Abstract
Objective
For the past several decades, the resins used in dental restorations have been plagued with numerous problems, including their implication in biofilm formation and secondary caries. The need for alternative resins is critical, and evaluation of biofilm formation on these resins is essential. The aim of this study was to evaluate in vitro biofilm formation on the surface of novel copper (I)-catalyzed azide-alkyne cycloaddition (CuAAC)-based resins and composites.
Methods
CuAAC-based resins/composites made from varying azide monomers and different copper concentrations were compared with BisGMA-TEGDMA resins/composites that served as the control. Biofilms were formed using a mono-species model containing a luciferase-expressing strain of Streptococcus mutans. Luciferase activity was measured and the number of viable bacteria was enumerated on biofilms associated with each resin and composite.
Results
A significant reduction (p < 0.05) in luciferase activity, and the number of viable bacteria recovered from biofilms on CuAAC-based resins and composites was observed in comparison to biofilms associated with the BisGMA-TEGDMA controls.
Significance
CuAAC-based resins do still allow for the formation of biofilms; however, the statistically significant reduction of growth that was associated with the CuAAC resin may enhance the longevity of restorations that incorporate CuAAC-based materials.
Keywords: Dental materials, biofilm, Streptococcus mutans, dental resin composites, Copper-catalyzed azide-alkyne polymerization
1. Introduction
The oral cavity is a complex environment where over 700 bacterial species have been detected in the oral common microbiota [1–4]. Within this diverse community of bacteria found in the mouth, Streptococcus mutans, due to its acidogenic nature and its ability to form biofilms on tooth surfaces, is one of the primary species associated with human dental caries and secondary caries formation [5, 6]. Recent studies have also indicated that numerous other oral bacteria, most notably those that are acid producing, work together to form polymicrobial biofilms that ultimately initiate and further develop tooth decay [7–10]. In fact, Lactobacillus acidophilus, another commonly found oral acid-producing bacterium, has been found in high numbers in both superficial and deep dental caries [11, 12], and its ability to form biofilms on tooth surfaces is enhanced by the presence of S. mutans, augmenting the ability to cause carious lesions [13, 14].
More than 100 million dental restorations are performed each year and over 50% of these restorations utilize resin-based composites over amalgams [15–17]. Resin-based composites have many benefits over amalgams, including improved aesthetics, adhesive strength, and filling capability [16–18]. However, resin-based restoratives frequently have limited longevity due to restoration failure, commonly caused by degradation or fracture of the restoration directly or by failure due to secondary caries formation at the margins around the restoration [17, 19–21]. Nearly all resin-based composite restoratives are methacrylate-based and consist of a comonomer mixture comprised from components such as 2,2-bis [4-(2-hydroxy-3-methacryloxypropoxy) phenyl] propane (BisGMA) and triethylene glycol dimethacrylate (TEGDMA) [18, 22] or related monomers. The functional integrity of these methacrylate restoratives relies on the polymerization of resin monomers, and their limited conversion leads not only deterioration of the restorative, but also to release of these monomers into the surrounding tissues [17, 18, 23, 24]. Additionally, numerous studies have demonstrated sensitivity of these restorations in general, and these methacrylate monomers specifically, to hydrolysis by salivary and bacterial esterases found in the oral cavity, resulting in biodegradation by-products (BBPs) [25–31]. The biodegradation of these restorations increases bacterial leakage between resin-dentin interfaces, leading to further damage to the tooth [32]. Additionally, residual monomers released from resin restorations and resulting BBPs, such as methacrylic acid (MA), bishydroxypropoxyphenyl-propane (Bis-HPPP), and triethylene glycol (TEG), have been thoroughly investigated and implicated in adverse manifestations in the host. In particular, these effects include disruption of immune function [33–37], cytotoxicity [38–41], microbiota shifts [42], and accelerated formation of biofilms [43–46].
Due to the various pitfalls associated with the currently used methacrylate-based composites, the development of new, longer-lasting polymers for dental restoration is of utmost importance and could have a significant positive impact on global oral health. Since bacterial accumulation and biofilms have been implicated in the deterioration of the current BisGMA-TEGDMA based composites, novel resins that have structural stability and also limit bacterial prevalence are desirable to prolong the longevity of dental resins. Recently, novel resins have been developed that specifically have antibacterial properties to resist biofilm formation and incorporate antimicrobial monomers such as novel quaternary ammonium methacrylates [47–49], methacryloxylethylcetyl dimethyl ammonium chloride [50], and 12-methacryloyloxydodecylpyridinium bromide (MDPB) [51–54]. Additional strategies have also included the inclusion of alternative antimicrobial agents such as fluoride [49, 55, 56], silver nanoparticles [57–60], and chlorhexidine [61–63], to name a few. While providing additional benefits over the currently used resin-based composite systems, many of the approaches listed above continue to rely on the existing methacrylate system and, as such, have been plagued with similar challenges.
Recently, the development and analysis of novel visible light-initiated copper-catalyzed azide-alkyne cycloaddition (CuAAC)-based resins that possess superior mechanical properties, significantly reduced shrinkage stress and suitable polymerization kinetics as compared to BisGMA-based polymers [64–70]. However, the ability of these resins to promote or restrict bacterial growth has not been evaluated. Therefore, the aim of this study was to evaluate in vitro biofilm formation on the surface of novel copper (I)-catalyzed azide-alkyne cycloaddition (CuAAC)-based resins.
