ABSTRACT
DEP domain containing 1 (DEPDC1) and M-phase phosphoprotein 1 (MPHOSPH1) are human cancer testis antigens that are frequently overexpressed in urinary bladder cancer. In a phase I/II clinical trial, a DEPDC1- and MPHOSPH1-derived short peptide vaccine demonstrated promising efficacy in preventing bladder cancer recurrence. Here, we aimed to identify long peptides (LPs) derived from DEPDC1 and MPHOSPH1 that induced both T-helper (Th) cells and tumor-reactive cytotoxic T lymphocytes (CTLs). Stimulation of peripheral blood mononuclear cells (PBMCs) from healthy donors with the synthetic DEPDC1- and MPHOSPH1-LPs predicted to bind to promiscuous human leukocyte antigen (HLA) class II molecules by a computer algorithm induced specific CD4+ T cells as revealed by interferon-γ enzyme-linked immunospot assays. Three of six LPs encompassed HLA-A2- or -A24-restricted CTL epitopes or both, and all six LPs stimulated DEPDC1- or MPHOSPH1-specific Th cells restricted by promiscuous and frequently observed HLA class II molecules in the Japanese population. Some LPs are naturally processed from the proteins in DCs, and the capacity of these LPs to cross-prime CTLs was confirmed in vivo using HLA-A2 or -A24 transgenic mice. The LP-specific and HLA class II-restricted T-cell responses were also observed in PBMCs from patients with bladder cancer. Repeated stimulation of PBMCs with DEPDC1-LPs and MPHOSPH1-LPs yielded clonal Th cells expressing specific T-cell receptor (TCR)-α and β genes. These DEPDC1- or MPHOSPH1-derived LPs may have applications in immunotherapy in patients with bladder cancer, and the TCR genes identified may be useful for monitoring of Th cells specific to LPs in vivo.
KEYWORDS: bladder cancer, cross presentation, cancer-testis antigen, DEP domain containing 1, helper T-cell epitope, M-phase phosphoprotein 1, T-cell receptor, cancer immunotherapy, tumor immunity
Introduction
Bladder cancer is the most common malignancy involving the genitourinary system. Platinum-based chemotherapy remains the standard of care for first-line treatment of advanced/metastatic urothelial carcinoma (UC).1,2 There is an estimated overall survival of 9–15 months in metastatic bladder cancer in those who receive the standard treatment with platinum-based chemotherapy; stage IV bladder cancer has a relative 5-year survival rate of about 15%.1,3 Until recently, there were few treatment options for patients who were refractory to chemotherapy or did not tolerate chemotherapy due to serious adverse reactions, and overall outcomes remained very poor.2 Currently, systemic immunotherapy using programmed cell death protein 1 (PD-1)- and PD-1 ligand (PD-L1)-specific monoclonal antibodies, i.e., Nivolumab/Pembrolizumab and Atezolizumab, respectively, has been approved for the treatment of urothelial bladder cancer to achieve significant improvement in response rates.2,4
Peptide vaccines are also expected to have applications in immunotherapy. Indeed, many studies have examined the roles of tumor-associated antigens (TAAs), which are commonly expressed in tumor cells and are recognized as foreign antigens by the host immune system because of dissimilarities between these substances and those expressed in normal cells. Therefore, in order to achieve effective antitumor immunity in patients with cancer, it is important to identify good TAAs as targets for cancer immunotherapy.
A recent report identified the cancer-testis antigens DEP domain containing 1 (DEPDC1) and M-phase phosphoprotein 1 (MPHOSPH1, also called KIF20B) as frequently overexpressed in bladder cancer, breast cancer, and several other malignancies using genome-wide cDNA microarray analysis.5-12 The growth of bladder cancer cells is significantly inhibited by knockdown of DEPDC1 or MPHOSPH1 expression using small interfering RNAs; thus, DEPDC1 and MPHOSPH1 are essential for tumor growth and survival, indicating the clinical significance of these antigens as targets for antigen-specific cancer immunotherapy. Subsequent reports further identified highly immunogenic DEPDC1- and MPHOSPH1-derived short peptides (SPs) that can induce HLA-A24 (A*24:02)-restricted CTLs from peripheral blood mononuclear cells (PBMCs) from patients with bladder cancer.5,13 In a phase I/II clinical trial, combination immunotherapy with intravesical BCG inoculation together with DEPDC1- and MPHOSPH1-derived SPs vaccination demonstrated promising clinical effects in preventing the recurrence of non-muscle invasive bladder cancer without adverse events and with good immunogenicity to induce CTLs.5 Another recent clinical trial showed that DEPDC1- and MPHOSPH1-derived SP vaccines administered to patients with progressive UC was safe and well tolerated, and that efficient activation of CTLs was associated with better survival of patients.13
However, vaccination with HLA-class I-restricted short CTL epitopes alone did not show dramatic antitumor effects in patients.5,13 One reason for the inadequate effects of SP vaccine therapies could be induction of T-cell tolerance or anergy because exogenously added SPs can bind directly to the HLA class I molecules expressed on all nucleated cells and stimulate T cells without so-called accessory signals.14,15
LPs do not bind directly to cell surface HLA-class I molecules because of their relatively closed peptide binding grooves, which can accommodate 8–10mer SPs in most cases. However, professional antigen-presenting cells (APCs), such as dendritic cells (DCs), can engulf and process LPs and present CTL epitopes on their HLA class I molecules by cross-presentation.14 Therefore, induction of the antitumor responses of CTLs by LPs through cross-presentation could overcome the drawbacks of SP vaccine therapy.
Tumor-specific CD4+ helper T (Th) cells, particularly Th type 1 (Th1) cells, play a critical role in efficient induction and maintenance of CTL-mediated antitumor immunity.16 Interferon (IFN)-γ and tumor necrosis factor (TNF)-α produced by Th1 cells are key molecules that induce and maintain long-lived CTL responses through multiple interactions.17-19 Th1 cells also mediate direct cytotoxic activity toward tumor cells and have anti-angiogenic effects.20 Furthermore, Th cells facilitate the entry of CTLs into the tumor site.21 Therefore, identification of Th-cell epitopes that can activate tumor-specific Th1 cells is important for induction of effective tumor immunity. Melief et al. recently reported that human papillomavirus-derived synthetic LPs naturally include CTL epitopes as an attractive vaccine compound for the treatment of vulvar and cervical cancers. 22 Following the injection of LPs, patient DCs take up LPs, process them, and present possible CTL and Th-cell epitopes in the context of the patient's HLA class I and class II molecules, respectively.14 In addition, recent clinical studies using a promiscuous telomerase-derived Th-cell epitope vaccine (GV1001) increased the survival time of patients with cancer when combined with radiotherapy and chemotherapy.23,24 Thus, an ideal peptide vaccine for cancer immunotherapy may be a single polypeptide containing both CTL and Th1-cell epitopes, which are naturally proximal to each other and can be induced simultaneously.22
Accordingly, in this study, we developed promiscuous HLA class II-binding LPs with or without known CTL-epitope sequences recognized by HLA-A2- or -A24-restricted CTLs to induce effective antitumor T cell responses. We demonstrated that the LPs induced specific Th1 responses in vitro from PBMCs of healthy individuals and some UC patients, suggesting the potential applications of these LPs in immunotherapy. Moreover, we found that repeated LP-stimulation yielded nearly clonal DEPDC1-LP- or MPHOSPH1-LP-specific T cells that expressed a convergent pair of TCRA and TCRB genes. Transduction of these TCR genes in the murine TCR-negative T cells restored the original LP- and HLA class II-specific responses, suggesting a potential use for gene-transfer therapy of the bladder cancer patients as well as the monitoring of antitumor Th cells specific to the LPs in vivo.
Results
Prediction and selection of potential promiscuous HLA class II-binding DEPDC1- and MPHOSPH-LPs
To identify potential promiscuous HLA-class II binding Th-cell epitopes of DEPDC1 and MPHOSPH1, we first examined the amino acid sequences of DEPDC1 and MPHOSPH1 using a public computer algorithm provided by IEDB (Fig. 1A, C, Table S1 and 2).25,26 Four DEPDC1 peptide fragments (191–213, 292–313, 301–329, and 613–634) and two MPHOSPH1 peptide fragments (272–298 and 326–351 with relatively lower percentile ranks, i.e., higher binding affinities to frequently observed HLA-class II molecules in the Japanese population (HLA-DR4, HLA-DR9, HLA-DR15, HLA-DP2, and HLA-DP5), were chosen for peptide synthesis. Three of these peptides, i.e., DEPDC1191–213 (DEPDC1-LP1), DEPDC1613–634 (DEPDC1-LP4), and MPHOSPH1326–351 (MPHOSPH1-LP2) did not include known CTL-epitope sequences (Fig. 1B and D). Another three regions, i.e., DEPDC1292–313 (DEPDC1-LP2), DEPDC1301–329 (DEPDC1-LP3), and MPHOSPH1272–298 (MPHOSPH1-LP1) included CTL epitopes recognized by HLA-A2- or -A24-restricted CTLs (Fig. 1B and D).5
Figure 1.
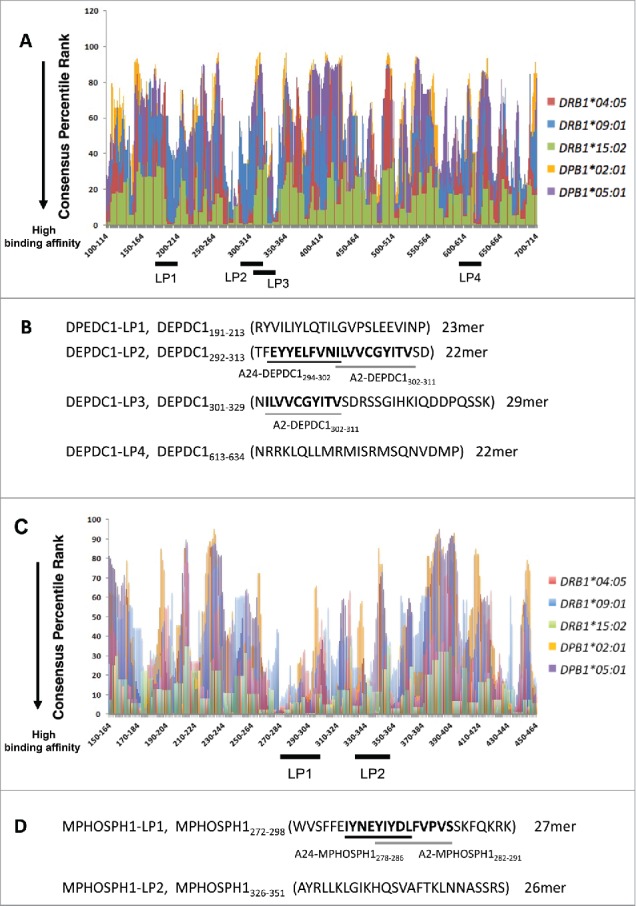
DEPDC1- and MPHOSPH1-derived promiscuous HLA class II-binding peptides predicted by a computer algorithm. (A, C) The amino acid sequences of human DEPDC1 and MPHOSPH1 proteins were analyzed using an algorithm (IEDB analysis resource, consensus method). Numbers on the X-axis indicate N-terminal amino acid positions of DEPDC1- and MPHOSPH1- derived 15mer peptides, respectively. A lower consensus percentile rank indicates higher binding affinity to HLA class-II molecules. Thick black bars below the X-axis indicate ranges of the predicted amino acid sequences of LPs, i.e., DEPDC1-LP1-4 (B), MPHOSPH1-LP1, and MPHOSPH1-LP2 (D), with lower consensus percentile ranks for multiple and frequently observed HLA class-II allelic products in the Japanese population (e.g., DRB1*04:05: red, DRB1*09:01: blue, DRB1*15:02: green, DRP1*02:01: yellow, DPB1*05:01: purple). Nonamer SPs (DEPDC1-A2302–311, DEPDC1-A24294–302, MPHOSPH1-A24278–286, and MPHOSPH1-A2282–291) that are recognized by HLA-A2- or HLA-A24-restricted CTLs are indicated with underlined bold letters.
