Abstract
37-kDa immature laminin receptor protein (iLRP), the precursor of 67-kDa laminin receptor protein (LRP), is overexpressed on the surface of most cancer cells and recognized as a universal tumor antigen. The role makes it a potential target for cancer immunotherapy, which has been well-studied. Our study aimed to produce high quality of human iLRP in bacteria so that the needs in research of its clinical application could be met. The powerful system for heterologous protein expression, pET system was used. Two types of DNA sequences encoding the same amino acid sequences were separately cloned into the vector pET30a(+). One of the resulting vectors includes the wild-type iLRP, and other one includes the codon-optimized iLRP. The expression by both genes was then compared in Escherichia coli BL21(DE3). Our results revealed that the performance of codon optimization was crucial for the expression of human iLRP in Escherichia coli. The yield was significantly enhanced up to 300 mg/L of bacterial culture by this approach.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1234-y) contains supplementary material, which is available to authorized users.
Keywords: Codon optimization, Immature laminin receptor protein, Heterologous expression, pET system
Introduction
As the incidence of cancer happens more frequently than before, more effective treatments are in urgent need. Over the past decades, one of the promising treatments, called as cancer immunotherapy, has been developed. For this approach, one of key factors is to select a well-studied, immune-reactive tumor antigen. The 37-kDa immature laminin receptor protein (iLRP) overexpressed on the surface of tumor cells, but not on normal tissues, is featured by its multifaceted functions in tumor growth and metastasis (Barsoum and Schwarzenberger 2014; Coggin et al. 1999). Together with the ability to induce autologous immunity in iLRP-positive cancer patients, it has been thoroughly studied both in pre-clinical and clinical trials as a target for immunotherapy (DiGiacomo and Meruelo 2016; McClintock et al. 2015). This suggested that it be promising in future clinics, thus leading to a requirement of large amount of iLRP. Although a way to produce high quality of iLRP was published for a clinical trial (Barsoum et al. 2009), the way can be improved. In the work, an additional TEV (Tobacco Etch Virus) protease site was inserted between the 6× His-tag and the N-terminus of iLRP, which was supposed to be cleaved off by TEV protease. Since the final products still carried the site, unexpected effects might occur, especially, on the host immune system after immunization. It is important to avoid unnecessary amino acid sequences in a recombinant protein to be used in clinical trials. With this regard, we constructed an expression vector similar with the one that they used except the TEV protease site being removed. Unexpectedly, the removal of the site extremely impacted the expression level to the extent that hardly detected when a small-scale expression test was performed. To solve this problem, a gene design strategy used frequently by other researchers was explored.
The expression of heterologous proteins originated from 1970s and has become basic biotechnology since then. However, for a specific protein, problems might occur frequently in expressing, such as insufficient product, lacking of function, or even nothing expressed (Burgess-Brown et al. 2008). Several factors can be attributed to these issues. The underlying point is that the DNA sequence used to encode a protein in one organism is often quite different from the sequence that would be used to encode the same protein in another organism. To obtain high expression level in the new host, hence, it is fairly important to change foreign DNA sequence into the sequence that can be readily recognized by it. In this process, GC content, relative frequency of codons, hairpins, and negative cis-acting elements, can all affect the expression level of the foreign protein (Chung and Lee 2012). Taking above parameters into consideration, we redesigned the human iLRP gene sequence by codon optimization technology referring to the codon usage frequency in Escherichia coli. The optimized sequence was then synthesized and cloned into the vector pET30a(+) for expression analysis. Results demonstrated that the iLRP expression level was significantly increased comparing with the wild gene sequence.
Materials and methods
Strains and plasmids
Escherichia coli DH5α was used as the host for recombinant DNA manipulation, and BL21(DE3) was used for gene expression. Plasmids pUC57 and pET30a(+) for cloning and expression are from ChinaPeptides, Shanghai, China. All hosts and plasmids were stored at − 80 °C in our lab.