2. Materials/Methods
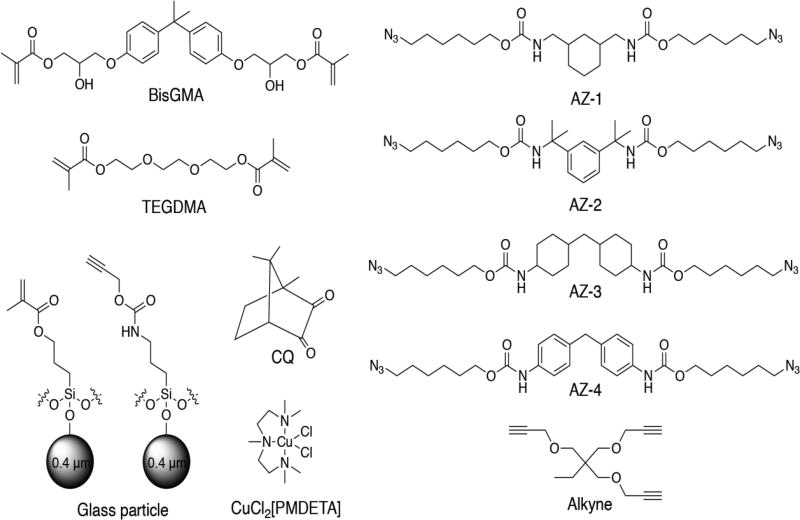
1,3-Bis(isocyanatomethyl)cyclohexane, 1,3-Bis(2-isocyanatopropan-2-yl)benzene, 4,4-methylenebis(cyclohexyl isocyanate), 4,4′-methylenebis(phenyl isocyanate), dibutyltin dilaurate, tetrahydrofuran, 6-chloro-1-hexanol, sodium azide, 1,1,1-tris(hydroxymethyl)propane, propargyl bromide, propargyl alcohol, 3-(triethoxysilyl)propyl isocyanate, copper(II) chloride, N,N,N′, N′, N ″-pentamethyldiethylenetriamine (PMDETA), camphorquinone (CQ), toluene, and acetonitrile were used as received from Sigma Aldrich. Propylamine, sodium hydroxide, dimethyl sulfoxide, dimethylformamide, methanol, and sodium sulfate were used as received from Fisher Scientific. The BisGMA/TEGDMA (70/30) comonomer mixture was used as donated from ESSTECH. Schott glass (mean particle size of 0.4 µm) with both untreated surface and surface treated with γ–methacryloxypropyltrimethoxysilane were used as received from ESSTECH. Syntheses of the azide monomers (AZ-1, AZ-2, AZ-3, AZ-4) and the alkyne monomer and alkyne silanization on glass microparticle (0.4 µm) were performed according to previously reported procedures [64, 66]. All synthetic procedures along with NMR predictions and particle functionalization are provided in the Supporting Information. Structures for BisGMA and TEGDMA monomers used for methacrylate polymerization, the various azide monomers (AZ-1, AZ-2, AZ-3, AZ-4) and the alkyne monomer used for CuAAC polymerizations, as well as the copper catalyst CuCl2[PMDETA] and photoinitiator CQ used in this study are shown in Fig. 1. Azides were synthesized according to the azide safety rules and handled with appropriate precaution when working with monomers, resins, and polymers in small quantities [71].
Fig. 1.
BisGMA and TEGDMA monomers used for methacrylate polymerization. Various structures of azide monomers, AZ-1, AZ-2, AZ-3, AZ-4, and alkyne monomer used for CuAAC polymerization. Copper catalyst CuCl2[PMDETA], photoinitiator CQ, and glass microparticle functionalized with methacrylates or alkynes are shown.
2.1 Preparation of BisGMA-TEGDMA and CuAAC resin and composite samples
2.1.1 BisGMA-TEGDMA and CuAAC resin formulation
A BisGMA:TEGDMA (70:30 weight ratio) mixture with 2 weight percentage of CQ was prepared by physical mixing. Stoichiometric mixtures of a diazide, trialkyne (a mole ratio of 1:1 to N3:alkyne), with different mole percentage of CuCl2[PMDETA] per functionality and 2 mole percentage of CQ were prepared, and methanol was added to homogenize the mixture and later removed in vacuo.
2.1.2 BisGMA-TEGDMA and CuAAC composite formulation
A BisGMA-TEGDMA (70:30 weight ratio) mixture with 2-weight percentage of CQ was prepared by physical mixing. 60-weight percentage of methacrylated microfillers (Schott, 0.4 µm) were added to the resin mixture and blended in a speedmixer (DAC 150 FVZ, Flakteck) to ensure uniform dispersion of the fillers. Stoichiometric mixtures of the AZ-2 diazide, trialkyne (a mole ratio of 1:1 to N3:alkyne), with 2 mole percentage of CuCl2[PMDETA] per functionality and 2 mole percentage of CQ were prepared. Methanol was added to homogenize the mixture and later removed in vacuo. 60-weight percentage of alkyne-functionalized microfillers (Schott, 0.4 µm) were added to the resin mixture and blended in a speedmixer (DAC 150 FVZ, Flakteck) to ensure uniform dispersion of fillers.
2.1.3 Photo-curing and thermal treatment
A round disc-shaped HDPP (high density polypropylene) mold (7mm in diameter and 1mm in thickness) was glued on a glass slide, which was previously treated with Rain-x (TM of Illinois Tool Works Inc.). Each resin/composite mixture was poured into the mold, and the top surface was covered with a glass slide. Each sample was irradiated from the top with 300 mW/cm2 of 455 nm light for 5–10 min and post-cured in the oven at 70 °C overnight. The sample was removed from the mold and further post-cured at 100 °C for 5 h to ensure the maximum degree of polymerization. This step was performed in order to eliminate any potential extractables/leachables that might otherwise influence the biofilm formation. It should be noted that a high-temperature post-curing protocol was utilized in the study, which is not intended to represent a practical approach for clinical implementation; however, the greater extent of polymerization can minimize potential extractables/leachables and substantially reduce free monomers/oligomers during the biofilm formation. In this manner, the biofilm formation should primarily be influenced by the surfaces rather than any residual, extractable monomers. An FT-IR spectrometer was utilized to monitor the functional group conversion of each system (6250-6096 cm−1 for methacrylate and 6538-6455 cm−1 for alkyne) before photopolymerization and after thermal treatment to verify quantitative conversion (see Fig. S1) of each functional group.
2.1.4 Surface roughness of resins and composites
A stylus profiler (Veeco Dektak 6M) was utilized to measure 1D surface features of four different formulations (CuAAC AZ-2 resin, CuAAC AZ-2 composite, BisGMA-TEGDMA resin, and BisGMA-TEGDMA composite). The tip (radius of 12.5 µm) was scanned across both top and bottom surfaces of each disc (7mm in diameter and 1mm in thickness) with a scan resolution of 0.513µm/sample. From the 1D surface feature of each formulation (see Fig. S2), smooth surfaces with less than 2% variation on the surface height were observed in all of formulations tested, indicating the influence of surface roughness on biofilm formation is likely to be negligible. However, it should be noted that while one side has a smooth surface, the other side of each disc has a mildly rough surface presumably due to contraction from volumetric shrinkage or small bubble formations. To avoid this effect, the smoother surface of each disc was utilized in biofilm formation.
2.2 Bacterial growth conditions
A marker-less firefly luciferase expressing derivative of S. mutans UA159 in which luciferase expression is under the control of the lactate dehydrogenase promoter (ldh) [72], referred to as S. mutans firefly herein, was grown either on Brain Heart Infusion (BHI) agar (BBL; Becton Dickenson, Cockeysville, MD, USA), in BHI broth, or in BHI broth supplemented with 1% sucrose at 37°C with 5% CO2.
