Significance
One-third of the mammalian genome encodes proteins that are transported by the secretory pathway. Coat protein complexes such as COPII generate carrier vesicles and recruit cargos, orchestrated by cognate small GTPases as “molecular switches.” How coat complexes select specific cargos remains incompletely understood. Here we applied proximity-dependent biotinylation, fusing the promiscuous biotin ligase BirA* with the SAR1B GTPase, to track the dynamics of COPII-mediated export from the endoplasmic reticulum. LMAN1, a cargo receptor for COPII, is transiently enriched and released by the coat complex. The enrichment requires a dimeric sorting signal, formed by two copies of LMAN1. Hence, the COPII coat undergoes a dynamic engaging–disengaging cycle to select unique sets of secretory cargos.
Keywords: COPII, cargo sorting, cargo receptor, LMAN1, proximity-dependent biotinylation
Abstract
The flow of cargo vesicles along the secretory pathway requires concerted action among various regulators. The COPII complex, assembled by the activated SAR1 GTPases on the surface of the endoplasmic reticulum, orchestrates protein interactions to package cargos and generate transport vesicles en route to the Golgi. The dynamic nature of COPII, however, hinders analysis with conventional biochemical assays. Here we apply proximity-dependent biotinylation labeling to capture the dynamics of COPII transport in cells. When SAR1B was fused with a promiscuous biotin ligase, BirA*, the fusion protein SAR1B-BirA* biotinylates and thus enables the capture of COPII machinery and cargos in a GTP-dependent manner. Biochemical and pulse–chase imaging experiments demonstrate that the COPII coat undergoes a dynamic cycle of engagement–disengagement with the transmembrane cargo receptor LMAN1/ERGIC53. LMAN1 undergoes a process of concentrative sorting by the COPII coat, via a dimeric sorting code generated by oligomerization of the cargo receptor. Similar oligomerization events have been observed with other COPII sorting signals, suggesting that dimeric/multimeric sorting codes may serve as a general mechanism to generate selectivity of cargo sorting.
The secretory pathway in eukaryotic cells delivers a broad array of cargos that differ in both quantity and quality (1, 2). A variety of cellular transport machineries, often governed by unique small GTPases, operate concertedly to maintain and regulate the flow of these cargos in space and time (3, 4). Among these machineries, the coat proteins play a central role in sculpturing carrier vesicles and recruiting cargo molecules (1).
The COPII coat generates transport vesicles that ferry newly synthesized proteins from the endoplasmic reticulum (ER) to the Golgi apparatus (5–7). Assembly of the COPII complex is initiated by the small GTPase SAR1, upon its activation by the guanine nucleotide exchange factor (GEF) SEC12. Activated SAR1 inserts its N-terminal amphipathic helix into the ER surface deforming the ER membrane, and subsequently recruits the inner coat complex, a heterodimer of SEC23 and SEC24. SEC23 also possesses GTPase-activating protein (GAP) activity with the capacity to inactivate SAR1. The SEC31/SEC13 outer coat builds upon the SAR1–SEC23/SEC24 prebudding complex to form caged vesicles. SEC31 subsequently stimulates the GAP activity of SEC23 and cycles SAR1 to the inactive form, thereby allowing the scission of cargo vesicles and subsequently a reset of the COPII machinery. Intriguingly, mutations in specific paralogs of the COPII genes, including SAR1B, SEC23A, and SEC23B, cause diverse genetic disorders in human patients, though the underlying mechanism remains unclear (7).
Direct interactions between SEC24 and transmembrane cargos/cargo receptors provide selectivity in ER export, a process involving specific sorting signals located in the cytosolic portion of cargos or receptors (8–10). A number of sorting signals that occupy specific sites on SEC24 have been characterized (11–14), most of which are short stretches of two to four amino acids. While simple, short sorting motifs may provide flexibility by allowing more cargos to be accommodated by COPII (3, 15), additional mechanisms may be necessary to ensure the specificity of their selection by SEC24.
Transmembrane cargo receptors are thought to accelerate the ER export of selective proteins by initially concentrating these cargos into COPII vesicles (8, 16, 17). Though a number of cargo receptors have been identified in yeast and mammals that mediate concentrative sorting of cargos such as the α-factor, these receptors often contain simple sorting motifs composed of only two amino acids, which seemingly contrasts the efficiency and selectivity of cargo concentration mediated by COPII recognition (3, 18–23). LMAN1/ERGIC53 was originally identified as a marker for the ER–Golgi intermediate compartment (ERGIC) (24). Human genetics and cell biology studies subsequently showed that LMAN1 functions as the cargo receptor for coagulation factors V and VIII (25–27), and potentially also for alpha1-antitrypsin and cathepsins C and Z (28, 29). LMAN1 is an oligomeric single-span type I transmembrane protein, with a large N-terminal luminal domain and a short (12 amino acids) cytosolic tail terminating with diphenylalanine (FF). The FF motif is required for LMAN1 ER export in COPII vesicles, with the adjacent dilysine (KK) necessary for retrograde transport in COPI vesicles (16, 30).
Here we adapt a biochemical assay based on proximity-dependent biotinylation with a promiscuous biotin ligase, BirA* [BioID (31)], to investigate interactions of the COPII complex with cargos in cells. A SAR1B-BirA* fusion protein efficiently biotinylates COPII subunits as well as transmembrane cargos transiently recruited to COPII vesicles. Biotin labeling of these proteins is dependent on GTP loading of SAR1B, and thus specific to transport events following COPII assembly. The proximity assay reveals that LMAN1, a prototype of COPII cargos, is selectively enriched by the coat complex. Pulse–chase imaging also confirms the concentrative sorting of LMAN1 by COPII. This enrichment required at least two copies of LMAN1 to present a dimeric FF/FF sorting signal for the COPII complex, a phenomenon that may be common for other hydrophobic amino acid-based sorting motifs for the COPII coat.
Results and Discussion
Proximity-Dependent Capture of COPII Vesicle Components with SAR1B-BirA*.