Identification of promiscuous DEPDC1- and MPHOSPH1-derived Th-cell epitopes
To examine the immunogenicity of these candidate peptides, we examined whether LP-specific CD4+ T-cells could be induced from PBMCs of healthy donors (HDs) stimulated with the LPs. HLA genotypes from the HDs are listed in Table 1, Table 2, and S3.
Table 1.
Identification of DEPDC1-derived and promiscuous HLA-class II-restricted CD4+ T-cell epitopes encompassing cytotoxic T lymphocyte (CTL) epitopes.
| Designaton of Long peptide (LP) | a.a. residue position | Sequence | a.a. length | T-cell donor | aDonors' HLA class-II alleles | Immune response | Restriction HLA class-II molecule |
|---|---|---|---|---|---|---|---|
| DEPDC1-LP1 | 191-213 | RYVILIYLQTILGVPSLEEVINP | 23 | HD1 | DRB1*04:05/09:01,(DR53) | Positive | DR53 (DRB4*01:03) |
| HD2 | DRB1*04:05/09:01 | Negative | |||||
| HD3 | DRB1*08:03/14:05 | Negative | |||||
| HD4 | DRB1*04:05/−,(DR53) | Positive | DR4 (DRB1*04:05) | ||||
| HD7 | DRB1*14:03/15:02 | Negative | |||||
| DEPDC1-LP2 | 292-313 | TFEYYELFVN ILVVCGYITV SD Underlined and bold sequences; bCTL epitope:A24-DEPDC1294–302 Boxed and bold sequences; bCTL epitope:A2-DEPDC1302–311 | 22 | HD1 | DRB1*04:05/09:01,(DR53) | Positive | DR4 (DRB1*04:05) |
| HD2 | DRB1*04:05/09:01 | Negative | |||||
| HD3 | DRB1*08:03/14:05 | Negative | |||||
| HD4 | DRB1*04:05/−,(DR53) | Negative | |||||
| HD6 | DRB1*09:01/13:02 | Negative | |||||
| HD7 | DRB1*14:03/15:02 | Negative | |||||
| DEPDC1-LP3 | 301-329 | N ILVVCGYITV SDRSSGIHKIQDDPQSSK | 29 | HD1 | DRB1*04:05/09:01,(DR53) | Negative | |
| Boxed and bold sequences; | HD3 | DPB1*02:02/05:01 | Positive | DP5 (DPB1*05:01) | |||
| bCTL epitope:A2-DEPDC1302–311 | HD4 | DRB1*04:05/−,(DR53) | Positive | DR4 (DRB1*04:05) | |||
| HD6 | DRB1*09:01/13:02 | Negative | |||||
| HD7 | DRB1*14:03/15:02 | Negative | |||||
| DEPDC1-LP4 | 613-634 | NRRKLQLLMRMISRMSQNVDMP | 22 | HD1 | DPB1*05:01/− | Positive | DP5 (DPB1*05:01) |
| HD2 | DRB1*04:05/09:01 | Negative | |||||
| HD3 | DRB1*08:03/14:05 | Negative | |||||
| HD4 | DRB1*04:05/−,(DR53) | Negative | |||||
| HD5 | DRB1*08:03/15:02 | Positive | DR15 (DRB1*15:02) | ||||
| HD7 | DPB1*05:01/09:01 | Positive | DP5 (DPB1*05:01) |
Summary of DEPDC1-LPs and the immune responses of Th cells reactive to DEPDC1-LPs in five HDs.
The underlined HLA-class II alleles encode HLA-class II molecules presenting DEPDC1-LPs to Th cells in HDs; details of donors' HLA alleles are shown in Table S3.
Underlined or boxed bold sequences are CTL epitopes; DEPDC1-LP1 and DEPDC1-LP4 sequences were selected based on high-affinity binding to HLA-class II molecules predicted by the algorithm;
DEPDC1-LP2 and DEPDC1-LP3 were selected based on the prediction of HLA class-II binding and the proximity to the known CTL epitopes. a.a.: amino acid, Negative: we could not obtain positive data and did not proceed further, LP: long peptide, HD: healthy donor.
Table 2.
Identification of MPHOSPH1-derived and promiscuous HLA-class II-restricted CD4+ T-cell epitopes encompassing cytotoxic T lymphocyte (CTL) epitopes.
| Designaton of Long peptide (LP) | a.a. residue position | Sequence | a.a. length | T-cell donor | aDonors' HLA class-II alleles | Immune response | Restriction HLA class-II molecule |
|---|---|---|---|---|---|---|---|
| MPHOSPH1-LP11 | 272-298 | WVSFFEIYNEY IYDLFVPVS SKFQKRK Underlined and bold sequences; bCTLepitope:A24-MPHOSPH1278–286 Boxed and bold sequences; bCTL epitope:A2-MPHOSPH1282–291 | 27 | HD1 | DRB1*04:05/09:01,(DR53) | Positive | DR9 (DRB1*09:01) |
| HD3 | DPB1*02:02/05:01 | Positive | DP5 (DPB1*05:01) | ||||
| HD4 | DRB1*04:05/−,(DR53) | Positive | DR4 (DRB1*04:05) | ||||
| HD5 | DRB1*08:03/15:02 | Negative | |||||
| HD6 | DRB1*09:01/13:02 | Negative | |||||
| HD7 | DRB1*14:03/15:02 | Negative | |||||
| MPHOSPH1-LP2 | 326-351 | AYRLLKLGIKHQSVAFTKLNNASSRS | 26 | HD1 | DRB1*04:05/09:01,(DR53) | Positive | DR4 (DRB1*04:05) |
| HD2 | DPB1*02:01/04:02 | Positive | DP2 (DPB1*02:01) | ||||
| HD3 | DRB1*08:03/14:05 | Negative | |||||
| HD4 | DRB1*04:05/−,(DR53) | Positive | DR4 (DRB1*04:05) |
Summary of MPHOSPH1-LPs and the immune responses of Th cells reactive to MPHOSPH1-LPs in four HDs.
The underlined HLA class-II alleles encode HLA class-II molecules presenting MPHOSPH1-LPs to Th cells in HDs; details of donors' HLA alleles are shown in Table S3.
Underlined or boxed bold sequences are CTL epitopes; MPHOSPH1-LP2 sequences were selected based on high-affinity binding to HLA class-II molecules predicted by the algorithm; MPHOSPH1-LP1 were selected based on the prediction of HLA class-II binding and the proximity to the known CTL epitopes. a.a.: amino acid, Negative: we could not obtain positive data and did not proceed further, LP: long peptide, HD: healthy donor.
As shown in Fig. 2A, after at least three rounds of stimulations, the Th cells established from an HLA-DR4- and HLA-DR53-positive HD (HD1) produced a significant amount of IFN-γ in response to DEPDC1-LP1-pulsed PBMCs in an HLA-DR-dependent manner as revealed by the enzyme-linked immunospot (ELISPOT) assays. The Th cells specifically recognized mouse fibroblast L cells expressing HLA-DR53 (DRB4*01:03; L-DR53) pulsed with DEPDC1-LP1 but did not recognize unpulsed L-DR53 cells, DEPDC1-LP1-pulsed L cells expressing HLA-DR4 (DRB1*04:05; L-DR4), or HLA-DR9 (DRB1*09:01; L-DR9). These results indicated that DEPDC1-LP1 was presented by HLA-DR53 and induced DEPDC1-LP1-specific Th-cell responses in an HLA-DR53-restricted manner. To investigate whether DEPDC1-LP1 could induce Th-cell responses restricted by other HLA class II molecules, CD4+ T cells from another HD (HD4) were tested. We confirmed that DEPDC1-LP1 induced HLA-DR4-restricted Th-cell responses (Fig. 2B). Thus, DEPDC1-LP1 bound to HLA-DR4 in addition to HLA-DR53 and induced peptide-specific Th-cell responses.
Figure 2.
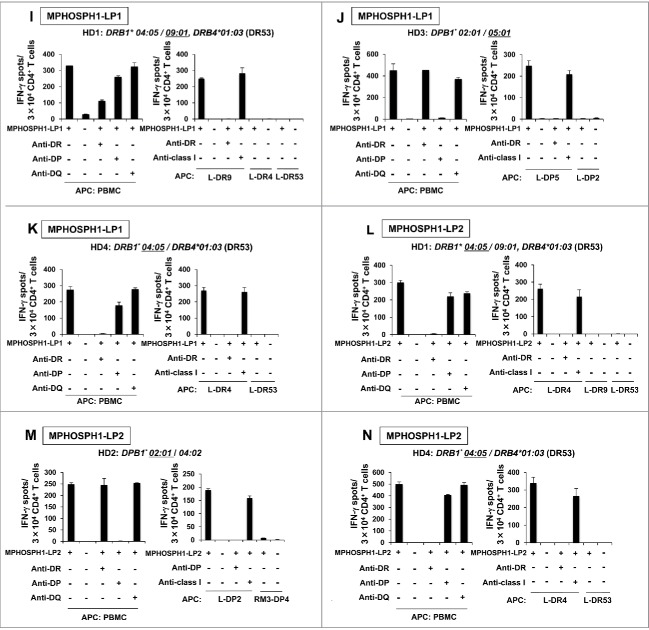
Induction of DEPDC1- or MPHOSPH1-specific Th cells from healthy donors. CD4+ T cells isolated from PBMCs of healthy donors were stimulated with autologous monocyte-derived DCs followed by irradiated PBMCs pulsed with DEPDC1-LPs (A–H) or MPHOSPH1-LPs (I-N). Generated Th cells were restimulated with autologous PBMCs or HLA class II-expressing L cells pulsed with DEPDC1- or MPHOSPH1-LPs in the absence or presence of the indicated blocking antibodies. The number of IFN-γ-producing Th cells was then analyzed by ELISPOT assays. Representative data from at least three independent experiments with similar results are shown. The HLA class II genotypes of the donors are indicated at the top of each panel. The underlines indicate HLA class II alleles encoding the restricting HLA class II molecules.