2.3 In vitro biofilm formation
To evaluate biofilm formation on novel CuAAC-based resins, biofilms were formed using a monospecies biofilm model containing S. mutans firefly. Briefly, BisGMA-TEGDMA and CuAAC-based resin discs (n=12 discs for each sample and control) were sterilized in 70% alcohol containing 0.5% triclosan for 30 min. Following sterilization, discs were rinsed in sterile saline and BHI broth prior to being used as a substrate for biofilm formation. In vitro monospecies S. mutans firefly biofilms were formed on novel and control resin discs similarly to that previously described [73]. Briefly, S. mutans firefly was grown in BHI broth overnight at 37°C with 5% CO2. Following incubation, the optical density at 600 nm (OD600) was measured, and cultures were diluted to an optical density OD600 = 0.05 into fresh BHI broth supplemented with 1% sucrose. Resin discs were transferred to sterile 24-well plates (Fisher Scientific, USA), 1 mL of the bacterial dilution was added to each well, and the plates were incubated with static conditions at 37°C with 5% CO2 for 24 hours to allow for mature biofilm formation. No bacteria added controls were also prepared where only BHI + 1% sucrose were added to the wells containing discs.
2.4 Quantification of bioluminescence by biofilm-associated S. mutans
To quantify the amount of viable bacteria attached to each resin disc, a non-disruptive luciferase assay was performed as previously described with modifications [73, 74]. Briefly, resin discs were rinsed to remove unattached planktonic bacteria and were transferred to sterile 24-well opaque white plates (Fisher Scientific, USA). To replenish ATP for luciferase activity measurement, discs were incubated in 1 mL BHI + 1% sucrose for 1 hour at 37°C with 5% CO2. Following incubation, 2 mM D-Luciferin substrate (Fisher Scientific, USA) suspended in 0.1 M citrate buffer (pH 6.0) was added, and bioluminescence was measured at 2-min intervals for up to 10 min using a luminometer (FilterMax F5, Molecular Devices, USA). Statistical analysis was performed using a Student’s t-test and ANOVA.
2.5 Quantification of viable bacteria released from biofilms
Following the measurement of bioluminescence on the discs, the number of viable bacteria on each disc was determined through disruption and subsequent plating. Briefly, each resin disc was transferred to a microcentrifuge tube containing 1 mL saline, and biofilm removal was facilitated through sonication, using a waterbath sonicator (Ultrasonic Bath 1.9L, Fisher Scientific, USA), for 10 min followed by agitation by vortexing for 3 min. An aliquot of the resulting bacterial suspension was diluted and plated in triplicate on BHI agar. The number of viable bacteria was determined by counting colonies. Statistical analysis was performed using a Student’s t-test and ANOVA.
3. Results
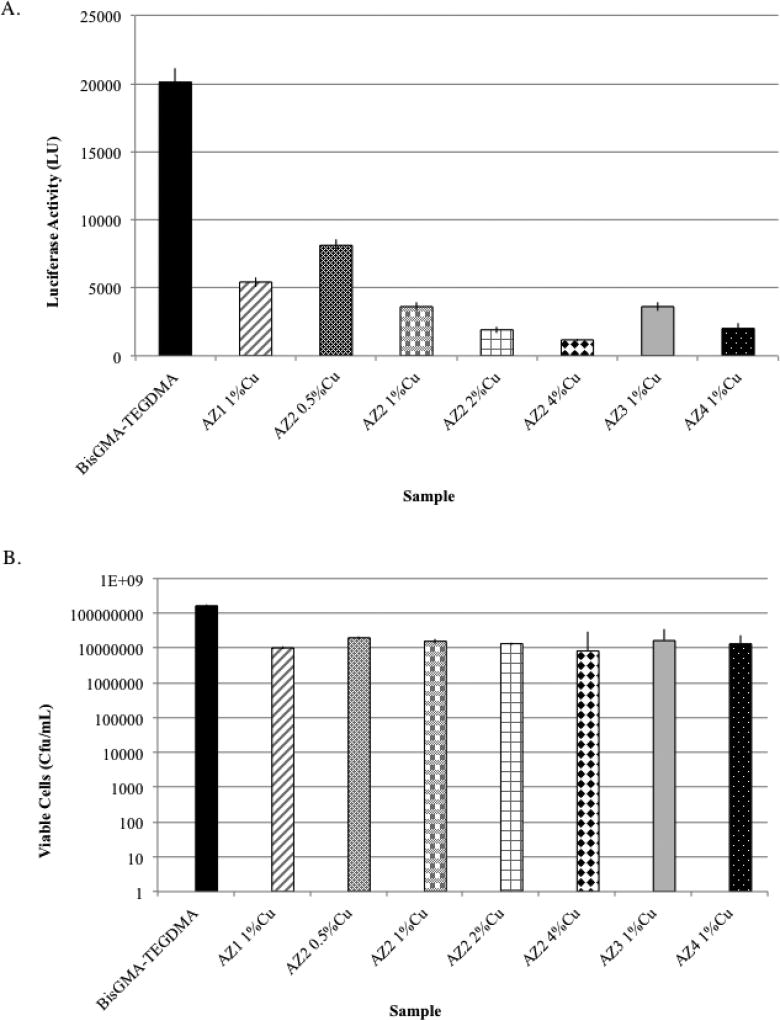
The bioluminescence and viable count analysis of bacteria associated with biofilms grown on CuAAC-based resins showed a significant reduction when compared to that quantified for the BisGMA-TEGDMA control resin discs. Fig. 2A displays the production of luciferase activity by S. mutans by non-disrupted biofilms associated with CuAAC-based resin discs and BisGMA-TEGDMA resin discs. Luciferase activity was measured at 2-min intervals, but only the 6-min interval is shown due to it having the highest bioluminescence emission. Bioluminescence analysis showed at least a 4-fold reduction in the metabolic activity of S. mutans associated with all forms of CuAAC.
Fig. 2.
Evaluation of bioburden on BisGMA-TEGDMA and novel CuAAC-based resins having varying Azide monomers and differing concentrations of CuCl2 in AZ-2 containing resins. Non-disruptive analysis of biofilm bioburden using luciferase measurements (A) indicated a statistically significant reduction (p <0.05) in the luminescence observed on the novel CuAAC-based discs compared to BisGMA-TEGDMA-based resins (n=12 discs), suggesting a reduction in the bioburden associated with the biofilms grown on the CuAAC resin-based discs. Disruptive analysis of biofilms (B) from BisGMA-TEGDMA and CuAAC resin-based discs (n=12) indicated a significant reduction (p <0.05) in the number of bacteria recovered from biofilms on each novel resin when compared to biofilms associated with the BisGMA-TEGDMA.
