Significance
Advances in technology are enabling evaluation for prevention and early detection of age-related chronic diseases associated with premature mortality, such as cancer and cardiovascular diseases. These diseases kill about one-third of men and one-quarter of women between the ages of 50 and 74 years old in the United States. We used whole-genome sequencing, advanced imaging, and other clinical testing to screen 209 active, symptom-free adults. We identified a broad set of complementary age-related chronic disease risks associated with premature mortality.
Keywords: precision medicine, screening, genomics, genome, magnetic resonance imaging
Abstract
Reducing premature mortality associated with age-related chronic diseases, such as cancer and cardiovascular disease, is an urgent priority. We report early results using genomics in combination with advanced imaging and other clinical testing to proactively screen for age-related chronic disease risk among adults. We enrolled active, symptom-free adults in a study of screening for age-related chronic diseases associated with premature mortality. In addition to personal and family medical history and other clinical testing, we obtained whole-genome sequencing (WGS), noncontrast whole-body MRI, dual-energy X-ray absorptiometry (DXA), global metabolomics, a new blood test for prediabetes (Quantose IR), echocardiography (ECHO), ECG, and cardiac rhythm monitoring to identify age-related chronic disease risks. Precision medicine screening using WGS and advanced imaging along with other testing among active, symptom-free adults identified a broad set of complementary age-related chronic disease risks associated with premature mortality and strengthened WGS variant interpretation. This and other similarly designed screening approaches anchored by WGS and advanced imaging may have the potential to extend healthy life among active adults through improved prevention and early detection of age-related chronic diseases (and their risk factors) associated with premature mortality.
The near-doubling of average human life expectancy over the last 150 y is a tribute to scientific advancements in medicine and public health (1). This success is largely the result of progress in control and prevention of infectious diseases, particularly in prevention of early childhood deaths. Eighty-five percent of children born now in the United States can expect to live to at least 65 y of age, and 42% will likely celebrate an 85th birthday (1). Partly because of this progress, the United States and many other parts of the world are facing a daunting and costly new and growing epidemic of age-related chronic diseases (1, 2).
Most age-related chronic diseases have substantial heritability (3, 4), often are slowly progressive with symptom-free onset (5), and are associated with common risk factors (2, 6). In 2015, the estimated US cumulative mortality risk among males 50–74 y of age was 39%; for women, the risk was lower but still substantial at 24% (6, 7). The causes of these deaths are similar across men and women, with neoplasms and cardiovascular disease accounting for about one-third each. Diabetes and related conditions, respiratory diseases, cirrhosis and other liver diseases, and neurologic disorders account for most of the remaining one-third.
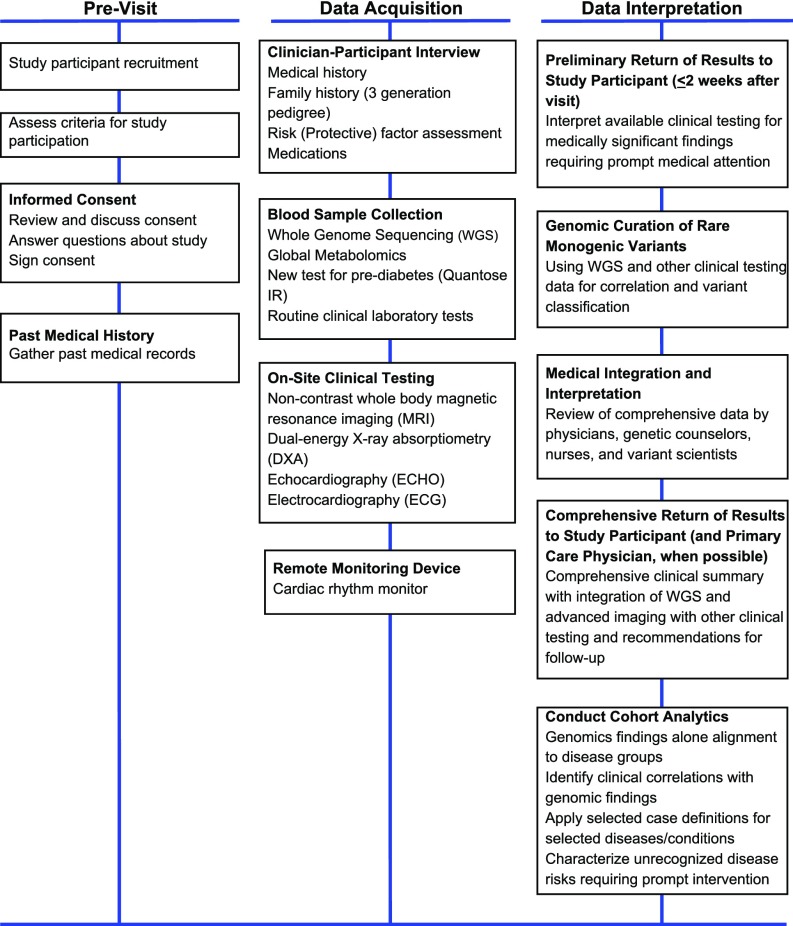
Few published examples show how genomics (8, 9) might be proactively incorporated into new models for medical practice and what infrastructure will be needed to support data generation and use (10–16). We used medical and family history and routine clinical testing in addition to clinical-grade whole-genome sequencing (WGS) (9), noncontrast whole-body magnetic resonance imaging (MRI) (17–19), dual-energy X-ray absorptiometry (DXA), global metabolomics (12, 20, 21) and a new blood test for prediabetes (Quantose IR) (22), echocardiography (ECHO), and ECG and 2-wk cardiac rhythm monitoring in an effort to identify age-related chronic disease risks associated with premature death (Fig. 1). Our objective for precision medicine screening of active, symptom-free adults was, in some ways, like successful newborn screening programs using advanced MS technologies for early simultaneous detection of multiple life-threatening conditions (23, 24). Age-related chronic diseases associated with premature mortality are much more common among active adults than diseases targeted in newborn screening, which make them good candidates for screening, but they require a broader set of specialized tools and technologies for identification of disease risk than any single modality, such as WGS. We evaluated whether active integration of routine and advanced clinical data with WGS has the potential to improve disease risk detection and strengthen WGS variant interpretation in support of precision medicine and discovery.