Figure 2.
(Continued)
Using similar experiments, we could generate IFN-γ-producing Th cells specific to DEPDC1-LP2 (Fig. 2C), DEPDC1-LP3 (Fig. 2D and E), DEPDC1-LP4 (Fig. 2F–H), MPHOSPH1-LP1 (Fig. 2I–K), and MPHOSPH1-LP2 (Fig. 2L–N). In summary, DEPDC1-LPs induced HLA-DR4 (HD1 and HD4)-, HLA-DR53 (HD1), HLA-DR15 (HD5)- and HLA-DP5 (HD1, HD3 and HD7)-restricted Th cells, and MPHOSPH1-LPs induced HLA-DR4 (HD1 and HD4)-, HLA-DR9 (HD1)-, HLA-DP2 (HD2)- and HLA-DP5 (HD3)-restricted Th cells. The data indicated that these LPs were promiscuous Th-cell epitopes presented by frequently observed HLA class II molecules in the Japanese population (Table S4) as expeted,27,28 and combination of these peptides would induce DEPDC1- or MPHOSPH1-specific Th-cell responses in PBMCs of many Japanese donors.
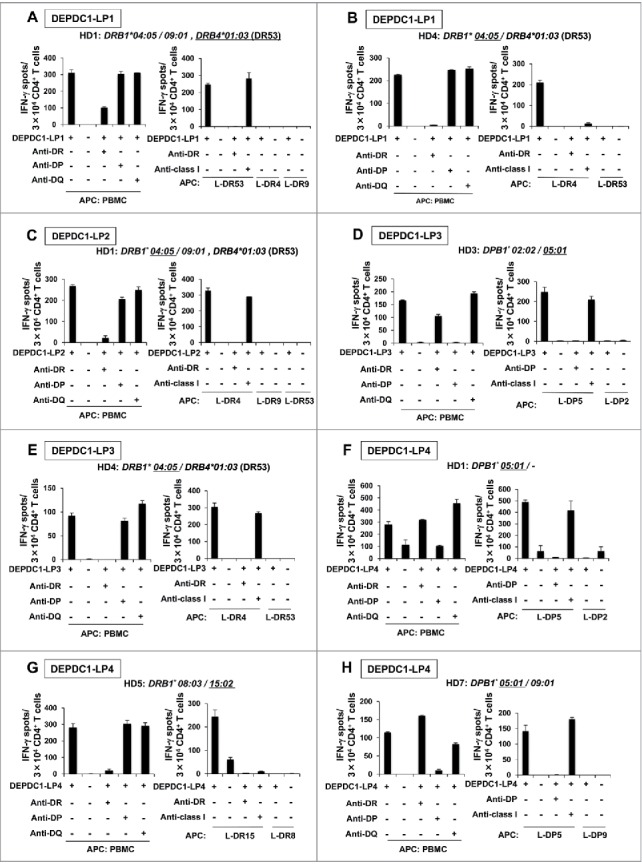
Some DEPDC1- and MPHOSPH1-LPs were naturally processed and presented by DCs
We next assessed whether DCs took up and processed DEPDC1- and MPHOSPH1-derived proteins to stimulate DEPDC1-LP- or MPHOSPH1-LP-specific Th cells. DCs loaded with recombinant DEPDC1 or MPHOSPH1 protein were prepared and used as APCs in IFN-γ ELISPOT assays.1,24 HLA-DR4-restricted DEPDC1-LP2-reactive Th cells efficiently recognized DCs loaded with DEPDC1 protein, but did not recognize those with control protein, indicating that this epitope was naturally processed and presented by DCs (Fig. 3A). Correspondingly, Th cells reactive to DEPDC1-LP3 (HD4, Fig. 3B), DEPDC1-LP4 (HD7, Fig. 3C), and MPHOSPH1-LP2 (HD4, Fig. 3D) efficiently recognized DCs loaded with DEPDC1 or MPHOSPH1 protein, but did not recognize those with the control protein. In summary, these results indicated that many of the LPs selected by IEDB prediction not only induced peptide-specific Th-cell responses but were also produced through natural processing of DEPDC1 or MPHOSPH1 protein by DCs and presented by various HLA class II molecules.
Figure 3.
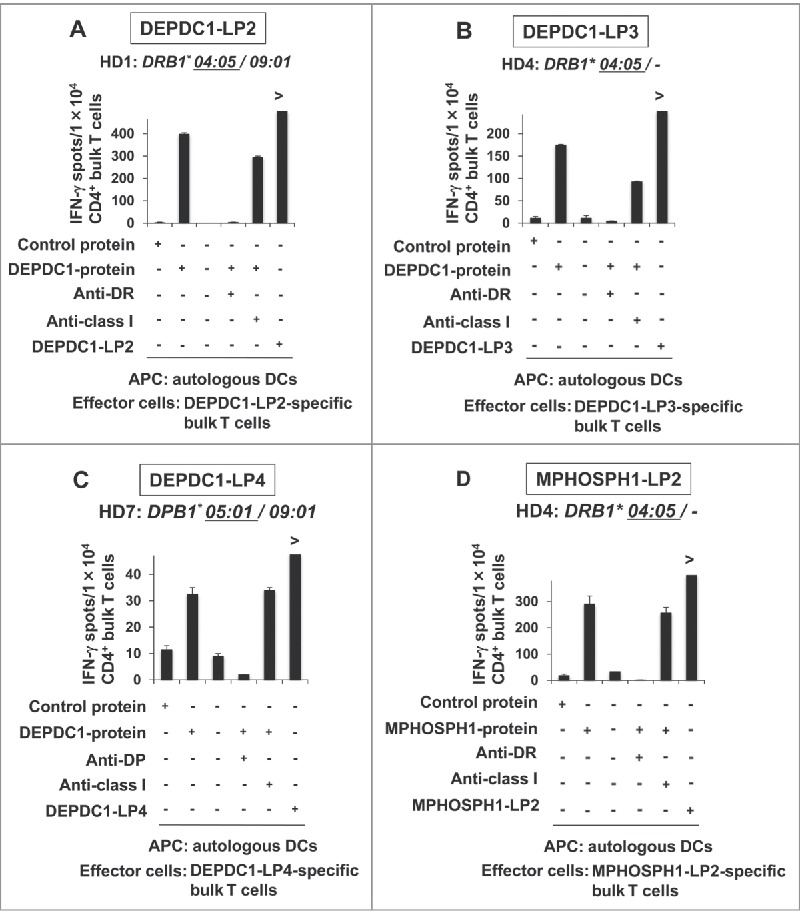
Natural processing and presentation of DEPDC1- or MPHOSPH1-LPs by DCs. Responses of HLA-DR4-restricted and DEPDC1-LP2-specific Th cells from HD1 (A), DEPDC1-LP3 specific Th cells from HD4 (B) and MPHOSPH1-LP2-specific Th cells from HD4 (D). Responses of HLA-DP5-restricted and DEPDC1-LP4-specific Th cells from HD7 (C). Anti-DR and Anti-DP: blocking antibodies to HLA-DR and HLA-DP, respectively. Anti-class I: blocking antibody to pan-HLA class I.
DEPDC1-LP- and MPHOSPH1-LP-induced Th cells produced Th1 cytokines
To characterize DEPDC1-LP- or MPHOSPH1-LP-specific Th cells, we measured various cytokines secreted by the Th cells in response to stimulation with autologous PBMCs pulsed with respective cognate peptide. Th1 cytokines, such as IFN-γ, TNF-α interleukin (IL)-2 and granulocyte macrophage colony-stimulating factor (GM-CSF) but relatively small amount of IL-4 and little IL-17, if any, were produced by DEPDC1-LP- and MPHOSPH1-LP-specific Th cells after restimulation with the cognate peptides (Fig. 4), suggesting that DEPDC1-LPs and MPHOSPH1-LPs had the capacity to induce Th1-polarized Th cells. Therefore, induction of Th cells by DEPDC1-LPs or MPHOSPH1-LPs could be useful for antitumor therapy.
Figure 4.
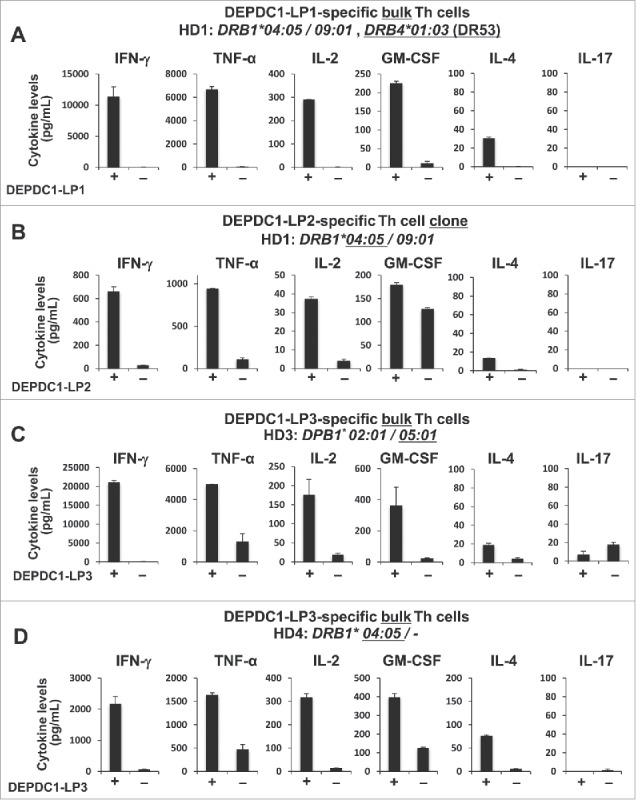
DEPDC1- or MPHOSPH1-LP-specific Th clones produced various Th1-type cytokines. (A–H) DEPDC1- or MPHOSPH1-LP-specific bulk Th cells or Th clones were stimulated with autologous PBMCs pulsed with or without cognate peptides. The culture supernatants were collected after 24 hours of stimulation and the concentrations of indicated cytokines were measured. Data are presented as the means ± standard deviations of triplicate assays.
Figure 4.
