression. Gene. 2017;601:21–26. doi: 10.1016/j.gene.2016.11.041. [DOI] [PubMed] [Google Scholar]
- 152.Jendrzejewski J, et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci USA. 2012;109:8646–8651. doi: 10.1073/pnas.1205654109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xu Y, et al. LncSubpathway: a novel approach for identifying dysfunctional subpathways associated with risk lncRNAs by integrating lncRNA and mRNA expression profiles and pathway topologies. Oncotarget. 2017;8:15453–15469. doi: 10.18632/oncotarget.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yan X, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sullenger BA, Nair S. From the RNA world to the clinic. Science. 2016;352:1417–1420. doi: 10.1126/science.aad8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127:761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zarnegar BJ, et al. irCLIP platform for efficient characterization of protein-RNA interactions. Nat Methods. 2016;13:489–492. doi: 10.1038/nmeth.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 159.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Quinn JJ, et al. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32:933–940. doi: 10.1038/nbt.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.West JA, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Olexiouk V, et al. sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2016;44:D324–D329. doi: 10.1093/nar/gkv1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kelley DR, Snoek J, Rinn JL. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016;26:990–999. doi: 10.1101/gr.200535.115. [DOI] [PMC free article] [PubMed] [Google Scholar]