Fig. 1.
Study process for precision medicine screening including WGS and advanced imaging as reported. Comprehensive return of results available within 10–12 wk of study visit.
Results
We enrolled 209 study participants with median age of 55 y old (range 20–98 y old, 34.5% female) between September 10, 2015 and May 16, 2016. There were 2 study participants 20–24 y old, 3 participants 25–29 y old, 8 participants 30–34 y old, 15 participants 35–39 y old, 19 participants 40–44 y old, 27 participants 45–49 y old, 31 participants 50–54 y old, 40 participants 55–59 y old, 28 participants 60–64 y old, 21 participants 65–69 y old, 10 participants 70–75 y old, 2 participants 75–79 y old, 1 participant 80–84 y old, 1 participant 85–89 y old, 0 participants 90–94 y old, and 1 participant 95–100 y old. Selected characteristics comparing study participants with an age- and sex-adjusted National Health and Nutrition Survey (NHANES) cohort, a US population-based sample, are shown in Table 1. Routine clinical laboratory testing was obtained on 90 study participants (43%); noncontrast whole-body MRI, DXA, and ECHO were conducted on all study participants. A specific MRI protocol to obtain body compartment-specific fat and muscle estimation was conducted on 126 participants (60%) (19). Global metabolomics and a new blood test for prediabetes (Quantose IR), including fasting blood glucose and other metabolites, were obtained on 208 participants (12, 20–22). ECG was performed on 202 study participants. Some portion of the intended 2-wk cardiac rhythm monitoring was completed on 140 (67%) participants; the median duration of monitoring was 5.9 d (range 0.8–14 d). Study participants who had the cardiac rhythm monitoring kit applied during their study visit had better use and duration of monitoring than those who applied the cardiac rhythm monitoring device at home. Abnormal findings for routine clinical laboratories, whole-body MRI, DXA, ECHO, ECG, and 2-wk cardiac rhythm monitoring are selectively summarized for participants with likely clinical correlations with genomic findings (Table 2 and Table S2), previously unrecognized age-related chronic disease risk requiring prompt (<30 d) medical attention (Table S3), and to apply case definitions for five diseases or conditions, including type 2 diabetes mellitus (diabetes) and diabetes risk, atherosclerosis or atherosclerosis risk, metabolic syndrome, nonalcoholic fatty liver disease (NAFLD), and nonalcoholic steatohepatitis (NASH) (Fig. 2). Seventy (34%) study participants were referred for follow-up imaging based on findings from noncontrast whole-body MRI.
Table 1.
Study participant characteristics and comparison with the NHANES
| Variables | Study participant | NHANES adult | Standardized incidence ratio (55) | 95% CI | P value |
| Characteristics | |||||
| Age, y | 4.43E-40* | ||||
| Median | 55 | 26 | |||
| Range | 20–98 | 0–80 | |||
| Sex | 4.84E-04* | ||||
| Male | 65.6% | 49.2% | |||
| Female | 34.4% | 50.8% | |||
| Measured BMI | |||||
| Median (25–75%) | 26 (23–29) | 24.7 (20–30) | |||
| Measured systolic blood pressure | |||||
| Median (25–75%) | 123.5 (115–133) | 116 (106–128) | |||
| Measured LDL | |||||
| Median (25–75%) | 114.5 (96–135) | 103 (81–127) | |||
| Diseases | |||||
| Neoplasms | |||||
| Ever told you had cancer or malignancy | 15.1% | 9.5% | 1.5 | 1.02–2.16 | 3.39E-02* |
| Cardiovascular | |||||
| Ever told you had coronary heart disease | 4.1% | 4.0% | 0.9 | 0.38–1.74 | 7.98E-01 |
| Chronic respiratory diseases | |||||
| Ever told you had COPD | 1.0% | 3.3% | 0.2 | 0.02–0.88 | 9.52E-02 |
| Diabetes, urogenital, blood, and endocrine diseases | |||||
| Doctor told you have diabetes | 4.6% | 7.5% | 0.3 | 0.13–0.54 | 9.63E-04* |
| Cirrhosis and other chronic liver diseases | |||||
| Ever told you had any liver condition | 6.1% | 4.1% | 1.1 | 0.55–1.89 | 7.75E-01 |
| Neurological disorders | |||||
| Blood relatives have Alzheimer’s disease | 13.2% | 13.3% | 1.0 | 0.63–1.44 | 1.00E+00 |
| Risk factors | |||||
| Alcohol use | |||||
| Had at least 12 alcoholic drinks per 1 y | 90.0% | 70.0% | 1.2 | 0.99–1.37 | 2.76E-02* |
| Tobacco smoking | |||||
| Smoked at least 100 cigarettes in life | 38.4% | 42.2% | 0.8 | 0.58–0.97 | 8.90E-02 |
| High LDL cholesterol | |||||
| Now taking prescribed medicine | 78.9% | 85.4% | 1.1 | 0.74–1.48 | 6.02E-01 |
| High blood pressure | |||||
| Ever told you had high blood pressure | 23.0% | 33.7% | 0.5 | 0.38–0.69 | 6.81E-06* |
| Taking prescription for hypertension | 73.8% | 83.6% | 0.8 | 0.54–1.14 | 2.44E-01 |
The NHANES information is at https://www.cdc.gov/nchs/nhanes/. BMI, body mass index; COPD, chronic obstructive pulmonary disease.
