Significance
Voltage-gated sodium channels are integral in electrical signaling within the human body and are key targets for anesthetics and antiepileptic compounds used in surgeries and the treatment of neurological disorders. We have used molecular simulations to determine where a number of these compounds bind inside the pore of a voltage-gated sodium channel to aid the design of new compounds for treating chronic pain, heart conditions, and epilepsy. We uncover two distinct binding sites inside the pore harnessed by neutral and charged drugs, respectively. This explains why so many anesthetic compounds have both neutral and charged forms: The neutral form more easily enters the pore, but the charged form binds more tightly to effectively block the pore and prevent electrical signaling.
Keywords: sodium channel, local anesthetic, molecular dynamics, pain, structural biology
Abstract
Voltage-gated sodium channels are essential for carrying electrical signals throughout the body, and mutations in these proteins are responsible for a variety of disorders, including epilepsy and pain syndromes. As such, they are the target of a number of drugs used for reducing pain or combatting arrhythmias and seizures. However, these drugs affect all sodium channel subtypes found in the body. Designing compounds to target select sodium channel subtypes will provide a new therapeutic pathway and would maximize treatment efficacy while minimizing side effects. Here, we examine the binding preferences of nine compounds known to be sodium channel pore blockers in molecular dynamics simulations. We use the approach of replica exchange solute tempering (REST) to gain a more complete understanding of the inhibitors’ behavior inside the pore of NavMs, a bacterial sodium channel, and NavPas, a eukaryotic sodium channel. Using these simulations, we are able to show that both charged and neutral compounds partition into the bilayer, but neutral forms more readily cross it. We show that there are two possible binding sites for the compounds: (i) a site on helix 6, which has been previously determined by many experimental and computational studies, and (ii) an additional site, occupied by protonated compounds in which the positively charged part of the drug is attracted into the selectivity filter. Distinguishing distinct binding poses for neutral and charged compounds is essential for understanding the nature of pore block and will aid the design of subtype-selective sodium channel inhibitors.
Voltage-gated sodium channels (Navs) are transmembrane proteins responsible for generating action potentials in nerve and muscle cells. By opening in response to a small stimulus, they facilitate the passage of ions into the cell, leading to rapid depolarization of the membrane potential. Mutations of sodium channels are responsible for a variety of disorders, including cardiac arrhythmias, epilepsy, and pain syndromes (1–5). Thus, they are key targets for a variety of therapeutic compounds (6, 7), including the broad family of “local anesthetics.” While current local anesthetics inhibit all nine sodium channel subtypes found in the human body, there is a great interest in developing subtype-selective compounds to treat conditions like chronic pain (4, 5) or to reduce the side effects of epilepsy and arrhythmic medications. While there has been some progress in finding subtype-selective channel inhibitors (8–12), this has not yet been translated into clinical success. Thus, there is a great desire to better characterize the mode of action of existing channel inhibitors to determine what makes them inhibit all human subtypes, to define the common binding sites and pharmacophores, to elucidate the potential for making subtype-selective variants, and to gain a broader understanding of basic sodium channel function.
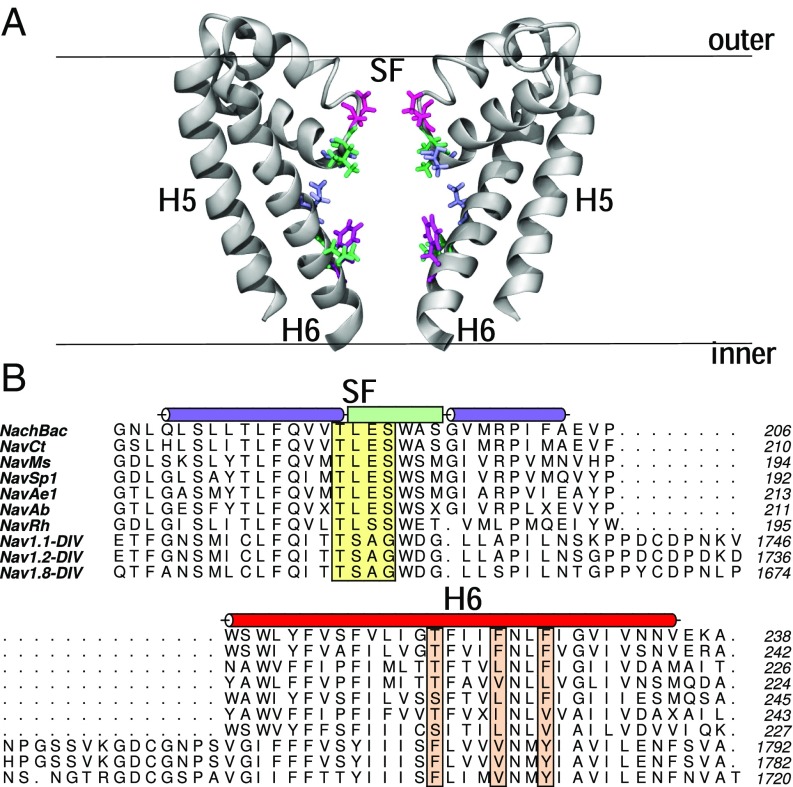
The publication of a number of bacterial Nav structures (13–22), as well as the most recent published structure of a eukaryotic channel (23), have opened the door to gaining detailed insight into the mechanism of action of sodium channel inhibitors. While bacterial channels have many differences from eukaryotic sodium channels, there are many similarities that make these a reasonable starting point for biophysical studies. Bacterial Navs consist of four identical subunits, while eukaryotic channels are one long sequence consisting of four homologous domains. However, the overall architecture of the channels is similar, with each subunit or domain containing six transmembrane helices (labeled S1–S6). The first four of these form an independent voltage-sensing domain, while the last two helices from each come together to form a central ion-conducting pore. The pore region itself contains: (i) a narrow selectivity filter at the extracellular end of the channel (Fig. 1, yellow box); (ii) a water-filled cavity in the center of the pore; (iii) a cytoplasmic activation gate that controls whether the channel is open or closed; and (iv) hydrophobic lateral fenestrations that extend from the center of the pore to the center of the bilayer (13–16, 23, 24). Mutagenesis studies have indicated that local anesthetics bind in the central cavity with key interactions with a number of polar and/or aromatic residues on helix S6 (25–32) (Fig. 1, orange boxes) and supported by a recent X-ray structure in which an inhibitor was partly resolved (16).
Fig. 1.
(A) Structure of the pore of the bacterial sodium channel NavMs. The backbone of the protein is shown in gray cartoon, and residues of interest that are highlighted in B are shown in licorice. The bilayer is represented by a black line. (B) Alignments of sequences of bacterial sodium channels and select eukaryotic channels, consisting of residues in the selectivity filter (SF) to the S6 helix (only domain IV of the eukaryotic channels is shown). The helices on each side of the selectivity filter are shown in a purple cylinder, with the selectivity filter shown as a light green box. The S6 helix is shown in a red cylinder. Residues of interest in the selectivity filter are highlighted in a yellow box, whereas residues on helix S6 are in orange boxes.
Local anesthetics are known to be able to inhibit sodium channels in at least two distinct ways. The first is “tonic block,” where the drug blocks resting channels. This is believed to occur when the compound enters the pore directly from the membrane via hydrophobic lateral fenestrations in the side of the pore (13, 19, 24, 33–36). Alternatively, anesthetic compounds can display “use-dependent” block (24, 37), in which the compounds only inhibit the channel after it has opened, presumably by first crossing the bilayer into the cytoplasm of the cell and entering through the open activation gate. Although close together, different residues contribute to the binding site of tonic and use-dependent block (30, 31), suggesting possible conformational changes occurring upon binding. Both pathways require the drug to first either enter or cross the bilayer. Interestingly, many local anesthetics (especially use-dependent blockers) have a pKa near 7, meaning that there are both neutral and charged forms at physiological pH. It is most likely the neutral state that crosses the bilayer, before regaining its charge in the cytoplasm and binding to the Nav protein; however, this has yet to be clearly shown. Simulations provide one way to examine which protonation state is likely to be present in each situation, but a comparative study of charged and neutral variants of local anesthetics has not yet been published.
The bilayer partitioning and energy barriers faced in crossing the membrane have been determined from simulations for a variety of both perpetually neutral and protonatable compounds. These include benzocaine (35, 38–40), lidocaine (41, 42), articaine (43), and phenytoin (35, 40). All show that, irrespective of charge, compounds prefer partitioning into the bilayer from the aqueous phase to sit just under the lipid head groups, and they all face a barrier to pass through the hydrophobic bilayer core. In addition, a number of molecular dynamics (MD) studies have been conducted to examine the interaction of perpetually neutral local anesthetic compounds with Nav channels. Direct simulations of drug–channel interactions have been conducted for benzocaine, phenytoin, sevoflurane, and isoflurane (33–36, 44). These studies indicated that the aforementioned compounds can enter the pore via the lateral fenestrations or through the activation gate, and they highlight potential binding sites inside the activation gate and at the internal mouth of the fenestrations. However, most of these studies have used unbiased simulations, which may oversample binding poses of the drug in metastable states. In addition, these same studies have only examined the behavior of at most two neutral compounds in the pore, making it difficult to determine the common mechanisms of action of this broad class of compounds and to examine the differences between the charged and neutral forms.