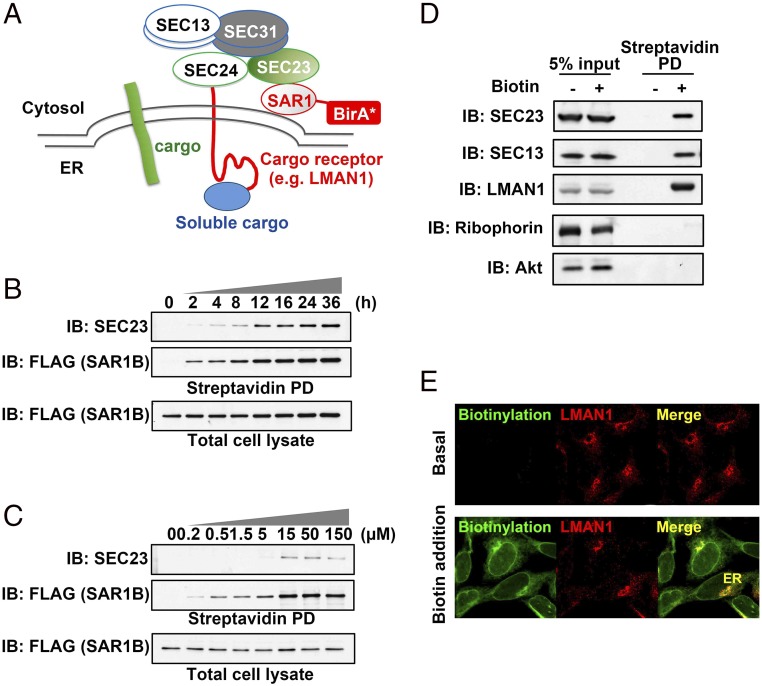
We hypothesized that actively assembling the COPII complex, though transient, could spatially enrich specific regulators and/or cargos for ER export within defined ER subdomains, thus allowing the proximity-dependent biotinylation technique to tag these dynamically recruited proteins and generate “permanent” marks to facilitate their tracking. To test this hypothesis, we placed the promiscuous biotin ligase BirA R118G (BirA*) at the C terminus of wild-type SAR1B that initiates COPII assembly when activated (Fig. 1A). Stably expressed SAR1B-BirA* biotinylated itself and endogenous SEC23 in a time- and biotin dose-dependent manner (Fig. 1 B and C). SAR1B-BirA* also efficiently labeled the outer COPII coat component SEC13 and the cargo receptor LMAN1, which does not directly interact with SAR1 but could be recruited by the COPII coats assembled by the GTPase. Little or no labeling by SAR1B-BirA* was observed for the control ER resident protein ribophorin I or the cytosolic protein Akt (Fig. 1D). Confocal microscopy demonstrated the distribution of SAR1B-BirA*–biotinylated proteins in the ER including the nuclear envelope, as well as post-ER organelles such as the ERGICs marked by LMAN1 staining (Fig. 1E), confirming that proximity-dependent biotinylation generates permanent labels to track proteins that were transiently recruited into COPII vesicles.
Fig. 1.
Development of a proximity-dependent biotinylation assay for COPII-mediated cargo transport. (A) Overall scheme of proximity-dependent biotinylation of the COPII machinery using SAR1-BirA*. Activated SAR1 initiates the assembly of the COPII coat, which recruits cargos and/or additional regulatory factors (e.g., the cargo receptor LMAN1) to defined microdomains, allowing biotinylation by BirA* fused to SAR1. (B) Time course of biotinylation of the COPII subunit SEC23 by SAR1B-BirA*. 293A cells stably expressing SAR1B-BirA* (with a FLAG tag at the C terminus of BirA*) were treated with 15 µM biotin for different time points as indicated. After cell lysis, biotinylated proteins were isolated by streptavidin bead pull-down (PD) and subjected to SDS/PAGE followed by immunoblotting (IB) with anti-SEC23 (Top) or anti-FLAG (recognizing SAR1B; Middle). Immunoblotting for SAR1B-FLAG in total cell lysates is shown (Bottom). The anti-SEC23 antibody recognizes both SEC23A and SEC23B. (C) Biotin dose dependence of SEC23 labeling by SAR1B-BirA*. 293A cells stably expressing SAR1B-BirA* were treated for 4 h with different doses of biotin as indicated. After cell lysis, biotinylated proteins were isolated by streptavidin beads and subjected to SDS/PAGE followed by immunoblotting with the indicated antibodies. (D) Capture of COPII cargos by SAR1B-BirA* in vivo. 293A cells stably expressing SAR1-BirA* were treated with 15 µM biotin for 4 h. After cell lysis, biotinylated proteins were isolated by streptavidin beads and subjected to SDS/PAGE followed by immunoblotting with the indicated antibodies; “5% input” indicates total cell lysate equivalent to 5% of the material used for streptavidin pull-down in lanes 3 and 4 (“Streptavidin PD”). (E) Subcellular localization of biotinylated proteins. 293A cells stably expressing SAR1B-BirA* were treated with 15 µM biotin for 4 h. After cell fixation, biotinylated proteins were visualized by immunostaining and confocal microscopy with an anti-LMAN1 antibody or Alexa Fluor-conjugated streptavidin.
The Assembly–Disassembly Cycle of COPII Coats Driven by SAR1B.
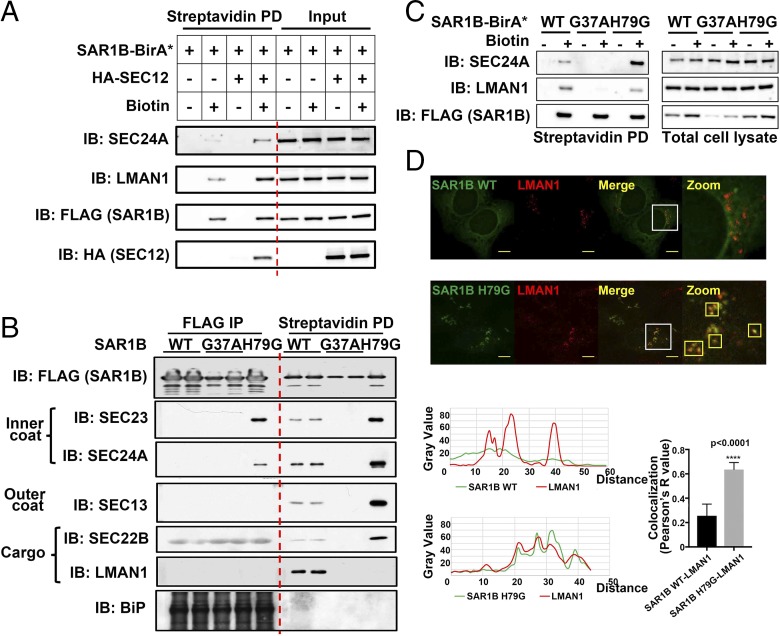
We first examined the regulatory steps of COPII function with the SAR1B-BirA* proximity assay. Overexpression of SEC12, the GEF that switches SAR1 to the active state (32), increased biotin labeling and capture of the COPII subunit SEC24A, as well as LMAN1 as a COPII cargo (Fig. 2A), confirming that SAR1B-BirA* could detect induction of COPII vesicle formation.
Fig. 2.