P ≤ 0.05.
Table 2.
Clinical correlates with rare monogenic variants by disease group and screening test
| Disease group | Screening test | |||||||
| Rare monogenic variants | Global metabolomics and Quantose IR | MRI | ECHO | ECG | Cardiac rhythm monitoring | Clinical laboratories | Medical and family history | |
| Neoplasms | 14 | 3 | 2 | 0 | 0 | 0 | 0 | 12 |
| Cardiovascular diseases | 15 | 0 | 0 | 8 | 5 | 5 | 4 | 14 |
| Diabetes, urogenital, blood, and endocrine diseases | 7 | 7 | 1 | 0 | 0 | 0 | 3 | 3 |
| Cirrhosis and other chronic liver diseases | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| Neurological disorders | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Other (metabolic) | 12 | 12 | 1 | 0 | 0 | 0 | 1 | 0 |
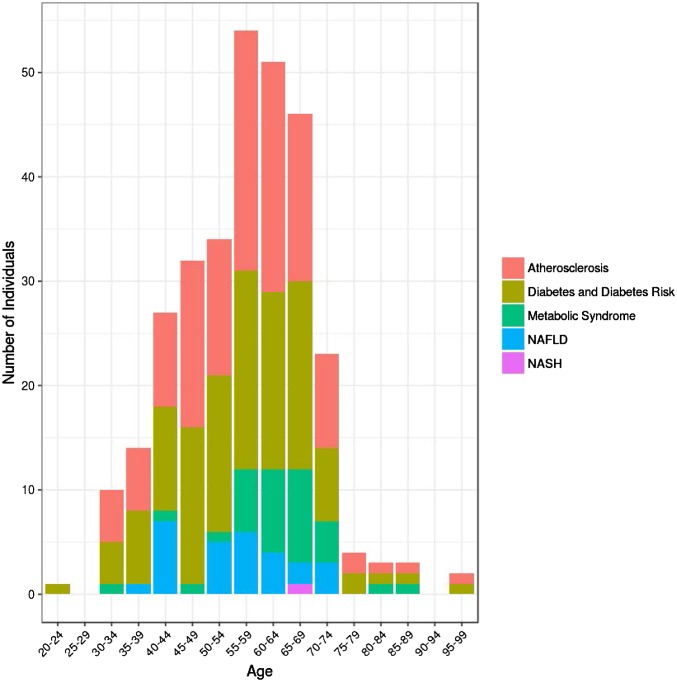
Fig. 2.
Frequency of five diseases or conditions identified by applying case definitions among our study participants by age group. The five diseases or conditions are type 2 diabetes mellitus (diabetes) and diabetes risk, atherosclerosis or atherosclerosis risk, metabolic syndrome, NAFLD, and NASH. Cohort denominators in Results show estimation of prevalence in the study cohort.
WGS was obtained on all 209 study participants. The median numbers of variants identified by predicted ethnicity were 3.60 million for European (n = 159, 76%), 3.65 million for admixture (n = 32, 15%), 3.59 million for east Asian (n = 10, 5%), 3.66 million for south central Asian (n = 4, 2%), 4.36 million for African (n = 2, 1%), and 3.75 million for Middle Eastern (n = 2, 1%). Twenty-one (10%) of the 209 participants were from seven families. WGS revealed 27,482,829 unique variants [21,761,709 single-nucleotide variants (SNVs) and 5,721,120 insertion–deletions (indels)]. Of these, 1,953,187 (1,769,795 SNVs and 183,392 indels) were observed only once in the our database; many of these occurred in 5′ and 3′ UTRs, which contain the largest enrichment of genome-wide association studies (GWAS)-implied disease associations (25) (Table S1).