Here, we have used MD simulations to better understand the binding of a range of known pore-blocking compounds to sodium channels. The compounds studied include three perpetually neutral compounds (benzocaine, lamotrigine, and carbamazepine) and three protonatable compounds in both neutral and charged states (PF-5215786, PF-6305591, and lidocaine). By studying a range of compounds, we are able to not only appreciate the common aspects of binding, we can tease out differences between protonatable and neutral compounds, as well as tonic and use-dependent blockers. Furthermore, to overcome the limitations in sampling the preferred locations of compounds in the pore, we used the approach of the replica exchange with solute scaling/tempering method (REST2) (45, 46). Using MD and REST2, we were able to (i) determine the ability of these compounds to enter and cross the bilayer, (ii) greatly increase sampling of the compounds inside the pore compared with unbiased simulations, and (iii) uncover distinct modes of binding for neutral and charged compounds. While they shared some interactions with the protein, charged compounds effectively blocked the pore by extending into the base of the selectivity filter, whereas neutral compounds resided lower in the central cavity, both in prokaryotic and eukaryotic channels. These findings support the hypothesis that the protonatable amine group is common to many Nav inhibitors, because it allows the compounds to lose their charge before they traverse the bilayer, only to regain the charge in the cytoplasm to more effectively bind to and block the channel via interactions with the selectivity filter.
Results
Parameterization and Validation of Compounds.
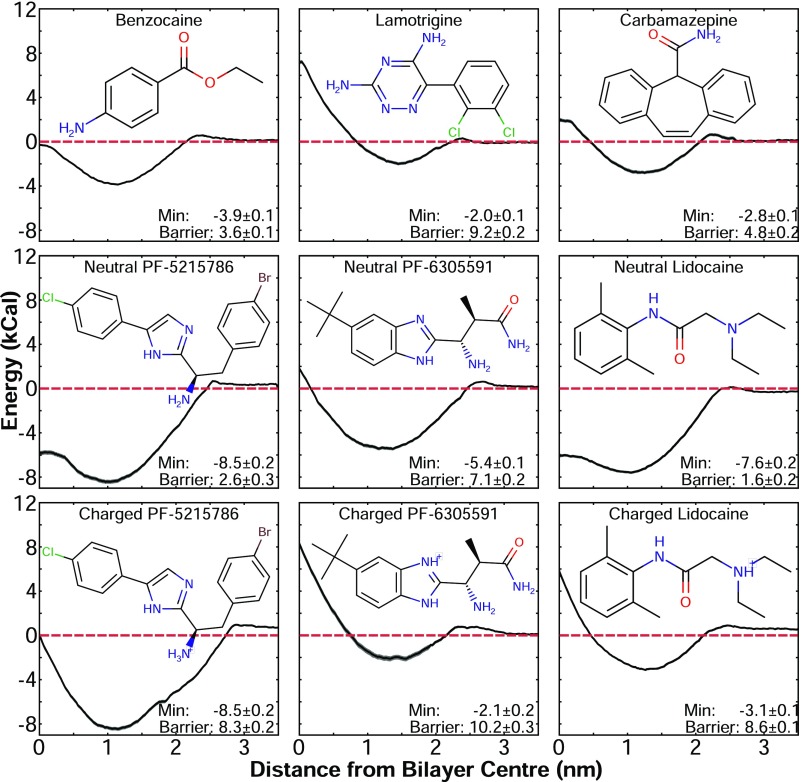
Nine compounds were initially modeled and parameterized for use in MD simulations [Fig. 2; structures are shown along with their corresponding potentials of mean force (PMFs)]. The validity of the parameters for each compound was tested by calculating the water/octanol and water/cyclohexane partition coefficients in simulations (logP) and comparing them to experimental values. Although experimental data were not available for all compounds (Table S1), the simulations accurately reproduced the trends seen in existing values for the experimental logPs. The main exception to this was lidocaine, for which we overestimated the water/octanol partition coefficient. However, we only predicted the value for the neutral species of lidocaine, due to difficulties in running free energy profile (FEP) calculations for non-charge-conserving processes, whereas the experimental value would include a contribution from the charged form of the molecule. This trend was expected to extend to PF-5215786 and PF-6305591, as their partitioning coefficients would also have contributions from both charged and neutral species. In addition to partitioning coefficients, we determined the PMF of each compound traversing a dipalmitoylphosphatidylcholine (DPPC) lipid bilayer to determine how the newly parameterized drugs would compare with previous studies of drug–lipid interactions and to gain new information about trends in sodium channel inhibitors. All compounds in this study (Fig. 2), irrespective of charge, had an energy minima between 1 and 1.5 nm away from the center of the bilayer. This position corresponded to the drug partitioning into the membrane, just below the lipid headgroups. In addition, each compound then faced an energy barrier when crossing the hydrophobic portion of the membrane and was in excellent agreement with simulation data for previously simulated compounds (35, 38–43). These PMFs showed that all of these compounds will partition into the membrane; however, both the proportion of each compound that enters the bilayer rather than remaining in the aqueous phase, and the rate at which they will cross the bilayer differed. While you may expect that neutral compounds will more readily partition into the bilayer, there was no clear trend in the depth of the energy minima presented in Fig. 2. The perpetually neutral compounds benzocaine, lamotrigine, and carbamazepine all had relatively small minima in the range of 2–4 kcal/mol, similar in depth to most of the charged versions of the protonatable compounds. Lamotrigine had a larger energy barrier to pass through the bilayer center than either carbamazepine or benzocaine, but this was not entirely surprising, given the presence of polar groups on this molecule. The neutral protonatable compounds, surprisingly, had deeper wells (in the range of 6–8 kcal/mol) than the perpetually neutral compounds, with lower energies as they inserted themselves into the hydrophobic core of the bilayer. In general, our results reflected the expected trend that larger and more polar compounds will have slower bilayer crossing rates. All of the charged protonatable compounds, however, had very large energy barriers to cross the bilayer in the charged form, which was significantly reduced when in the neutral form. This strongly suggests that the protonatable molecules are more likely to permeate the bilayer core in the neutral form.
Fig. 2.
PMF for lipid partitioning of the compounds used in this study, along with their corresponding molecular structure. The dashed red line is at an energy of 0 kcal/mol. The value of the energy minima (Min) and the barrier to cross the bilayer are indicated.
Understanding Anesthetics Behavior in the Pore.
Increased sampling of benzocaine inside pore.
To best study the likely binding sites of local anesthetics in the sodium channel, we used REST2 (45) as implemented in NAMD (46). Replica-exchange methods have been shown to more efficiently improve conformational sampling than alternative methods such as metadynamics or accelerated MD (34, 46, 47), and the solute tempering method allows the approach to be used efficiently in large systems. To show that replica exchange solute tempering (REST) simulations indeed increase sampling of a compound in the pore, we chose a test system that has been well-characterized in simulation: the structure of NavAb [Protein Data Bank (PDB) ID code 3rvy] with benzocaine inside the pore (34–36), embedded in a pure 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) bilayer. This system was simulated for 100 ns by using both unbiased and REST simulations. To measure sampling efficiency, and indirectly convergence, we chose to track the conformational sampling of the drug as measured by the physical volume visited by the center of mass of benzocaine. As seen in SI Appendix, Fig. S1A, the volume visited by the center of mass of benzocaine increased rapidly at the beginning of the REST simulations (purple line), compared with the unbiased simulation (orange line), indicative of more rapid sampling. In addition, the volume asymptoted toward a much lower value in the unbiased simulations than with REST, indicating that it is unlikely that even an extremely long unbiased simulation would achieve the same conformational sampling obtained by using REST. Energy landscapes obtained from both the unbiased and REST simulations (SI Appendix, Fig. S1 B and C, respectively) supported the conclusions from the volume sampling. The energy landscape from the unbiased simulation showed that benzocaine only sampled the known binding site near the fenestrations, and did not explore the cytoplasmic activation gate where benzocaine is also known to bind. However, when using REST, both the known binding sites were sampled. In addition, when cluster analysis was done on both the unbiased and REST simulation (SI Appendix, Fig. S1 D and E, respectively), the four most populous clusters associated with the unbiased simulations were all close together and near the same binding site. In the REST simulations, these clusters represented both the activation gate and fenestration binding sites, agreeing well with previous simulations on NavAb with benzocaine (34, 36). All these results indicated that REST not only increased sampling in the pore, it also identified binding modes that may not be seen in unbiased simulations.
Simulations determine two distinct binding modes of PF-5215786 with NavMs.