Codon optimization
It has been a valuable tool to produce proteins as therapeutic agents or research reagents in heterologous host. With the availability of vast amount of genomics data, along with increased knowledge of protein expression, function and structure relationships, gene expression levels can be improved significantly through codon optimization (Gao et al. 2015). A variety of critical factors involved in different stages of protein expression, such as GC content, codon adaptability, mRNA structure, and various cis-elements in transcription and translation can be taken into consideration for this process (Gvritishvili et al. 2010). In this study, the human iLRP gene (NM_002295) was optimized according to the above points in order to over express it in Escherichia coli using a commercial proprietary algorithm, NG® Codon Optimization Technology (Synbio Technologies, NJ, USA).
Construction of expression vectors
The optimized iLRP gene with a 6× His-tag at its 5′-end was synthesized by ChinaPeptides, Shanghai, China, and was cloned into vector pET30a(+) between NdeI and EcoRV. The wild-type cDNA of human iLRP gene was purchased from Vigene Biosciences, Shandong, China as an insert in the plasmid pENTER. The cDNA was then constructed into pET30a(+) between NdeI and XhoI with an added 6× His-tag same as the optimized construct at the N-terminus of iLRP. Resulted vectors were, respectively, named as pET30-OP-iLR for the optimized insert and pET30-WT-iLR for the wild insert and transformed into Escherichia coli DH5α for plasmid preparation using PureYield™ Plasmid Miniprep System (Promega Biotech, Beijing, China). Purified plasmids were confirmed by enzyme digestion first and then sent to Sangon Biotech, Shanghai, China for further confirmation by DNA sequencing using T7 promoter and T7 terminator universal primers. Constructs confirmed by both methods were used for subsequent assessment of protein expression.
Analysis of protein expression
Vectors containing the wild-type iLRP gene and the optimized iLRP gene were both transformed into Escherichia coli BL21 (DE3) for protein expression. Three independent transformants for both vectors were tested for the expression at a small scale. The most expressed clone was selected and expanded for the storage. To analyze the expression level, a fresh Terrific Broth (TB) agar plate containing 50 µg/ml kanamycin was inoculated using the selected clone. Next day, one isolate was picked from the plate and cultured overnight in 3 ml TB containing 50 µg/ml kanamycin. Overnight cultures were transferred to 100 ml fresh TB and incubated at 37 °C until the OD600 = 1.0. Each culture was then divided into 4 aliquots, one as a negative control, and other three parts were induced by addition of isopropyl-thio-β-D-galactoside (IPTG) to a final concentration of 0.3, 0.5 or 1.0 mM, respectively, at 37 °C. In order to find the optimal time course for high level iLRP expression, the above cultures with different IPTG concentrations were sampled at time points 6, 10, or 16 h for each. The final OD600 values were measured and equivalent amounts of culture were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were stained by Coomassie Brilliant Blue R-250 (ThermoFisher Scientific, Shanghai, China).
Large-scale expression of iLRP
Escherichia coli BL21 (DE3) transformant that had been tested from above step harboring the optimized gene were used to express the recombinant iLRP at a large-scale level. A single clone from an overnight TB agar plate at 37 °C was picked and cultured in 3 ml TB medium containing 50 µg/ml kanamycin. Culture was then transferred to 30 ml TB medium with kanamycin and grew for 2 h. The bacteria were expanded in 500 ml of TB medium in the presence of 50 µg/ml kanamycin to an OD600 of 1.0 at 37 °C in a 2 L flask. Expression was then induced by adding IPTG at a final concentration of 0.5 mM and the cells were continually grown at 37 °C for 6 h. Cells were harvested using high speed centrifugation at 4500 g for 10 min after induction. Cell pellets were washed with 10 volumes of buffer (50 mM Tris–HCl, 150 mM NaCl, PH 8.0) and stored at − 20 °C. Once 4 batches of culture were available, we put them together and proceeded to the next step, protein purification.