Rare monogenic variant findings were identified with standardized and phenotype-based queries using an internal version of Human Longevity, Inc. (HLI) Open Search (9) and the American College of Medical Genetics (ACMG) criteria for interpretation (26, 27). A total of 310 unique medically significant risk alleles in 231 genes were identified, a median of 2 per study participant (range, 0–7); 25 study participants had none. Of these, we classified the inheritance of 261 alleles in 190 genes as autosomal recessive (AR), we classified the inheritance of 38 alleles in 33 genes as autosomal dominant (AD), we classified the inheritance of 14 alleles in 9 genes as AR/AD, and we classified the inheritance of one allele as X-linked dominant. The most commonly affected genes (number of variants) were BTD (six), HFE (two), SERPINA1 (three), ABCA4 (five), and GJB2 (three) for AR; F5 (one), F2 (one), ALDH2 (one), NBN (two), and PPP1R3A (two) for AD; and CFTR (four), SPINK1 (one), VWF (three), ALPL (two), and F11 (one) for AR/AD. Table 3 shows the distribution of these AD, AD/AR, and homozygous AR genes and related variants classified as pathogenic and likely pathogenic by major age-related chronic disease using Global Burden Disease groupings with the number of study participants impacted (6, 7). There were 9 genes and 10 alleles with neoplasm-associated risk, 4 genes and 4 alleles with cardiovascular-associated risk, 6 genes and 8 alleles with diabetes-associated risk, 1 gene and 1 allele with cirrhosis-associated risk, and 6 cases with the homozygous APOE 4 c.388T > C allele.
Table 3.
Rare monogenic variants associated with age-related chronic disease risks
| Disease group (56) | Rare monogenic variants | |
| Genes (variants) | Participants impacted (%) | |
| Neoplasms | 9 (10) | 16 (7.7) |
| Cardiovascular diseases | 4 (4) | 4 (1.9) |
| Chronic respiratory diseases | — | — |
| Diabetes, urogenital, blood, and endocrine diseases | 6 (8) | 12 (5.7) |
| Cirrhosis and other chronic liver diseases | 1 (1) | 1 (0.5) |
| Neurological disorders | 1 (1) | 6 (2.9) |
| Other | 8 (8) | 23 (11.0) |
| Totals | 29 (32) | 62 (29.7) |
Using our full range of screening tests, we identified clinical correlations with genomic findings among 43 (21%, 1:5) study participants. A summary of sources for clinical correlation with genomic findings by age-related chronic disease group is shown in Table 2. Detailed data describing clinical correlations by specific gene, variant, mode of inheritance, and zygosity are provided in Table S2.
Through exploration of rare monogenic variants and associated global metabolomic results, we identified 10 unique alleles in 14 study participants with metabolic signatures consistent with penetrance (Table S2). Metabolic pathways impacted by the allelic differences included fatty acid beta oxidation, fatty acid synthesis, urea cycle, and signatures associated with oxidative stress. Strong metabolic signatures were observed for two polymorphisms matching the genes’ function. Two heterozygous variants in the ACADS gene, c.1510G > A and c.1030C > T, coding for the short-chain acyl-CoA dehydrogenase (SCAD) were detected in one case. In another case, the heterozygous ACADM variant c.1456C > T coding for medium-chain acyl-CoA dehydrogenase (MCAD) was detected, and interestingly, both enzymes participate in fatty acid beta oxidation by reducing different fatty acid chain length (28). SCAD specifically acts on the short-chain fatty acid butyryl-CoA, and MCAD reduces acyl-CoA chains containing 6–12 carbons. In the absence of SCAD activity, by-products of butyryl-CoA, including butyrylcarnitine and ethylmalonate, accumulate (29). Greatly elevated levels of butyrylcarnitine and ethylmalonate (Z scores above the 97.5th percentile) were observed in the plasma, suggestive of combined metabolic penetrance of these variants.
Moreover, greatly elevated medium-chain acylcarnitines (hexanoylcarnitine, octanoylcarnitine, and decanoylcarnitine; Z scores above 97.5 the percentile) were detected, suggestive of reduced MCAD activity. Large GWAS combined with metabolic profiling have previously identified associations between ACADS and MCAD and their respective metabolic substrates, lending support to the metabolic penetrance observed on an individual basis in this study (30–32). We previously reported on additional metabolomic/genetic variants, which are heterozygotes for known recessively inherited disorder (12, 20). These studies established that “carrier” disease state does not reflect carrier for individual metabolic variation. The number of adult cases of metabolic penetrance will continue to expand using this approach.
Metabolomic analysis also detected xanthinuria in an individual with early-onset (20s) recurrent renal stones (six episodes) as well as the drug effect of xanthine oxidase inhibitors in three other individuals.
Although hypoxanthine and especially, xanthine levels were elevated in both cases, normal urate and elevated orotate and orotidine levels, due to perturbed pyrimidine synthesis (33), were only observed in individuals taking xanthine oxidase inhibitors (allopurinols) for their gout conditions.