Following the measurement of luciferase activity, biofilms associated with CuAAC-based and BisGMA-TEGDMA resins were disrupted via sonication and agitation; the number of viable bacteria was determined. Fig. 2B shows the number of viable cells (cfu/mL) released from each novel resin and control discs. The results reveal a statistically significant reduction (p <0.05) where between a 1–2-log reduction was observed in the number of viable bacteria released from CuAAC-based resins in comparison to BisGMA-TEGDMA.
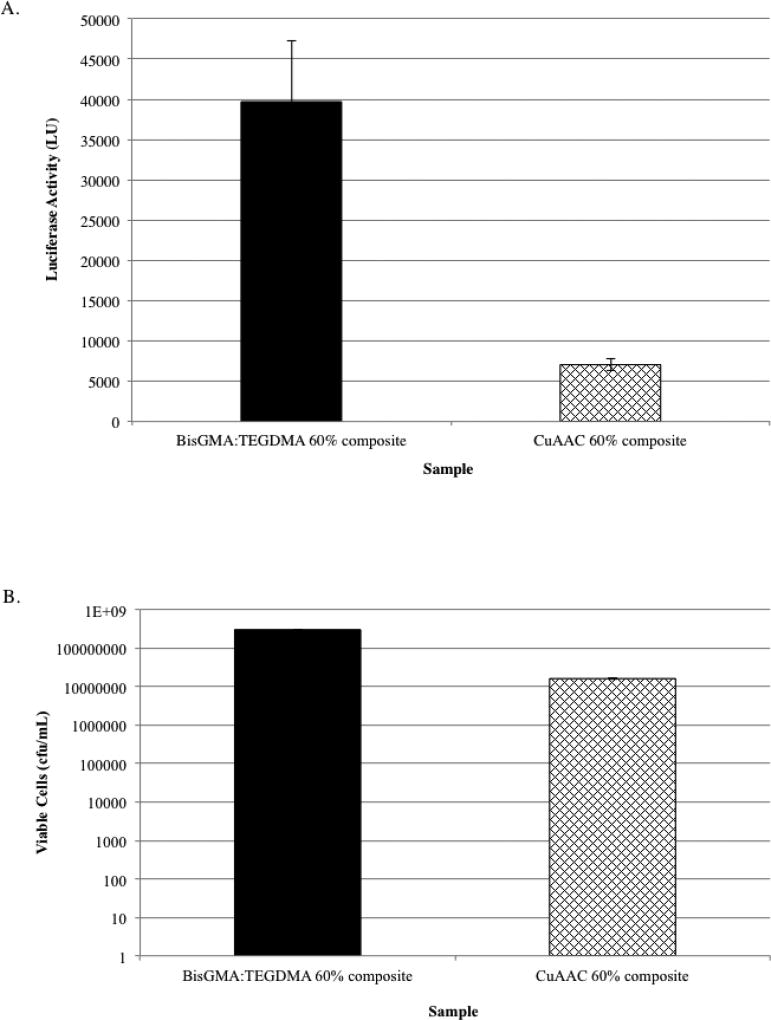
In addition to evaluating the resins for biofilm formation, we also measured luciferase activity and the number of viable bacteria associated with biofilms that formed on CuAAC-based composites. For this measurement, a formulation containing the AZ-2 diazide (Fig. 1), trialkyne (a mole ratio of 1:1 to N3:alkyne), with 2 mole percentage of CuCl2[PMDETA] per functionality and 2 mole percentage of CQ was used over the other CuAAC formulations because its formulation has ease of preparation, mixing, and kinetics for composite preparation in addition to having superior mechanical properties (data not shown). Both luciferase activity and the number of viable bacteria quantified showed a statistically significant reduction for biofilms grown on CuAAC-based composites in comparison to BisGMA-TEGDMA composites. There was over a 5-fold reduction in bioluminescence measured from CuAAC-based composite-associated biofilms (Fig. 3A) in comparison to biofilms grown on BisGMA-TEGDMA composites. Similarly, nearly a 2-log reduction was observed in the number of viable cells released from biofilms grown on CuAAC-based composites (Fig. 3B).
Fig. 3.
Evaluation of bioburden on novel CuAAC-based and BisGMA-TEGDMA-based composites using non-disruptive (A) and disruptive analyses (B). Nondisruptive luciferase analysis (A) of S. mutans firefly biofilm grown on CuAAC 60% composite discs (containing AZ-2 diazide, trialkyne (a mole ratio of 1:1 to N3:alkyne), with 2 mole percentage of CuCl2[PMDETA] per functionality, 2 mole percentage of CQ and 60-weight percentage of alkyne-functionalized microfillers (Schott, 0.4 µm)) and on BisGMA-TEGDMA 60% composite discs (n=12) (containing BisGMA-TEGDMA (70:30 weight ratio) with 2-weight percentage of CQ having 60-weight percentage of methacrylated microfillers (Schott, 0.4 µm)) indicated a statistically significant reduction (p >0.05) in luminescence, indicating a reduced bioburden on CuAAC composite discs. Disruptive analysis (B) of CuAAC 60% composite discs and BisGMA-TEGDMA 60% composite discs also showed a significant reduction (p <0.05), having nearly a 2-log reduction in bioburden on CuAAC based composites.
4. Discussion
For this study, we evaluated the ability of a monospecies biofilm to form on novel CuAAC-based resins. S. mutans is readily used as a model organism to evaluate dental restorative materials, particularly their antibacterial effects. Recently, Esteban Florez et al [73] reported a non-disruptive evaluation of S. mutans biofilm formation on resin-based composites and showed correlation between luciferase activity and viable bacteria recovered from biofilms on the composites. In this study, we used a similar approach and utilized a marker-less luciferase expressing strain of S. mutans (S. mutans firefly) [72] to investigate antibacterial effects of novel CuAAC resin and resin-based composites. In both non-disruptive and disruptive assays, we showed a reduction in the bioburden in biofilms associated with CuAAC-based materials. While this study used a mono-species biofilm model, initial studies were performed using a defined multi-species biofilm assay similar to that described by Arthur et al [75] in which oral bacteria including Streptococcus mutans, Streptococcus sanguinis, Streptcococcus salivarius, and Lactobacillus casei were utilized. The results were comparable to that observed using the single-species biofilm assay; a statistically significant reduction in bioburden was observed (Fig. S3). Further evaluation of the effect of CuAAC materials on biofilm formation in the oral cavity is needed. To better simulate the oral environment, a dental plaque microcosm biofilm model will be utilized in future experiments; a similar plaque microcosm model was previously used to evaluate the effect of dental adhesives on biofilm formation [76]. Alternatively, future evaluation of S. mutans biofilm formation on CuAAC dental materials will be performed in a CDC biofilm reactor, using the widely accepted standardized protocol [77].