Assembly–disassembly cycle of COPII revealed by the proximity assay. (A) The SAR1 activator SEC12 promotes labeling of COPII subunits and cargos. 293A cells transfected with the indicated DNA constructs were treated with 15 µM biotin for 4 h. After cell lysis, biotinylated proteins were isolated by streptavidin beads and subjected to SDS/PAGE followed by immunoblotting with the indicated antibodies. (B) GTP-dependent cycling of the COPII coats revealed by coimmunoprecipitation (Left) and biotinylation (Right). 293A cells stably expressing the indicated SAR1B-BirA*-FLAG constructs were treated with 15 µM biotin for 4 h. After cell lysis, proteins were isolated by mouse anti-FLAG beads (FLAG IP) or streptavidin beads (Streptavidin PD) and subjected to SDS/PAGE followed by immunoblotting with the indicated antibodies. Mouse antibodies against BiP yielded strong nonspecific signal with the mouse anti-FLAG IgG used for IP. (C) GTP-locked SAR1B (H79G) decreases LMAN1 biotinylation. 293A cells transfected with the indicated DNA constructs were treated with 15 µM biotin for 4 h. After cell lysis, biotinylated proteins were isolated by streptavidin beads and subjected to SDS/PAGE followed by immunoblotting with the indicated antibodies. (D) GTP-locked SAR1B (H79G) traps LMAN1 on the ER. 293A cells stably expressing SAR1-BirA* or the H79G mutant were fixed and visualized by immunostaining and confocal microscopy with the indicated antibodies. Smaller boxes outlined in yellow indicate colocalization of LMAN1 and SAR1B. (Scale bars, 8 μm.) (D, Bottom) Quantification of the colocalization of green (SAR1B) and red (LMAN1) fluorophores. A total of 10 cells was analyzed for SAR1B H79G and for SAR1B WT. Error bars represent SEM.
Activated SAR1 is switched off by the GAP activity of SEC23, a required step in COPII transport that results in a transient interaction between SAR1 and the COPII coat (33). To dissect this dynamic cycle of COPII, we generated stable cell lines expressing BirA*-FLAG fused with either wild-type SAR1B, an inactive mutant (G37A), or a constitutively active mutant (H79G) that cannot be inactivated by SEC23 (34). Only the constitutively active SAR1B H79G mutant formed a stable complex with SEC23/SEC24 identifiable by FLAG coimmunoprecipitation (co-IP) (Fig. 2B, Left), with the absence of such signals for the wild-type SAR1B, likely due to rapid termination of SAR1–SEC23 interaction. In contrast, SEC23 and SEC24A were efficiently labeled by BirA* fused to wild-type SAR1B. This labeling was substantially increased by SAR1B H79G but was absent for SAR1B G37A (Fig. 2B, Right), demonstrating the role of the SAR1 GDP/GTP cycle in COPII assembly. Similar biotinylation was observed for the outer coat proteins SEC13, and not detected in the co-IP experiments (Fig. 2B). Moreover, the proximity assay also captured the COPII cargo SEC22B in a SAR1-GTP–dependent manner, an indirect interaction not detected by standard co-IP, though the light chains of anti-FLAG IgG give nonspecific signal across the IP samples. Taken together, these data suggest that the SAR1B-BirA* proximity assay faithfully captures multilayer protein interactions during the assembly of the COPII complex, despite their transient or indirect nature.
We also observed that LMAN1 was effectively biotinylated by SAR1B WT-BirA* but not by the inactive SAR1B G37A-BirA*, consistent with the requirement for SAR1 activation in COPII assembly and cargo recruitment. Surprisingly, SAR1B H79G-BirA* failed to label LMAN1 (Fig. 2B, Far Right). Similarly, LMAN1 showed decreased labeling by SAR1B H79G-BirA* compared with SAR1B WT-BirA* with transient overexpression (Fig. 2C), ruling out potential clonal effects with stable cell lines. LMAN1 contains a short cytosolic tail of 12 amino acids, including the only two lysine residues available for biotinylation (Fig. 3A). The constitutively active SAR1B H79G could potentially freeze the LMAN1–COPII interaction, masking the dilysine motif from biotinylation. Consistent with this notion, though LMAN1 displayed classic ERGIC staining with little colocalization with wild-type SAR1B on the ER surface, LMAN1 in SAR1B H79G-expressing cells primarily localized to concentrated punctae on the ER, which were also marked by SAR1 H79G (Fig. 2D). ER-trapped LMAN1 in the SAR1 H79G-expressing cells also displayed significant colocalization with SEC24A (Fig. S1), further indicating that the “stalled” LMAN1–COPII interaction may prevent the exposure of the lysine residues in LMAN1 tails for biotinylation. Taken together, the itinerary of LMAN1 exemplifies the assembly–disassembly cycle of COPII regulated by the GDP/GTP switch of SAR1.
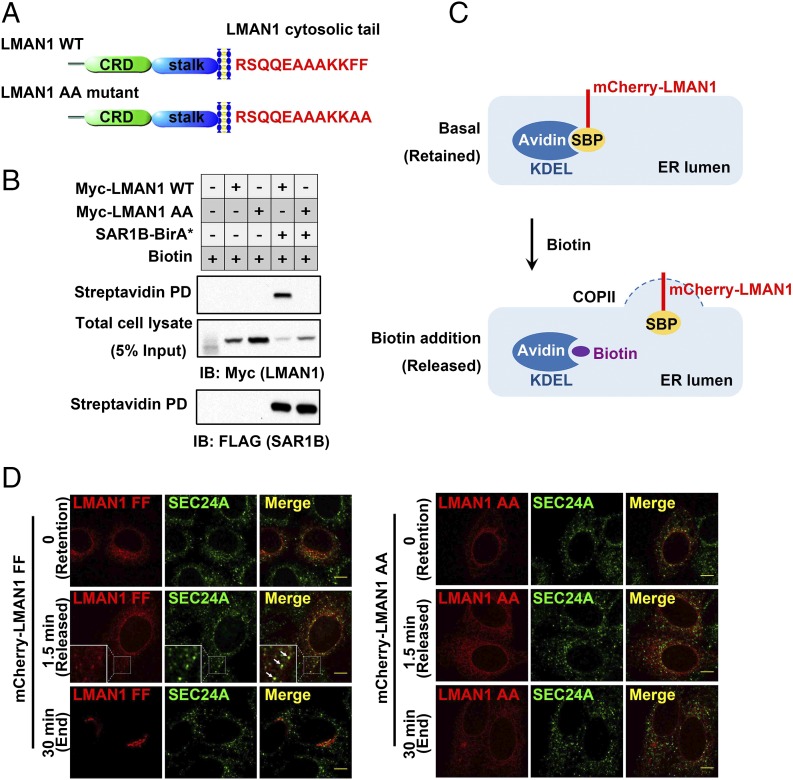
Fig. 3.