Protein purification using nickel–nitrilotriacetic acid (Ni–NTA) column
The cell pellets harvested from the large-scale expression were weighted and resuspended in 1/25 volume (80 ml/2 L of culture) of buffer (50 mM Tris–HCl, 150 mM NaCl, pH 8.0). Cell suspension was subjected to sonication with 20 s pulse-on and pulse-off time each, using an ultrasonicator (Ningbo Scientz Biotechnology Co. LTD, China). During sonication, the beaker with sample was kept in ice-bath to avoid over-heating. The inclusion bodies were then isolated by centrifugation at 16,000g for 20 min at 4 °C. After washing twice with 50 ml ice-cold buffer (1 M guanidine-HCl, 50 mM Tris–HCl, 150 mM NaCl, pH 8.0), the inclusion bodies were dissolved in 50 ml of solubilization buffer (10 mM Tris–HCl, 100 mM sodium phosphate, 6 M guanidine-HCl, 10 mM imidazole, 2 mM 2-mercaptoethanol, pH 8.0). The dissolved protein solution was centrifuged at 16,000g for 30 min at 4 °C. The supernatant was then transferred to an Ni–NTA column, and unbound proteins were removed by washing the column with the solubilization buffer. Elution buffers including different concentrations of imidazole (10 mM Tris–Cl, 100 mM sodium phosphate, 2 mM mercaptoethanol, 20, or 50, or 100, or 200, or 500 mM imidazole, pH 8.0) was used to elute the His-tagged protein. The eluted solutions were detected by SDS-PAGE for the presence of iLRP. The concentration of final products was determined by scanning densitometry on a SDS-PAGE gel using bovine serum albumin standards (ThermoFisher Scientific, Shanghai, China) as a reference.
SDS-PAGE and Western blotting analysis
All protein samples were boiled in 1× reducing SDS-PAGE loading buffer at 95 °C for 10 min and subjected to 12% denaturing SDS-PAGE. Gels were stained by Coomassie Brilliant Blue R-250 to identify the expected protein bands. For Western blotting analysis, gels were blotted to nitrocellulose membrane. The membrane was blocked with 2.5% w/v skimmed milk powder in phosphate-buffered saline (PBS, pH 7.2) for 2 h at room temperature (RT), and successively incubated with mouse anti-iLRP monoclonal antibody (Abcam, Shanghai, China) at a dilution of 1:5000 at 4 °C overnight. After washing, the membrane was incubated with a 1:10,000 dilution of horseradish peroxidase (HRP) conjugated anti-mouse IgG secondary antibody at RT for 2 h. Following the final step of washing, the blotted membrane was incubated with ECL reagent (ThermoFisher Scientific, Shanghai, China), and protein bands were detected by ChemiDoc™ MP imaging System (Bio-Rad Laboratories Co. LTD, Shanghai, China).
Results
Codon optimization
Codon optimization can refer to several meanings as, avoiding rare codons with low utilization, simplifying mRNA’s secondary structure after gene transcription, removing the motif which is not conducive to efficient expression and add the helpful one, adjusting the GC content and other methods to re-design genes. In the present study, GC content, codon adaptation index (CAI), mRNA structure and cis-acting elements in the wild iLRP gene were optimized to match the host requirements for expression in Escherichia coli by NG® Codon Optimization Technology. A more smooth curve of GC content was obtained (Online Resource Fig S1), meanwhile, the stable hairpin structures were minimized to 1 from 11. These allow ribosomes to translate more efficiently. Furthermore, codon usage and distribution of the gene became more fit to the host genome, and the CAI was improved from 0.60 to 0.84 after replacing rare codons (Online Resource Fig S2). The negative cis-acting elements and killer sequences were eliminated also. Finally, the gene optimization model evaluated the integrated scores that correlate with gene expression level negatively. For the wild iLRP gene, the scores are 1,632,435, and for the optimized gene, the scores were reduced to 776,954, which decreased the value by 47%. The DNA sequence changed after performing the optimization was aligned with the wild-type gene (Online Resource Fig S3). The alignment clearly showed that both sequences encode a protein with same amino acid sequences.