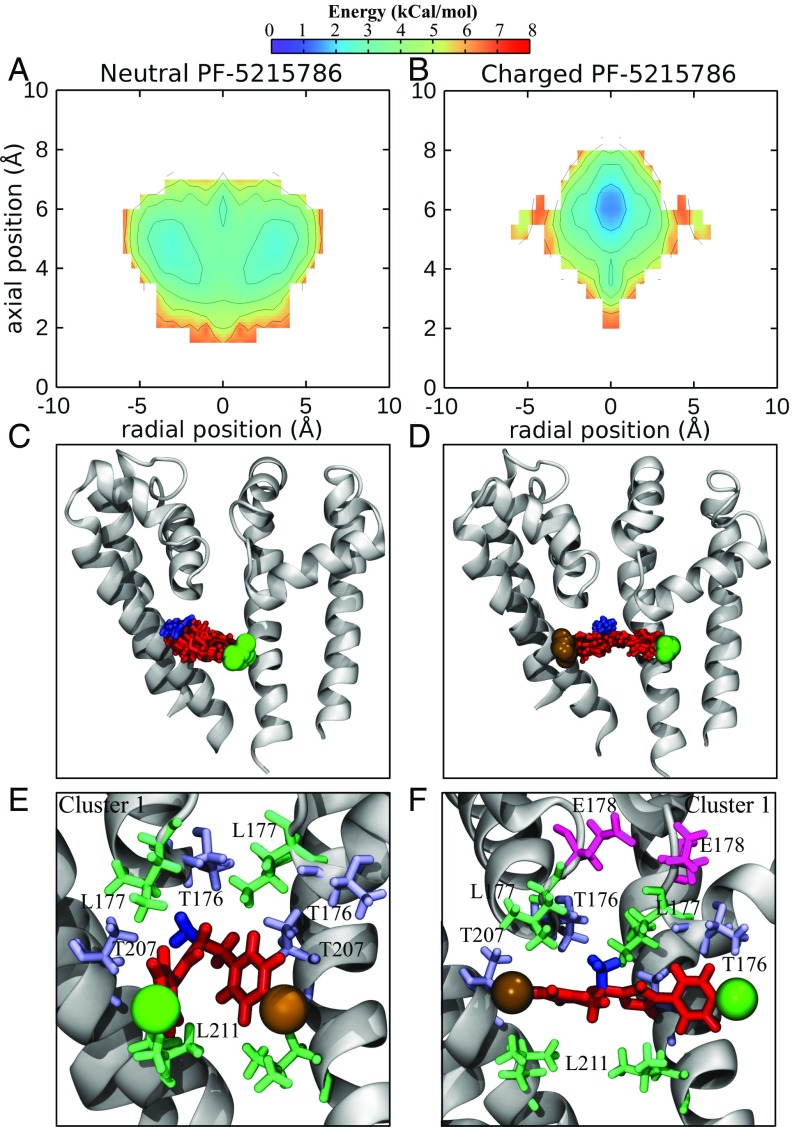
To gain a direct comparison between the simulation results and experimental data, a system was constructed with NavMs (PDB ID code 4p9o) embedded in POPC, with the brominated compound PF-5215786 inside the pore. The structure of NavMs with this compound bound has previously been solved, and the electron density of the bromine atom is visible near the internal opening of the fenestrations (16). When both the neutral and charged states of PF-5215786 were simulated with NavMs, different energy landscapes were produced (Fig. 3 A and B; neutral and charged respectively), indicative of distinct binding modes of the neutral and charged versions. While both neutral and charged PF-5215786 bound in the central cavity, the free energy surfaces showed that the neutral version of the molecule bound lower in the cavity (z 4 Å) and away from the central axis (r 3 Å) (Fig. 3A). In contrast, the charged version sat higher in the pore (z 6 Å) with its center of mass close to the pore axis (r 0 Å) (Fig. 3B). This difference in the binding modes is visualized in Fig. 3 C and D, by looking at the most populous binding pose. Neutral PF-5215786 preferred to bind close to helix S6, where it interacted with residues on both the S6 helix (T207 and L211; Fig. 3E) and at the bottom of the selectivity filter (T176 and L177; Fig. 3E). In this position, the amine (blue, Fig. 3E) pointed into a lateral fenestration. Charged PF-5215786, in contrast, spanned the width of the pore (Fig. 3D), with the halide (colored in brown and green for Br and Cl, respectively) on either end of the molecule inserting itself into the entrance of a fenestration. The protonated amine (Fig. 3D, blue) pointed toward the negatively charged selectivity filter, preferentially interacting with T176 and L177 (Fig. 3F), while the other parts of charged PF-5215786 were interacting with T207 and L211. As a final indication of the different binding modes of the neutral and charged compounds, we plotted the average interaction energy of each compound with each protein residue in SI Appendix, Fig. S2. While the neutral compound had a fairly strong interaction for the pore chamber, the charged compound had stronger interactions, especially with residues in the selectivity filter. In short, while both the neutral and charged compounds interacted with residues on S6 at positions found to be important in eukaryotic channels (30), the charged compounds had additional interactions with the selectivity filter, which was likely to result in stronger binding.
Fig. 3.
Binding of PF-5215789 to the NavMs pore. (A and B) Energy landscapes (in kcal/mol) of the centers of mass of neutral PF-5215786 (A) and charged PF-5215786 (B) inside the channel. (C and D) Snapshots from the most populated clusters for neutral PF-5215786 (C) and charged PF-5215786 (D) are shown, with the channel in gray cartoon and the compound in red licorice. (E and F) A representative frame from the cluster shown in C and D of both neutral PF-5215786 (E) and charged PF-5215786 (F) is shown as red licorice, with the bromine atom shown in a brown sphere, the chlorine atom shown as a green sphere, and the protonatable amine highlighted in blue. The channel is shown in gray cartoon, and surrounding residues (T176, L177, E178, T207, and L211) are shown in licorice, with the threonines shown in light blue, the leucines in green, and the glutamatic acid in pink.
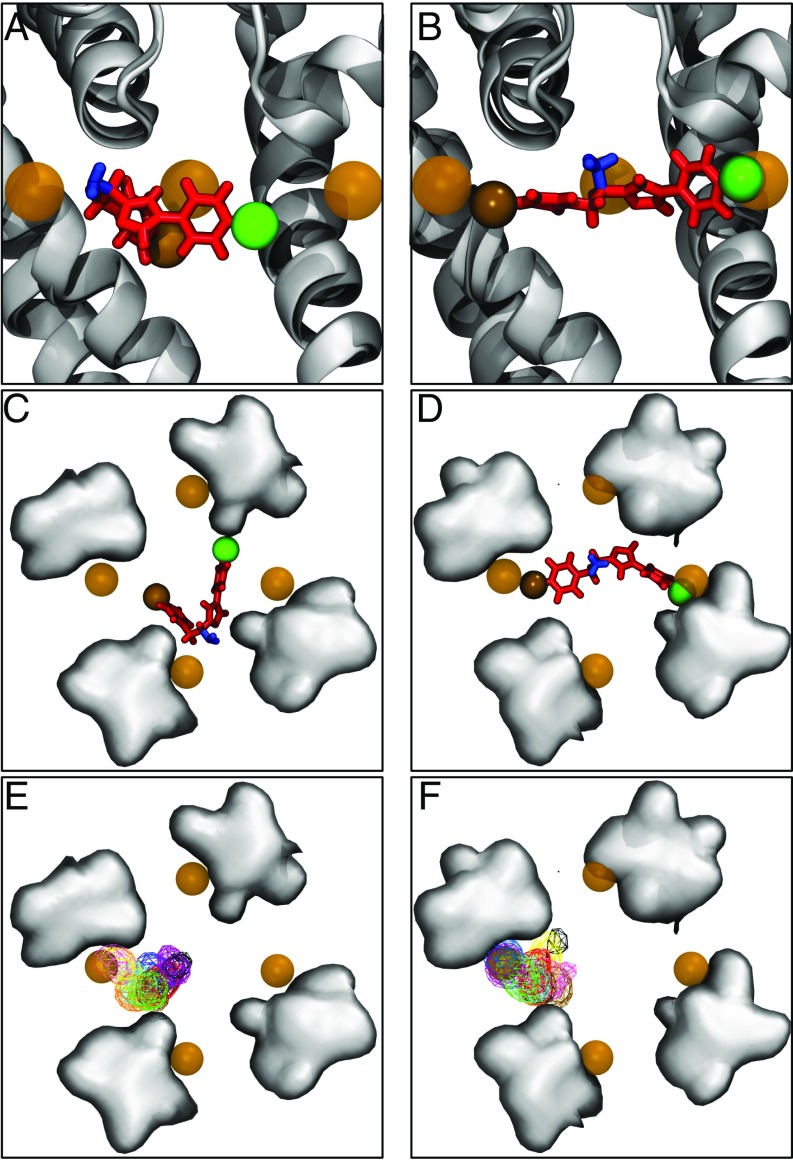
To compare our predicted binding poses with the crystallographic structure containing resolved bromines [PDB ID code 4p9o (16)], we aligned representative structures from the most populous binding poses with this crystal structure. As seen in Fig. 4 A and C, the bromine atom of neutral PF-5215786 did not overlap with any of the crystallographic bromines. Instead, the amine group was closer in position to the crystallographic bromines than either the bromine or chlorine atom on either end of the molecule. In contrast, the bromine and chlorine atoms on charged PF-5215786’s were in close proximity to the crystallographic bromine (Fig 4 B and D). The same pattern of binding for both protonation states was seen for all of the 10 most populated clusters, as demonstrated in SI Appendix, Figs. S3E and S4F, in which we plotted the positions of the bromine atom for all clusters. For neutral PF-5215786, there was very little overlap in the positions of either halide with the crystallographic bromine (SI Appendix, Fig. S5 A and C). In the case of charged PF-5215786, the bromine atom often was near or overlapped with one of the bromine positions seen in the crystal structure (SI Appendix, Fig. S5 B and D). In addition, the chlorine atom sat near to another crystallographic bromine position, either in the opposite or adjacent fenestration, suggesting that the chlorine atoms could be contributing to the halide density seen in the crystal structures. This observation was supported by the occupancy of both the bromine and chlorine atoms for each cluster, as shown in SI Appendix, Fig. S5. The closer agreement between both the bromine and chloride position in our simulations of the charged compound with those in the crystal structure, as opposed to the neutral compound, strongly suggests that the compound Bagneris et al. (16) cocrystallized with the pore of NavMs was in the charged state.
