Abstract
In multicellular organisms, regulation of telomere length in pluripotent stem cells is critical to ensure organism development and survival. Telomeres consist of repetitive DNA that are progressively lost with each cellular division. When telomeres become critically short, they activate a DNA damage response that results in cell cycle arrest. To counteract telomere attrition pluripotent stem cells are equipped with telomere elongation mechanisms that ensure prolonged proliferation capacity and self-renewal capacity. Excessive telomere elongation can also be deleterious and is counteracted by a rapid telomere deletion mechanism termed telomere trimming. While the consequences of critically short telomeres are well established, we are only beginning to understand the mechanisms that counteract excessive telomere elongation. The balance between telomere elongation and shortening determine the telomere length set point in pluripotent stem cells and ensures sustained proliferative potential without causing chromosome instability.
Keywords: Telomere, Stem Cells, Shelterin, TZAP, ZBTB48, Telomere Trimming
Telomere Length Regulation
Telomeres are essential nucleoprotein structures required to cap and protect chromosome ends. In mammals, telomeres consist of repetitive [TTAGGG]n sequences that represent the binding site of a protective protein complex called shelterin (de Lange, 2005). Shelterin is a six-protein complex containing two double-stranded binding proteins TRF1 and TRF2 that specifically recruit the rest of the complex (TIN2-TPP1-POT1 and RAP1) to chromosome ends. The shelterin complex shapes telomeric DNA into a lasso–like secondary “t-loop” that results from invasion of the single-stranded 3′ telomeric overhang into the double stranded telomeric region(Doksani et al., 2013; Griffith et al., 1999). T-loop structures as well as binding of the shelterin complex ensure chromosome end protection from nucleolytic degradation and activation of the DNA damage response (Denchi, 2009; Denchi and de Lange, 2007; Doksani et al., 2013; Karlseder et al., 2004; Okamoto et al., 2013; Sfeir and de Lange, 2012). Gradual telomere shortening occurs with each cellular division due to the inability of DNA polymerases to completely replicate a linear template, the so-called “end replication problem” (Olovnikov, 1973; Watson, 1972). As a result, replicating cells undergo progressive telomere attrition that, if not counteracted by telomere elongating mechanisms, results in critically short telomeres that do not recruit sufficient shelterin complex. Telomere length is a major determinant of the proliferation potential in cells that lack telomere lengthening mechanisms such as human somatic cell lines.
Telomere elongation is ensured by telomerase, an enzyme composed of the reverse transcriptase TERT and an RNA template, TERC, as well as associating proteins such as the ribonucleoprotein Dyskerin (Cohen et al., 2007). Telomerase performs de novo addition of TTAGGG repeats to chromosome ends, allowing replenishment of terminal sequences lost due to the end replication problem (Blackburn, 1997) (Figure 1). As a result, telomere elongation in germ cells and stem cells is critical to ensure sufficient cellular divisions for development, tissue turnover and tissue regeneration. In long-lived mammals such as humans, telomerase expression is repressed in most somatic tissues (Gomes et al., 2011). A set of human diseases associated with defective telomere elongation are collectively called Telomere Biology Disorders (TBD), which highlight the importance of proper telomere length regulation (Savage, 2014). Patients affected by these diseases have critically short telomeres and, depending on the severity of the disease, display symptoms associated with defective cellular proliferation(Savage, 2014).
Figure 1.