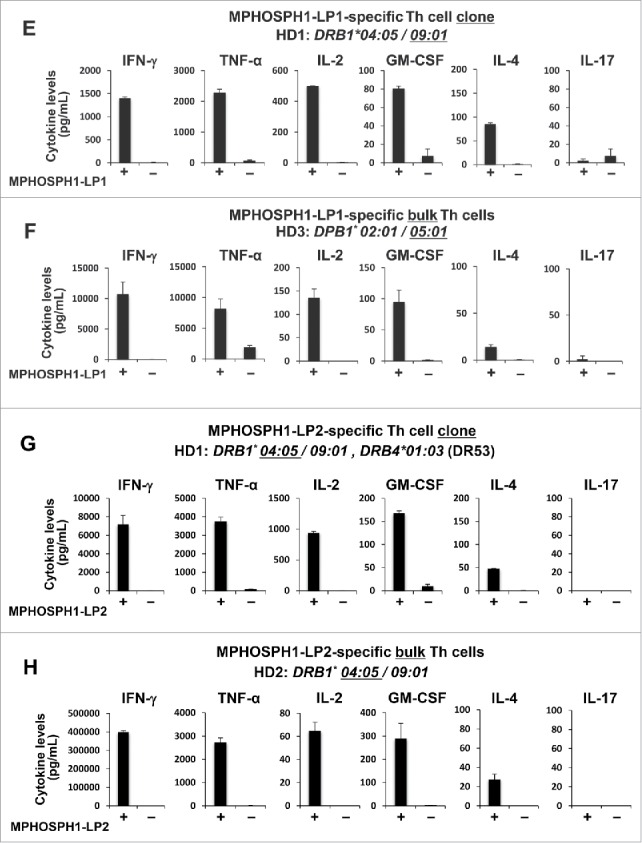
(Continued)
DEPDC1-LP2- or MPHOSPH1-LP1-specific bulk Th cells expressed converged TCR-α and -β genes
The DEPDC1-LP2- or MPHOSPH1-LP1-specific T-cell repertoire was analyzed by deep cDNA sequencing of TCR-α and -β genes using next-generation sequencing. After several stimulations of bulk Th cells, frequency of a particular pair of TCR-α and -β gene usage was increased in both bulk Th cells. DEPDC1-LP2- and MPHOSPH1-LP1-specific T-cell clones established from the bulk T cells by limiting dilution expressed the same pair of TRVA and TRVB genes as the most converged pair of TCR genes observed in the bulk Th cells (a in Fig. 5A-D). Introduction of the full-length TCR-α and -β genes prepared from the T-cell clones into the murine TCR-negative T-cell line TG40 successfully acquired the original antigenic peptide specificity and HLA restriction of the parental T cells, as revealed by up-regulation of CD69 and CD137 detected by flow cytometric analysis (Fig. 5E). These observations confirmed the clonality of the established T cells and specificity of these pairs of TCR-α and -β chains.
Figure 5.

TCR gene analysis in DEPDC1-LP2- or MPHOSPH1-LP1-specific Th cells. (A–D) The DEPDC-LP2- or MPHOSPH1-LP1-specific T-cell repertoires were analyzed by deep cDNA sequencing of T-cell receptor α and β genes using next-generation sequencing. TCR-α is presented in the top panels, and TCR-β is presented in the lower panels. The Y-axis indicates the number of Th cell stimulations with DCs pulsed with the cognate peptide. Cloning of DEPDC1-LP2-specific bulk Th cells was performed after five stimulations of bulk Th cells with irradiated PBMCs pulsed with the cognate peptide. Cloning of MPHOSPH1-LP1-specific bulk Th cells was performed after four stimulations of bulk Th cells with irradiated PBMCs pulsed with the cognate peptide. For both, the total number of stimulation was nine times. The X-axis indicates the top 10 most frequent V-(D)-J-CDR3 sequences (details are shown in the right boxes), the Z-axis indicates the frequency of individual V-J-CDR3 sequences. (E) The TCR-negative murine T-cell line TG40 expressing full-length TCR-α and -β genes isolated from DEPDC1-LP2- and MPHOSPH1-LP1-specific T-cell clones specifically responded to HLA-DR expressing L cells that were pulsed with cognate peptides, as revealed by the cell-surface expression of CD69 and CD137.
Cross-presentation of DEPDC1- and MPHOSPH1-derived LPs efficiently primed SP-specific CD8+ T cells in vivo
The capacity of DEPDC1-LP2 to prime DEPDC1-A2- and DEPDC1-A24-specific CTLs was examined by an ex vivo IFN-γ ELISPOT assay. HLA-A2 and HLA-A24 transgenic mice (Tgm) were immunized with DEPDC1-LP2 emulsified in incomplete Freund's adjuvant (IFA). The CD8+ T cells isolated from the lymph nodes of vaccinated HLA-A2 or HLA-A24 Tgm specifically produced IFN-γ in response to stimulation with bone marrow (BM)-derived DCs pulsed with DEPDC1-A2-SPs or DEPDC1-A24-SPs (Fig. 6A and B). Similar results were obtained from HLA-A2 or HLA-A24 Tgm immunized with MPHOSPH1-LP1 (Fig. 6C and D). These results suggested that after uptake of DEPDC1-LP2 and MPHOSPH1-LP1, BM-DCs cross-primed A2- or A24-DEPDC1-SPs, and A2- or A24-MPHOSPH1-SPs -specific CTLs in vivo in the Tgm immunized with DEPDC1-LP2 and MPHOSPH1-LP1, respectively, which would be advantageous for the induction of antitumor T-cell responses.
Figure 6.
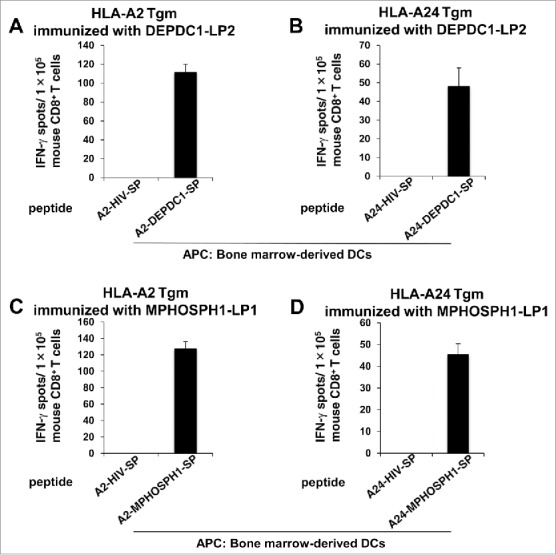
DEPDC1-LP2 and MPHOSPH1-LP1 induced cross-priming of CTLs in vivo. HLA-A2 or HLA-A24 Tgm were immunized with DEPDC1-LP2 or MPHOSPH1-LP1 emulsified in IFA. After vaccination, CD8+ T cells isolated from the inguinal lymph nodes were stimulated ex vivo with BM-DCs pulsed with DEPDC1-A2302–311 (A), DEPDC1-A24294–302 (B), MPHOSPH1-A2282–291 (C), MPHOSPH1-A24278–286 (D), or HIV-A2 or -A24 SPs. The number of IFN-γ-producing murine CD8+ T cells was analyzed by ex vivo ELISPOT assays. Representative data from three independent experiments (two or three mice in each group) performed in duplicate or triplicate are shown.
PBMCs isolated from patients with UC recognized some DEPDC1-LPs and MPHOSPH1-LPs
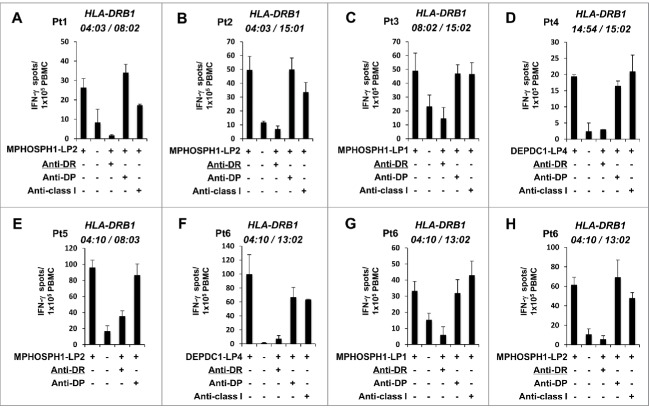
To examine the induction of DEPDC1-LP- and MPHOSPH1-LP-specific Th-cell responses in patients with UC, we stimulated PBMCs isolated from patients with advanced-stage UC with LPs in vitro. Patient characteristics are indicated in Table S5. After stimulation, the frequency of individual DEPDC1-LP- and MPHOSPH1-LP-specifc T cells was measured. DEPDC1-LP4, MPHOSPH1-LP1, and MPHOSPH1-LP2 induced peptide-specific T cells from six patients. Peptide-specific IFN-γ production by T cells was significantly inhibited by anti-HLA-DR monoclonal antibody (mAb) but not by anti-HLA-DP mAb or anti-HLA class I mAb (Fig. 7). These results demonstrated that IFN-γ was produced by DEPDC1-LP- and MPHOSPH1-LP-specific and HLA-DR-restricted CD4+ T cells, indicating that DEPDC1-LP4, MPHOSPH1-LP1, and MPHOSPH1-LP2 induced antigen-specific Th cells not only in HDs but also in UC patients with various HLA-DR alleles.
Figure 7.
The presence of DEPDC1- and MPHOSPH1-LP-specific Th cells in the PBMCs of UC patients. PBMCs isolated from patients with UC were stimulated with a mixture of DEPDC1-LP1, DEPDC1-LP2, DEPDC1-LP3, DEPDC1-LP4, MPHOSPH1-LP1, and MPHOSPH1-LP2 plus IL-2 and IL-7 in vitro, and the frequencies of individual DEPDC1- and MPHOSPH1-LP-specific T cells were assessed using ELISPOT assays. Th-cell responses specific to DEPDC1-LP4, MPHOSPH1-LP1, and MPHOSPH1-LP2 were observed in six patients (A–H). HLA class II restriction of DEPDC1- and MPHOSPH1-LP-specific Th cells was determined by blocking assays using monoclonal antibodies specific to HLA-DR or HLA-DP.