Fig. 4.
Binding modes of neutral and charged PF-5215786 compared with the crystal structure. (A–D) Side views of a representative snapshot from the most populous clusters of neutral PF-5215786 (A) and charged PF-5215786 (B), along with top views of neutral PF-5215786 (C) and charged PF-5215786 (D), when aligned with PDB ID code 4p9o. PF-5215786 is shown in red licorice, with the bromine atom as a solid brown sphere, the chlorine atom as a solid green sphere, and the protonatable amine group as blue licorice. NavMs is shown in cartoon (light gray for the simulation structure, dark gray for the crystal structure). Bromine atoms from the crystal structure are shown in brown transparent spheres. (E and F) Average occupancies of the bromine atoms for each cluster is shown for the neutral (E) and charged PF-5215786 (F), shown in wire mesh and colored according to cluster. For C–F, the positions of the S6 helices are shown by the gray surface, with the fenestrations occupying the space between these surfaces.
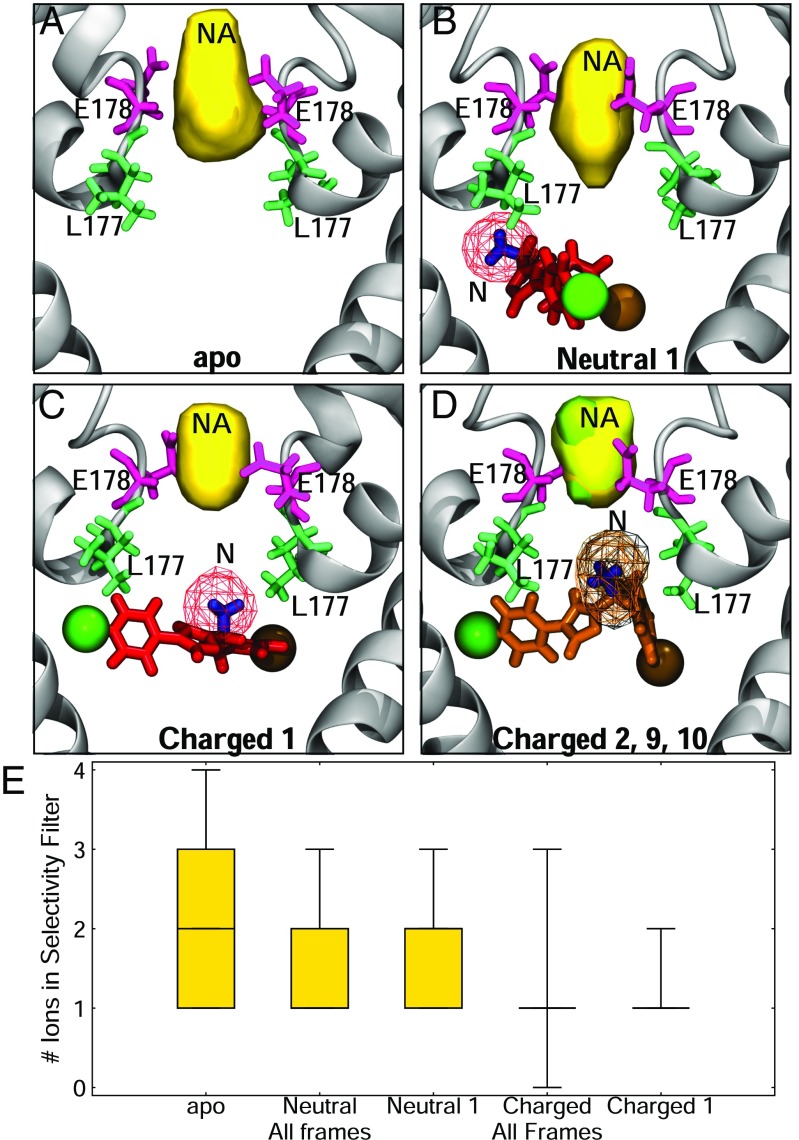
Our simulations also suggested that the presence of the inhibitor in the pore alters the position and number of in the selectivity filter. The charged form of the compound partially inserted itself into the selectivity filter via the protonated amine group, occluding the pore and influencing ion permeation. To highlight this point in Fig. 5E, we showed that in an unbiased simulation without the compound present, the number of ions inside the filter fluctuated between one, two, and three. The average density was spread throughout the filter (Fig. 5A), in agreement with previous simulations of the channel (48–50). When the neutral PF-5215786 was present, it lowered the number of sodium ions in the filter to between one and two (Fig. 5E), both in the whole simulation and in the most populous binding pose. Despite this, the distribution of the sodium density in the selectivity filter was similar to the apo form (Fig. 5B). In contrast, for charged PF-5215786, there was a marked decrease in the number of ions present in the filter, with the protonated amine effectively replacing a sodium ion. This reduced the most frequently occurring number of ions in the pore to one (Fig. 5 C and D) and significantly altered the distribution of in the filter for both the whole simulation and for the most populous cluster (Fig. 5E). This observation correlated with the lack of electron density seen in the selectivity filter between apo and PF-5215786 crystals, indicating that something was occluding the selectivity filter (16). Bagneris et al. (16) used molecular docking to predict a binding pose of PF-5215786 in which the bromine atom overlapped the crystallographic bromine and the chlorine atom pointed into the selectivity filter, where it was proposed to alter the ion densities and occlude the pore. We did not see this binding pose in any of our simulations, and think it unlikely that the electronegative atom would either be attracted to the filter (given repulsive electrostatic interactions) or displace resident (given the attractive electrostatic interactions). We suggest that the lack of density seen in the selectivity filter in the crystal structure was not caused by the chlorine-containing end of PF-5215786 blocking the filter, but rather by the protonated amine group in the middle of the molecule interacting with the selectivity filter residues.
Fig. 5.
Drug binding influences in the channel. (A–D) Densities of (yellow) and the protonatable amine (mesh) are shown in both the absence (A) and presence (B) of neutral PF-5215786 and charged PF-5215786 cluster 1 (C) and 2, 9, and 10 (D). The protein is shown in gray cartoon, L177 is shown in green licorice, and E178 is shown in pink licorice. (E) The average number of sodium ions present in the selectivity filter are shown for simulations containing no drug (apo) as well as neutral and charged compounds using box-and-whiskers plots. Results are shown for all frames of each compound as well as for just those in the most populous cluster.
Binding of other protonatable compounds.
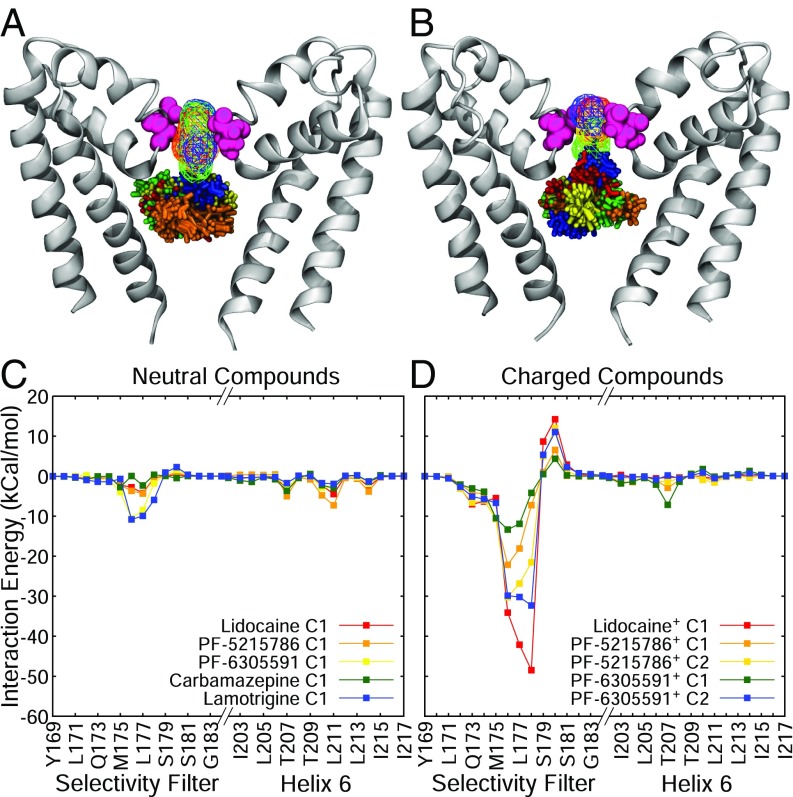
To see if the different binding modes of the neutral and charged forms of PF-5215786 are also seen for other compounds, two more compounds with a protonatable amine group (lidocaine and PF-6305591) were simulated by using REST. Lidocaine is a commonly used local anesthetic, and PF-6305591 is a newer compound reported to display selectivity for Nav1.8 and suspected to also bind inside the pore. Both lidocaine and PF-6305591 have a protonatable amine group, similar to PF-5215786, and both displayed similar behaviors as PF-5215786 in our simulations. The energy landscapes of the neutral states of all protonatable compounds (SI Appendix, Fig. S6 A, C, and E) displayed preferred binding modes lower in the pore than the charged states. For the charged states (SI Appendix, Fig. S6 B, D, and F), all three had multiple binding positions, but included one at the base of the selectivity filter (z 8 Å), as seen for charged PF-5215786. When these different binding modes were compared between compounds, the neutral compounds preferred to bind in approximately the same place in the pore (Fig. 6A) and occluded the pore, although a relatively large density of sodium ions was still present in the selectivity filter. In contrast, some clusters in the charged compounds were seen entering the selectivity filter site, with a noticeably smaller density of sodium ions in the selectivity filter (Fig. 6B). A plot of the average interaction energy of each compound with each protein residue also highlighted the difference between the neutral and charged compounds (Fig. 6 C and D). While both compounds interacted with S6 and the selectivity filter, the strength of interaction with the filter was much larger for the charged compounds than the neutral ones. As seen in SI Appendix, Fig. S8, while the neutral forms of each compound had a small influence on the number of in the filter, the effect was much greater for all of the charged compounds. In the latter case, the charged protonated portion of the compound effectively replaced a sodium ion in the filter. Overall, these data reinforce that there are distinct binding modes for neutral and charged compounds, with the latter interacting more strongly with the selectivity filter.