The antimicrobial nature of copper is well known. While an essential trace element in living organisms, found in more than 30 types of proteins, copper ions can also be fairly toxic through the generation of reactive hydroxyl radicals and through the depletion of sulfhydryls within cells [78, 79]. Numerous studies investigating the antimicrobial activity of copper have shown that bacteria, yeasts, and viruses are killed through contact with copper and copper alloy surfaces, showing nearly complete killing of all microbes present upon contact; however, copper surfaces are most effective as an antimicrobial when having between 50–75% copper content [80–83]. In this study, we showed between a 1–2 log reduction that was slightly dose dependent with an increased amount of copper added, observing the most bacterial reduction in biofilms formed on CuAAC resins containing 4% copper chloride (CuCl2). While we observed a reduction of bacterial prevalence with increased CuCl2, resins containing 4% or greater CuCl2 are challenging to prepare and photo-cure, which makes the possibility of further increasing the Cu2+ content impossible. Additionally, too high of a Cu2+ and its possible dissociation could result in harmful effects to the host. Biocompatibility analysis was performed on the described CuAAC-based resins and CuAAC-based composites to ensure that there is low cytotoxicity of these materials and no toxic effects were observed (data not shown). A similar study investigating the effects of ≥100 ppm silver nanoparticles (AgNPs) showed a 2.3-log reduction in S. mutans biofilm bioburden and any amount of AgNPs greater than 10 ppm showed a significant cytotoxic effect on fibroblasts; the authors concluded that the anti-biofilm activity of AgNPs would be effective [84]. While the biological relevance of the 1–2 log reduction of bioburden observed on the CuAAC-based dental materials requires further investigation, scanning electron microscopy of biofilms formed on CuAAC-based materials and BisGMA-TEGDMA controls showed a distinct difference in the appearance of the structure of the biofilms formed on the CuAAC-based materials. In specific, while confluent growth was observed on both the CuAAC materials and the controls, a distinct reduction or complete lack of microcolony formation was observed on the CuAAC-based materials (data not shown). Further evaluation of the live-dead percentage, thickness, and general effect of CuAAC materials on the biofilm is necessary.
Alternatively, while the dental materials described herein utilize CuCl2, the incorporation of copper nanoparticles in dental resins is also a probable and valid solution for reducing bioburden on dental materials; various studies have shown the use of copper nanoparticles is highly effective against S. mutans at lower amounts [85, 86]. The incorporation of copper nanoparticles into these CuAAC resins is outside of the scope of the current study. While the CuAAC described herein possess minimal bactericidal activity, with the increased mechanical properties of the CuAAC resin over the currently used BisGMA-TEGDMA [64–66], in conjunction with a reduction in biofilm formation, these resins represent a viable replacement for currently used dental restorative materials. Because it is unknown if this bactericidal effect of CuAAC is a prolonged effect, future evaluations will measure bioburden after prolonged exposure to CuAAC-based composites and will additionally investigate the effect that biofilm formation has on the various composite formulations.
5. Conclusions
Oral biofilms have been implicated in the failure of currently used dental materials; therefore, a dental material having improved mechanical performance and a reduction in biofilm formation may enhance the longevity of dental restoratives. In this study, we noted a statistically significant reduction in S. mutans biofilm bioburden on CuAAC resins and CuAAC-based microfilled composites, which in combination with CuAAC-based materials’ superior mechanical properties and significantly reduced shrinkage stress in comparison to BisGMA-based polymers [64–66], makes CuAAC-based resins a promising alternative.
Supplementary Material
Highlights.
Biofilm formation on the surface of CuAAC-based resins and composites was assessed.
A significant reduction in biofilm bioburden was observed on CuAAC-based materials.
Longevity of dental restorations that contain CuAAC-based materials may be enhanced.
Acknowledgments
This study was supported in part by National Institutes of Health/NIDCR grant U01DE023774 and U01DE023777 We would like to thank Dr. Justin Merritt for kindly providing the luciferase expressing S. mutans that we used in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hojo K, Nagaoka S, Ohshima T, Maeda N. Bacterial interactions in dental biofilm development. J Dent Res. 2009;88:982–90. doi: 10.1177/0022034509346811. [DOI] [PubMed] [Google Scholar]
- 3.Faveri M, Mayer MP, Feres M, de Figueiredo LC, Dewhirst FE, Paster BJ. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol Immunol. 2008;23:112–8. doi: 10.1111/j.1399-302X.2007.00397.x. [DOI] [PubMed] [Google Scholar]
- 4.Yasunaga H, Takeshita T, Shibata Y, Furuta M, Shimazaki Y, Akifusa S, et al. Exploration of bacterial species associated with the salivary microbiome of individuals with a low susceptibility to dental caries. Clin Oral Investig. 2017;21:2399–406. doi: 10.1007/s00784-016-2035-5. [DOI] [PubMed] [Google Scholar]
- 5.Forssten SD, Bjorklund M, Ouwehand AC. Streptococcus mutans, caries and simulation models. Nutrients. 2010;2:290–8. doi: 10.3390/nu2030290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–80. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munson MA, Banerjee A, Watson TF, Wade WG. Molecular analysis of the microflora associated with dental caries. J Clin Microbiol. 2004;42:3023–9. doi: 10.1128/JCM.42.7.3023-3029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon-Soro A, Belda-Ferre P, Cabrera-Rubio R, Alcaraz LD, Mira A. A tissue-dependent hypothesis of dental caries. Caries Res. 2013;47:591–600. doi: 10.1159/000351663. [DOI] [PubMed] [Google Scholar]