We identified 164 (78%, >3:4) participants with evidence of age-related chronic disease or risk factors. One hundred eighteen study participants (56%) had evidence of diabetes or risk for diabetes: 15 (7%) had type 2 diabetes, 80 (38%) had prediabetes, and 23 (11%) had insulin resistance suggesting prediabetes risk (based on Quantose IR). Only 19 (9%) reported a history of type 2 diabetes or prediabetes. One hundred twenty-four participants (59%) had evidence of atherosclerotic disease or risk. Thirty-three (16%) had evidence of metabolic syndrome. Twenty-eight participants (13%) met a screening definition for NAFLD, and one had suspected NASH. Many participants had multiple overlapping conditions, including 29 with prediabetes and atherosclerotic disease or risk; 19 with prediabetes, atherosclerotic disease or risk, and metabolic syndrome; and 13 with insulin resistance and atherosclerotic disease or risk. When diabetes, prediabetes, and insulin resistance were considered as a group of diseases and conditions, 28 (11%) had all four of the common diseases and conditions (diabetes and diabetes risk, atherosclerosis or atherosclerosis risk, metabolic syndrome, and NAFLD). As expected, there was a strong effect of age on the prevalence of these conditions, with exception of NAFLD (Fig. 2).
We identified 17 study participants (8%) with previously unrecognized age-related chronic disease risk requiring prompt (<30 d) medical attention after confirmation of screening findings. This includes 4 with early-stage neoplasias (thymoma, renal cell carcinoma, and two high-grade prostate neoplasms all initially suspected on MRI and confirmed through biopsy), 1 with enlarged aortic root, 2 with newly recognized atrial fibrillation cases, 2 with medically significant arrhythmias, 1 with third degree heart block, 1 with primary biliary cholangitis, and 1 with xanthinuria (Table S3).
Discussion
We used a precision medicine screening approach anchored by WGS and noncontrast whole-body MRI along with other screening tests among active, symptom-free adults to identify age-related chronic disease risks associated with premature mortality. We hypothesized that, by doing this, we may accelerate identification of age-related chronic disease risk, allowing for a range of earlier interventions and potentially, better health outcomes. We found that WGS alone identified possible age-related chronic disease risks associated with premature mortality (19% of participants), including neoplasms (8%), cardiovascular diseases (2%), diabetes and related diseases (6%), cirrhosis and other chronic liver disease (<1%), and neurologic disorders (3%). Combining WGS with advanced imaging and other testing strengthened guideline-driven WGS variant interpretation (26, 27). As shown in Table 2, a broad range of our imaging and other screening testing was useful in strengthening WGS variant interpretation, and many of our study participants had multiple lines of supporting clinical evidence (Table S2).
Additionally, we could correlate alterations in global metabolomics levels (a phenotype) with 15 heterozygous AR alleles. This is a relatively unexplored realm of human biology and clinical application, particularly among adults, but our data suggest that this may be a relatively common phenomenon (12, 20). In total, we could identify likely clinical (or phenotypic) correlations in one-fifth of our study participants. This is an encouraging baseline for clinical utility given that we could characterize only a miniscule fraction of the total WGS variation that we identified in this cohort.
We looked at two other risk perspectives in our study to more fully characterize the likely potential of this screening approach to identify age-related chronic disease risks associated with premature mortality.
Identifying risk includes not only prevention opportunities but also early detection of these diseases and risks associated with these diseases. We used case definitions to identify four common diseases or conditions that are age-related chronic diseases associated with premature mortality (diabetes and diabetes risk) or are risk factors for these diseases (atherosclerosis for cardiovascular diseases, metabolic syndrome for diabetes and cardiovascular diseases, and NAFLD for cirrhosis) (Fig. 1) (34–36). More than three-quarters of our study participants had at least one of these diseases or conditions, and 28 (11%) had all four of these diseases or conditions. The overall prevalence of these diseases or conditions increased with age, except for NAFLD, which was relatively constant by age, although the cohort is relatively small. The other risk perspective that we highlight for early detection is that 17 (8%) participants who we identified as having previously unrecognized age-related chronic disease risk required prompt (<30 d) medical attention, including 4 (2%) with early-stage neoplasms. Surprisingly given our overall data, we did not identify high-risk rare monogenic variants in any of these individuals; this emphasizes the important of advanced imaging and our other clinical tests as a complement WGS for screening. Overall, WGS was useful in explaining past medical history and possible future individual (and familial) disease risk for prevention, while advanced imaging and other testing were most useful for (early) detection of active disease.
There is warranted concern about testing performance whenever screening is undertaken in medical practice. False positives may expose people to unnecessary risks, anxiety, costs, and inconvenience (37). The traditional medical approach to minimizing false positives is to rely on occurrence of symptoms to increase pretest probabilities, although this is poorly understood by most physicians (38). Targeting age-related chronic diseases associated with premature mortality as we have offers the potential to mitigate some negative aspects of screening through (i) the high prevalence and life-threatening nature of these conditions, (ii) use of low to no risk technologies, and (iii) convergent approaches to strengthen interpretation, particularly for WGS variant data.
We recommended follow-up imaging studies for slightly more than one-third of our study participants. Some of this is the nature of screening, which drives the need for more definitive imaging studies better suited to specific abnormalities. Other instances of referral were intended to identify change over a specified time period, which might be suggestive of cancer, such as finding a cystic pancreatic lesion (39), or instability of a vascular lesion, such an intracranial aneurysm (40). In some instances, data are lacking to confidently predict the natural course of these findings, and thus, the findings may cause unnecessary anxiety and unneeded surgery (39, 40). Additional research with longer follow-up periods will be required to resolve outcomes associated with follow-up imaging. However, the life-threatening consequences and relatively high prevalence of diseases associated with these lesions suggest that early recognition is likely to be beneficial for most individuals.