Protein expression and detection
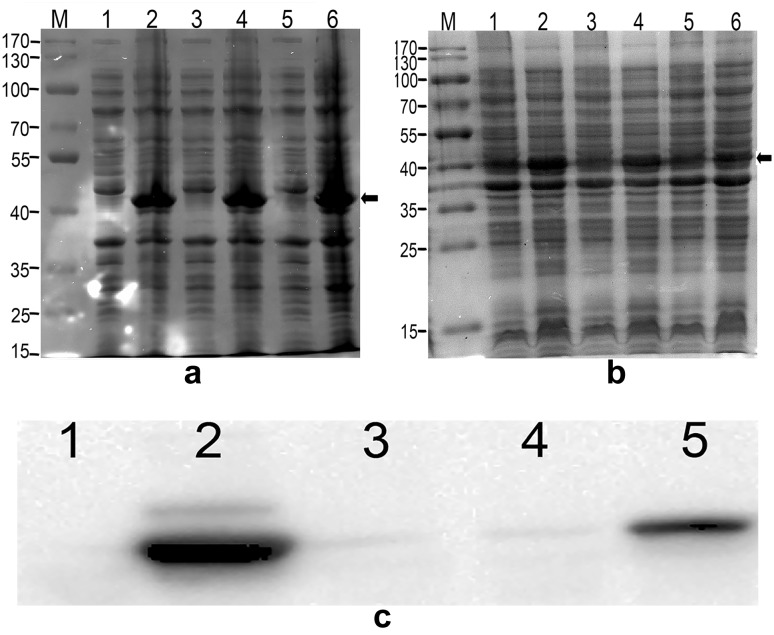
To meet the need of high quality of recombinant human iLRP protein in clinics, we made a change comparing with the previous work by removing the extra TEV protease site in the expressing vector. Unexpectedly, when an expression test was performed for this vector without the TEV protease site, it was difficult to detect the target protein on an SDS-PAGE gel after being dyed with Coomassie Brilliant Blue (data not shown). We then tried various conditions to improve the expression level but failed. The analysis by Western Blotting could not reveal the expression either (Fig. 1c, lanes 3 and 4). As a disappointing result, we turned to the codon optimization technique according to the codon usage bias in Escherichia coli and synthesized the optimized iLRP sequence and then constructed it into the expression vector pET30a(+), named as pET30-OP-iLR. The construct was then transformed into Escherichia coli BL21 (DE3) for the expression analysis. Parameters that might influence the expression were explored, including time duration and the concentration of IPTG. The induced products were analyzed by SDS-PAGE. Heavily dark bands with electrophoretic mobility of about 37 kDa could be found at lanes loaded with samples induced by different concentrations of IPTG on the SDS-PAGE gel, with the darkest band from 0.5 mM IPTG (Fig. 1a, lanes 2, 4 and 6). From the result, we also observed that induced products reached to the peak level at the time point 6 h after induction (Fig. 1b, lane 2). Hence, we applied the optimal conditions for iLRP expression in our study were as follows. The overnight culture from a freshly inoculated plate would be expanded in a 2-l flask until an OD600 of 1.0 at 37 °C, then the expanded bacteria would be induced by 0.5 mM IPTG and continually cultured at 37 °C for 6 h.
Fig. 1.
iLRP expression and detection. a Various IPTG concentrations were evaluated for the induction of iLRP expression. Lanes 1, 3, and 5 were samples of cell lysates before IPTG induction, and lanes 2,4, and 6 were samples of cell lysates after IPTG induction at concentrations of 0.3, 0.5, and 1.0 mM, respectively. The target bands were indicated by a solid arrow. A darker band could be observed on lane 4, consequently, the corresponding concentration of 0.5 mM was selected for the induction. M: molecular weight marker. b To find an optimal time course for high level expression of iLRP, we sampled at time points 6, 10, and 16 h after induction by 0.5 mM IPTG. The lane 2 showed the darkest band at the right size of 37 kDa indicated by a solid arrow, which meant the corresponding time point at 6 h was the appropriate point for the induction. Lanes 1, 3 and 5 were samples before inducing. Lane 2, 4, and 6 were