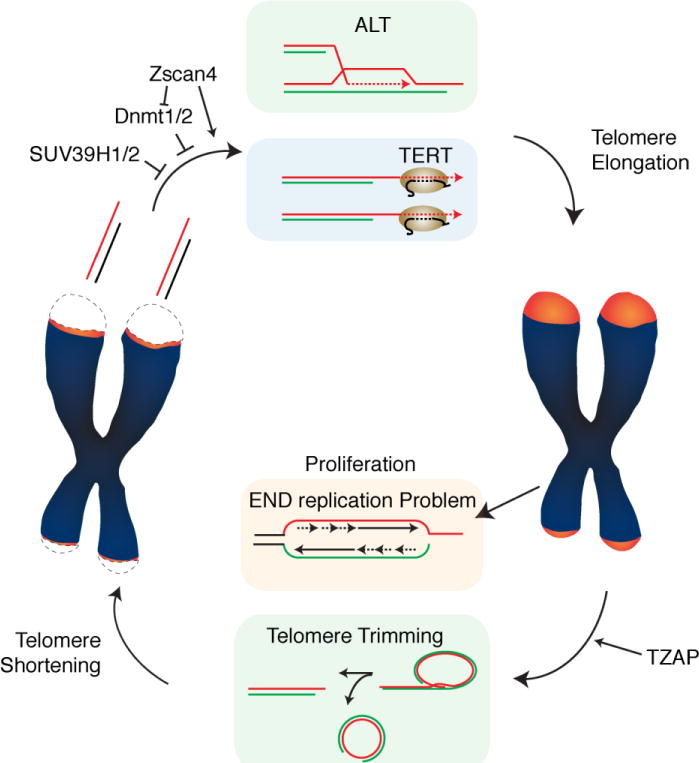
Balance between telomere elongation and telomere shortening maintain telomere length set-point in pluripotent stem cells. Telomere elongation by telomerase or ALT maintain the lower threshold of telomere length to sustain self-renewal capacity. Telomere trimming by t-loop excision maintains the upper threshold of telomere length.
Telomeres can also be extended in a telomerase-independent manner by a recombination-based process termed alternative lengthening of telomeres (ALT) (Bryan et al., 1997) (Figure 1). A significant fraction of cancer cells (approx. 15%) do not express telomerase and maintain telomeres using the ALT pathway. ALT engages the homologous recombination machinery to use telomeric sequences as a template for telomere extension (Dunham et al., 2000). Interestingly, ALT has also been reported to occur in non-transformed mouse somatic cells (Neumann et al., 2013). Furthermore, it has been reported that ALT-like mechanisms are active during early stages of embryogenesis (Liu et al.). In these cases, ALT-like features co-occur with telomerase expression. It is currently unclear to what extent these telomerase-independent mechanisms contribute to telomere elongation in normal cells. Finally, telomere elongation is balanced by a process called telomere trimming, which negatively regulates telomere length by actively eliminating excessively long telomeres harmful for genome stability(Pickett et al., 2009) (Figure 1). While telomere elongation mechanisms that maintain the lower limit of telomere length have been well studied for decades, how the upper limit of telomere length is determined by telomere trimming remains poorly understood. Here we provide an overview of the recent advancements of telomere length control with a particular emphasis on the balance between elongation and trimming in pluripotent stem cells.
Telomere elongation in Pluripotent Stem Cells
Pluripotent stem cells such as embryonic stem cells (ESC) are able to self-renew and give rise to virtually any type of somatic cell. ESCs were the first pluripotent stem cells that could be isolated and cultured in vitro (Evans and Kaufman, 1981; Martin, 1981). Mouse ESCs revolutionized the field of mouse genetics based on the fact that they are immortal, can be genetically modified and used to generate any mouse even after months of in vitro culture. Recently, establishment of human ESCs cultures as well as the ability to create “induced” Pluripotent Stem Cells (iPSCs) from somatic cells represent the promise of new transplantation therapies (Takahashi and Yamanaka, 2006; Thomson et al., 1998). ESCs and iPSCs can elongate their telomeres and proliferate indefinitely while maintaining pluripotency and genome stability. Highlighting the importance of telomere homeostasis in these cells is the finding that ESCs with short telomeres show reduced pluripotency and display differentiation defects (Pucci et al., 2013).
Multiple mechanisms ensure telomere elongation in ESCs, including elevated levels of telomerase activity (Thomson et al., 1998). Interestingly, ALT-like activity has been observed in ESCs (Liu et al., 2007). However, given that depletion of telomerase activity ESCs results in critical telomere shortening (Huang et al., 2011; Niida et al., 1998; Pucci et al., 2013) the contribution of ALT-like mechanism to telomere homeostasis in ESCs remains to be established.