Fig. 6.
General behavior of neutral and charged anesthetics. (A) Most populous clusters of neutral lidocaine (red licorice), neutral PF-5215786 (orange licorice), neutral PF-6305591 (yellow licorice), carbamazepine (green licorice), and lamotrigine (blue licorice). (B) Clusters of charged lidocaine (red licorice), charged PF-5215786 (orange licorice), and charged PF-6305591 (yellow licorice). For both structures, the channel is shown in silver cartoon, and residue E178 is shown in pink spheres for reference. (C and D) Drug–protein interaction energies of neutral (C) and charged (D) compounds. For both C and D, energies are in kcal/mol, and each cluster is shown as a separate line, and only residues from the selectivity filter (left half of each graph) and the S6 helix (right half of each graph) are shown.
Perpetually neutral drugs.
As a comparison with the protonatable compounds, two drugs which are always in a neutral state were simulated with NavMs by using REST: carbamazepine and lamotrigine. Both had a relatively similar energy landscape to the neutral protonatable compounds (protonatable: SI Appendix, Fig. S6; and carbamazepine/lamotrigine: SI Appendix, Fig. S9 A and B), in that they had preferred binding modes at a z coordinate between 4 and 8 Å. However, lamotrigine was more likely to venture toward the selectivity filter than carbamazepine. This could also be seen by snapshots of the most likely positions of each compound (SI Appendix, Fig. S9 C and D). Carbamazepine appeared to have a preference of binding to helix 6, but the most populous cluster of lamotrigine sat higher, with the nitrogen-containing moieties pointing toward the selectivity filter. Despite these differences, both compounds interacted with a similar set of protein residues: M175, T176, L177, 207T, and L211 (SI Appendix, Fig. S9 E and F and Fig. S12). These are the same residues that the protonatable compounds in this study prefer to bind to in their neutral state, and compounds that are perpetually neutral that have been previously studied (benzocaine and lidocaine).
Extension to eukaryotic channels.
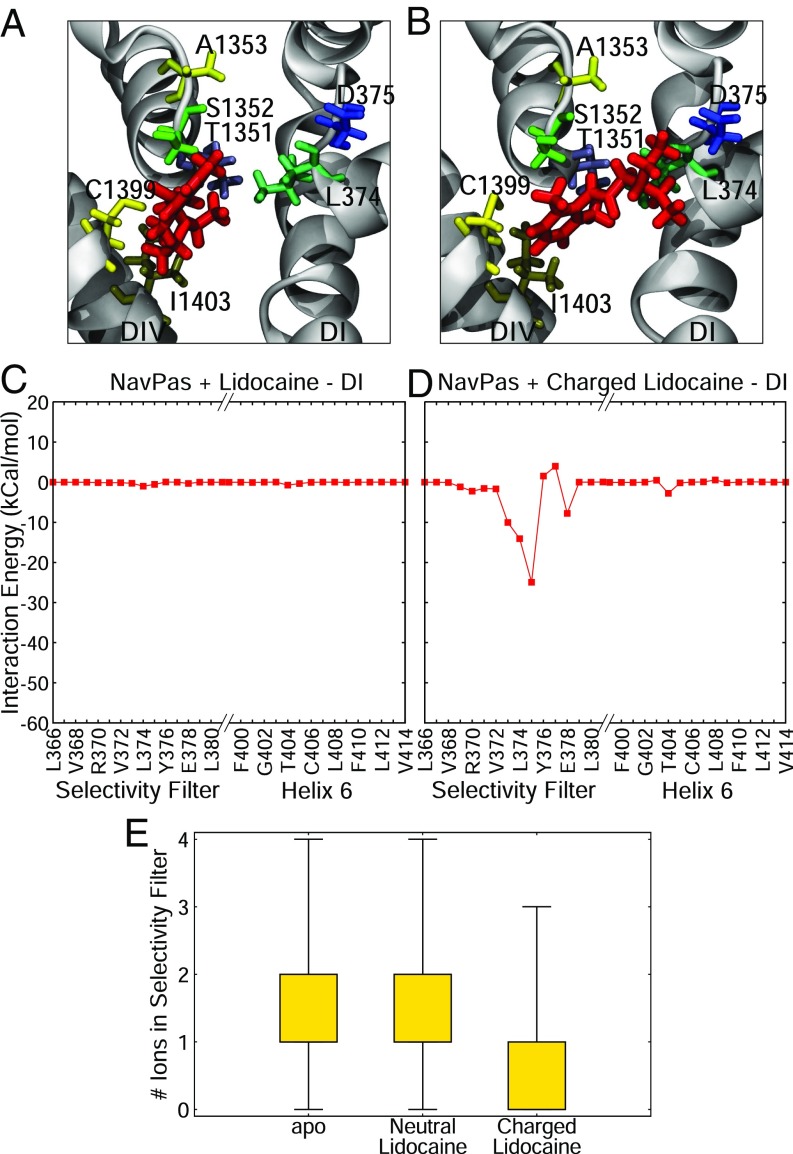
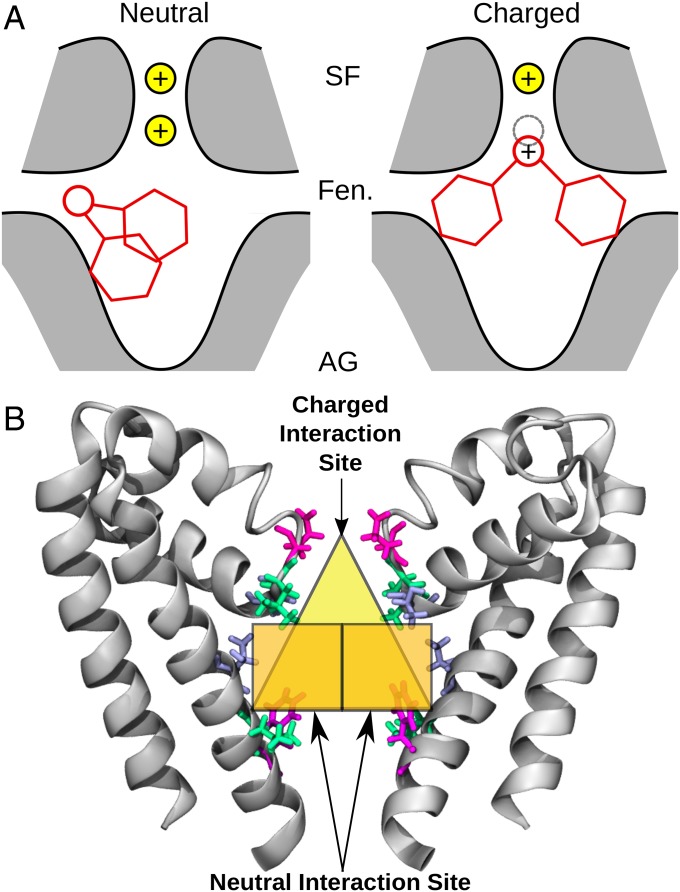
Although we have identified distinct binding sites for a range of neutral and charged compounds in bacterial channels, it is not obvious whether these same sites will be present in eukaryotic channels. In particular, the amino acid sequence of the selectivity filter is distinctly different in prokaryotic and eukaryotic sodium channels, so it is important to confirm if charged molecules can interact with the filter of eukaryotic channels in the same way we found for the bacterial channels. To determine this, we ran REST simulation of both protonation states of lidocaine inside the pore region of the most recent structure of a sodium channel from the American cockroach, denoted NavPas (23). When inside the pore, the neutral form of lidocaine did not interact with the selectivity filter of NavPas and preferred to interact lower in the pore with the S6 helix of DIII and DIV (Fig. 7A), in a relatively similar position to the one seen in the bacterial channels. However, the charged form of lidocaine preferentially interacted with the selectivity filter of NavPas, specifically the aspartic acid residue in the “DEKA” ring (Fig. 7B). The difference in the binding pose of the neutral and charged forms of lidocaine was highlighted by the fact that there was a large interaction energy between the charged version of lidocaine and the aspartic acid residue, whereas there was virtually no interaction when the neutral state of lidocaine was in the pore (Fig. 7 C and D). This was a remarkable result, as charged lidocaine displayed the same binding position in NavMs, which has a glutamic acid “EEEE” ring, rather than the DEKA ring present in eukaryotic channels. Charged lidocaine’s affinity for the aspartic acid residue in NavPas resulted in a binding pose where the compound blocked ion conduction by both physically occluding the filter and by electrostatic repulsion, as seen by the difference in the number of sodium ions in the selectivity filter (Fig. 7E) in the presence of charged lidocaine. The results from simulations with both eukaryotic and prokaryotic channels point to two different binding sites for local anesthetics, depending on their charge state (Fig. 8A). The neutral compounds preferred to bind in the central cavity close to helix S6 and had their primary interactions with the hydrophobic or aromatic residues in this region (neutral binding site, Fig. 8 A, Left). The charged compounds could also have the same hydrophobic interactions with S6, but preferentially sat higher in the pore, exhibiting strong interactions with the negatively charged selectivity filter residues (charged binding site, Fig. 8 A, Right). Both the neutral and charged compounds shared common interacting residues, but only the charged compounds ventured toward the selectivity filter (Fig. 8B).