Concentrative sorting of LMAN1 by COPII coats. (A) Schematics of LMAN1. The luminal domains and amino acid sequences of the cytosolic tails in wild-type LMAN1 and the AA mutant are shown. CRD, carbohydrate-binding domain. The stalk domain mediates LMAN1 oligomerization (51). (B) Enrichment of wild-type LMAN1, but not the LMAN1-AA mutant, by COPII revealed by SAR1B-BirA*. 293A cells stably expressing SAR1B-BirA* were transfected with the indicated LMAN1 constructs and treated with 15 µM biotin for 24 h. After cell lysis, biotinylated proteins were isolated by streptavidin beads and subjected to SDS/PAGE followed by immunoblotting with the indicated antibodies. (C) Schematics of the pulse–chase experiment using the RUSH imaging strategy. In the basal state (no biotin), SBP-mCherry-LMAN1 is retained in the ER by streptavidin-KDEL. Addition of biotin releases SBP-mCherry-LMAN1 from streptavidin-KDEL, allowing LMAN1 to be exported from the ER. (D) Wild-type LMAN1, but not the LMAN1-AA mutant, is enriched on COPII-coated puncta before ER export. HeLa cells expressing the indicated SBP-mCherry-LMAN1 constructs and streptavidin-KDEL were fixed at different time points following biotin treatment and subjected to immunostaining with an anti-SEC24A antibody, followed by confocal microscopy. Arrows indicate the colocalization of LMAN1 and SEC24A on the ER surface. (Scale bars, 8 µm.)
Concentrative Sorting of LMAN1 by COPII.
The cycling of the LMAN1–COPII interaction may enable efficient processing of cargos. Biotinylation, as a covalent modification, should permanently mark LMAN1 when it is “processed” by the assembled COPII. We therefore used the efficiency of LMAN1 biotinylation as a quantitative readout of its recruitment by the COPII coat in vivo, and observed that ∼50% of total wild-type LMAN1 is labeled by biotin at 24 h (Fig. 3B). The two phenylalanine residues in the C terminus of LMAN1 are required for COPII interaction (35, 36). In contrast to the efficient biotinylation of wild-type LMAN1, biotin labeling was undetectable with an LMAN1 mutant in which the C-terminal FF motif was replaced by AA (Fig. 3B), suggesting that, in the absence of specific sorting signals, incorporation of LMAN1-AA into COPII by general bulk flow is inefficient.
To directly visualize the enrichment of LMAN1 by COPII, we performed a pulse–chase experiment using the RUSH (retention using selective hooks) system (37). Either LMAN1 or LMAN1-AA was fused with a streptavidin-binding peptide (SBP), which can be selectively retained in the ER by streptavidin tagged with the ER retention signal KDEL (Fig. 3C). Addition of biotin disrupts the SBP–streptavidin interaction, thereby allowing synchronized release of LMAN1 from the ER (Movie S1). Before release, LMAN1 displayed a diffuse, reticular localization throughout the ER. Upon biotin-induced release, LMAN1 was quickly concentrated in ER punctations (in ∼1 to 2 min), which were marked by the COPII subunit SEC24A. By 30 min, LMAN1 can be seen in the ERGIC and clearly separated from SEC24A punctations (Fig. 3D, Left), further supporting a dynamic engagement–disengagement cycle between the cargo receptor and COPII. In contrast to wild-type LMAN1, this concentration and subsequent transport were not observed with LMAN1-AA, which displayed a diffuse ER localization throughout the recording (Fig. 3D, Right and Movie S2). Taken together, the biochemical and pulse–chase imaging data demonstrate concentrative sorting of LMAN1 by the COPII coat and the subsequent release of this cargo receptor, with the former process requiring a specific sorting signal based on the FF motif in the LMAN1 cytosolic tail.
An LMAN1 Dimer Represents the Minimal Unit for ER Exit.
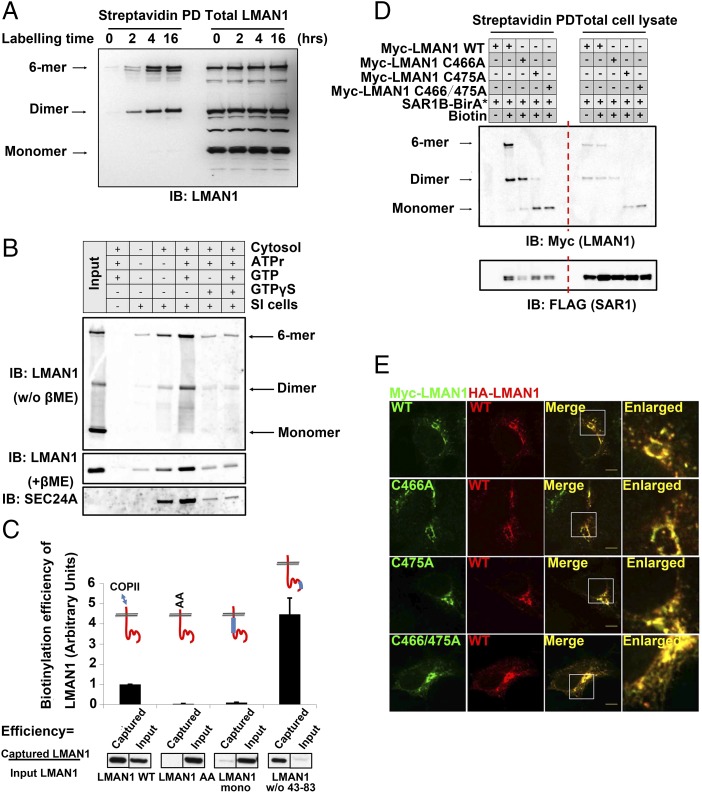
The efficient concentration of LMAN1 is surprising, compared with the apparent simplicity of its FF motif required for sorting, leading us to search for additional mechanisms that account for the sorting specificity by COPII. Analysis by nondenaturing SDS/PAGE, preserving LMAN1 oligomers, demonstrated that only dimeric or higher oligomers of LMAN1 were biotinylated by SAR1-BirA* (Fig. 4A), while monomeric LMAN1 displayed no such labeling. These data suggest that the LMAN1 dimer constitutes the minimal unit to exit the ER via COPII. Consistent with this idea, cell-free reconstitution assays also demonstrated the recruitment of LMAN1 dimers and oligomers to COPII-coated vesicles formed in vitro, while LMAN1 monomers were excluded from the vesicle fraction (Fig. 4B).
Fig. 4.