Genomics has been disappointing in its ability to unravel the estimated heritability of most age-related chronic diseases and other common diseases (41–43). First, we expect and are increasingly seeing evidence of the recognition of rare variants with large effect sizes (3, 9, 44). Combining these findings with advancements in the regulatory genome (45); study of genomic essentiality (46); monogenic and polygenic methodologies to assess causation, including Mendelian randomization methods (47); extension of GWAS to create hazard models (48); and continued exploration of pleiotropy (49) will increase clinical utility. Second, increasingly detailed mapping of molecular pathways and mechanisms associated with diseases and risk factors will provide a much-needed improved capability to link genotype and phenotype data (12, 43, 50). In our study, we could show the use of global metabolomics in mapping to genomic variation. This integration will strengthen with additional automation of analysis. Third, we are working to quantitatively integrate genomics with advanced imaging data and other clinical data to create point-of-care clinical decision support (48, 51, 52). The version of HLI Open Search that we are using internally can query individual genomes (and families) to facilitate rapid exploration of genotype–phenotype associations.
The traditional symptom-driven medical model is clearly inadequate for early recognition of age-related chronic diseases associated with premature mortality, many of which are preventable. The sequelae of these diseases represent most of the current total US Medicare expenditure (2, 53). For nationally sanctioned proactive single-disease adult screening programs, there are robust long-term evaluations of test performance in the context of clinical harms and benefits and costs—at the population level—although it is now increasingly well-recognized that individual risk varies widely for these conditions (54). Single-disease approaches are problematic in clinical use, because many individuals have risk for or are suffering from multiple rather than single diseases, and clear clinical guidance in these real world situations is lacking. Symptom-driven medicine and single disease-based approaches to prevention have advanced health but are likely to become anachronistic with the introduction of genomics and other new science and technologies (e.g., advanced imaging and metabolomics) to medicine, particularly when combined with the rapid demographic and epidemiologic changes underway in the United States and globally. A major promise of genomics and precision medicine is to more tightly link curative (to identify pathology) and preventive (to identify risk) medical disciplines by creating health care platforms to personalize disease risk and longitudinal care. Our data suggest a route to creating such an approach, initially focusing on prevention of premature deaths among active adults associated with age-related chronic diseases and then expanding to other causes of disability and additional life stages.
Materials and Methods
We enrolled active adults ≥18 y old (without acute illness, activity-limiting unexplained illness or symptoms, or known active cancer) able to come for 6–8 h of onsite data collection who were able to undergo MRI without sedation; in the case of women, were not pregnant or attempting to become pregnant; and were interested in undergoing a precision medicine screening approach for disease risk detection, including genomics and other testing, as part of an institutional review board (IRB)-approved clinical research protocol. Study results were returned to study participants (within 10–12 wk after visit), who were encouraged to involve their primary care physicians.
Participants underwent a verbal review of the IRB-approved consent (Western IRB) and were given time to ask and receive answers to questions during a 0.5- to 1-h session conducted by a health professional. We received permission from the IRB to collect up to $25,000 for participation in this study. Study participants underwent standardized activities related to data collection and return of results in previsit, data acquisition, and data interpretation during a 1-y study period. Readers interested in access to data, associated protocols, code, and/or other materials that may not be included in this manuscript or SI Materials and Methods should contact the corresponding authors.
Supplementary Material
Acknowledgments
We acknowledge the individuals who participated in this precision medicine screening study, without whom the findings would not be possible. Julie Ellison and Natalie Schenker-Ahmed provided medical writing assistance, and Anna Georgalis provided editorial assistance. We also acknowledge staff involved in the conduct of this study: Amy Reed, Ana Sanchez, Athena Hutchinson, Carina Sarabia, Cheryl Buffington, Cheryl Greenberg, Christina Bonas, Daniel Jones, Diana Cardin Escobedo, Emily Smith, Frank, Song, Genelle Olsen, Greg Olson, Heidi Millard, Helen Messier, Keisha Robinson, Laura Edwards, Nicole Boramanand, Nolan Tengonciang, Patrick Jamieson, Saints Dominguez, Samantha Punsalan, and William Herrera.
Footnotes
Conflict of interest statement: Some authors are employees of Human Longevity, Inc. as indicated by institution affiliation, and the study was funded by Human Longevity, Inc.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706096114/-/DCSupplemental.