A distinguishing feature of ESCs is represented by an “open” telomeric chromatin structure with reduced levels of the heterochromatin markers H3K9me3 and H4K20me3. Lack of these markers suggests that in ESCs telomeric chromatin is “decompacted”, a state that has been linked with increased telomerase-mediated elongation. Indeed, reduction of H3K9me3 by depletion of SUV39H1/H2 or reduction of H4K20me3 by depletion of Suv4-20h both induce telomere elongation (Benetti et al., 2007; Gomes et al., 2011). Similarly, during mouse development, expression of Zscan4 results in telomere elongation through the reduction of DNA methylation (Dan et al., 2017; Zalzman et al., 2010). In mouse embryonic stem cells, loss of the DNA methyl transferases (Dnmt1, Dnmt3a/3b) results in telomere elongation (Gonzalo et al., 2006). However, in human cells loss of DNA methyltransferase has a different outcome in terms of telomere length. Patients deficient of DNMT3b have very short telomeres and suffer from the Immunodeficiency, Centromeric instability and Facial anomalies (ICF)-syndrome (Yehezkel et al., 2008). This discrepancy could be due to specific differences between mouse and human cells in terms of telomere length regulation or by additional modifier factors that contribute to telomere shortening in ICF patients.
Interestingly, reduced methylation levels have also been associated with ALT-like activities such as an increase in telomere sister chromatid exchange (TSCE) (Dan et al., 2017; Zalzman et al., 2010). In agreement with this observation, low levels of H3K9me3 and H4K20me3 have also been reported in cancer cells that use the ALT pathway to elongate telomeres (Episkopou et al., 2014). In cancer cells ALT is correlated with mutations in the ATRX/DAXX remodeling complex and in the histone variant H3.3 (Heaphy et al., 2011; Lovejoy et al., 2012). The ATRX/DAXX complex acts as a chaperone that deposits histone H3.3 at pericentric heterochromatin, telomeres, as well as heterochromatic sites throughout the genome (Voon et al., 2015). Collectively, these data show a clear connection between chromatin status and telomere elongation mechanisms, a topic that has been extensively covered by previous reviews (for further detail see (O’Sullivan and Almouzni, 2014)).
Evidence of an upper threshold of telomere length in yeast
While the need to maintain a lower threshold of telomere length has been well established, whether an upper threshold of telomere length is important to maintain genome stability is less clear. Early clues of an upper limit of telomere length came from the analysis of S. cerevisiae strain carrying mutant alleles of Rap1 (Kyrion et al., 1992). Telomeres in this mutant strain were elongated up to 4kb from the normal size of ~300bp (Kyrion et al., 1992; Lustig and Petes, 1986). Strikingly, these elongated telomeres were found to be highly unstable, causing elevated rates of chromosome loss. Analysis of the fate of cells carrying these hyper-elongated telomeres revealed a process termed Telomere Rapid Deletions (TRD) that could reset excessively long telomeres back to WT length. Telomere Rapid Deletions occurred through a homologous recombination pathway, mechanistically distinct from the gradual loss of telomeres known as telomere attrition (Li and Lustig, 1996). Genetically, TRD require the homologous recombination factors Mre11 and Rad50 and are suppressed by the non-homologous-end-joining (NHEJ) factor Ku70 (Bucholc et al., 2001; Li and Lustig, 1996; Lustig, 2003). It was later shown that, in S. cerevisiae, meiotic cells undergo high rates of precise deletion to wild-type telomere size in a process that resembles TRD (Joseph et al., 2005). In this study, TRDs were 30 fold to 70 fold greater in meiotic cells compared to mitotic cells, suggesting that the control of the upper limit of telomere length is particularly important in this stage in development. In S. pombe, depletion of the telomere associated protein Taz1 leads to massive telomere elongation and defects in telomere replication as well as frequent telomere entanglements (Cooper et al., 1997; Miller and Cooper, 2003). Interestingly, loss of taz1 leads to rapid loss of telomeres due to problems with semiconservative replication of telomeric DNA that can be compensated by telomerase-mediated telomere elongation (Miller et al., 2006).