LMAN1 dimers constitute the minimal unit for export from the ER. (A) ER export of LMAN1 dimers and oligomers revealed by the proximity assay. 293A cells stably expressing SAR1B-BirA* were treated with 15 µM biotin for the indicated time points. After cell lysis, biotinylated proteins were isolated by streptavidin beads and subjected to SDS/PAGE without reducing agents followed by immunoblotting with an anti-LMAN1 antibody. (B) Cell-free “budding” assay to reconstitute ER export of LMAN1 dimers/oligomers. Semiintact (SI) cells were mixed with cytosol and the indicated reagents to catalyze in vitro vesicle formation from the ER. The isolated vesicles were subjected to immunoblotting without reducing agent (Top) or with reducing agent (Middle and Bottom) with the indicated antibodies. No dimers or hexamers of LMAN1 were observed in the reducing condition. ATPr, ATP regeneration system. GTPγS is the nonhydrolyzable analog of GTP, and blocks COPII budding from the ER. (C) Biotinylation efficiency of LMAN1 mutants in the proximity assay. 293A cells stably expressing SAR1B-BirA* were transfected with the indicated Myc-LMAN1 mutants and treated with 15 µM biotin for 4 h. After cell lysis, biotinylated proteins were isolated by streptavidin beads and subjected to SDS/PAGE followed by immunoblotting with an anti-Myc antibody. (C, Top) LMAN1 is depicted in red, with blue bars representing deleted regions within the LMAN1 protein. Error bars represent SEM. (D) Dimeric LMAN1 is biotinylated by SAR1B-BirA*. 293A cells stably expressing SAR1B-BirA* were transfected with the indicated Myc-LMAN1 cysteine-to-alanine mutants and treated with 15 µM biotin for 4 h. After cell lysis, biotinylated proteins were isolated by streptavidin beads and subjected to native SDS/PAGE followed by immunoblotting with an anti-Myc antibody. Different oligomerization states of LMAN1 are depicted (Left). (E) Dimeric LMAN1 is targeted to the ERGICs. Cos-1 cells expressing the indicated LMAN1 constructs were fixed and subjected to immunostaining with the indicated antibodies, followed by confocal microscopy. (Scale bars, 8 µm.)
To further dissect the molecular basis for LMAN1 sorting, we employed additional mutants of the cargo receptor (38). A truncated LMAN1 lacking the luminal dimerization domain (“stalk” in Fig. 3A) but still containing one FF motif in the cytoplasmic tail largely failed to be incorporated into COPII vesicles (Fig. 4C, column 3, “LMAN1 mono”), demonstrating that the monomeric FF sorting signal is not sufficient for COPII interaction. This is consistent with previous reports using LMAN1 fragments fused with glycosylation sequences (39). Of note, an N-terminal β1-domain–truncated LMAN1 mutant showed increased recruitment by COPII (Fig. 4C, column 4). This mutant fails to interact with the auxiliary protein MCFD2 but retains carbohydrate interaction (38), suggesting that LMAN1 luminal domains may influence the LMAN1–COPII interaction from across the membrane.
To differentiate the involvement of the LMAN1 dimer and hexamer in ER exiting, we mutated the cysteines in the juxtamembrane regions. Consistent with a previous report (39), native SDS/PAGE revealed that single C466A or C475A or the double mutant of LMAN1 failed to form covalent hexamers but existed in covalent or noncovalent dimers (Fig. 4D). These dimers can all be labeled by SAR1B-BirA* (Fig. 4D). Taken together with the higher stoichiometric labeling of WT LMAN1 dimers over hexamers in Fig. 4A, the data demonstrated dimeric LMAN1 as the minimal unit for ER exporting via the COPII complex. When examined by confocal microscopy, single or double C/A mutants of LMAN1 can also be targeted to the ERGICs, similar to the wild-type cargo receptor (Fig. 4E).
Dimeric Sorting Codes Enable COPII-Mediated ER Exit.
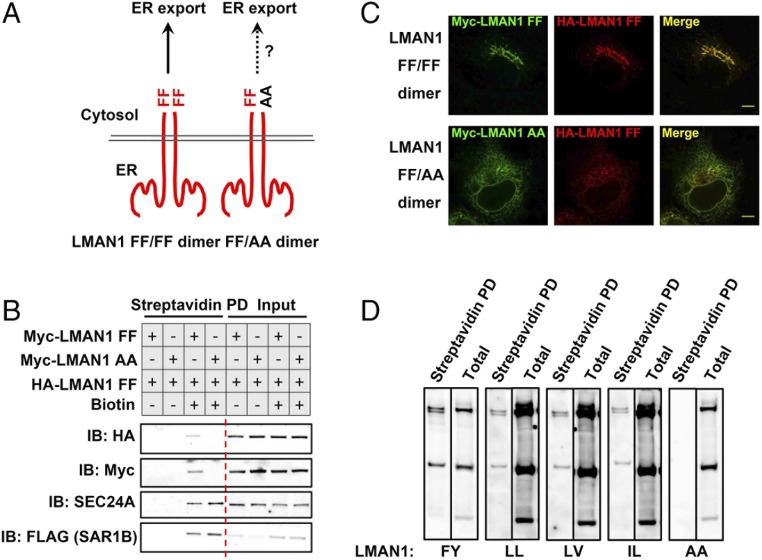
The apparent failure of monomeric LMAN1 to be exported from the ER led us to hypothesize that a dimeric FF–FF sorting code is required for interaction with the COPII machinery. In this case, FF–AA heterodimers would fail to be recruited by COPII. To test this hypothesis, we coexpressed HA-tagged LMAN1 WT, with either Myc-tagged LMAN1 WT or LMAN1-AA, to examine their recruitment by the COPII coat using the proximity assay (Fig. 5A). LMAN1-AA failed to be labeled by SAR1B-BirA*, consistent with the data in Fig. 3B. However, coexpression of LMAN1-AA also inhibited the recruitment of LMAN1 WT by the COPII machinery, as measured by SAR1B-BirA* biotinylation (Fig. 5B). Similar inhibitory effects were observed for LMAN1 dimers or hexamers under nondenaturing conditions (Fig. S3). Furthermore, coexpression of LMAN1-AA resulted in retention of wild-type LMAN1 in the ER, in contrast to the ERGIC punctae observed with control wild-type LMAN1 expression (Fig. 5C). LMAN1-AA has been reported to function as a dominant-negative mutant when coexpressed with the wild-type cargo receptor, though the mechanism was previously unknown (35, 40, 41). Our data suggest that a dimeric FF–FF motif is required for LMAN1 interaction with COPII, and that LMAN1-AA acts as a dominant negative by “poisoning” wild-type LMAN1 through the formation of a nonfunctional heterodimer and disrupting the dimeric sorting signal.
Fig. 5.