samples from time points at 6, 10, and 16 h after induction. M: molecular weight marker. c Western Blotting analysis. The result not only showed that the wild iLRP gene expressed very little, but also confirmed the protein identity for the optimized gene product. Lane 1: cell lysates from pET-OP-iLR before IPTG induction; lane 2: cell lysates from pET-OP-iLR after 0.5 mM IPTG induction; lane 3: cell lysates from pET-WT-iLR before induction; lane 4: cell lysates from pET-WT-iLR after 0.5 mM IPTG induction; lane 5: Purified iLRP by Ni–NTA column
Protein purification
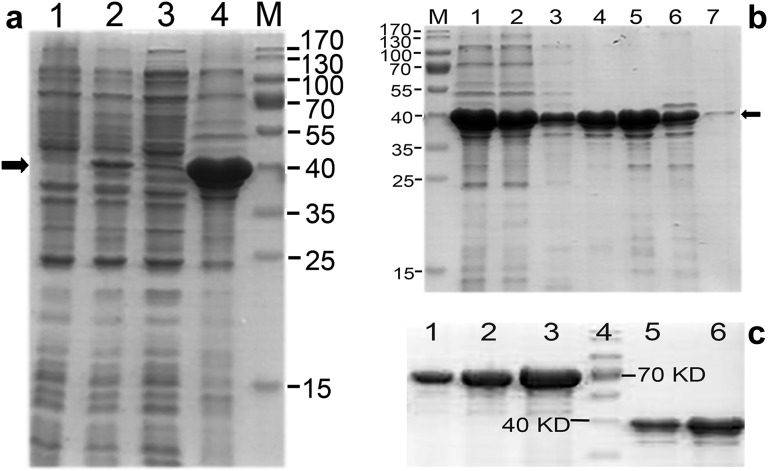
The cell pellets collected from 2 L of bacterial culture induced by IPTG were sonicated according to the procedure above. Resulting cell lysates were fractionated by centrifuging at 16,000g for 20 min and analyzed by SDS-PAGE. The data indicated that the target protein iLRP was expressed as a form of inclusion body (Fig. 2a). Subsequently, the fraction with the inclusion bodies was dissolved in the solubilization buffer and the clarified supernatant was applied to an Ni–NTA column for purification of His-tagged protein. To determine an optimal concentration of imidazole for eluting His-tagged target, we performed a small-scale purification. Briefly, the pre-equilibrated column was loaded by the clarified supernatant, and washed by the washing buffer first, and then consecutively eluted by 2 CV of each of elution buffers in which different concentrations of 20, 50, 100, 200 and 500 mM imidazole, were added. The eluates were individually collected to detect the presence of iLRP by SDS-PAGE. The data demonstrated that a part of recombinant protein from the solubilized inclusion bodies flowed through the column at washing. A small amount of protein was eluted at the concentrations of 20 and 200 mM imidazole. The majority of protein was eluted at the concentrations of 50 and 100 mM. The eluates at 500 mM could only detect a very weak band with a size of 37 kDa around (Fig. 2b). Consequently, we chose the concentration of 100 mM as the elution condition for the subsequent protein purification. The eluted solutions at this concentrations were collected and dialyzed against 3 M Guanidine-HCl in phosphate-buffered saline (pH 7.4) firstly, then against Guanidine-free buffer at 4 °C. The final product from 2 L of bacterial culture was concentrated in 60 ml of phosphate-buffered saline. The diluted solution by 10 times was used to determine the yield by scanning densitometry (Fig. 2c). From the result, the estimated concentration for the diluted sample was about 1 µg/µL, which meant the total yield in 2 L of culture was about 600 mg. Hence, the expression level in our study reached up to 300 mg/L of culture medium evaluated right after purification by Ni–NTA.
Fig. 2.