References
- 1.Olshansky SJ. Articulating the case for the longevity dividend. Cold Spring Harb Perspect Med. 2016;6:a025940. doi: 10.1101/cshperspect.a025940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer UE, Briss PA, Goodman RA, Bowman BA. Prevention of chronic disease in the 21st century: Elimination of the leading preventable causes of premature death and disability in the USA. Lancet. 2014;384:45–52. doi: 10.1016/S0140-6736(14)60648-6. [DOI] [PubMed] [Google Scholar]
- 3.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 4.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray CJ, et al. US Burden of Disease Collaborators The state of US health, 1990-2010: Burden of diseases, injuries, and risk factors. JAMA. 2013;310:591–608. doi: 10.1001/jama.2013.13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benziger CP, Roth GA, Moran AE. The global burden of disease study and the preventable burden of NCD. Glob Heart. 2016;11:393–397. doi: 10.1016/j.gheart.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 8.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Telenti A, et al. Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci USA. 2016;113:11901–11906. doi: 10.1073/pnas.1613365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashley EA, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Garay ML, McGuire AL, Pereira S, Caskey CT. Personalized genomic disease risk of volunteers. Proc Natl Acad Sci USA. 2013;110:16957–16962. doi: 10.1073/pnas.1315934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo L, et al. Plasma metabolomic profiles enhance precision medicine for volunteers of normal health. Proc Natl Acad Sci USA. 2015;112:E4901–E4910. doi: 10.1073/pnas.1508425112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caskey CT, Gonzalez-Garay ML, Pereira S, McGuire AL. Adult genetic risk screening. Annu Rev Med. 2014;65:1–17. doi: 10.1146/annurev-med-111212-144716. [DOI] [PubMed] [Google Scholar]
- 14.Ashley EA. Towards precision medicine. Nat Rev Genet. 2016;17:507–522. doi: 10.1038/nrg.2016.86. [DOI] [PubMed] [Google Scholar]
- 15.Green RC, et al. CSER Consortium Clinical sequencing exploratory research consortium: Accelerating evidence-based practice of genomic medicine. Am J Hum Genet. 2016;99:246. doi: 10.1016/j.ajhg.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel CJ, et al. Whole genome sequencing in support of wellness and health maintenance. Genome Med. 2013;5:58. doi: 10.1186/gm462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holland D, Brewer JB, Hagler DJ, Fennema-Notestine C, Dale AM. Alzheimer’s Disease Neuroimaging Initiative Subregional neuroanatomical change as a biomarker for Alzheimer’s disease. Proc Natl Acad Sci USA. 2009;106:20954–20959, and erratum (2010) 107:6551. doi: 10.1073/pnas.0906053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunsing RL, et al. Restriction spectrum imaging: An evolving imaging biomarker in prostate MRI. J Magn Reson Imaging. 2017;45:323–336. doi: 10.1002/jmri.25419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West J, et al. Feasibility of MR-based body composition analysis in large scale population studies. PLoS One. 2016;11:e0163332. doi: 10.1371/journal.pone.0163332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long T, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet. 2017;49:568–578. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- 21.Evans AMBB, et al. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics. 2014;4:132. [Google Scholar]
- 22.Cobb J, et al. A novel fasting blood test for insulin resistance and prediabetes. J Diabetes Sci Technol. 2013;7:100–110. doi: 10.1177/193229681300700112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans JP, Berg JS, Olshan AF, Magnuson T, Rimer BK. We screen newborns, don’t we?: Realizing the promise of public health genomics. Genet Med. 2013;15:332–334. doi: 10.1038/gim.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CDC Impact of expanded newborn screening—United States, 2006. Morb Mortal Wkly Rep. 2008;57:1012–1015. [PubMed] [Google Scholar]
- 25.Schork AJ, et al. Tobacco and Genetics Consortium; Bipolar Disorder Psychiatric Genomics Consortium; Schizophrenia Psychiatric Genomics Consortium All SNPs are not created equal: Genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. doi: 10.1371/journal.pgen.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards S, et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalia SS, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 28.Jethva R, Bennett MJ, Vockley J. Short-chain acyl-coenzyme A dehydrogenase deficiency. Mol Genet Metab. 2008;95:195–200. doi: 10.1016/j.ymgme.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corydon MJ, et al. Ethylmalonic aciduria is associated with an amino acid variant of short chain acyl-coenzyme A dehydrogenase. Pediatr Res. 1996;39:1059–1066. doi: 10.1203/00006450-199606000-00021. [DOI] [PubMed] [Google Scholar]
- 30.Gieger C, et al. Genetics meets metabolomics: A genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suhre K, et al. CARDIoGRAM Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin SY, et al. Multiple Tissue Human Expression Resource (MuTHER) Consortium An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beardmore TD, Kelley WN. Mechanism of allopurinol-mediated inhibition of pyrimidine biosynthesis. J Lab Clin Med. 1971;78:696–704. [PubMed] [Google Scholar]
- 34.Chamberlain JJ, Rhinehart AS, Shaefer CF, Jr, Neuman A. Diagnosis and management of diabetes: Synopsis of the 2016 American Diabetes Association Standards of Medical Care in Diabetes. Ann Intern Med. 2016;164:542–552. doi: 10.7326/M15-3016. [DOI] [PubMed] [Google Scholar]