Discussion
Previous phase I/II clinical trials have shown that vaccination with a DEPDC1-SP and an MPHOSPH1-SP alone did not induce dramatic antitumor effects.5,13 Therefore, we attempted to identify DEPDC1- and MPHOSPH1-derived LPs capable of inducing both CTLs and Th1 cells in order to further develop better peptide-based cancer vaccine therapies. Because HLA class II molecules are highly polymorphic, in order to clinically apply LPs activating HLA class II-restricted Th cells to cancer vaccine therapy, it should be noted that the peptides are capable of binding to multiple HLA class II molecules and have a wider range of adaptation.27,29,30 We have demonstrated herein that DEPDC1- and MPHOSPH1-LPs were presented by HLA-DR4, HLA-DR9, HLA-DR15, HLA-DR53, HLA-DP2, and HLA-DP5, suggesting that vaccination with a combination of DEPDC1- and MPHOSPH1-LPs could cover the majority of the Japanese population (Table S4). Although both HD1 and HD4 are DR4-positive, Th cells generated from HD4 was restricted by HLA-DR4 in respect to IFN-γ production upon stimulation with DEPDC1-LP1, but Th cells generated from HD1 was restricted by DR53, resulting in a difference in HLA restriction of Th cells specific to the same DEPDC1-LP1. It is likely that healthy donors have not been previously sensitized with DEPDC1-LP1 itself. Therefore, the T cell response to the LP in healthy donors might be due to cross-reaction of T cells specific to certain other antigens previously exposed. Since the exposure process varies depending on each individual environment and history, HLA restriction of the T cells specific to LP could be different between HD1 and HD4. As a result, we observed the presence of DEPDC1-LP- or MPHOSPH1-LP-specific Th cells in six patients with UC, suggesting that DEPDC1- and MPHOSPH1-LPs may be broadly useful in many of the patients. In some patients, DEPDC1-LP and MPHOSPH1-LP-specific T cell reactivity is inhibited by anti-HLA class I antibody. In the case of UC patients, due to the limitation of available amounts of the patients' blood, PBMCs were directly stimulated with the peptides without sorting of CD4+ T cells and CTLs must be present in the culture. Therefore, observed inhibition by the addition of anti-HLA class I antibody may indicate the presence of peptide-specific CTL responses. Additional studies with more patients harboring various HLA-class II types should be investigated.
When using LPs harboring Th-cell epitopes for cancer immunotherapy, clinicians should consider whether antitumor Th1 cells could be induced. The phenotype of Th cells is affected not only by the surrounding cytokine milieu at the differentiation stage but also by the affinity/avidity of TCR and MHC-peptide complexes.28,31-33 In other words, there is a tendency for CD4+ T cells to differentiate into Th1 cells based on the high density of peptide/HLA class II complexes inducing strong TCR-mediated signals. Conversely, Th cells tend to differentiate into Th2 cells. The DEPDC1- and MPHOSPH1-LP-specific Th cells induced from HDs in this study were all Th1-type cells that mainly produced INF-γ, TNF-α, IL-2 and GM-CSF, although the production of IL-2 was less compared to those of IFN-γ and TNF-α. Therefore, the DEPDC1-LPs and MPHOSPH1-LPs may preferentially trigger Th1-type Th cell differentiation, possibly due to the higher affinity binding to HLA class II molecules, which could produce high-density LP-HLA class II complexes to stimulate durable and strong TCR-mediated activation signals.
A clinical trial demonstrated that vaccination with a human epidermal growth factor receptor 2 (HER-2)/neu-derived LP encompassing an HLA-A2-binding CTL epitope could induce both HER-2/neu-specific Th-cell responses and HLA-A2-binding epitope-specific CTL responses in patients with metastatic breast cancer.34 Furthermore, Melief et al. reported that LPs encompassing CTL epitopes had superior CTL induction efficiency compared with an SP with minimal CTL epitopes because of the increased duration of antigen cross-presentation exclusively by DCs.14,15 The DEPDC1-LP2 and MPHOSPH1-LP1 encompassing known CTL epitopes induced CTL epitope-specific CTLs via the cross-presentation pathway in HLA-A2 or -A24 Tgm in vivo. Therefore, vaccination with these peptides may induce DEPDC1- or MPHOSPH1-LP1-specific Th cells and CTLs simultaneously in patients with cancer. Thus, further analyses are necessary to elucidate which LPs or combination with SPs would induce stronger DEPDC1- and MPHOSPH1-specific antitumor responses in vivo, with the goal of conducting clinical trials.
Although we did not observe the cross-presentation of soluble DEPDC1-LP2 and MPHOSPH1-LP1 by in vitro culture with human DCs, we could observe sufficient cross-priming in vivo using HLA-A2 and HLA-A24 Tgm vaccinated with those LPs. One possible reason for this discrepancy between humans and mice may be the use of IFA as an adjuvant in mouse in vivo experiments. IFA not only promotes antigen persistence at the injection site and prolongs the duration of antigen incorporation by DCs but also increases antigen presentation of lower amounts of antigenic peptides by DCs.35 Therefore, IFA-embedded DEPDC1- and MPHsOSPH1-LPs could be eventually incorporated by DCs and cross-prime CTLs in vivo in HLA-A2 and HLA-A24 Tgm.
DEPDC1-LP1, DEPDC1-LP4, and MPHOSPH1-LP2 efficiently induced LP-specific Th-cell responses; however, they did not contain known HLA class I-restricted CTL epitopes. Thus, we asked whether these were inferior to DEPDC1-LP2, DEPDC1-LP3, and MPHOSPH1-LP1, which contained HLA class I epitopes, to induce antitumor T-cell responses. A recent study reported that the epitope spreading phenomenon is critical for induction of effective antitumor immune responses.36,37 In fact, patients who received therapeutic cancer vaccines (containing minimal CTL epitope peptides) exhibited clinical responses and long-term survival. In those patients, both Th-cell and CTL responses to other epitopes of the same proteins or several unrelated antigens not included in the vaccines were observed. This is because the vaccination induced CTLs specific to the target antigens, and as a result of damaging tumor cells, other TAAs were released, taken up by APCs, and presented to induce other TAA-specific T-cell responses. If this was the case, vaccination with CTL epitope-free DEPDC1-LP1, DEPDC1-LP4, and MPHOSPH1-LP2 would induce LP-specific Th1 responses together with other antitumor Th1 and CTL responses. Furthermore, if the epitopes spread to include neoantigens, i.e., the aberrant peptides derived from proteins encoded by somatic missense mutant genes generated in cancer cells, the antitumor T-cell responses would be more efficient since those neoantigens were non-self. In fact, recent studies have shown that there are many T cells specific to neoantigens, suggesting that the genes encoding for the antigens have mutations and that the spread epitopes may also include neoantigens. Cancer antigen vaccine therapies using neoantigens have attracted much attention.38,39 Beside the induction of the epitope spreading, to induce and activate as many TAA-specific T cells as possible, which is key to the success of cancer vaccine therapy, the vaccine formulation of DEPDC1- and MPHOSPH1-LPs and -SPs may be promising.
Rather than inducing various TAA-specific Th cells and CTLs by vaccination, direct introduction of genetically engineered T cells specific to TAA into patients with cancer may be another effective antitumor therapy. In clinical trials, Rosenberg et al. introduced autologous T cells retrovirally transduced with TCR genes of NY-ESO-1-reactive CTLs into patients with metastatic synovial cell sarcoma and melanoma, exhibiting objective clinical responses.40 In our study, TCRs on DEPDC1-LP- and MPHOSPH1-LP-specific Th cells converged to a combination of TCRA and TCRB genes, respectively, after repeated stimulation. Introduction of each pair of TCR genes into the murine TCR-negative T-cell line TG40 produced cells with the original T-cell specificity to their cognate ligands, suggesting that transfer of patients' autologous T cells expressing those TCR genes could be an effective therapy. The convergence of the TCR repertoire of DEPDC1- and MPHOSPH1-LP-specific Th cells into those with particular complementarity determining region (CDR) sequences seemed to be similar to recent observations by Dash et al. and Glanville et al., in which antigen-specific TCRs shared specifically enriched short sequence motifs in CDRs compared with random TCR repertoires that had not been selected for binding to the particular antigenic peptide.41,42 It is interesting to investigate TCR repertoire usages of CTLs induced by SPs incorporated in LPs, and this is remained to be analyzed in a future study.
In conclusion, we identified four DEPDC1-derived LPs and two MPHOSPH1-derived LPs that could induce promiscuous HLA-class II-restricted Th cells, three of which included HLA-A2- and A24-restricted CTL epitopes. Our results suggested that these LPs could represent a useful tool for propagation of both DEPDC1- and MPHOSPH1-specific Th1 cells and CTLs. These findings provided a basis for clinical trials of DEPDC1-LP- and MPHOSPH1-LP-based immunotherapy in various types of cancer. In addition, converged TCRs expressed on LPs-specific Th cells may be useful for monitoring of LP-specific Th cells in vitro and possibly in vivo.
Materials and methods
Cells
The research protocol for collecting and using PBMCs from HDs and patients with cancer was approved by the Institutional Review Board of Kumamoto University. PBMCs were obtained from HDs and patients with cancer after obtaining written informed consent. Human monocyte-derived DCs and murine BM-derived DCs were generated as previously reported.43-48
Mouse fibroblast cell lines (L cells), genetically engineered to express DR4 (DRB1*04:05, L-DR4), DR9 (DRB1*09:01, L-DR9), DR15 (DRB1*15:02, L-DR15), DR53 (DRB4*01:03, L-DR53), DP2 (DPA1*01:03/DPB1*02:01, L-DP2), DP4 (DPA1*01:03/DPB1*04:01, RM3-DP4), DP5 (DPA1*02:02/DPB1*05:01, L-DP5), and DP9 (DPB1*09:01, L-DP9) were provided by Dr. Alessandro Sette (La Jolla Institute for Allergy and Immunology, CA, USA).49
Prediction and selection of HLA class II-binding peptides
To predict potential promiscuous HLA-class II binding human DEPDC1- or MPHOSPH1-derived peptides, the amino acid sequences of human DEPDC1 or MPHOSPH1 protein were analyzed by a computer algorithm (IEDB analysis resource, consensus method, http://tools.immuneepitope.org/analyze/html/mhc_II_binding.html).26,25 The program ranks overlapping 15-amino acid-long sequences that encompass the entire protein of interest according to the inverse of binding affinities (consensus percentile rank) to particular HLA molecules. The 22–29-amino acid-long peptides DEPDC1191–213LP (DEPDC1-LP1; RYVILIYLQTILGVPSLEEVINP), DEPDC1292–313-LP (DEPDC1-LP2; TFEYYELFVNILVVCGYITVSD), DEPDC1301–329-LP (DEPDC1-LP3; NILVVCGYITVSDRSSGIHKIQDDPQSSK), and DEPDC1613–634-LP (DEPDC1-LP4; NRRKLQLLMRMISRMSQNVDMP), which showed relatively lower consensus percentile ranks for multiple HLA class II molecules encoded by HLA-DRB1*04:05, HLA-DRB1*09:01, HLA-DRB1*15:02, HLA-DPB1*02:01 and HLA-DPB1*05:01 alleles were selected for synthesis (Fig. 1 and Table 1). DEPDC1-LP2 and -LP3 included either DEPDC1-derived 9mer or 10mer CTL epitopes (A24-DEPDC1294–302 and A2-DEPDC1302–311 in DEPDC1-LP2 and A2-DEPDC1302–311 in DEPDC1-LP3).