- 9.Simon-Soro A, Guillen-Navarro M, Mira A. Metatranscriptomics reveals overall active bacterial composition in caries lesions. J Oral Microbiol. 2014;6:25443. doi: 10.3402/jom.v6.25443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon-Soro A, Mira A. Solving the etiology of dental caries. Trends Microbiol. 2015;23:76–82. doi: 10.1016/j.tim.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Hahn CL, Falkler WA, Jr, Minah GE. Microbiological studies of carious dentine from human teeth with irreversible pulpitis. Arch Oral Biol. 1991;36:147–53. doi: 10.1016/0003-9969(91)90077-8. [DOI] [PubMed] [Google Scholar]
- 12.Mei ML, Chu CH, Low KH, Che CM, Lo EC. Caries arresting effect of silver diamine fluoride on dentine carious lesion with S. mutans and L. acidophilus dual-species cariogenic biofilm. Med Oral Patol Oral Cir Bucal. 2013;18:e824–31. doi: 10.4317/medoral.18831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen ZT, Yates D, Ahn SJ, Burne RA. Biofilm formation and virulence expression by Streptococcus mutans are altered when grown in dual-species model. BMC Microbiol. 2010;10:111. doi: 10.1186/1471-2180-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinberg I. A mixed-bacteria ecological approach to understanding the role of the oral bacteria in dental caries causation: an alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit Rev Oral Biol Med. 2002;13:108–25. doi: 10.1177/154411130201300202. [DOI] [PubMed] [Google Scholar]
- 15.Correa MB, Peres MA, Peres KG, Horta BL, Barros AD, Demarco FF. Amalgam or composite resin? Factors influencing the choice of restorative material. J Dent. 2012;40:703–10. doi: 10.1016/j.jdent.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 16.Demarco FF, Correa MB, Cenci MS, Moraes RR, Opdam NJ. Longevity of posterior composite restorations: not only a matter of materials. Dent Mater. 2012;28:87–101. doi: 10.1016/j.dental.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Drummond JL. Degradation, fatigue, and failure of resin dental composite materials. J Dent Res. 2008;87:710–9. doi: 10.1177/154405910808700802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferracane JL. Resin composite--state of the art. Dent Mater. 2011;27:29–38. doi: 10.1016/j.dental.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Mjor IA, Moorhead JE, Dahl JE. Reasons for replacement of restorations in permanent teeth in general dental practice. Int Dent J. 2000;50:361–6. doi: 10.1111/j.1875-595x.2000.tb00569.x. [DOI] [PubMed] [Google Scholar]
- 20.Bernardo M, Luis H, Martin MD, Leroux BG, Rue T, Leitao J, et al. Survival and reasons for failure of amalgam versus composite posterior restorations placed in a randomized clinical trial. J Am Dent Assoc. 2007;138:775–83. doi: 10.14219/jada.archive.2007.0265. [DOI] [PubMed] [Google Scholar]
- 21.Soncini JA, Maserejian NN, Trachtenberg F, Tavares M, Hayes C. The longevity of amalgam versus compomer/composite restorations in posterior primary and permanent teeth: findings From the New England Children's Amalgam Trial. J Am Dent Assoc. 2007;138:763–72. doi: 10.14219/jada.archive.2007.0264. [DOI] [PubMed] [Google Scholar]
- 22.Cramer NB, Stansbury JW, Bowman CN. Recent advances and developments in composite dental restorative materials. J Dent Res. 2011;90:402–16. doi: 10.1177/0022034510381263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durner J, Spahl W, Zaspel J, Schweikl H, Hickel R, Reichl FX. Eluted substances from unpolymerized and polymerized dental restorative materials and their Nernst partition coefficient. Dent Mater. 2010;26:91–9. doi: 10.1016/j.dental.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Ferracane JL. Resin-based composite performance: are there some things we can't predict? Dent Mater. 2013;29:51–8. doi: 10.1016/j.dental.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bourbia M, Ma D, Cvitkovitch DG, Santerre JP, Finer Y. Cariogenic bacteria degrade dental resin composites and adhesives. J Dent Res. 2013;92:989–94. doi: 10.1177/0022034513504436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaviz Y, Finer Y, Santerre JP. Biodegradation of resin composites and adhesives by oral bacteria and saliva: a rationale for new material designs that consider the clinical environment and treatment challenges. Dent Mater. 2014;30:16–32. doi: 10.1016/j.dental.2013.08.201. [DOI] [PubMed] [Google Scholar]
- 27.Finer Y, Santerre JP. The influence of resin chemistry on a dental composite's biodegradation. J Biomed Mater Res A. 2004;69:233–46. doi: 10.1002/jbm.a.30000. [DOI] [PubMed] [Google Scholar]
- 28.Finer Y, Santerre JP. Influence of silanated filler content on the biodegradation of bisGMA/TEGDMA dental composite resins. J Biomed Mater Res A. 2007;81:75–84. doi: 10.1002/jbm.a.31004. [DOI] [PubMed] [Google Scholar]
- 29.Jaffer F, Finer Y, Santerre JP. Interactions between resin monomers and commercial composite resins with human saliva derived esterases. Biomaterials. 2002;23:1707–19. doi: 10.1016/s0142-9612(01)00298-8. [DOI] [PubMed] [Google Scholar]
- 30.Lin BA, Jaffer F, Duff MD, Tang YW, Santerre JP. Identifying enzyme activities within human saliva which are relevant to dental resin composite biodegradation. Biomaterials. 2005;26:4259–64. doi: 10.1016/j.biomaterials.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Santerre JP, Shajii L, Leung BW. Relation of dental composite formulations to their degradation and the release of hydrolyzed polymeric-resin-derived products. Crit Rev Oral Biol Med. 2001;12:136–51. doi: 10.1177/10454411010120020401. [DOI] [PubMed] [Google Scholar]
- 32.Kermanshahi S, Santerre JP, Cvitkovitch DG, Finer Y. Biodegradation of resin-dentin interfaces increases bacterial microleakage. J Dent Res. 2010;89:996–1001. doi: 10.1177/0022034510372885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuan YH, Huang FM, Lee SS, Li YC, Chang YC. Bisgma stimulates prostaglandin E2 production in macrophages via cyclooxygenase-2, cytosolic phospholipase A2, and mitogen-activated protein kinases family. PLoS One. 2013;8:e82942. doi: 10.1371/journal.pone.0082942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckhardt A, Muller P, Hiller KA, Krifka S, Bolay C, Spagnuolo G, et al. Influence of TEGDMA on the mammalian cell cycle in comparison with chemotherapeutic agents. Dent Mater. 2010;26:232–41. doi: 10.1016/j.dental.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Krifka S, Petzel C, Hiller KA, Frank EM, Bosl C, Spagnuolo G, et al. Resin monomer-induced differential activation of MAP kinases and apoptosis in mouse macrophages and human pulp cells. Biomaterials. 2010;31:2964–75. doi: 10.1016/j.biomaterials.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Krifka S, Spagnuolo G, Schmalz G, Schweikl H. A review of adaptive mechanisms in cell responses towards oxidative stress caused by dental resin monomers. Biomaterials. 2013;34:4555–63. doi: 10.1016/j.biomaterials.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 37.Schweikl H, Widbiller M, Krifka S, Klement J, Petzel C, Bolay C, et al. Interaction between LPS and a dental resin monomer on cell viability in mouse macrophages. Dent Mater. 2016;32:1492–503. doi: 10.1016/j.dental.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 38.de Souza Costa CA, Hebling J, Hanks CT. Effects of light-curing time on the cytotoxicity of a restorative resin composite applied to an immortalized odontoblast-cell line. Oper Dent. 2003;28:365–70. [PubMed] [Google Scholar]