Fig. 7.
Binding of lidocaine to NavPas. (A and B) Representative snapshots from the most populous cluster with neutral (A) and charged (B) lidocaine. Lidocaine is shown in red licorice, the backbone of the protein is in gray cartoon, and relevant residues are shown in licorice of varying colors. (C and D) Associated drug–protein interaction energies for neutral (C) and charged (D) lidocaine for DI are shown. (E) The number of ions present in the selectivity filter for apo, NavPas + neutral lidocaine, and NavPas + charged lidocaine REST simulations are shown as a boxplot.
Fig. 8.
(A) Schematic of the binding modes of neutral (Left) and charged (Right) compounds. The protein is shown in gray shading with black outline, the drug is shown in red lines, and the sodium ions are shown as yellow spheres, with a gray dotted line outlining a sodium ion site. (B) Neutral and charged interaction sites of local anesthetics. The sodium channel pore is shown in gray cartoon. Select residues involved in binding are shown in blue, green, and pink licorice, and each binding site is highlighted by orange (neutral site) or yellow (charged site) regions. AG, activation gate; Fen, fenestration; SF, selectivity filter.
Discussion
In this study, we have taken a number of steps to improve our understanding of sodium channel inhibition. First, we have rigorously parameterized and characterized a range of pore-blocking compounds and were successful in replicating trends seen in drug-solvent partitioning experiments. Such parameterization is essential before using these compounds in MD simulations with the protein. Second, we determined the energy profiles for partitioning of each compound from the aqueous phase into the lipid bilayer. This indicated that even the charged compounds were likely to move into the membrane from the aqueous phase, but, as expected, the neutral forms were more likely to pass across it. Third, we used an improved sampling method to determine binding positions of the compounds in the pore (REST) and demonstrated that this approach improved the sampling of benzocaine in a previously characterized sodium channel system. This knowledge allowed us to be more confident that we could find the most likely binding position for the newly simulated compounds in the pore. Finally, we characterized the binding of a range of sodium channel inhibitors inside the sodium channel pore. As an MD study investigating a large number of both neutral and charged compounds, we are able to compare our results with a large amount of previous experimental and computational data, and determine trends within our own data, to shed light on the characteristics of different sodium channel inhibitors. Notably, we show that neutral and positively charged compounds have distinct binding modes in the pore.
Interactions between all inhibitors in this study and the NavMs sodium channel pore involved both polar and nonpolar interactions, with pore-lining S6 residues and residues at the base of the selectivity filter. This was mainly with key leucines and threonines (T176, L177, T207, and L211), and, in a few cases, there were hydrophobic interactions with F214. These residues were the dominating interaction partners of both the neutral states of PF-5215786, PF-6305591, and lidocaine, as well as the perpetually neutral compounds, carbamazepine and lamotrigine. All of the neutral compounds remained in the central pore chamber, interacting primarily with residues on helix S6. Only benzocaine entered the fenestrations, and none of the compounds ventured into the selectivity filter. Carbamazepine and lamotrigine had more significant interactions with the selectivity filter, with some clusters having their amine groups interacting with T176 and L177. This behavior was also seen in a docking study (51) in the presence of a sodium ion in the filter. This could be because the amines in carbamazepine and lamotrigine made them more strongly attracted to the positive charge of the sodium ion, encouraging the amines to point up toward the selectivity filter. The interaction of carbamazepine and lamotrigine with this sodium ion would prevent sodium currents, with the ion playing a similar role to the protonatable amine of the charged compounds.
In contrast to the neutral compounds, the protonated states of PF-5215786, PF-6305591, and lidocaine had much stronger interactions with residues in the selectivity filter. This was clearly seen in the most populated poses of PF-5215786. In these, the compound spanned the width of the pore, with the halides on the ends of the molecule occupying either opposite or adjacent fenestrations, and the protonated amine in the center of the pore pointed up toward the selectivity filter. These positively charged amines effectively replaced a sodium ion at the bottom of the filter and formed strong interactions with residues T176 and L177. The compound thus prevented a positive sodium ion from passing through the filter by both physical occlusion and electrostatics. The binding position seen for this compound agreed with the position of the bromine atoms seen in the crystal structure in which PF-5215786 was cocrystallized with NavMs. However, the specific binding pose predicted by Bagneris et al. (16), in which the chlorine occludes the selectivity filter, was not seen in any of the frames of our simulation. As chlorine is electronegative, we think it is unlikely that chlorine would be attracted to the negatively charged selectivity filter. It is more likely that PF-5215786 spans the pore with both halides occupying fenestrations, providing density for the halide seen in the crystal structure, while the positively charged protonated amine points toward the selectivity filter, altering the sodium ion density.
Similar binding poses were seen for the other protonated compounds, charged PF-6305591 and charged lidocaine, as described above for PF-5215786. For all these compounds, the protonated amine formed strong interactions with residues in the selectivity filter: T176, L177, and E178. In addition to this, all these compounds interacted strongly with T207 on the S6 helix, one of the key interaction partners for the neutral compounds. As suggested in a recent docking study, it is possible that charged compounds form aromatic interactions in the center of the pore while extending their positively charged ends into the selectivity filter (51). The other residues that were implicated in binding neutral compounds (L211 and F214) did not have such significant interactions when the compounds were charged. Interestingly, T207 in NavMs corresponds to one of two residues that were seen as being important for tonic and use-dependent block in a rat sodium channel: F1764 (30, 31). Taking into account this present work, and Ragsdale’s previous study (31), this residue at this particular region could act as a scaffold and/or major interacting partner for the compounds in this study, and this residue could be key in understanding sodium channel block. The other residue highlighted by Ragsdale et al. (31), as important for use-dependent block, Y1771 (corresponding to F214 in NavMs), was not found as a major interacting partner in this study. Given that we have not used the structure of an inactivated channel, it could be that this residue becomes available due to conformational changes happening as the channel changes from the resting or open state studied here, to the inactivated state which is the principle target of use-dependent blockers. Indeed, the limited interactions of the compounds studied here with F214 agree with the observation of Ragsdale et al. (30, 31) that mutations of this residue had little influence on tonic block.
Our study used the crystal structures of two bacterial channels (NavAb and NavMs) and one eukaryotic channel (NavPas). While previous studies have indicated that binding of compounds to NavMs (16) and NavAb (52) can reflect binding in eukaryotic channels, there is potential for significant differences to arise. Remarkably, in light of potential differences, lidocaine bound in a similar fashion in both prokaryotic (NavMs) and eukaryotic (NavPas) channels. The neutral state of lidocaine bound lower in the pore and interacted with polar residues, while the charged state used these polar residues as a scaffold to position the charged end of lidocaine next to the aspartic acid residue in the selectivity filter of NavPas, which occluded the pore. Since lidocaine behaved the same way in both prokaryotic and eukaryotic channels, it is likely that the other two protonatable compounds (PF-5215786 and PF-6305591) would exhibit similar behavior when simulated with eukaryotic channels. As charge seems to be important in determining the interaction sites within the pore, it is important to consider whether the compounds will be protonated while in the pore. At physiological pH, all three protonatable compounds studied here will largely be in their protonated state when they are in the aqueous phase. While our lipid-partitioning data indicated that these compounds are much more likely to cross the bilayer in the neutral state, at physiological pH, we can expect a majority of each compound to return to the charge state in the cytoplasm. If these compounds enter through the open activation gate as suspected, then we would expect that they can enter and bind while protonated. This claim is supported by the close agreement of the halide positions with those observed in a recent crystal structure for the charged state (16) of the charged state of PF-5215786, but not the neutral state. The pKa of the compounds may change inside the pore, but the net negative charge on the pore walls used to attract into the channel may make it more likely for the compounds to protonate in this environment.