Similarly, the 27- and 26-amino acid-long peptides MPHOSPH1272–298-LP (MPHOSPH1-LP1; WVSFFEIYNEYIYDLFVPVSSKFQKRK) and MPHOSPH1326–351-LP (MPHOSPH1-LP2; AYRLLKLGIKHQSVAFTKLNNASSRS) were selected for synthesis (Fig. 1 and Table 2). MPHOSPH1-LP1 included the MPHOSPH1-derived 9–10mer CTL epitopes A24-MPHOSPH1278–286 and A2-MPHOSPH1282–291.
Peptide synthesis and preparation of recombinant proteins
Four human DEPDC1- and MPHOSPH1-derived SPs presented by HLA-A2 (A2-DEPDC1302–311 and A2-MPHOSPH1282–291)6 or HLA-A24 (A24-DEPDC1294–302 and A24-MPHOSPH1278–286),7 six LPs (DEPDC1-LP1–4, MPHOSPH1-LP1, and MPHOSPH1-LP2) and HLA-A2-binding human immunodeficiency virus (HIV)-SP as a negative control6,50 were synthesized by MBL (Nagoya, Japan; purity > 95%; Fig. 1B). A promiscuous HIV-derived LP were synthesized and used as the LP-negative control.51 Peptides were dissolved in dimethylsulfoxide at 10 μg/μL and stored at −80°C until use.
Recombinant human whole DEPDC1 and MPHOSPH1 proteins were generated in Escherichia coli BL21 strain transformed with pET28a vectors (69864–3; Novagen, Buenos Aires, AR) encoding DEPDC1 and MPHOSPH1, respectively, according to the manufacturer's instructions. The molecular sizes of both DEPDC1 and MPHOSPH1 proteins were assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and purified by HisTrap FF columns (GE Healthcare, Little Chalfont, Buckinghamshire, UK). In assays for specific T-cell responses to DEPDC1 protein, MPHOSPH1 protein was used as a negative control, and vice versa.
Generation of antigen-specific CD4+ T-cells from HDs
A fraction of PBMCs obtained from seven HDs was subjected to genotyping of HLA-A, DRB1, and DPB1 alleles at the HLA Laboratory (Kyoto, Japan; Table S3). Induction of antigen-specific CD4+ T cells from the PBMCs was performed as described previously using monocyte-derived DCs as APCs.50-52 The CD4+ T cells were purified by positive selection using magnetic microbeads (Miltenyi Biotec, Auburn, California, USA).53
Mixed 6 peptides were used from 1st stimulation until 3rd stimulation. The specificity of CD4+ Tcells was assessed by IFN-γ ELISPOT assays (BD Biosciences, Franklin Lakes, New Jersey, USA) as described previously,50 and the stimulation from the 4th time onward was done using a specific LP.
Assessment of T-cell responses to peptides
The immune responses of CD4+ cells were assessed by IFN-γ ELISPOT assays (BD Biosciences), as described previously.50 Briefly, the number of IFN-γ-producing peptide-specific CD4+ T cells per 3 × 104 bulk CD4+ T cells upon stimulation with peptide-pulsed irradiated PBMCs (3 × 104) or per 1 × 104 bulk CD4+ T cells upon stimulation with peptide-pulsed HLA-DR-expressing L cells (5 × 104/well) was analyzed. To determine the restricting HLA molecules, blocking of antigen-induced IFN-γ production was investigated by adding anti-HLA-DR mAbs (L243; BioLegend, San Diego, California, USA), anti-HLA-DP mAbs (B7/21; Abcam, Cambridge, UK), anti-HLA-DQ mAbs (SPV-L3; Abcam), or anti-HLA class I mAbs (W6/32; Abcam). All mAbs were used at a final concentration of 5 μg/mL. All assessments of IFN-γ ELISPOT assays were carried out in triplicate or duplicate, and results are presented as means ± standard deviations.
Assessment of DEPDC1- and MPHOSPH1-LP-specific CD4+ T-cell responses in patients with UC
PBMCs obtained from patients with six UC were cultured in the presence of a mixture of DEPDC1-LPs or MPHOSPH1-LPs (20 μg/mL) in a final volume of 2 mL AIM-V medium (Invitrogen, Carlsbad, California, USA) supplemented with 5% human decomplemented plasma at 37°C (2 × 106/well, 24-well plates). IL-2 (20 U/mL) and IL-7 (5 ng/mL) were added on days 0 and 2. After 7 days of cell culture, the cells were collected, washed, and transferred in ELISPOT plates (5–10 × 104/well) with DEPDC1-LP, MPHOSPH1-LP, or control LPs for 18 h. The numbers of DEPDC1- or MPHOSPH1-LP-specific PBMCs were counted as described above.
In vivo cross-priming assay
HLA-A2 and HLA-A24 (HHD) Tgm were kindly provided by Dr. F.A. Lemonnier.54,55 This study was approved by the animal research committee of Kumamoto University. The mice were maintained at the Center for Animal Resources and Development of Kumamoto University, and were handled in accordance with the animal care guidelines of Kumamoto University. The DEPDC1-LP2 or MPHOSPH1-LP1 emulsified in IFA were intradermally injected at the tail base of the Tgm at 7-day intervals. Seven days after the second vaccination, CD8+ T cells were isolated from inguinal lymph nodes by positive selection with magnetic microbeads (Miltenyi Biotec). The number of IFN-γ-producing CD8+ T cells (1 × 105/well) upon stimulation with A2-DEPDC1302–311SP-, A24-DEPDC1294–302SP-, A2-MPHOSPH1282–291SP-, or A24-MPHOSPH1278–286-SP-pulsed BM-DCs (2 × 104/well) was counted by ex vivo ELISPOT assays.52,56
Cytokine analysis
DEPDC1- or MPHOSPH1-LP-specific bulk Th cells or Th clones (1 × 104/well) were cultured with autologous PBMCs (3 × 104/well) pulsed with cognate peptides in 96-well plates. Using the Bio-Plex system (Bio-Rad, Hercules, CA, USA), cytokine levels in the culture supernatants after 24 h were measured according to the manufacturer's instructions.
TCR sequencing
T cells were cloned by limiting dilution for further studies as described previously.57 After 5th stimulation with DEPDC1-LP or after 4th stimulation with MPHOSPH1-LP, Th cells were stimulated with specific single peptide. A DEPDC1- LP2-specific Th cell clone was established from five times stimulated bulk Th cells. A MPHOSPH1-LP1-specific Th cell clone was established from four times stimulated bulk Th cells. In both of them, the final stimulation counts were 9 times.
Total RNAs were isolated from DEPDC1-LP2- or MPHOSPH1-LP1-specific T cells using an RNeasy mini kit (Qiagen, Valencia, CA, USA). Sequencing libraries for TRAV and TRBV were prepared as described previously,58,59 with some modifications, and then subjected to sequencing on the Illumina Miseq platform using 600 cycles Miseq Reagent Kit V3 (Illumina, Inc., San Diego, CA, USA). Sequence reads were then analyzed with the algorithm described previously.58 To identify V-(D)-J segments in individual TRAV and TRBV sequencing reads, each of the sequence reads in FASTQ files were mapped to the reference sequences derived from IMGT/GENE-DB60 using Bowtie2 aligner (Version 2.1.0).61,62 To define amino acid sequences of CDR3 in TRAV and TRBV, a conserved cysteine encoded in the 3′ portion of the V segment and a conserved phenylalanine encoded in the 5′ portion of the J segments that formed the boundaries of CDR3 were identified.58 The nucleotide sequences of the conserved cysteine and phenylalanine were extracted to determine the amino acid sequence of the CDR3 region.
TCR cloning and expression in TG40 cells
TCR cDNAs were amplified from T cells using one-step multiplex reverse transcription-polymerase chain reaction (PCR), as described previously.63 The DNA sequences of the amplified PCR products were determined by direct sequencing. To analyze the antigen-specificity of the obtained TCR cDNAs, expression vectors for the TCRs were constructed and then transferred to Plat-E cells to produce the recombinant retroviruses, as described previously.64-66 The produced TCR genes were transferred to TG40 cells67 as described previously.65 Next, 1 × 105 cells of TCR-transferred TG40 cells were cocultured with 1 × 105 cells of restricting HLA-DR-expressing L cells in the presence of 10 μg/mL of antigenic peptides overnight at 37°C in an atmosphere containing 5% CO2, and the expression of CD69 and CD137 on the cell surface was analyzed with a flow cytometer.
Statistical analysis
We compared data by two-tailed Student's t-tests (bar graphs) or by nonparametric Mann-Whitney U tests (scatter-dot graph). Differences with P values of less than 0.05 were considered statistically significant for all tests.
Supplementary Material
Funding Statement
This research was supported by a MEXT Grant-in-Aid for Scientific Research on Innovative Areas (grant number 22133005); JSPS KAKENHI (grant numbers 23650609, 24300334, 15H04311 and 16H06498 for Scientific Research on Innovative Area “Neo-self”); the Project for Cancer Research And Therapeutic Evolution (P-CREATE) from the Japan Agency for Medical Research and Development, AMED; and OncoTherapy Science, Inc.
Disclosure of potential conflict of interests
Isao Fukuda, Poh Yi Yew, and Sachiko Yoshimura are current employees of Onco Therapy Science, Inc. and Cancer Precision Medicine, Inc. Yasuharu Nishimura is supported by funding from Onco Therapy Science, Inc.
Acknowledgments
We would like to thank Dr. Hirotake Tsukamoto of Kumamoto University for helpful advices on this study. We also would like to thank Drs. Tokunori Ikeda, Yusuke Tomita, Akira Yuno and Taku Kojima of Kumamoto University for their cooperation in this study.