iLRP purification and quantification. a iLRP was expressed as an insoluble form of inclusion body. An induced band could be observed clearly on lane 2, but the most induced products were revealed on lane 4 loaded with inclusion bodies. Lane 1: cell lysates from the sample without IPTG induction; lane 2: sample from induced bacterial culture by 0.5 mM IPTG at 37 °C for 6 h; lane 3: supernatant from the same sample as in lane 2 lysed by ultrasonication, then centrifuging at 16,000g; lane 4: inclusion body pellet from the same sample as in lane 3 after centrifugation. The solid arrow pointed at the proper position of target protein. M: molecular weight marker. b Elution profile of Ni–NTA column loaded with solubilized inclusion bodies. Lane 1: clarified supernatant from dissolved inclusion bodies; lane 2: flow through collected from sample loading; lane 3: eluted fraction collected at 20 mM imidazole; lane 4: eluted fraction at 50 mM imidazole; lane 5: eluted fraction at 100 mM imidazole; lane 6: eluted fraction at 200 mM imidazole; lane 7: eluted fraction at 500 mM imidazole. M: molecular weight marker. The solid arrow indicated the position of target protein with a size of around 37 kDa. c Quantification by scanning densitometry. Bovine serum albumin (BSA) standards were used to determine the yield of recombinant protein. Lane 1: 1 µg of BSA was loaded; lane 2: 2 µg of BSA was loaded; lane 3: 4 µg of BSA was loaded; lane 4: molecular weight marker; lane 5: purified iLRP in 1 µL of phosphate-buffered saline; lane 6: purified iLRP in 2 µL of phosphate-buffered saline. Lane 1 and 2 had similar band density with lane 5 and 6, respectively, which meant the amount of loaded protein in lane 5 was close to 1 µg and close to 2 µg in lane 6
Western blot analysis
To validate the final product was the protein we designed, Western blotting analysis was performed using purchased monoclonal antibody (mAb) whose specificity to iLRP has been confirmed by Abcam. At the position of approximate molecular weight of 37 kDa, positive bands were shown clearly not only for the sample from induced cell culture lysates (Fig. 1c, lane 2), but also for the purified products post Ni–NTA column (Fig. 1c, lane 5). Nonetheless, samples from vector pET-WT-iLR whether induced by IPTG or not showed hardly detectable weak bands (Fig. 1c, lanes 3 and 4). The immunoreactivity of samples above to anti-iLRP mAb revealed that the recombinant products we expressed and purified were the protein we designed, and further testified that the wild-type iLRP gene could not satisfactorily produce proteins in Escherichia coli BL21 (DE3).
Discussion
Overexpression of 37-kDa iLRP is prevalent in human cancers and correlates with poor patient prognosis. It plays a central role in tumor invasion and metastasis and is also vital for tumor cell proliferation, survival and protein translation (Poon et al. 2011). More importantly, documents have demonstrated that iLRP is a strong tumor rejection antigen capable of autologously inducing immune response in cancer patients (Friedrichs et al. 2008; Rohrer et al. 2006). These data suggest that iLRP could be a suitable candidate target for cancer immunotherapy. To fast forward its application in future clinics, obtaining large amount of the protein becomes the first issue that needs to be resolved. In this study, we adopted the most powerful system yet developed for recombinant protein expression in Escherichia coli, pET system, to produce iLRP. In order to accomplish this work, we optimized the human iLRP gene referring to the genome of Escherichia coli by NG® Codon Optimization Technology.
NG® Codon Optimization Technology is one of commercialized algorithms for heterologous protein expression, which possesses several advantages, such as, elimination of codon usage bias, fine-tuning secondary structure of mRNA, and removal of negative cis-acting elements (http://www.synbio-tech.com/). By using this technology, the human iLRP gene was optimized and cloned into the vector pET30a(+) so that it could be expressed in the pET system. Comparing the expression of codon-optimized gene with the wild-type gene, big difference was revealed. The expression level reached up to 300 mg/L after performing the optimization, however, the expression could hardly be detected for the wild gene even with Western blotting analysis (Fig. 1c, lanes 3 and 4). Since the gene optimization changed the genetic constitution of iLRP a lot by changing the GC content, improving the CAI, stabilizing the mRNA structures, and reducing the integrated scores negatively correlated with the protein expression (Online Resource Figs S1–S3), we inferred that the modified genetic constitution made combinatorial contribution to the improved iLRP products in the bacterial host. According to the report (Hanson and Coller 2018), heterologous expression of rare codon-containing genes is likely to exhaust the endogenous pools of the analogous tRNAs, which leading to growth inhibition, premature termination of transcription and/or translation, decreased mRNA stability, and increased frame-shifts, deletions and misincorporations. Therefore, it is feasible to improve the iLRP expression through the technique of codon optimization by modifying the genetic constitution in terms of the host. With the development of gene synthesis, direct modifications of any coding sequences can be easily synthesized. Combining gene synthetic design with codon optimization, some other proteins were overexpressed in Escherichia coli for various purposes (Nguyen et al. 2017; Shi et al. 2018; Song et al. 2014).