- 39.Hanks CT, Strawn SE, Wataha JC, Craig RG. Cytotoxic effects of resin components on cultured mammalian fibroblasts. J Dent Res. 1991;70:1450–5. doi: 10.1177/00220345910700111201. [DOI] [PubMed] [Google Scholar]
- 40.Geurtsen W. Biocompatibility of resin-modified filling materials. Crit Rev Oral Biol Med. 2000;11:333–55. doi: 10.1177/10454411000110030401. [DOI] [PubMed] [Google Scholar]
- 41.Geurtsen W, Lehmann F, Spahl W, Leyhausen G. Cytotoxicity of 35 dental resin composite monomers/additives in permanent 3T3 and three human primary fibroblast cultures. J Biomed Mater Res. 1998;41:474–80. doi: 10.1002/(sici)1097-4636(19980905)41:3<474::aid-jbm18>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 42.Paolantonio M, D'Ercole S, Perinetti G, Tripodi D, Catamo G, Serra E, et al. Clinical and microbiological effects of different restorative materials on the periodontal tissues adjacent to subgingival class V restorations. J Clin Periodontol. 2004;31:200–7. doi: 10.1111/j.0303-6979.2004.00472.x. [DOI] [PubMed] [Google Scholar]
- 43.Khalichi P, Singh J, Cvitkovitch DG, Santerre JP. The influence of triethylene glycol derived from dental composite resins on the regulation of Streptococcus mutans gene expression. Biomaterials. 2009;30:452–9. doi: 10.1016/j.biomaterials.2008.09.053. [DOI] [PubMed] [Google Scholar]
- 44.Singh J, Khalichi P, Cvitkovitch DG, Santerre JP. Composite resin degradation products from BisGMA monomer modulate the expression of genes associated with biofilm formation and other virulence factors in Streptococcus mutans. J Biomed Mater Res A. 2009;88:551–60. doi: 10.1002/jbm.a.31879. [DOI] [PubMed] [Google Scholar]
- 45.Sadeghinejad L, Cvitkovitch DG, Siqueira WL, Merritt J, Santerre JP, Finer Y. Mechanistic, genomic and proteomic study on the effects of BisGMA-derived biodegradation product on cariogenic bacteria. Dent Mater. 2016 doi: 10.1016/j.dental.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sousa RP, Zanin IC, Lima JP, Vasconcelos SM, Melo MA, Beltrao HC, et al. In situ effects of restorative materials on dental biofilm and enamel demineralisation. J Dent. 2009;37:44–51. doi: 10.1016/j.jdent.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Antonucci JM, Zeiger DN, Tang K, Lin-Gibson S, Fowler BO, Lin NJ. Synthesis and characterization of dimethacrylates containing quaternary ammonium functionalities for dental applications. Dent Mater. 2012;28:219–28. doi: 10.1016/j.dental.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng Y, Howard L, Guo X, Chong VJ, Gregory RL, Xie D. A novel antibacterial resin composite for improved dental restoratives. J Mater Sci Mater Med. 2012;23:1553–61. doi: 10.1007/s10856-012-4629-z. [DOI] [PubMed] [Google Scholar]
- 49.Xu X, Wang Y, Liao S, Wen ZT, Fan Y. Synthesis and characterization of antibacterial dental monomers and composites. J Biomed Mater Res B Appl Biomater. 2012;100:1151–62. doi: 10.1002/jbm.b.32683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F, Chen J, Chai Z, Zhang L, Xiao Y, Fang M, et al. Effects of a dental adhesive incorporating antibacterial monomer on the growth, adherence and membrane integrity of Streptococcus mutans. J Dent. 2009;37:289–96. doi: 10.1016/j.jdent.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Imazato S. Bio-active restorative materials with antibacterial effects: new dimension of innovation in restorative dentistry. Dent Mater J. 2009;28:11–9. doi: 10.4012/dmj.28.11. [DOI] [PubMed] [Google Scholar]
- 52.Imazato S, Ebi N, Tarumi H, Russell RR, Kaneko T, Ebisu S. Bactericidal activity and cytotoxicity of antibacterial monomer MDPB. Biomaterials. 1999;20:899–903. doi: 10.1016/s0142-9612(98)00247-6. [DOI] [PubMed] [Google Scholar]
- 53.Imazato S, Kinomoto Y, Tarumi H, Ebisu S, Tay FR. Antibacterial activity and bonding characteristics of an adhesive resin containing antibacterial monomer MDPB. Dent Mater. 2003;19:313–9. doi: 10.1016/s0109-5641(02)00060-x. [DOI] [PubMed] [Google Scholar]
- 54.Zhang K, Cheng L, Imazato S, Antonucci JM, Lin NJ, Lin-Gibson S, et al. Effects of dual antibacterial agents MDPB and nano-silver in primer on microcosm biofilm, cytotoxicity and dentine bond properties. J Dent. 2013;41:464–74. doi: 10.1016/j.jdent.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu X, Burgess JO. Compressive strength, fluoride release and recharge of fluoride-releasing materials. Biomaterials. 2003;24:2451–61. doi: 10.1016/s0142-9612(02)00638-5. [DOI] [PubMed] [Google Scholar]
- 56.Xu X, Ling L, Wang R, Burgess JO. Formulation and characterization of a novel fluoride-releasing dental composite. Dent Mater. 2006;22:1014–23. doi: 10.1016/j.dental.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 57.Cheng L, Zhang K, Melo MA, Weir MD, Zhou X, Xu HH. Anti-biofilm dentin primer with quaternary ammonium and silver nanoparticles. J Dent Res. 2012;91:598–604. doi: 10.1177/0022034512444128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melo MA, Cheng L, Zhang K, Weir MD, Rodrigues LK, Xu HH. Novel dental adhesives containing nanoparticles of silver and amorphous calcium phosphate. Dent Mater. 2013;29:199–210. doi: 10.1016/j.dental.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng L, Zhang K, Weir MD, Melo MA, Zhou X, Xu HH. Nanotechnology strategies for antibacterial and remineralizing composites and adhesives to tackle dental caries. Nanomedicine (Lond) 2015;10:627–41. doi: 10.2217/nnm.14.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Markowska K, Grudniak AM, Wolska KI. Silver nanoparticles as an alternative strategy against bacterial biofilms. Acta Biochim Pol. 2013;60:523–30. [PubMed] [Google Scholar]