References
- 1.von der Maase H, Sengelov L, Roberts JT, Ricci S, Dogliotti L, Oliver T, Moore MJ, Zimmermann A, Arning M. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol. 2005;23(21):4602–4608. doi: 10.1200/JCO.2005.07.757. [DOI] [PubMed] [Google Scholar]
- 2.Gupta S, Gill D, Poole A, Agarwal N. Systemic immunotherapy for urothelial cancer: current trends and future directions. Cancers. 2017;9(2):E15. doi: 10.3390/cancers9020015. PMID:28134806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Santis M, Bellmunt J, Mead G, Kerst JM, Leahy M, Maroto P, Gil T, Marreaud S, Daugaard G, Skoneczna I, et al.. Randomized phase II/III trial assessing gemcitabine/carboplatin and methotrexate/carboplatin/vinblastine in patients with advanced urothelial cancer who are unfit for cisplatin-based chemotherapy: EORTC study 30986. J Clin Oncol. 2012;30(2):191–199. doi: 10.1200/JCO.2011.37.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zichi C, Tucci M, Leone G, Buttigliero C, Vignani F, Pignataro D, Scagliotti GV, Di Maio M. Immunotherapy for patients with advanced urothelial cancer: current evidence and future perspectives. Biomed Res Int. 2017;2017:5618174. PMID:28680882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obara W, Ohsawa R, Kanehira M, Takata R, Tsunoda T, Yoshida K, Takeda K, Katagiri T, Nakamura Y, Fujioka T. Cancer peptide vaccine therapy developed from oncoantigens identified through genome-wide expression profile analysis for bladder cancer. Jpn J Clin Oncol. 2012;42(7):591–600. doi: 10.1093/jjco/hys069. PMID:22636067 [DOI] [PubMed] [Google Scholar]
- 6.Kanehira M, Harada Y, Takata R, Shuin T, Miki T, Fujioka T, Nakamura Y, Katagiri T. Involvement of upregulation of DEPDC1 (DEP domain containing 1) in bladder carcinogenesis. Oncogene. 2007;26(44):6448–6455. doi: 10.1038/sj.onc.1210466. PMID:17452976 [DOI] [PubMed] [Google Scholar]
- 7.Harada Y, Kanehira M, Fujisawa Y, Takata R, Shuin T, Miki T, Fujioka T, Nakamura Y, Katagiri T. Cell-permeable peptide DEPDC1-ZNF224 interferes with transcriptional repression and oncogenicity in bladder cancer cells. Cancer Res. 2010;70(14):5829–5839. doi: 10.1158/0008-5472.CAN-10-0255. PMID:20587513 [DOI] [PubMed] [Google Scholar]
- 8.Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10(1):15. doi: 10.1186/1476-4598-10-15. PMID:21314937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishiu M, Yanagawa R, Nakatsuka S, Yao M, Tsunoda T, Nakamura Y, Aozasa K. Microarray analysis of gene-expression profiles in diffuse large B-cell lymphoma: identification of genes related to disease progression. Jpn J Cancer Res. 2002;93(8):894–901. doi: 10.1111/j.1349-7006.2002.tb01335.x. PMID:12716467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanehira M, Katagiri T, Shimo A, Takata R, Shuin T, Miki T, Fujioka T, Nakamura Y. Oncogenic role of MPHOSPH1, a cancer-testis antigen specific to human bladder cancer. Cancer Res. 2007;67(7):3276–3285. doi: 10.1158/0008-5472.CAN-06-3748. PMID:17409436 [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Zhou Y, Liu X, Peng A, Gong H, Huang L, Ji K, Petersen RB, Zheng L, Huang K. MPHOSPH1: a potential therapeutic target for hepatocellular carcinoma. Cancer Res. 2014;74(22):6623–6634. doi: 10.1158/0008-5472.CAN-14-1279. PMID:25269478 [DOI] [PubMed] [Google Scholar]
- 12.Miyata Y, Kumagai K, Nagaoka T, Kitaura K, Kaneda G, Kanazawa H, Suzuki S, Hamada Y, Suzuki R. Clinicopathological significance and prognostic value of Wilms' tumor gene expression in colorectal cancer. Cancer Biomark. 2015;15(6):789–797. doi: 10.3233/CBM-150521. PMID:26406403 [DOI] [PubMed] [Google Scholar]
- 13.Obara W, Eto M, Mimata H, Kohri K, Mitsuhata N, Miura I, Shuin T, Miki T, Koie T, Fujimoto H, et al.. A phase I/II study of cancer peptide vaccine S-288310 in patients with advanced urothelial carcinoma of the bladder. Ann Oncol. 2017;28(4):798–803. PMID:27998971 [DOI] [PubMed] [Google Scholar]
- 14.Melief CJ, van der Burg SH. Immunotherapy of established (pre)malignant disease by synthetic long peptide vaccines. Nat Rev Cancer. 2008;8(5):351–360. doi: 10.1038/nrc2373. PMID:18418403 [DOI] [PubMed] [Google Scholar]
- 15.Bijker MS, van den Eeden SJ, Franken KL, Melief CJ, van der Burg SH, Offringa R. Superior induction of anti-tumor CTL immunity by extended peptide vaccines involves prolonged, DC-focused antigen presentation. Eur J Immunol. 2008;38(4):1033–1042. doi: 10.1002/eji.200737995. PMID:18350546 [DOI] [PubMed] [Google Scholar]
- 16.Chamoto K, Tsuji T, Funamoto H, Kosaka A, Matsuzaki J, Sato T, Abe H, Fujio K, Yamamoto K, Kitamura T, et al.. Potentiation of tumor eradication by adoptive immunotherapy with T-cell receptor gene-transduced T-helper type 1 cells. Cancer Res. 2004;64(1):386–390. doi: 10.1158/0008-5472.CAN-03-2596. PMID:14729649 [DOI] [PubMed] [Google Scholar]
- 17.Bevan MJ. Helping the CD8(+) T-cell response. Nat Rev Immunol. 2004;4(8):595–602. doi: 10.1038/nri1413. PMID:15286726 [DOI] [PubMed] [Google Scholar]
- 18.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300(5617):337–339. doi: 10.1126/science.1082305. PMID:12690201 [DOI] [PubMed] [Google Scholar]
- 19.Anders K, Buschow C, Herrmann A, Milojkovic A, Loddenkemper C, Kammertoens T, Daniel P, Yu H, Charo J, Blankenstein T. Oncogene-targeting T cells reject large tumors while oncogene inactivation selects escape variants in mouse models of cancer. Cancer Cell. 2011;20(6):755–767. doi: 10.1016/j.ccr.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Street SE, Cretney E, Smyth MJ. Perforin and interferon-gamma activities independently control tumor initiation, growth, and metastasis. Blood. 2001;97(1):192–197. doi: 10.1182/blood.V97.1.192. PMID:11133760 [DOI] [PubMed] [Google Scholar]
- 21.Bos R, Sherman LA. CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res. 2010;70(21):8368–8377. doi: 10.1158/0008-5472.CAN-10-1322. PMID:20940398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenter GG, Welters MJ, Valentijn AR, Lowik MJ, Berends-van der Meer DM, Vloon AP, Essahsah F, Fathers LM, Offringa R, Drijfhout JW, et al.. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N Engl J Med. 2009;361(19):1838–1847. doi: 10.1056/NEJMoa0810097. PMID:19890126 [DOI] [PubMed] [Google Scholar]
- 23.Brunsvig PF, Kyte JA, Kersten C, Sundstrom S, Moller M, Nyakas M, Hansen GL, Gaudernack G, Aamdal S. Telomerase peptide vaccination in NSCLC: a phase II trial in stage III patients vaccinated after chemoradiotherapy and an 8-year update on a phase I/II trial. Clin Cancer Res. 2011;17(21):6847–6857. doi: 10.1158/1078-0432.CCR-11-1385. PMID:21918169 [DOI] [PubMed] [Google Scholar]
- 24.Kyte JA, Gaudernack G, Dueland S, Trachsel S, Julsrud L, Aamdal S. Telomerase peptide vaccination combined with temozolomide: a clinical trial in stage IV melanoma patients. Clin Cancer Res. 2011;17(13):4568–4580. doi: 10.1158/1078-0432.CCR-11-0184. PMID:21586625 [DOI] [PubMed] [Google Scholar]
- 25.Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4(4):e1000048. doi: 10.1371/journal.pcbi.1000048. PMID:18389056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. PMID:21092157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabowska AK, Kaufmann AM, Riemer AB. Identification of promiscuous HPV16-derived T helper cell epitopes for therapeutic HPV vaccine design. Int J Cancer. 2015;136(1):212–224. doi: 10.1002/ijc.28968. PMID:24824905 [DOI] [PubMed] [Google Scholar]
- 28.Constant S, Pfeiffer C, Woodard A, Pasqualini T, Bottomly K. Extent of T cell receptor ligation can determine the functional differentiation of naive CD4+ T cells. J Exp Med. 1995;182(5):1591–1596 doi: 10.1084/jem.182.5.1591. PMID:7595230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi H, Song Y, Hoon DS, Appella E, Celis E. Tumor-reactive T helper lymphocytes recognize a promiscuous MAGE-A3 epitope presented by various major histocompatibility complex class II alleles. Cancer Res. 2001;61(12):4773–4778. PMID:11406551 [PubMed] [Google Scholar]
- 30.Zarour HM, Maillere B, Brusic V, Coval K, Williams E, Pouvelle-Moratille S, Castelli F, Land S, Bennouna J, Logan T, et al.. NY-ESO-1 119-143 is a promiscuous major histocompatibility complex class II T-helper epitope recognized by Th1- and Th2-type tumor-reactive CD4+ T cells. Cancer Res. 2002;62(1):213–218. PMID:11782380 [PubMed] [Google Scholar]
- 31.Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J Exp Med. 2005;202(6):793–804. doi: 10.1084/jem.20051304. PMID:16172258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Panhuys N, Klauschen F, Germain RN. T-cell-receptor-dependent signal intensity dominantly controls CD4(+) T cell polarization in vivo. Immunity. 2014;41(1):63–74. doi: 10.1016/j.immuni.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosken NA, Shibuya K, Heath AW, Murphy KM, O'Garra A. The effect of antigen dose on CD4+ T helper cell phenotype development in a T cell receptor-alpha beta-transgenic model. J Exp Med. 1995;182(5):1579–1584. doi: 10.1084/jem.182.5.1579. PMID:7595228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Disis ML, Wallace DR, Gooley TA, Dang Y, Slota M, Lu H, Coveler AL, Childs JS, Higgins DM, Fintak PA, et al.. Concurrent trastuzumab and HER2/neu-specific vaccination in patients with metastatic breast cancer. J Clin Oncol. 2009;27(28):4685–4692. doi: 10.1200/jco.2008.20.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson D, Bundell C, Robinson B. In vivo cross-presentation of a soluble protein antigen: kinetics, distribution, and generation of effector CTL recognizing dominant and subdominant epitopes. J Immunol. 2000;165(11):6123–6132. doi: 10.4049/jimmunol.165.11.6123. PMID:11086045 [DOI] [PubMed] [Google Scholar]
- 36.Inderberg-Suso EM, Trachsel S, Lislerud K, Rasmussen AM, Gaudernack G. Widespread CD4+ T-cell reactivity to novel hTERT epitopes following vaccination of cancer patients with a single hTERT peptide GV1001. OncoImmunology. 2012;1(5):670–686. doi: 10.4161/onci.20426. PMID:22934259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corbiere V, Chapiro J, Stroobant V, Ma W, Lurquin C, Lethe B, van Baren N, Van den Eynde BJ, Boon T, Coulie PG. Antigen spreading contributes to MAGE vaccination-induced regression of melanoma metastases. Cancer Res. 2011;71(4):1253–1262. doi: 10.1158/0008-5472.CAN-10-2693. PMID:21216894 [DOI] [PubMed] [Google Scholar]