Dimeric sorting motifs mediate ER export via the COPII coat. (A) Schematics of a homodimeric FF–FF motif or a heterodimeric FF–AA motif. (B) LMAN1-AA inhibits ER export of the wild-type LMAN1 revealed by the proximity assay. 293A cells stably expressing SAR1B-BirA* were transfected with the indicated LMAN1 mutants and treated with 15 µM biotin for 4 h. After cell lysis, biotinylated proteins were isolated by streptavidin beads and subjected to SDS/PAGE followed by immunoblotting with the indicated antibodies. (C) LMAN1-AA traps wild-type LMAN1 in the ER. HeLa cells expressing the indicated LMAN1 constructs were fixed and subjected to immunostaining with the indicated antibodies, followed by confocal microscopy. (Scale bars, 8 µm.) (D) Dimeric FY, LL, LV, and IL sorting motifs in ER export. 293A cells stably expressing SAR1-BirA* were transfected with Myc-LMAN1 mutants with the indicated sorting motifs and treated with 15 µM biotin for 4 h. After cell lysis, biotinylated proteins were isolated by streptavidin beads and subjected to SDS/PAGE followed by immunoblotting with an anti-Myc antibody.
To test whether other di-amino acid sorting motifs also undergo dimerization/oligomerization to interact with the COPII machinery, we replaced FF in LMAN1 with FY, or other hydrophobic di-amino acid-sorting motifs, such as LL, LV, or IL. When analyzed with nondenaturing SDS/PAGE, all three of these additional motifs exhibited dimer/oligomer-dependent biotinylation, though with variable efficiency, while the monomeric forms failed to be labeled by SAR1-BirA* (Fig. 5D), suggesting dimerization/oligomerization may be a common phenomenon in COPII–cargo interactions. Consistent with this notion, we observed several biotinylated high-molecular-weight bands under nondenaturing conditions that were lost following denaturation (Fig. S2), implying the presence of other unidentified oligomeric proteins in the proximity of SAR1B-BirA*.
Mechanisms governing the specificity of COPII-mediated transport remain incompletely understood (5–7). We report here concentrative sorting of the cargo receptor LMAN1, enabled by a dimeric sorting signal formed by two copies of the transmembrane protein. Such concentrative sorting could provide a mechanism to segregate a specific subset of COPII cargo receptors and their cargos from the bulk flow of other secreted proteins (3, 16). Concentrative selection requires specific interactions between COPII and cargo/cargo receptors, via obligatory sorting signals (3, 15, 42). The FF–FF dimer of LMAN1 may increase selectivity over the monomeric FF signal, potentially explaining the highly specific LMAN1–COPII interaction. In addition to LMAN1, other cargo receptors, such as Rer1p in yeast (43) and the p24 family of proteins, also undergo dimerization or oligomerization (17), raising the possibility that dimeric or oligomeric sorting motifs may serve as a general mechanism to form specific interactions with COPII. Such dimers in trans could provide greater complexity than one monomer or linear repeats in cis, with the potential to generate “3D” sorting codes for COPII recognition. Formation of oligomers may also provide another level of regulation during cargo recruitment, particularly from within the ER lumen. Although LMAN1 dimers are stabilized by disulfide bonds, luminal calcium fluctuations and oxidative states have been shown to influence LMAN1 ER exit (28, 41, 44), while other noncovalently linked oligomers may be more prone to regulation. Intriguingly, previous work on the yeast heteromeric cargo receptor complex Erv41–Erv46 has shown that Erv41 relies on an IL motif for COPII recognition and Erv46 uses an FY motif to stimulate vesicle formation, though it is unclear whether these motifs also require dimerization for these functions (45). Further study, likely with the aid of structural biology, will ultimately illustrate the molecular architecture of COPII in complex with these high-order, multimeric signals, and may identify other examples of dimeric/oligomeric cargos of COPII.
Materials and Methods
DNA Constructs, Chemicals, and Antibodies.
LMAN1 deletion mutants have been described previously (38) and were subcloned into pKH3 or pK-Myc vectors (46). Gateway vectors expressing humanized BirA* bearing R118G cDNA were kind gifts from A. Gingras, Lunenfeld-Tanenbaum Research Institute, Toronto (47). Both human SAR1A and SAR1B were placed in the N terminus of BirA*-FLAG, and SAR1B was primarily used in this study. The RUSH constructs (streptavidin-KDEL-IRES-SBP-mCherry-MAN II) were obtained from Addgene, and MAN II was replaced by LMAN1 cDNA. Other expression constructs have been described previously (48). All constructs were confirmed by complete DNA sequencing. Chemicals were purchased from Sigma-Aldrich.
The anti-SEC24A antibody has been previously described (48). Rabbit anti-FLAG antibody, mouse anti-actin antibody, and rabbit anti-SEC23 antibody were purchased from Sigma-Aldrich. Rabbit anti-LMAN1 antibodies were purchased from Stressgen. Preparation of anti-SAR1, anti-ribophorin, and anti-SEC22B antibodies has been described previously (49).
Cell Culture, Transfection, and Immunostaining.
293A and HeLa cells were grown in Dulbecco’s modified Eagle’s medium (GIBCO) containing 10% FBS (Sigma-Aldrich) and 1% penicillin-streptomycin (GIBCO) at 37 °C in the presence of 5% CO2. Cells were transfected with polyethylenimine (EMD Millipore) according to the manufacturer’s instructions for 16 to 24 h before the following experiments. The Flp-In System (Invitrogen) was used to generate 293A cells stably expressing SAR1B-BirA* proteins as previously described (47).
For immunostaining experiments, cells were grown on glass coverslips and washed with PBS before fixation. After fixation with methanol at −20 °C for 3 min, cells were rehydrated in PBS and then blocked with 1% BSA and 1% chicken albumin. Monoclonal Myc and HA antibodies (Santa Cruz) were used at 1:500 dilution; rabbit anti-SEC24A antibody was used at 1:100 dilution. Other antibodies have been previously described (46, 50). Alexa Fluor-conjugated goat anti-mouse/rabbit secondary antibodies, Alexa Fluor-conjugated streptavidin, and Vectashield mounting medium were from Molecular Probes. Confocal images were obtained using an inverted Olympus X81 confocal microscope operated with the MetaMorph program (Molecular Devices). Digital images were finally processed with the entire microscopic field using ImageJ software (NIH); colocalization of different fluorophores was performed with ImageJ’s colocalization plugin (Coloc 2) to determine Pearson’s R value (no threshold).
Immunoprecipitation, Streptavidin Pull-Down, and Immunoblotting.