Rapid Telomere Deletion in mammalian cells
A process similar to the yeast TRD was first reported in mammalian cells expressing an allele of the telomere binding protein TRF2 lacking the N-terminus basic domain: TRF2ΔBasic (Wang et al., 2004). Binding of the TRF2ΔB to telomeres triggered the deletion of large portions of telomeric repeats resulting in rapid telomere shortening. The excised telomeric repeats where found to be extrachromosomal telomeric circles (t-circles), suggesting that they resulted from the excision of t-loops. Genetically, these rapid deletion of telomeric repeats depends on the homologous recombination proteins XRCC3 and NBS1. XRCC3 is a resolvase acting at Holliday junctions (Liu et al., 2004) while NBS1 is a component of the MRE11-Rad50-NBS1 (MRN) complex (Stracker and Petrini, 2011; Tauchi et al., 2002). These results suggested that, similarly to what is observed in yeast, mammalian cells are also capable of resetting telomere length through rapid deletion events. This notion was corroborated by the analysis of cells with hyper-elongated telomeres. High levels of telomerase activity induced by concomitant overexpression of the catalytic component, hTERT, and the RNA component, hTR, resulted in progressive telomere elongation (Cristofari and Lingner, 2006). However, when cancer cells with high levels of telomerase were kept in culture, telomere elongation eventually halted and cells accumulated c-circles, an indication of rapid telomere depletion (Pickett et al., 2009). In this setting, telomere depletion did not cause telomere dysfunction, suggesting a regulated process involved in telomere length regulation, a process that was termed “telomere trimming”. A similar process was recently described in human embryonic stem cells (hESC) as well as in induced pluripotent stem cells (iPCS) with upregulated levels of telomerase activity (Rivera et al., 2017). Thus, hyper-elongated telomeres both in human cancer cells as well as in pluripotent stem cells reset telomere length through telomere trimming.
Telomere trimming was found to play a physiological role in germ cells and activated T cells, two conditions in which telomerase activity is upregulated (Pickett et al., 2011). In germline cells telomerase is activated at high levels prior to fertilization to elongate telomeres and ensure proper telomere length in the offspring. Similarly, telomerase induction in activated T cells is required to sustain the massive cellular proliferation required during the immune response. In both conditions, c-circles are detected, suggesting that telomere trimming ensures a balancing mechanism to prevent excessive telomere elongation. Here, t-circle formation was shown to be dependent on XRCC3.
What are the differences between short and long telomeres?
The observation that hyper-elongated telomeres can trigger a rapid deletion event(s) suggests that differences between short and long telomeres can set an upper limit to telomere length (Figure 2). One potential mechanism is represented by the dilution of telomere specific factors upon abnormal telomere elongation. Indeed, it has been shown that in mammalian cells the abundance of the shelterin complex does not change with difference in telomere length expression. As a result, the density of shelterin at telomeres is inversely proportional to the length of telomeres, with short telomeres displaying a higher density of shelterin (Takai et al., 2010). In agreement with this observation, mice with hyper-elongated telomeres show similar levels of shelterin proteins at individual telomeres (Varela et al., 2016).
Figure 2.
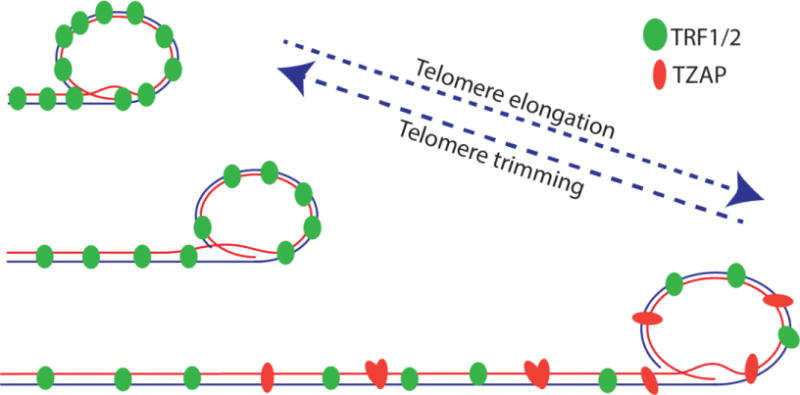
Binding competition between shelterin and TZAP regulates telomere trimming. Telomere elongation beyond the optimal upper threshold results in reduced shelterin binding, allowing TZAP to bind telomeres to reset the upper limit of telomere length.
Another difference between short and long telomeres is represented by the chromatin modifications. In mammals, telomeric chromatin is usually in a “silenced” repressive state, bound by the heterochromatin protein 1 (HP1) and with high levels of heterochromatin marks H3K9me3 and H4K20me3. As discussed above, changes in chromatin markers have been reported to affect telomerase mediated elongation (Benetti et al., 2007; Gonzalo et al., 2006). These data suggest that the heterochromatic state influences the ability of telomerase to act on telomeres and it is possible to envision that these changes could also mediate the ability of cells to engage in rapid telomere depletion events aimed at resetting telomere length. Strong data in support of a role for chromatin modification in telomere length regulation comes from the characterization of the ALT pathway. Cells that maintain telomeres using this pathway have lower levels of H3K9me3 and H4K20me3 and lower nucleosome density (Episkopou et al., 2014).