While this study has helped to highlight two distinct binding positions for charged and neutral pore blockers, there are a number of questions it cannot address. First, while the crystal structure of NavMs used in this study is claimed to be in the open state, in the absence of the C terminus (not resolved in the crystal structure), the S6 helices rapidly closed the pore during our simulations. Although it is difficult to be sure exactly what functional state this represents, it is unlikely to be the inactivated state to which most of the compounds studied bind most strongly. As such, we believe that these binding positions most likely represent tonic-like binding, something backed up by the lack of interaction with F214. Further studies will be required to ascertain the differences in binding of these compounds to the inactivated state. Second, this study only hints at why most of the compounds are use-dependent rather than tonic blockers. Benzocaine, the primary tonic blocker examined here, was the only compound in our study seen to enter the fenestrations. Thus, it is likely that it was simply the small size of benzocaine that allowed it to pass through the fenestrations to block the resting state of the channel. Previous work has concluded that compounds with the maximum width of a benzene ring (i.e., benzocaine) are able to fit through the fenestrations (49, 53). Apart from benzocaine, all of the drugs studied here are wider than the width of a benzene ring, making it unlikely that the compounds would pass through the fenestrations on their way to block the pore. Finally, future work could use the NavPas structure as a basis for studying human sodium channels, to further understand the interactions of pore-blocking compounds with eukaryotic sodium channels and open the door for more targeted sodium channel therapies.
Materials and Methods
Parameterization of Compounds and Validation.
Parameterization of the drug models was performed with the aid of Force Field Toolkit (FFTK) (54) implemented in VMD (55). FFTK does not support the parameterization of Lennard–Jones parameters and the Urey–Bradley term, and these parameters were obtained by analogy from the CGenFF program (https://cgenff.paramchem.org/) (56, 57).
The Gaussian09 package was used for all quantum mechanical (QM) calculations. Force-field parameter optimizations were done after the molecular geometries of drugs were optimized at the MP2/6-31G∗ (for drugs with 40 atoms) or B3LYP/6-31G** level of theory (for drugs with 40 atoms). Atomic charges were assigned based on the minimum interaction energy and distance between one water and all hydrogen-bond donors and acceptors, optimized at the HF/6-31G* level of theory. Of note, aliphatic and aromatic nonpolar hydrogens in CHARMM have predetermined charges of 0.09 and 0.15, while aliphatic and aromatic carbons not adjacent to a heteroatom are assigned -0.18 (or -0.27 for terminal group) and −0.15 respectively (58). Equilibrium bonds and angle values were optimized through comparison with the QM-optimized drug geometries, while the force constants were derived from the Hessian calculated in Gaussian. The optimizations of dihedral parameters were done by using a simulated annealing protocol developed by Guvench and MacKerell (59) and implemented in FFTK.
To validate the parameters for each drug, the octanol–water and cyclohexane–water partitioning free energies for neutral drugs were determined by using the Bennett acceptance ratio approach (60), as implemented in GROMACS4.5.4 (61), and compared with available experimental logP values. The simulations were carried out at a constant isotropic pressure of 1 atm and a temperature of 300 K. A series of 51 windows with the equally spaced between 0 and 1 were constructed for both water and octanol or cyclohexane phases. Each window was simulated for 1 ns, and the first 0.2-ns simulation was discarded for equilibration. A soft core potential with = 1, = 0.3 nm and -power of 1 was used to avoid singularities. Results presented used the CHARMM general force field (58) for the octanol and cyclohexane partitioning simulations. The TIP3P model (62) was used for water. Further details are given in SI Appendix.
Bilayer Partitioning.
The method of umbrella sampling was used to determine the PMF to move the compounds from bulk aqueous solution into the interior of a DPPC bilayer. Starting configurations for umbrella sampling were generated by using steered MD. The drug was pulled along the z axis (perpendicular to the bilayer/water interface) at a rate of 0.01 nm/ps and with a force constant of 1,000 kJ/mol. Starting points for umbrella sampling were selected every 0.1 nm along the z axis. In each umbrella sampling window, we carried out 50-ns umbrella sampling simulations. During umbrella sampling simulations (63), a biased harmonic potential with a force constant of 3,000 kJ/molnm−2 was used to confine the drug within the sampling window. The FEPs and error estimates were determined by using the weighted histogram analysis method (64). The first 20 ns within each window was discarded to allow for equilibration at the new solute position, and the FEP was determined from the remaining 30 ns. Further details of the simulation methods are described in SI Appendix.
Pore and Drug Simulations.
NavAb (PDB ID code 3RVY), NavMs (PDB ID code 4P9O), and NavPas (PDB ID code 5XOM) proteins were embedded into pure POPC bilayers, with explicit TIP3P solvent and 150 mM NaCl. One drug molecule was randomly placed at the center of the pore, and constraints were placed on the protein backbone with a force constant of 0.1 kcal/molÅ−2 to prevent excessive conformational changes in the high-temperature REST simulations. The compound was prevented from exiting the pore by a spherical potential with a radius of 37 Å from the pore center, and force constant of 20 kcal/molÅ−2. For the REST2 (46) simulations, the patch was compiled with NAMD2.10 (65). A total of 21 parallel simulations for NavMs (25 for NavPas) were run in an NPT ensemble, with 21 and 25 being the number of replicas predicted by a temperature generator for REMD simulations (66) (folding.bmc.uu.se/remd/). Only the protein and the drug were used as the “hot” parts of the system, while the lipids, waters, and ions were used as the “cold” parts of the system, with a temperature range from 310 to 410 K. Exchanges were attempted every 2 ps, yielding an average swap acceptance ratio of 0.4. All other simulation details are described in detail in SI Appendix. Cluster analysis was performed to identify distinct binding poses and to select representative snapshots. This was done by using the measure cluster tool in VMD (55), with 10 clusters and a 2.5-Å cutoff used for all compounds in the study. Energy landscape analysis and volume sampling were performed with locally written scripts.
Supplementary Material
Acknowledgments
This research was undertaken with the assistance of Australian Research Council Grant FT130100781 and resources provided at the National Computational Infrastructure National Facility at the Australian National University (ANU) and the Pawsey Center through the National Merit Allocation Scheme supported by the Australian Government and through the ANU Partner Scheme.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 3512.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714131115/-/DCSupplemental.
References
- 1.Meisler MH, Kearney JA. Sodium channel mutations in epilepsy and other neurological disorders. J Clin Invest. 2005;115:2010–2017. doi: 10.1172/JCI25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roden DM, George AL. The cardiac ion channels: Relevance to management of arrhythmias. Annu Rev Med. 1996;47:135–148. doi: 10.1146/annurev.med.47.1.135. [DOI] [PubMed] [Google Scholar]
- 3.Ruan Y, Liu N, Priori SG. Sodium channel mutations and arrhythmias. Nat Rev Cardiol. 2009;6:337–348. doi: 10.1038/nrcardio.2009.44. [DOI] [PubMed] [Google Scholar]
- 4.Waxman SG, Dib-Hajj S, Cummins TR, Black JA. Sodium channels and pain. Proc Natl Acad Sci USA. 1999;96:7635–7639. doi: 10.1073/pnas.96.14.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waxman SG, Hains BC. Fire and phantoms after spinal cord injury: Na+ channels and central pain. Trends Neurosci. 2006;29:207–215. doi: 10.1016/j.tins.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Catterall WA. From ionic currents to molecular mechanisms: The structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 7.Fozzard HA, Sheets MF, Hanck DA. The sodium channel as a target for local anesthetic drugs. Front Pharmacol. 2011;2:1–6. doi: 10.3389/fphar.2011.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarvis MF, et al. A-803467, a potent and selective Nav1.8 sodium channel blocker, attenuates neuropathic and inflammatory pain in the rat. Proc Natl Acad Sci USA. 2007;104:8520–8525. doi: 10.1073/pnas.0611364104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kort ME, et al. Subtype-selective Nav1.8 sodium channel blockers: Identification of potent, orally active nicotinamide derivatives. Bioorg amd Med Chem Lett. 2010;20:6812–6815. doi: 10.1016/j.bmcl.2010.08.121. [DOI] [PubMed] [Google Scholar]
- 10.Scanio MJC, et al. Discovery and biological evaluation of potent, selective, orally bioavailable, pyrazine-based blockers of the Nav1.8 sodium channel with efficacy in a model of neuropathic pain. Bioorg Med Chem Lett. 2010;18:7816–7825. doi: 10.1016/j.bmc.2010.09.057. [DOI] [PubMed] [Google Scholar]
- 11.McCormack K, et al. Voltage sensor interaction site for selective small molecule inhibitors of voltage-gated sodium channels. Proc Natl Acad Sci USA. 2013;110:E2724–E2732. doi: 10.1073/pnas.1220844110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corry BA. Physical basis of specificity and delayed binding of a subtype selective sodium channel inhibitor. Sci Rep. 2017;8:1356. doi: 10.1038/s41598-018-19850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahuja S, et al. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science. 2015;350:aac5464. doi: 10.1126/science.aac5464. [DOI] [PubMed] [Google Scholar]