- 61.Jedrychowski JR, Caputo AA, Kerper S. Antibacterial and mechanical properties of restorative materials combined with chlorhexidines. J Oral Rehabil. 1983;10:373–81. doi: 10.1111/j.1365-2842.1983.tb00133.x. [DOI] [PubMed] [Google Scholar]
- 62.Riggs PD, Braden M, Patel M. Chlorhexidine release from room temperature polymerising methacrylate systems. Biomaterials. 2000;21:345–51. doi: 10.1016/s0142-9612(99)00187-8. [DOI] [PubMed] [Google Scholar]
- 63.Leung D, Spratt DA, Pratten J, Gulabivala K, Mordan NJ, Young AM. Chlorhexidine-releasing methacrylate dental composite materials. Biomaterials. 2005;26:7145–53. doi: 10.1016/j.biomaterials.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 64.Baranek A, Song HB, McBride M, Finnegan P, Bowman CN. Thermomechanical formation-structure-property relationships in photopolymerized copper-catalyzed azide-alkyne (CuAAC) networks. Macromolecules. 2016;49:1191–200. doi: 10.1021/acs.macromol.6b00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song HB, Sowan N, Shah PK, Baranek A, Flores A, Stansbury JW, et al. Reduced shrinkage stress via photo-initiated copper(I)-catalyzed cycloaddition polymerizations of azide-alkyne resins. Dent Mater. 2016;32:1332–42. doi: 10.1016/j.dental.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song HB, Wang X, Patton JR, Stansbury JW, Bowman CN. Kinetics and mechanics of photo-polymerized triazole-containing thermosetting composites via the copper(I)-catalyzed azide-alkyne cycloaddition. Dent Mater. 2017;33:621–9. doi: 10.1016/j.dental.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adzima BJ, Tao Y, Kloxin CJ, DeForest CA, Anseth KS, Bowman CN. Spatial and temporal control of the alkyne-azide cycloaddition by photoinitiated Cu(II) reduction. Nat Chem. 2011;3:256–59. doi: 10.1038/nchem.980. [DOI] [PubMed] [Google Scholar]
- 68.El-Zaatari BM, Shete AU, Adzima BJ, Kloxin CJ. Towards understanding the kinetic behaviour and limitations in photo-induced copper(i) catalyzed azide-alkyne cycloaddition (CuAAC) reactions. Phys Chem Chem Phys. 2016;18:25504–11. doi: 10.1039/c6cp04950h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shete AU, El-Zaatari BM, French JM, Kloxin CJ. Blue-light activated rapid polymerization for defect-free bulk Cu(i)-catalyzed azide-alkyne cycloaddition (CuAAC) crosslinked networks. Chem Commun (Camb) 2016;52:10574–7. doi: 10.1039/c6cc05095f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gong T, Adzima BJ, Baker NH, Bowman CN. Photopolymerization reactions using the photoinitiated copper (I)-catalyzed azide-alkyne cycloaddition (CuAAC) reaction. Adv Mater. 2013;25:2024–8. doi: 10.1002/adma.201203815. [DOI] [PubMed] [Google Scholar]
- 71.Brase S, Gil C, Knepper K, Zimmermann V. Organic azides: an exploding diversity of a unique class of compounds. Angew Chem Int Ed Engl. 2005;44:5188–240. doi: 10.1002/anie.200400657. [DOI] [PubMed] [Google Scholar]
- 72.Merritt J, Senpuku H, Kreth J. Let there be bioluminescence: development of a biophotonic imaging platform for in situ analyses of oral biofilms in animal models. Environ Microbiol. 2016;18:174–90. doi: 10.1111/1462-2920.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Esteban Florez FL, Hiers RD, Smart K, Kreth J, Qi F, Merritt J, et al. Real-time assessment of Streptococcus mutans biofilm metabolism on resin composite. Dent Mater. 2016;32:1263–9. doi: 10.1016/j.dental.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Merritt J, Kreth J, Qi F, Sullivan R, Shi W. Non-disruptive, real-time analyses of the metabolic status and viability of Streptococcus mutans cells in response to antimicrobial treatments. J Microbiol Methods. 2005;61:161–70. doi: 10.1016/j.mimet.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 75.Arthur RA, Waeiss RA, Hara AT, Lippert F, Eckert GJ, Zero DT. A defined-multispecies microbial model for studying enamel caries development. Caries Res. 2013;47:318–24. doi: 10.1159/000347050. [DOI] [PubMed] [Google Scholar]
- 76.Zhang N, Melo MA, Chen C, Liu J, Weir MD, Bai Y, et al. Development of a multifunctional adhesive system for prevention of root caries and secondary caries. Dent Mater. 2015;31:1119–31. doi: 10.1016/j.dental.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mah TF, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–9. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 78.Karlin KD. Metalloenzymes, structural motifs, and inorganic models. Science. 1993;261:701–8. doi: 10.1126/science.7688141. [DOI] [PubMed] [Google Scholar]
- 79.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A. 2009;106:8344–9. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Prado JV, Esparza MM, Vidal AR, Duran TC. Adherence to copper and stainless steel metal coupons of common nosocomial bacterial strains. Rev Med Chil. 2013;141:291–7. doi: 10.4067/S0034-98872013000300002. [DOI] [PubMed] [Google Scholar]
- 81.Prado JV, Vidal AR, Duran TC. Application of copper bactericidal properties in medical practice. Rev Med Chil. 2012;140:1325–32. doi: 10.4067/S0034-98872012001000014. [DOI] [PubMed] [Google Scholar]
- 82.Quaranta D, Krans T, Espirito Santo C, Elowsky CG, Domaille DW, Chang CJ, et al. Mechanisms of contact-mediated killing of yeast cells on dry metallic copper surfaces. Appl Environ Microbiol. 2011;77:416–26. doi: 10.1128/AEM.01704-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Warnes SL, Keevil CW. Inactivation of norovirus on dry copper alloy surfaces. PLoS One. 2013;8:e75017. doi: 10.1371/journal.pone.0075017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perez-Diaz MA, Boegli L, James G, Velasquillo C, Sanchez-Sanchez R, Martinez-Martinez RE, et al. Silver nanoparticles with antimicrobial activities against Streptococcus mutans and their cytotoxic effect. Mater Sci Eng C Mater Biol Appl. 2015;55:360–6. doi: 10.1016/j.msec.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 85.Ramazanzadeh B, Jahanbin A, Yaghoubi M, Shahtahmassbi N, Ghazvini K, Shakeri M, et al. Comparison of Antibacterial Effects of ZnO and CuO nanoparticles coated brackets against Streptococcus mutans. J Dent (Shiraz) 2015;16:200–5. [PMC free article] [PubMed] [Google Scholar]
- 86.Gutierrez MF, Malaquias P, Matos TP, Szesz A, Souza S, Bermudez J, et al. Mechanical and microbiological properties and drug release modeling of an etch-and-rinse adhesive containing copper nanoparticles. Dent Mater. 2017;33:309–20. doi: 10.1016/j.dental.2016.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.