An upper threshold of telomere length may also be triggered by replication stalling at the repetitive TTAGGG repeats. Telomeres consist of tandem TTAGGG repeats that have the inherent tendency to form G-quadruplex, which acts as a structural barrier that causes replication fork collapse if unresolved. Indeed, telomeric DNA is prone to replication defects if not aided by telomere associated proteins(Miller et al., 2006; Sfeir et al., 2009). It is therefore likely that longer telomeres are more likely to incur replication stalling a signal that could mediate the engagement in rapid telomere deletions. This notion is supported by numerous studies that link replication stress induction at telomeres with the accumulation of c-circles. For instance, depletion of ASF1 the histone chaperone responsible for proper nucleosome assembly during replication leads to the accumulation of c-circles (O’Sullivan et al., 2014). Similarly, depletion of SMARCAL1 results in the induction of C-circles in telomerase positive cancer cells (Poole et al., 2015). SMARCAL1 is a member of the SWI/SNF-related family of chromatin remodelers with helicase and ATPase activity that allows proper progression of the replication fork during DNA replication(Flaus et al., 2006). These data suggest that replication fork stalling acts as a trigger for telomere trimming activity.
TZAP: a telomere associated protein involved in telomere trimming
Recently we and others have identified and characterized the zinc finger protein ZBTB48 as a novel specific telomere associated protein. To highlight the specificity of this factor we renamed it TZAP for Telomeric Associated Zinc finger Protein (TZAP) (Jahn et al., 2017; Li et al., 2017; Zhao et al., 2018). TZAP directly binds TTAGGG repeats independently from the shelterin complex. As shown both in a cellular context as well as in vitro, the binding of TZAP to chromosome ends is mediated by direct interaction between the terminal 3 zinc finger domains of TZAP and double-stranded telomeric repeats. TZAP binding to telomeres can be displaced by TRF1 and TRF2 overexpression, suggesting that TZAP and the shelterin complex compete for binding to telomeres. As a result of this competition, TZAP preferentially binds to long telomeres that have a low density of the shelterin complex (Takai et al., 2010). TZAP localization at telomeres resulted in the induction of rapid loss of telomeric sequences and concomitant accumulation of c-circles, suggesting a role for TZAP in telomere trimming. In agreement with this, depletion of TZAP results in reduced levels of c-circles and increased telomere length in mouse ESCs (Li et al., 2017).
How does the localization of TZAP promote trimming? TZAP does not contain any enzymatic domains, so two possible scenarios can be envisioned: TZAP could physically recruit resolvases able to dislodge the t-loop resulting in t-loop excision and telomere trimming (Figure 3). Potential candidates include the SLX4-SLX1-Mus81 resolvases or the Bloom-TopoIIIa-Rmi1 dissolvases. Alternatively, it is possible that TZAP might induce or stabilize DNA structures that act as substrates for these potential downstream factors. In these scenarios, TZAP would counteract the action of the basic domain of TRF2 that has been shown to bind three-way junctions at the base of the t-loop to prevent t-loop cleavage by HJ resolves (Schmutz et al., 2017; Wang et al., 2004). In line with this later model, the upper telomere length threshold would be determined by the balance between TZAP and TRF2 binding. Additional experiments are required to assess the mechanism of action of TZAP and its potential ability to bind and stabilize telomeres DNA structures.
Figure 3.