- 15.Bagnéris C, et al. Role of the C-terminal domain in the structure and function of tetrameric sodium channels. Nat Commun. 2013;4:2465. doi: 10.1038/ncomms3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagnéris C, et al. Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism. Proc Natl Acad Sci USA. 2014;111:8428–8433. doi: 10.1073/pnas.1406855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCusker EC, et al. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naylor CE, et al. Molecular basis of ion permeability in a voltage-gated sodium channel. EMBO J. 2016;35:820–830. doi: 10.15252/embj.201593285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Payandeh J, El-din TMG, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaya D, et al. Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J Mol Biol. 2014;426:467–483. doi: 10.1016/j.jmb.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai Cj, et al. Two alternative conformations of a voltage-gated sodium channel. J Mol Biol. 2013;425:4074–4088. doi: 10.1016/j.jmb.2013.06.036. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen H, et al. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science. 2017;355:1–12. doi: 10.1126/science.aal4326. [DOI] [PubMed] [Google Scholar]
- 24.Hille B. Local anesthetics: Hydrophilic and hydrophobic pathways for the drug-receptor reaction. J Gen Physiol. 1977;69:497–515. doi: 10.1085/jgp.69.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahern CA, Eastwood AL, Dougherty DA, Horn R. Electrostatic contributions of aromatic residues in the local anesthetic receptor of voltage-gated sodium channels. Circulations Res. 2008;102:86–94. doi: 10.1161/CIRCRESAHA.107.160663. [DOI] [PubMed] [Google Scholar]
- 26.Gingrich KJ, Beardsley D, Yue DT. Ultra-deep blockade of Na+ channels by a quaternary ammonium ion: Catalysis by a transition-intermediate state? J Physiol. 1993;471:319–341. doi: 10.1113/jphysiol.1993.sp019903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimbrough JT, Gingrich KJ. Quaternary ammonium block of mutant Na+ channels lacking inactivation: Features of a transition-intermediate mechanism. J Physiol. 2000;529:93–106. doi: 10.1111/j.1469-7793.2000.00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNulty MM, et al. Charge at the lidocaine binding site residue Phe-1759 affects permeation in human cardiac voltage-gated sodium channels. J Physiol. 2007;581:741–755. doi: 10.1113/jphysiol.2007.130161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pless SA, Galpin JD, Frankel A, Ahern CA. Molecular basis for clas Ib anti-arrhythmic inhibition of cardiac sodium channels. Nat Commun. 2011;2:351–359. doi: 10.1038/ncomms1351. [DOI] [PubMed] [Google Scholar]
- 30.Ragsdale DS, Mcphee JC, Scheuer T, Catterall WA. Molecular determinants of state-dependent block of nal channels by local anesthetics. Science. 1994;265:1724–1728. doi: 10.1126/science.8085162. [DOI] [PubMed] [Google Scholar]
- 31.Ragsdale DS, Mcphee JC, Scheuer T, Catterall WA. Common molecular determinants of local anesthetic, antiarrhythmic, and anticonvulsant block of voltage-gated Na + channels. Proc Natl Acad Sci USA. 1996;93:9270–9275. doi: 10.1073/pnas.93.17.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zamponi GW, Doyle DD, French RJ. Fast lidocaine block of cardiac and skeletal muscle sodium channels: One site with two routes of access. Biophys J. 1993;65:80–90. doi: 10.1016/S0006-3495(93)81042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raju SG, Barber AF, Lebard DN, Klein ML, Carnevale V. Exploring volatile general anesthetic binding to a closed membrane-bound bacterial voltage-gated sodium channel via computation. PLoS Comput Biol. 2013;9:e1003090. doi: 10.1371/journal.pcbi.1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin LJ, Corry B. Locating the route of entry and binding sites of benzocaine and phenytoin in a bacterial voltage gated sodium channel. PLoS Comput Biol. 2014;10:e1003688. doi: 10.1371/journal.pcbi.1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boiteux C, et al. Local anesthetic and antiepileptic drug access and binding to a bacterial voltage-gated sodium channel. Proc Natl Acad Sci USA. 2014;111:13057–13062. doi: 10.1073/pnas.1408710111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith NE, Corry B. Mutant bacterial sodium channels as models for local anesthetic block of eukaryotic proteins. Channels. 2016;10:225–237. doi: 10.1080/19336950.2016.1148224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hille B. Ion Channels of Excitable Membranes. Sinauer Associates, Inc.; Sunderland, MA: 2001. [Google Scholar]
- 38.Porasso RD, Bennett WFD, Lo JJ. Study of the benzocaine transfer from aqueous solution to the interior of a biological membrane. J Phys Chem B. 2009;113:9988–9994. doi: 10.1021/jp902931s. [DOI] [PubMed] [Google Scholar]
- 39.Cascales JJL, et al. Thermodynamic study of benzocaine insertion into different lipid bilayers. J Chem Phys. 2011;135:135103. doi: 10.1063/1.3643496. [DOI] [PubMed] [Google Scholar]
- 40.Martin LJ, Chao R, Corry B. Molecular dynamics simulation of the partitioning of benzocaine and phenytoin into a lipid bilayer. Biophysical Chem. 2014;185:98–107. doi: 10.1016/j.bpc.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Högberg Cj, Maliniak A, Lyubartsev AP. Dynamical and structural properties of charged and uncharged lidocaine in a lipid bilayer in a lipid bilayer. Biophys Chem. 2007;125:416–424. doi: 10.1016/j.bpc.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Högberg CJ, Lyubartsev AP. Effect of local anesthetic lidocaine on electrostatic properties of a lipid bilayer. Biophysical J. 2008;94:525–531. doi: 10.1529/biophysj.107.104208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mojumdar EH, Lyubartsev AP. Biophysical chemistry molecular dynamics simulations of local anesthetic articaine in a lipid bilayer. Biophys Chem. 2010;153:27–35. doi: 10.1016/j.bpc.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Barber AF, Liang Q, Amaral C, Treptow W, Covarrubias M. Molecular mapping of general anesthetic sites in a voltage-gated ion channel. Biophys J. 2011;101:1613–1622. doi: 10.1016/j.bpj.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Friesner RA, Berne BJ. Replica exchange with solute scaling: A more efficient version of replica exchange with solute tempering ( REST2 ) J Phys Chem B. 2011;115:9431–9438. doi: 10.1021/jp204407d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jo S, Jiang W. A generic implementation of replica exchange with solute tempering (REST2) algorithm in NAMD for complex biophysical simulations. Computer Phys Commun. 2015;197:304–311. [Google Scholar]
- 47.Walczewska-Szewc K, Deplazes E, Corry B. Comparing the ability of enhanced sampling molecular dynamics methods to reproduce the behavior of fluorescent labels on proteins. J Chem Theory Comput. 2015;11:3455–3465. doi: 10.1021/acs.jctc.5b00205. [DOI] [PubMed] [Google Scholar]
- 48.Corry B, Thomas M. Mechanism of ion permeation and selectivity in a voltage-gated sodium channel. JACS. 2012;134:1840–1846. doi: 10.1021/ja210020h. [DOI] [PubMed] [Google Scholar]
- 49.Boiteux C, Vorobyov I, Allen TW. Ion conduction and conformational flexibility of a bacterial voltage-gated sodium channel. Proc Natl Acad Sci USA. 2014;111:1840–1846. doi: 10.1073/pnas.1320907111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furini S, Domene C. On conduction in a bacterial sodium channel. PLoS Comput Biol. 2012;8:e1002476. doi: 10.1371/journal.pcbi.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tikhonov DB, Zhorov BS. Mechanism of sodium channel block by local anaesthetics, antiarrhythmics, and anticonvulsants. J Gen Physiol. 2017;149:1–17. doi: 10.1085/jgp.201611668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee S, Goodchild SJ, Ahern CA. Local anesthetic inhibition of a bacterial sodium channel. J Gen Physiol. 2012;139:507–516. doi: 10.1085/jgp.201210779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaczmarski JA, Corry B. Investigating the size and dynamics of voltage-gated sodium channel fenestrations. Channels. 2014;8:264–277. doi: 10.4161/chan.28136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayne CG, Saam J, Schulten K, Tajkhorshid E, Gumbart JC. Rapid parameterization of small molecules using the Force Field Toolkit. J Chem Theory Comput. 2014;34:1–28. doi: 10.1002/jcc.23422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 56.Vanommeslaeghe K, Mackerell AD. Automation of the CHARMM general force field (CGenFF) I: Bond perception and atom typing. J Chem Inf Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanommeslaeghe K, Prabhu Raman E, MacKerell AD., Jr Automation of the CHARMM General Force Field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J Chem Inf Model. 2012;52:3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanommeslaeghe K, et al. CHARMM general force field (CGenFF): A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guvench O, MacKerell AD., Jr Automated conformation energy fitting for force-field development. J Mol Model. 2008;14:667–679. doi: 10.1007/s00894-008-0305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bennett CH. Efficient estimation of free energy differences from Monte Carlo data. J Comput Phys. 1976;22:245–268. [Google Scholar]
- 61.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Inf Model. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 62.Jorgensen WL, et al. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 63.Torrie GM, Valleau JP. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J Comput Phys. 1977;23:187–199. [Google Scholar]
- 64.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 65.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patriksson A, van der Spoel. A temperature predictor for parallel tempering simulations. Phys Chem Chem Phys. 2008;10:2073–2077. doi: 10.1039/b716554d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.