- 38.Kreiter S, Vormehr M, van de Roemer N, Diken M, Lower M, Diekmann J, Boegel S, Schrors B, Vascotto F, Castle JC, et al.. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520(7549):692–696. doi: 10.1038/nature14426. PMID:25901682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linnemann C, van Buuren MM, Bies L, Verdegaal EM, Schotte R, Calis JJ, Behjati S, Velds A, Hilkmann H, Atmioui DE, et al.. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med. 2015;21(1):81–85. doi: 10.1038/nm.3773. PMID:25531942 [DOI] [PubMed] [Google Scholar]
- 40.Robbins PF, Kassim SH, Tran TL, Crystal JS, Morgan RA, Feldman SA, Yang JC, Dudley ME, Wunderlich JR, Sherry RM, et al.. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res. 2015;21(5):1019–1027. doi: 10.1158/1078-0432.CCR-14-2708. PMID:25538264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dash P, Fiore-Gartland AJ, Hertz T, Wang GC, Sharma S, Souquette A, Crawford JC, Clemens EB, Nguyen THO, Kedzierska K, et al.. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547(7661):89–93. doi: 10.1038/nature22383. PMID:28636592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, Ji X, Han A, Krams SM, Pettus C, et al.. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547(7661):94–98. doi: 10.1038/nature22976. PMID:28636589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomita Y, Harao M, Senju S, Imai K, Hirata S, Irie A, Inoue M, Hayashida Y, Yoshimoto K, Shiraishi K, et al.. Peptides derived from human insulin-like growth factor-II mRNA binding protein 3 can induce human leukocyte antigen-A2-restricted cytotoxic T lymphocytes reactive to cancer cells. Cancer Sci. 2011;102(1):71–78. doi: 10.1111/j.1349-7006.2010.01780.x. PMID:21087352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomita Y, Yuno A, Tsukamoto H, Senju S, Kuroda Y, Hirayama M, Irie A, Kawahara K, Yatsuda J, Hamada A, et al.. Identification of promiscuous KIF20A long peptides bearing both CD4+ and CD8+ T-cell epitopes: KIF20A-specific CD4+ T-cell immunity in patients with malignant tumor. Clin Cancer Res. 2013;19(16):4508–4520. doi: 10.1158/1078-0432.CCR-13-0197. PMID:23714729 [DOI] [PubMed] [Google Scholar]
- 45.Tomita Y, Yuno A, Tsukamoto H, Senju S, Yoshimura S, Osawa R, Kuroda Y, Hirayama M, Irie A, Hamada A, et al.. Identification of CDCA1-derived long peptides bearing both CD4+ and CD8+ T-cell epitopes: CDCA1-specific CD4+ T-cell immunity in cancer patients. Int J Cancer. 2014;134(2):352–366. doi: 10.1002/ijc.28376. PMID:24734272 [DOI] [PubMed] [Google Scholar]
- 46.Tomita Y, Yuno A, Tsukamoto H, Senju S, Kuroda Y, Hirayama M, Imamura Y, Yatsuda J, Sayem MA, Irie A, et al.. Identification of immunogenic LY6K long peptide encompassing both CD4+ and CD8+ T-cell epitopes and eliciting CD4+ T-cell immunity in patients with malignant disease. OncoImmunology. 2014;3:e28100. doi: 10.4161/onci.28100. PMID:25340007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sayem MA, Tomita Y, Yuno A, Hirayama M, Irie A, Tsukamoto H, Senju S, Yuba E, Yoshikawa T, Kono K, et al.. Identification of glypican-3-derived long peptides activating both CD8+ and CD4+ T cells; prolonged overall survival in cancer patients with Th cell response. OncoImmunology. 2016;5(1):e1062209. doi: 10.1080/2162402X.2015.1062209. PMID:26942076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirayama M, Tomita Y, Yuno A, Tsukamoto H, Senju S, Imamura Y, Sayem MA, Irie A, Yoshitake Y, Fukuma D, et al.. An oncofetal antigen, IMP-3-derived long peptides induce immune responses of both helper T cells and CTLs. OncoImmunology. 2016;5(6):e1123368. doi: 10.1080/2162402X.2015.1123368. PMID:27471607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKinney DM, Southwood S, Hinz D, Oseroff C, Arlehamn CS, Schulten V, Taplitz R, Broide D, Hanekom WA, Scriba TJ, et al.. A strategy to determine HLA class II restriction broadly covering the DR, DP, and DQ allelic variants most commonly expressed in the general population. Immunogenetics. 2013;65(5):357–370. doi: 10.1007/s00251-013-0684-y. PMID:23392739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomita Y, Imai K, Senju S, Irie A, Inoue M, Hayashida Y, Shiraishi K, Mori T, Daigo Y, Tsunoda T, et al.. A novel tumor-associated antigen, cell division cycle 45-like can induce cytotoxic T-lymphocytes reactive to tumor cells. Cancer Sci. 2011;102(4):697–705. doi: 10.1111/j.1349-7006.2011.01865.x. PMID:21231984 [DOI] [PubMed] [Google Scholar]
- 51.Ramduth D, Day CL, Thobakgale CF, Mkhwanazi NP, de Pierres C, Reddy S, van der Stok M, Mncube Z, Nair K, Moodley ES, et al.. Immunodominant HIV-1 Cd4+ T cell epitopes in chronic untreated clade C HIV-1 infection. PLoS One. 2009;4(4):e5013. doi: 10.1371/journal.pone.0005013. PMID:19352428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harao M, Hirata S, Irie A, Senju S, Nakatsura T, Komori H, Ikuta Y, Yokomine K, Imai K, Inoue M, et al.. HLA-A2-restricted CTL epitopes of a novel lung cancer-associated cancer testis antigen, cell division cycle associated 1, can induce tumor-reactive CTL. Int J Cancer. 2008;123(11):2616–2625. doi: 10.1002/ijc.23823. PMID:18770861 [DOI] [PubMed] [Google Scholar]
- 53.Inoue M, Senju S, Hirata S, Ikuta Y, Hayashida Y, Irie A, Harao M, Imai K, Tomita Y, Tsunoda T, et al.. Identification of SPARC as a candidate target antigen for immunotherapy of various cancers. Int J Cancer. 2010;127(6):1393–1403. doi: 10.1002/ijc.25160. PMID:20063317 [DOI] [PubMed] [Google Scholar]
- 54.Firat H, Garcia-Pons F, Tourdot S, Pascolo S, Scardino A, Garcia Z, Michel ML, Jack RW, Jung G, Kosmatopoulos K, et al.. H-2 class I knockout, HLA-A2.1-transgenic mice: a versatile animal model for preclinical evaluation of antitumor immunotherapeutic strategies. Eur J Immunol. 1999;29(10):3112–3121. doi: 10.1002/(SICI)1521-4141(199910)29:10%3c3112::AID-IMMU3112%3e3.0.CO;2-Q. PMID:10540322 [DOI] [PubMed] [Google Scholar]
- 55.Jung KO, Khan AM, Tan BY, Hu Y, Simon GG, Nascimento EJ, Lemonnier F, Brusic V, Miotto O, Tan TW, et al.. West Nile virus T-cell ligand sequences shared with other flaviviruses: a multitude of variant sequences as potential altered peptide ligands. J Virol. 2012;86(14):7616–7624. doi: 10.1128/JVI.00166-12. PMID:22573867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inoue M, Senju S, Hirata S, Irie A, Baba H, Nishimura Y. An in vivo model of priming of antigen-specific human CTL by Mo-DC in NOD/Shi-scid IL2rgamma(null) (NOG) mice. Immunol Lett. 2009;126(1-2):67–72. doi: 10.1016/j.imlet.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Tabata H, Kanai T, Yoshizumi H, Nishiyama S, Fujimoto S, Matsuda I, Yasukawa M, Matsushita S, Nishimura Y. Characterization of self-glutamic acid decarboxylase 65-reactive CD4+ T-cell clones established from Japanese patients with insulin-dependent diabetes mellitus. Hum Immunol. 1998;59(9):549–560. doi: 10.1016/S0198-8859(98)00050-0. PMID:9757911 [DOI] [PubMed] [Google Scholar]
- 58.Fang H, Yamaguchi R, Liu X, Daigo Y, Yew PY, Tanikawa C, Matsuda K, Imoto S, Miyano S, Nakamura Y. Quantitative T cell repertoire analysis by deep cDNA sequencing of T cell receptor alpha and beta chains using next-generation sequencing (NGS). OncoImmunology. 2014;3(12):e968467. doi: 10.4161/21624011.2014.968467. PMID:25964866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choudhury NJ, Kiyotani K, Yap KL, Campanile A, Antic T, Yew PY, Steinberg G, Park JH, Nakamura Y, O'Donnell PH. Low T-cell receptor diversity, high somatic mutation burden, and high neoantigen load as predictors of clinical outcome in muscle-invasive bladder cancer. Eur Urol Focus. 2016;2(4):445–452. doi: 10.1016/j.euf.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 60.Giudicelli V, Chaume D, Lefranc MP. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33(Database issue):D256–261. doi: 10.1093/nar/gki010. PMID:15608191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. PMID:22388286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lefranc MP, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D, Ginestoux C, Clement O, Chaume D, Lefranc G. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2005;33(Database issue):D593–597. doi: 10.1093/nar/gki065. PMID:15608269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamana H, Shitaoka K, Kishi H, Ozawa T, Muraguchi A. A novel, rapid and efficient method of cloning functional antigen-specific T-cell receptors from single human and mouse T-cells. Biochem Biophys Res Commun. 2016;474(4):709–714. doi: 10.1016/j.bbrc.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 64.Mou Z, Li J, Boussoffara T, Kishi H, Hamana H, Ezzati P, Hu C, Yi W, Liu D, Khadem F, et al.. Identification of broadly conserved cross-species protective Leishmania antigen and its responding CD4+ T cells. Sci Transl Med. 2015;7(310):310ra167. doi: 10.1126/scitranslmed.aac5477. PMID:26491077 [DOI] [PubMed] [Google Scholar]
- 65.Kobayashi E, Mizukoshi E, Kishi H, Ozawa T, Hamana H, Nagai T, Nakagawa H, Jin A, Kaneko S, Muraguchi A. A new cloning and expression system yields and validates TCRs from blood lymphocytes of patients with cancer within 10 days. Nat Med. 2013;19(11):1542–1546. doi: 10.1038/nm.3358. PMID:24121927 [DOI] [PubMed] [Google Scholar]
- 66.Miyama T, Kawase T, Kitaura K, Chishaki R, Shibata M, Oshima K, Hamana H, Kishi H, Muraguchi A, Kuzushima K, et al.. Highly functional T-cell receptor repertoires are abundant in stem memory T cells and highly shared among individuals. Sci Rep. 2017;7(1):3663. doi: 10.1038/s41598-017-03855-x. PMID:28623251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohno H, Ushiyama C, Taniguchi M, Germain RN, Saito T. CD2 can mediate TCR/CD3-independent T cell activation. J Immunol. 1991;146(11):3742–3746. PMID:1674520 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.