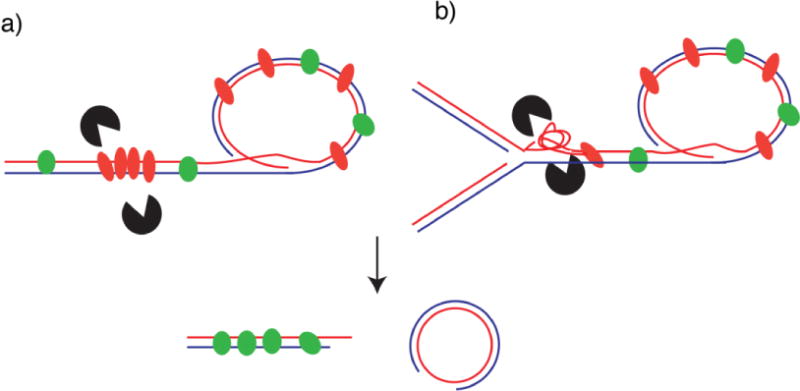
Potential mechanisms of TZAP-induced telomere trimming: (a) When bound to telomeres TZAP recruits directly nucleases involved in telomere trimming, or (b) when bound to telomeres TZAP facilitates formation of secondary DNA structures that are recognized by resolvases leading to telomere trimming. In both scenarios binding of TZAP to telomeres results in rapid telomere shortening and the release of extrachromosomal telomeric DNA (e.g. t-circles).
Consequences of telomere length de-regulation
In all model systems tested, defective telomere elongation ultimately causes severe proliferative/developmental defects. In humans, defect in telomerase-dependent telomere elongation is associated with dyskeratosis congenita (DC), a disorder that can affect different proliferative tissues such as the epidermis and the hematopoietic system. People with DC are vulnerable to disorders that impair bone marrow function (Armanios et al., 2007; Kirwan and Dokal, 2009; Shay and Wright, 1999; Tsakiri et al., 2007). In addition to developmental abnormalities, critically short telomeres coupled to an inappropriate DNA damage response also facilitates rounds of breakage-fusion-bridge cycles that drive genomic instability and tumorigenesis. Indeed, short telomeres have been linked to an increased risk for developing tumors of highly proliferative tissues in the gastrointestinal tract, head, and neck (Zhu et al., 2016). Similar phenotypes have been reported in late generation telomerase knockout mice, that display defects in tissue regeneration as well as increased predisposition to cancer development (Blasco et al., 1997; Rudolph et al., 1999). In contrast, whether over elongated telomeres have harmful or beneficial physiological consequences remain controversial. Based on the fact that telomere length represents a barrier against unlimited proliferation it is expected that excessive telomere length would increase the probability of tumor development, in particular in organisms that display a tight control of telomerase expression such as primates (Gomes et al., 2011). Evidence in support of this notion comes from the association between genetic determinants of long telomeres and increased overall cancer risk (Rode et al., 2016). Interpretation of these data is complicated by the fact that genetic predisposition to enhanced telomere maintenance may act exclusively as a survival advantage for cancerous cells rather than increasing the proliferation potential of pre-cancerous cells. Studies performed on genetically engineered mice have shown that increased telomerase activity driven in epithelial cells can be beneficial by slowing down the ageing process and increasing lifespan (Tomas-Loba et al., 2008). Similarly, mice generated from ES with hyper-elongated telomeres show reduced level of DNA damage and reduced sign of aging (Varela et al., 2016). However, mice are not the ideal system to assess the impact of hyper-elongated telomeres on tumor development given that in mice, telomere elongation is dispensable for tumor development (Blasco et al., 1997).
Highlights.
Regulation of telomere length in stem cells
Upper Limit of Telomere Length
Mechanisms of telomere trimming
Acknowledgments
We apologize to our colleagues whose work was not included in this review due to limited space and the expansive nature of this research area. The E.L.D. laboratory is supported by the NIH National Institute of General Medical Sciences (1R01GM122987) and the American Cancer Society (RSG-14-186-DMC).