Immunoprecipitation or streptavidin pull-down was carried out with cellular proteins extracted at 4 °C for 30 min with buffer A (100 mM Tris, pH 7.5, 1% Nonidet P-40, 10% glycerol, 130 mM sodium chloride, 5 mM magnesium chloride, 1 mM sodium vanadate, 1 mM sodium fluoride, and 1 mM EDTA) supplemented with protease inhibitor tablets (Roche), according to the manufacturer’s instructions. Cell lysates were then incubated with 10 µL M2 agarose (Sigma-Aldrich) or 20 µL streptavidin agarose (Sigma-Aldrich) for 4 h at 4 °C before washing four times with buffer A. Immune complexes were then solubilized with 1× SDS/PAGE sample buffer (Invitrogen) with or without (when indicated) 5% β-mercaptoethanol (β-ME), and separated with 3 to 15% Tris-acetate SDS/PAGE (Invitrogen). For streptavidin pull-down experiments, a final concentration of 1 mM biotin was included in the SDS/PAGE sample buffer, as previously described (31). Following SDS/PAGE, proteins were transferred onto nitrocellulose membranes (Bio-Rad) and immunoblotting was performed as previously described (50). Quantification of immunoblotting data was performed using ImageJ.
In Vitro Budding.
Cell-free reconstitution experiments using permeabilized cells were performed as previously described (49) with slight modification. Three plates of 293A cells grown to ∼80 to 90% confluency were collected by trypsinization and sedimented at 750 × g for 5 min at 4 °C. Cells were resuspended in 6 mL B88 buffer (20 mM Hepes, pH 7.2, 250 mM sorbitol, and 150 mM KOAc) and permeabilized with 40 µg/mL digitonin on ice for 5 min. Permeabilization was stopped by adding 8 mL ice-cold B88 buffer, and permeabilized cells were sedimented at 750 × g for 5 min at 4 °C. After washing twice with 6 mL B88 buffer at 4 °C, permeabilized cells were resuspended in 1 mL B88 buffer and centrifuged at 10,000 × g for 1 min at 4 °C. Permeabilized cells were then resuspended in 1 mL B88 buffer supplemented with 1 M MgCl2 for 10 min on ice to wash off cytosolic protein associated with cellular membranes. Permeabilized cells were then washed three times with 1 mL B88 buffer after being centrifuged at 10,000 × g for 20 s, and finally resuspended in 0.2 to 0.3 mL B88 buffer. Budding reactions (100 µL) were assembled in nonstick Eppendorf tubes on ice with 20 µL (OD600 0.1 to 0.2) semiintact cells as donor membranes, 10 µL 10× ATP regeneration system (10 mM ATP, 400 mM creatine phosphate, 2 mg/mL creatine phosphokinase, and 5 mM MgOAc in B88 buffer), 1.5 µL 10 mM GTP, and rat liver cytosol at 4 mg/mL final concentration. Reactions were performed at 30 °C for 60 min and stopped by centrifuging at 10,000 × g for 10 min at 4 °C. Seventy-five microliters of supernatant was centrifuged in a TLA100 rotor at 100,000 × g for 10 min at 4 °C. The supernatants were discarded by pipetting with gel-loading tips, and the high-speed pellet fractions (including COPII vesicles) were thoroughly resuspended in 20 µL of 1× SDS sample buffer (Invitrogen) supplemented with or without 5% β-ME and heated at 55 °C for 20 min before SDS/PAGE.
Pulse–Chase Imaging Using RUSH and Live-Cell Imaging.
For live-cell imaging, HeLa cells grown on confocal dishes were transfected with either streptavidin-KDEL-IRES-SBP-mCherry-LMAN1 FF or streptavidin-KDEL-IRES-SBP-mCherry-LMAN1 AA mutant constructs. Twenty-four hours after transfection, cells with ER-localized red fluorescence (mCherry-LMAN1) were identified. Prewarmed cell-culture medium containing 80 µM biotin was introduced into the dish cautiously to double the culture medium. Cells were imaged under a 63× oil lens at 10-s intervals for about 20 min at 37 °C with 5% CO2. For fixed-cell imaging, transfected HeLa cells treated with 40 µM biotin for two different time points as indicated before fixation proceeded to immunostaining. Images were processed by MetaMorph.
Supplementary Material
Acknowledgments
The authors thank R. Schekman, R. Khoriaty, and H. Cai for helpful comments. The work is supported by National Science Foundation of China Grants 31571213 and 31521062, National Science and Technology Support Project 2014BAI02B01, the Young 1000 Talents Plan (X.-W.C.), and National Institute of Health Grants R01 HL039693, P01 HL057346, and R35HL135793 (to D.G.). D.G. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704639115/-/DCSupplemental.
References
- 1.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 2.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 3.Barlowe C, Helenius A. Cargo capture and bulk flow in the early secretory pathway. Annu Rev Cell Dev Biol. 2016;32:197–222. doi: 10.1146/annurev-cellbio-111315-125016. [DOI] [PubMed] [Google Scholar]
- 4.Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat Rev Mol Cell Biol. 2013;14:382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gürkan C, Stagg SM, Lapointe P, Balch WE. The COPII cage: Unifying principles of vesicle coat assembly. Nat Rev Mol Cell Biol. 2006;7:727–738. doi: 10.1038/nrm2025. [DOI] [PubMed] [Google Scholar]
- 7.Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2011;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- 8.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 9.Miller E, Antonny B, Hamamoto S, Schekman R. Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 2002;21:6105–6113. doi: 10.1093/emboj/cdf605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller EA, et al. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- 11.Bickford LC, Mossessova E, Goldberg J. A structural view of the COPII vesicle coat. Curr Opin Struct Biol. 2004;14:147–153. doi: 10.1016/j.sbi.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Mancias JD, Goldberg J. The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol Cell. 2007;26:403–414. doi: 10.1016/j.molcel.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Mancias JD, Goldberg J. Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO J. 2008;27:2918–2928. doi: 10.1038/emboj.2008.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mossessova E, Bickford LC, Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- 15.Barlowe C. Signals for COPII-dependent export from the ER: What’s the ticket out? Trends Cell Biol. 2003;13:295–300. doi: 10.1016/s0962-8924(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 16.Baines AC, Zhang B. Receptor-mediated protein transport in the early secretory pathway. Trends Biochem Sci. 2007;32:381–388. doi: 10.1016/j.tibs.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Dancourt J, Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- 18.Malkus P, Jiang F, Schekman R. Concentrative sorting of secretory cargo proteins into COPII-coated vesicles. J Cell Biol. 2002;159:915–921. doi: 10.1083/jcb.200208074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowe T, et al. COPII vesicles derived from mammalian endoplasmic reticulum microsomes recruit COPI. J Cell Biol. 1996;135:895–911. doi: 10.1083/jcb.135.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thor F, Gautschi M, Geiger R, Helenius A. Bulk flow revisited: Transport of a soluble protein in the secretory pathway. Traffic. 2009;10:1819–1830. doi: 10.1111/j.1600-0854.2009.00989.x. [DOI] [PubMed] [Google Scholar]