As expected, a significant higher iLRP expression level was obtained after gene optimization. Nevertheless, the wild-type sequence expressed few proteins hardly detected, which was inconsistent with the previous work (Barsoum et al. 2009). In their work, high-quality recombinant iLRP was successfully produced without performance of codon optimization in the same pET expression system. The only difference from our study was that they inserted a TEV protease site between the His-tag and iLRP, which was removed hereof. The totally different expression profile between the vector with TEV protease site and the one without it showed that this extra fragment apparently affected the expression of iLRP. To find out the reason, we can design a set of vectors with or without a certain tag, then evaluate the influence on expression. The DNA sequences of tags should also be accounted for explaining the difference since they can make significant contribution to the whole gene in GC contents, mRNA stability, and CAI.
In the pET system, target genes under control of strong bacteriophage T7 transcription signals can be induced by providing a source of T7 RNA polymerase in the host cell. T7 RNA polymerase is so selective and active that the desired product can comprise more than 50% of the total cell protein a few hours after induction (Mierendorf et al. 1998). By optimizing culture condition and/or induction environment, the expression level can increase even more. Documentation reported that the induced product reached up to a level of 7 g/L of cell culture (Shin et al. 1997). In the present study, induction parameters, temperature, concentration of IPTG, and time points, were assessed. The optimal conditions for iLRP expression are revealed in Fig. 1a, b and depicted in detail in the section of “Results”. By applying the optimized parameters to inducing expression, the yield for the optimized iLRP gene reached up to 300 mg/L, which was significantly higher than the average yield of 80 mg/L in a previous work (Barsoum et al. 2009). This appreciable output will be of great value for developing on its therapeutic application. The work showed further an example of taking advantages of codon optimization and pET system in heterologous protein overexpression.
In summary, the present study has shown that codon optimization is a key for the successful expression of human iLRP in Escherichia coli. With codon optimization by the algorithm, NG®Codon Optimization Technology, we replaced the codons that are rare for the host with more frequent ones, and minimized any unfavorable mRNA structures during the translation. Furthermore, we found that the TEV protease site fused to the N-terminus of iLRP was very important for efficient expression by the wild iLRP gene. After the deletion of TEV protease site from the construct, the wild iLRP gene expressed few of products hardly detected by Western blotting. Nonetheless, the expression was remarkably improved after performing the codon optimization. Combining the codon optimization and the removal of TEV protease site, we obtained substantial iLRP product with the powerful protein expression system, pET system. This strategy will undoubtedly fast forward studies to validate therapeutic utility of this tumor antigen in clinical trials.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant no. 81560276), and the Joint Funds of Science and Technology Department of Guizhou Province of China (Grant No. LKZ[2013]02), and the Foundation for Returnees from the Department of Human Resources and Social Security of Guizhou Province of China (Grant no. [2014]08).
Author contributions
Dr. BL conceived and designed the study and performed the data analysis and wrote the manuscript; QK designed and performed the experiments and data analysis; DZ, and LY helped perform the analysis with constructive discussions.
Compliance with ethical standard
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
On behalf of all authors, Dr. Bainan Liu states that there is no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1234-y) contains supplementary material, which is available to authorized users.
Bainan Liu and Qianqian Kong made equal contributions to the study.