References
- Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA, 3rd, Lansdorp PM, Greider CW, Loyd JE. Telomerase mutations in families with idiopathic pulmonary fibrosis. The New England journal of medicine. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. The Journal of cell biology. 2007;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. The telomere and telomerase: nucleic acid-protein complexes acting in a telomere homeostasis system. A review. Biochemistry Biokhimiia. 1997;62:1196–1201. [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nature medicine. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- Bucholc M, Park Y, Lustig AJ. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Molecular and cellular biology. 2001;21:6559–6573. doi: 10.1128/MCB.21.19.6559-6573.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science (New York, NY) 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. The EMBO journal. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J, Rousseau P, Hardikar S, Veland N, Wong J, Autexier C, Chen T. Zscan4 Inhibits Maintenance DNA Methylation to Facilitate Telomere Elongation in Mouse Embryonic Stem Cells. Cell reports. 2017;20:1936–1949. doi: 10.1016/j.celrep.2017.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes & development. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Denchi EL. Give me a break: how telomeres suppress the DNA damage response. DNA repair. 2009;8:1118–1126. doi: 10.1016/j.dnarep.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nature genetics. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Episkopou H, Draskovic I, Van Beneden A, Tilman G, Mattiussi M, Gobin M, Arnoult N, Londono-Vallejo A, Decottignies A. Alternative Lengthening of Telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic acids research. 2014;42:4391–4405. doi: 10.1093/nar/gku114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic acids research. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NM, Ryder OA, Houck ML, Charter SJ, Walker W, Forsyth NR, Austad SN, Venditti C, Pagel M, Shay JW, Wright WE. Comparative biology of mammalian telomeres: hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging cell. 2011;10:761–768. doi: 10.1111/j.1474-9726.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nature cell biology. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, Offerhaus GJ, McLendon R, Rasheed BA, He Y, Yan H, Bigner DD, Oba-Shinjo SM, Marie SK, Riggins GJ, Kinzler KW, Vogelstein B, Hruban RH, Maitra A, Papadopoulos N, Meeker AK. Altered telomeres in tumors with ATRX and DAXX mutations. Science (New York, NY) 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang F, Okuka M, Liu N, Ji G, Ye X, Zuo B, Li M, Liang P, Ge WW, Tsibris JC, Keefe DL, Liu L. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell research. 2011;21:779–792. doi: 10.1038/cr.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn A, Rane G, Paszkowski-Rogacz M, Sayols S, Bluhm A, Han CT, Draskovic I, Londono-Vallejo JA, Kumar AP, Buchholz F, Butter F, Kappei D. ZBTB48 is both a vertebrate telomere-binding protein and a transcriptional activator. EMBO reports. 2017;18:929–946. doi: 10.15252/embr.201744095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph I, Jia D, Lustig AJ. Ndj1p-dependent epigenetic resetting of telomere size in yeast meiosis. Current biology : CB. 2005;15:231–237. doi: 10.1016/j.cub.2005.01.039. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS biology. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan M, Dokal I. Dyskeratosis congenita, stem cells and telomeres. Biochimica et biophysica acta. 2009;1792:371–379. doi: 10.1016/j.bbadis.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G, Boakye KA, Lustig AJ. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Molecular and cellular biology. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Lustig AJ. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes & development. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- Li JS, Miralles Fuste J, Simavorian T, Bartocci C, Tsai J, Karlseder J, Lazzerini Denchi E. TZAP: A telomere-associated protein involved in telomere length control. Science (New York, NY) 2017;355:638–641. doi: 10.1126/science.aah6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, Blasco MA, Keefe DL. Telomere lengthening early in development. Nature cell biology. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- Liu Y, Masson JY, Shah R, O’Regan P, West SC. RAD51C is required for Holliday junction processing in mammalian cells. Science (New York, NY) 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, Rosenbluh J, Zwang Y, Weir BA, Hatton C, Ivanova E, Macconaill L, Hanna M, Hahn WC, Lue NF, Reddel RR, Jiao Y, Kinzler K, Vogelstein B, Papadopoulos N, Meeker AK. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS genetics. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nature reviews Genetics. 2003;4:916–923. doi: 10.1038/nrg1207. [DOI] [PubMed] [Google Scholar]