- 21.Warren G, Mellman I. Bulk flow redux? Cell. 1999;98:125–127. doi: 10.1016/s0092-8674(00)81006-5. [DOI] [PubMed] [Google Scholar]
- 22.Wieland FT, Gleason ML, Serafini TA, Rothman JE. The rate of bulk flow from the endoplasmic reticulum to the cell surface. Cell. 1987;50:289–300. doi: 10.1016/0092-8674(87)90224-8. [DOI] [PubMed] [Google Scholar]
- 23.Nishimura N, et al. A di-acidic (DXE) code directs concentration of cargo during export from the endoplasmic reticulum. J Biol Chem. 1999;274:15937–15946. doi: 10.1074/jbc.274.22.15937. [DOI] [PubMed] [Google Scholar]
- 24.Schindler R, Itin C, Zerial M, Lottspeich F, Hauri HP. ERGIC-53, a membrane protein of the ER-Golgi intermediate compartment, carries an ER retention motif. Eur J Cell Biol. 1993;61:1–9. [PubMed] [Google Scholar]
- 25.Nichols WC, et al. Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell. 1998;93:61–70. doi: 10.1016/s0092-8674(00)81146-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang B, et al. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nat Genet. 2003;34:220–225. doi: 10.1038/ng1153. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B, et al. Mice deficient in LMAN1 exhibit FV and FVIII deficiencies and liver accumulation of α1-antitrypsin. Blood. 2011;118:3384–3391. doi: 10.1182/blood-2011-05-352815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Appenzeller C, Andersson H, Kappeler F, Hauri HP. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat Cell Biol. 1999;1:330–334. doi: 10.1038/14020. [DOI] [PubMed] [Google Scholar]
- 29.Nyfeler B, et al. Identification of ERGIC-53 as an intracellular transport receptor of alpha1-antitrypsin. J Cell Biol. 2008;180:705–712. doi: 10.1083/jcb.200709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itin C, Foguet M, Kappeler F, Klumperman J, Hauri HP. Recycling of the endoplasmic reticulum/Golgi intermediate compartment protein ERGIC-53 in the secretory pathway. Biochem Soc Trans. 1995;23:541–544. doi: 10.1042/bst0230541. [DOI] [PubMed] [Google Scholar]
- 31.Roux KJ, Kim DI, Burke B. BioID: A screen for protein-protein interactions. Curr Protoc Protein Sci. 2013;74:Unit 19.23. doi: 10.1002/0471140864.ps1923s74. [DOI] [PubMed] [Google Scholar]
- 32.Barlowe C, Schekman R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- 33.Yoshihisa T, Barlowe C, Schekman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- 34.Fromme JC, et al. The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev Cell. 2007;13:623–634. doi: 10.1016/j.devcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itin C, Schindler R, Hauri HP. Targeting of protein ERGIC-53 to the ER/ERGIC/cis-Golgi recycling pathway. J Cell Biol. 1995;131:57–67. doi: 10.1083/jcb.131.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B, Kaufman RJ, Ginsburg D. LMAN1 and MCFD2 form a cargo receptor complex and interact with coagulation factor VIII in the early secretory pathway. J Biol Chem. 2005;280:25881–25886. doi: 10.1074/jbc.M502160200. [DOI] [PubMed] [Google Scholar]
- 37.Boncompain G, et al. Synchronization of secretory protein traffic in populations of cells. Nat Methods. 2012;9:493–498. doi: 10.1038/nmeth.1928. [DOI] [PubMed] [Google Scholar]
- 38.Zheng C, Liu HH, Yuan S, Zhou J, Zhang B. Molecular basis of LMAN1 in coordinating LMAN1-MCFD2 cargo receptor formation and ER-to-Golgi transport of FV/FVIII. Blood. 2010;116:5698–5706. doi: 10.1182/blood-2010-04-278325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nufer O, Kappeler F, Guldbrandsen S, Hauri HP. ER export of ERGIC-53 is controlled by cooperation of targeting determinants in all three of its domains. J Cell Sci. 2003;116:4429–4440. doi: 10.1242/jcs.00759. [DOI] [PubMed] [Google Scholar]
- 40.Moussalli M, et al. Mannose-dependent endoplasmic reticulum (ER)-Golgi intermediate compartment-53-mediated ER to Golgi trafficking of coagulation factors V and VIII. J Biol Chem. 1999;274:32539–32542. doi: 10.1074/jbc.274.46.32539. [DOI] [PubMed] [Google Scholar]
- 41.Nufer O, et al. Role of cytoplasmic C-terminal amino acids of membrane proteins in ER export. J Cell Sci. 2002;115:619–628. doi: 10.1242/jcs.115.3.619. [DOI] [PubMed] [Google Scholar]
- 42.Barlowe C. Membrane trafficking: ER export encounters dualism. Curr Biol. 2015;25:R151–R153. doi: 10.1016/j.cub.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 43.Sato K, Sato M, Nakano A. Rer1p, a retrieval receptor for ER membrane proteins, recognizes transmembrane domains in multiple modes. Mol Biol Cell. 2003;14:3605–3616. doi: 10.1091/mbc.E02-12-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klumperman J, et al. The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J Cell Sci. 1998;111:3411–3425. doi: 10.1242/jcs.111.22.3411. [DOI] [PubMed] [Google Scholar]
- 45.Otte S, Barlowe C. The Erv41p-Erv46p complex: Multiple export signals are required in trans for COPII-dependent transport from the ER. EMBO J. 2002;21:6095–6104. doi: 10.1093/emboj/cdf598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen XW, et al. Exocyst function is regulated by effector phosphorylation. Nat Cell Biol. 2011;13:580–588. doi: 10.1038/ncb2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lambert JP, Tucholska M, Go C, Knight JD, Gingras AC. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J Proteomics. 2015;118:81–94. doi: 10.1016/j.jprot.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen XW, et al. SEC24A deficiency lowers plasma cholesterol through reduced PCSK9 secretion. eLife. 2013;2:e00444. doi: 10.7554/eLife.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, et al. Biogenesis of gamma-secretase early in the secretory pathway. J Cell Biol. 2007;179:951–963. doi: 10.1083/jcb.200709012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen XW, et al. A Ral GAP complex links PI 3-kinase/Akt signaling to RalA activation in insulin action. Mol Biol Cell. 2011;22:141–152. doi: 10.1091/mbc.E10-08-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng C, et al. Structural characterization of carbohydrate binding by LMAN1 protein provides new insight into the endoplasmic reticulum export of factors V (FV) and VIII (FVIII) J Biol Chem. 2013;288:20499–20509. doi: 10.1074/jbc.M113.461434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.