References
- Barsoum AL, Schwarzenberger PO. Oncofetal antigen/immature laminin receptor protein in pregnancy and cancer. Cell Mol Biol Lett. 2014;19:393–406. doi: 10.2478/s11658-014-0203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum AL, Liu B, Rohrer JW, Coggin JH, Jr, Tucker JA, Pannell LK, Schwarzenberger PO. Production, safety and antitumor efficacy of recombinant oncofetal antigen/immature laminin receptor protein. Biomaterials. 2009;30:3091–3099. doi: 10.1016/j.biomaterials.2009.02.022. [DOI] [PubMed] [Google Scholar]
- Burgess-Brown NA, Sharma S, Sobott F, Loenarz C, Oppermann U, Gileadi O. Codon optimization can improve expression of human genes in Escherichia coli: a multi-gene study. Protein Expr Purif. 2008;59:94–102. doi: 10.1016/j.pep.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Chung BK, Lee DY. Computational codon optimization of synthetic gene for protein expression. BMC Syst Biol. 2012;6:134. doi: 10.1186/1752-0509-6-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggin JH, Jr, Barsoum AL, Rohrer JW. 37 kiloDalton oncofetal antigen protein and immature laminin receptor protein are identical, universal T-cell inducing immunogens on primary rodent and human cancers. Anticancer Res. 1999;19:5535–5542. [PubMed] [Google Scholar]
- DiGiacomo V, Meruelo D. Looking into laminin receptor: critical discussion regarding the non-integrin 37/67-kDa laminin receptor/RPSA protein. Biol Rev Camb Philos Soc. 2016;91:288–310. doi: 10.1111/brv.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichs B, et al. Humoral immune responses against the immature laminin receptor protein show prognostic significance in patients with chronic lymphocytic leukemia. J Immunol. 2008;180:6374–6384. doi: 10.4049/jimmunol.180.9.6374. [DOI] [PubMed] [Google Scholar]
- Gao CY, Xu TT, Zhao QJ, Li CL. Codon optimization enhances the expression of porcine beta-defensin-2 in Escherichia coli. Genet Mol Res. 2015;14:4978–4988. doi: 10.4238/2015.May.12.1. [DOI] [PubMed] [Google Scholar]
- Gvritishvili AG, Leung KW, Tombran-Tink J. Codon preference optimization increases heterologous PEDF expression. PLoS ONE. 2010;5:e15056. doi: 10.1371/journal.pone.0015056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol. 2018;19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock SD, et al. Monoclonal antibodies specific for oncofetal antigen–immature laminin receptor protein: effects on tumor growth and spread in two murine models. Cancer Biol Ther. 2015;16:724–732. doi: 10.1080/15384047.2015.1026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierendorf RC, Morris BB, Hammer B, Novy RE. Expression and purification of recombinant proteins using the pET system. Methods Mol Med. 1998;13:257–292. doi: 10.1385/0-89603-485-2:257. [DOI] [PubMed] [Google Scholar]
- Nguyen AN, et al. Prokaryotic soluble expression and purification of bioactive human fibroblast growth factor 21 using maltose-binding protein. Sci Rep. 2017;7:16139. doi: 10.1038/s41598-017-16167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon SL, Klausen C, Hammond GL, Leung PC. 37-kDa laminin receptor precursor mediates GnRH-II-induced MMP-2 expression and invasiveness in ovarian cancer cells. Mol Endocrinol. 2011;25:327–338. doi: 10.1210/me.2010-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer JW, Barsoum AL, Coggin JH., Jr Identification of oncofetal antigen/immature laminin receptor protein epitopes that activate BALB/c mouse OFA/iLRP-specific effector and regulatory T cell clones. J Immunol. 2006;176:2844–2856. doi: 10.4049/jimmunol.176.5.2844. [DOI] [PubMed] [Google Scholar]
- Shi Y, Halperin SA, Lee SF. Expression, purification, and functional analysis of an antigen-targeting fusion protein composed of CD40 ligand and the C-terminal fragment of ovalbumin. Protein Expr Purif. 2018;142:37–44. doi: 10.1016/j.pep.2017.09.015. [DOI] [PubMed] [Google Scholar]
- Shin CS, Hong MS, Bae CS, Lee J. Enhanced production of human mini-proinsulin in fed-batch cultures at high cell density of Escherichia coli BL21(DE3)[pET-3aT2M2] Biotechnol Prog. 1997;13:249–257. doi: 10.1021/bp970018m. [DOI] [PubMed] [Google Scholar]
- Song H, Li G, Mai W, Huang G, Chen K, Zhou Y, Chen H. Codon optimization enhances protein expression of Bombyx mori nucleopolyhedrovirus DNA polymerase in E. coli. Curr Microbiol. 2014;68:293–300. doi: 10.1007/s00284-013-0476-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.