- 35.Grundy SM, et al. American Heart Association; National Heart, Lung, and Blood Institute Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute scientific statement. Circulation. 2005;112:2735–2752. doi: 10.1161/CIRCULATIONAHA.105.169404. [DOI] [PubMed] [Google Scholar]
- 36.Angulo P, et al. The NAFLD fibrosis score: A noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology. 2007;45:846–854. doi: 10.1002/hep.21496. [DOI] [PubMed] [Google Scholar]
- 37.Weiner C. Anticipate and communicate: Ethical management of incidental and secondary findings in the clinical, research, and direct-to-consumer contexts (December 2013 report of the Presidential Commission for the Study of Bioethical Issues) Am J Epidemiol. 2014;180:562–564. doi: 10.1093/aje/kwu217. [DOI] [PubMed] [Google Scholar]
- 38.Manrai AK, Bhatia G, Strymish J, Kohane IS, Jain SH. Medicine’s uncomfortable relationship with math: Calculating positive predictive value. JAMA Intern Med. 2014;174:991–993. doi: 10.1001/jamainternmed.2014.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konings IC, et al. Dutch Research Group on Pancreatic Cancer Surveillance in High-Risk Individuals Prevalence and progression of pancreatic cystic precursor lesions differ between groups at high risk of developing pancreatic cancer. Pancreas. 2017;46:28–34. doi: 10.1097/MPA.0000000000000725. [DOI] [PubMed] [Google Scholar]
- 40.Etminan N, Rinkel GJ. Unruptured intracranial aneurysms: Development, rupture and preventive management. Nat Rev Neurol. 2017;13:126. doi: 10.1038/nrneurol.2017.14. [DOI] [PubMed] [Google Scholar]
- 41.Katsanis N. The continuum of causality in human genetic disorders. Genome Biol. 2016;17:233. doi: 10.1186/s13059-016-1107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manrai AK, Ioannidis JP, Kohane IS. Clinical genomics: From pathogenicity claims to quantitative risk estimates. JAMA. 2016;315:1233–1234. doi: 10.1001/jama.2016.1519. [DOI] [PubMed] [Google Scholar]
- 43.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: From polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marouli E, et al. EPIC-InterAct Consortium; CHD Exome+ Consortium; ExomeBP Consortium; T2D-Genes Consortium; GoT2D Genes Consortium; Global Lipids Genetics Consortium; ReproGen Consortium; MAGIC Investigators Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.di Iulio J, et al. 2016. The human functional genome defined by genetic diversity, bioRxiv:10.1101/082362.
- 46.Telenti A, Perkins BA, Venter JC. Dynamics of an aging genome. Cell Metab. 2016;23:949–950. doi: 10.1016/j.cmet.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Ference BA, et al. Variation in PCSK9 and HMGCR and risk of cardiovascular disease and diabetes. N Engl J Med. 2016;375:2144–2153. doi: 10.1056/NEJMoa1604304. [DOI] [PubMed] [Google Scholar]
- 48.Desikan RS, et al. 2016. Personalized genetic assessment of age associated Alzheimers disease risk. bioRxiv:10.1101/074864.
- 49.Ellinghaus D, et al. International IBD Genetics Consortium (IIBDGC); International Genetics of Ankylosing Spondylitis Consortium (IGAS); International PSC Study Group (IPSCSG); Genetic Analysis of Psoriasis Consortium (GAPC); Psoriasis Association Genetics Extension (PAGE) Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: A complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hibar DP, et al. Novel genetic loci associated with hippocampal volume. Nat Commun. 2017;8:13624. doi: 10.1038/ncomms13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, Huang H, Shen D. Alzheimer’s Disease Neuroimaging Initiative Integrative analysis of multi-dimensional imaging genomics data for Alzheimer’s disease prediction. Front Aging Neurosci. 2014;6:260. doi: 10.3389/fnagi.2014.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.CMS . Chronic Conditions among Medicare Beneficiaries, Chartbook. Department of Health and Human Services; Baltimore: 2012. [Google Scholar]
- 54.Shieh Y, et al. Athena Breast Health Network Investigators Breast cancer screening in the precision medicine era: Risk-based screening in a population-based trial. J Natl Cancer Inst. 2017;109:djw290. doi: 10.1093/jnci/djw290. [DOI] [PubMed] [Google Scholar]
- 55.Tripepi G, Jager KJ, Dekker FW, Zoccali C. Stratification for confounding–part 2: Direct and indirect standardization. Nephron Clin Pract. 2010;116:c322–c325. doi: 10.1159/000319591. [DOI] [PubMed] [Google Scholar]
- 56.GBD 2015 Mortality and Causes of Death Collaborators Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: A method for quantifying insulin secretion and resistance. Am J Physiol. 1979;237:E214–E223. doi: 10.1152/ajpendo.1979.237.3.E214. [DOI] [PubMed] [Google Scholar]
- 58.Ferrannini E, et al. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes. 2013;62:1730–1737. doi: 10.2337/db12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gall WE, et al. RISC Study Group Alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One. 2010;5:e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tripathy D, et al. A novel insulin resistance index to monitor changes in insulin sensitivity and glucose tolerance: The ACT NOW study. J Clin Endocrinol Metab. 2015;100:1855–1862. doi: 10.1210/jc.2014-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lloyd-Jones DM, et al. Estimating longitudinal risks and benefits from cardiovascular preventive therapies among medicare patients: The million hearts longitudinal ASCVD risk assessment tool: A special report from the American Heart Association and American College of Cardiology. J Am Coll Cardiol. 2017;69:1617–1636. doi: 10.1016/j.jacc.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Landrum MJ, et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.