- Lustig AJ, Petes TD. Identification of yeast mutants with altered telomere structure. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:1398–1402. doi: 10.1073/pnas.83.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Cooper JP. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Molecular cell. 2003;11:303–313. doi: 10.1016/s1097-2765(03)00041-8. [DOI] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- Neumann AA, Watson CM, Noble JR, Pickett HA, Tam PP, Reddel RR. Alternative lengthening of telomeres in normal mammalian somatic cells. Genes & development. 2013;27:18–23. doi: 10.1101/gad.205062.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Matsumoto T, Satoh H, Shiwa M, Tokutake Y, Furuichi Y, Shinkai Y. Severe growth defect in mouse cells lacking the telomerase RNA component. Nature genetics. 1998;19:203–206. doi: 10.1038/580. [DOI] [PubMed] [Google Scholar]
- O’Sullivan RJ, Almouzni G. Assembly of telomeric chromatin to create ALTernative endings. Trends Cell Biol. 2014;24:675–685. doi: 10.1016/j.tcb.2014.07.007. [DOI] [PubMed] [Google Scholar]
- O’Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G, Karlseder J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nature structural & molecular biology. 2014;21:167–174. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, 3rd, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. Journal of theoretical biology. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. The EMBO journal. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett HA, Henson JD, Au AY, Neumann AA, Reddel RR. Normal mammalian cells negatively regulate telomere length by telomere trimming. Human molecular genetics. 2011;20:4684–4692. doi: 10.1093/hmg/ddr402. [DOI] [PubMed] [Google Scholar]
- Poole LA, Zhao R, Glick GG, Lovejoy CA, Eischen CM, Cortez D. SMARCAL1 maintains telomere integrity during DNA replication. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:14864–14869. doi: 10.1073/pnas.1510750112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci F, Gardano L, Harrington L. Short telomeres in ESCs lead to unstable differentiation. Cell stem cell. 2013;12:479–486. doi: 10.1016/j.stem.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera T, Haggblom C, Cosconati S, Karlseder J. A balance between elongation and trimming regulates telomere stability in stem cells. Nature structural & molecular biology. 2017;24:30–39. doi: 10.1038/nsmb.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode L, Nordestgaard BG, Bojesen SE. Long telomeres and cancer risk among 95 568 individuals from the general population. International journal of epidemiology. 2016;45:1634–1643. doi: 10.1093/ije/dyw179. [DOI] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, DePinho RA. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- Savage SA. Human telomeres and telomere biology disorders. Progress in molecular biology and translational science. 2014;125:41–66. doi: 10.1016/B978-0-12-397898-1.00002-5. [DOI] [PubMed] [Google Scholar]
- Schmutz I, Timashev L, Xie W, Patel DJ, de Lange T. TRF2 binds branched DNA to safeguard telomere integrity. Nature structural & molecular biology. 2017;24:734–742. doi: 10.1038/nsmb.3451. [DOI] [PubMed] [Google Scholar]
- Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science (New York, NY) 2012;336:593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Mutant dyskerin ends relationship with telomerase. Science (New York, NY) 1999;286:2284–2285. doi: 10.1126/science.286.5448.2284. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nature reviews Molecular cell biology. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. The Journal of biological chemistry. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, vanHeems D, Ito E, Nakamura A, Sonoda E, Takata M, Takeda S, Matsuura S, Komatsu K. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–98. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science (New York, NY) 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tomas-Loba A, Flores I, Fernandez-Marcos PJ, Cayuela ML, Maraver A, Tejera A, Borras C, Matheu A, Klatt P, Flores JM, Vina J, Serrano M, Blasco MA. Telomerase reverse transcriptase delays aging in cancer-resistant mice. Cell. 2008;135:609–622. doi: 10.1016/j.cell.2008.09.034. [DOI] [PubMed] [Google Scholar]
- Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, Rosenblatt RL, Shay JW, Garcia CK. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela E, Munoz-Lorente MA, Tejera AM, Ortega S, Blasco MA. Generation of mice with longer and better preserved telomeres in the absence of genetic manipulations. Nature communications. 2016;7:11739. doi: 10.1038/ncomms11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voon HP, Hughes JR, Rode C, De La Rosa-Velazquez IA, Jenuwein T, Feil R, Higgs DR, Gibbons RJ. ATRX Plays a Key Role in Maintaining Silencing at Interstitial Heterochromatic Loci and Imprinted Genes. Cell reports. 2015;11:405–418. doi: 10.1016/j.celrep.2015.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Watson JD. Origin of concatemeric T7 DNA. Nature: New biology. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Yehezkel S, Segev Y, Viegas-Pequignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Human molecular genetics. 2008;17:2776–2789. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, Wersto RP, Ko MS. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang G, He C, Mei Y, Shi Y, Li F. The 11th C2H2 zinc finger and an adjacent C-terminal arm are responsible for TZAP recognition of telomeric DNA. Cell research. 2018;28:130–134. doi: 10.1038/cr.2017.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Han W, Xue W, Zou Y, Xie C, Du J, Jin G. The association between telomere length and cancer risk in population studies. Scientific reports. 2016;6:22243. doi: 10.1038/srep22243. [DOI] [PMC free article] [PubMed] [Google Scholar]


