Significance
Bacterial viruses (bacteriophages) append a variety of molecules, including sugars, amino acids, and polyamines, to the nucleobases of their genomic DNA to circumvent the endonuclease-based defenses of their hosts. These DNA hypermodifications are formed through bacteriophage-encoded biosynthetic pathways, with steps occurring before and after replication of bacteriophage DNA. We report here the discovery of two thymidine hypermodifications: 5-(2-aminoethoxy)methyluridine replacing 40% of thymidine nucleotides in the Salmonella phage ViI and 5-(2-aminoethyl)uridine replacing 30% of thymidine in the DNA of the Pseudomonas phage M6. Additionally, we show in vitro reconstitution of 5-(2-aminoethyl)uridine biosynthesis from five recombinantly expressed proteins. These findings reveal an expanded diversity in the types of naturally occurring DNA modifications and their biosynthetic pathways.
Keywords: DNA modification, bacteriophage, hypermodification, pyrimidine
Abstract
Certain viruses of bacteria (bacteriophages) enzymatically hypermodify their DNA to protect their genetic material from host restriction endonuclease-mediated cleavage. Historically, it has been known that virion DNAs from the Delftia phage ΦW-14 and the Bacillus phage SP10 contain the hypermodified pyrimidines α-putrescinylthymidine and α-glutamylthymidine, respectively. These bases derive from the modification of 5-hydroxymethyl-2′-deoxyuridine (5-hmdU) in newly replicated phage DNA via a pyrophosphorylated intermediate. Like ΦW-14 and SP10, the Pseudomonas phage M6 and the Salmonella phage ViI encode kinase homologs predicted to phosphorylate 5-hmdU DNA but have uncharacterized nucleotide content [Iyer et al. (2013) Nucleic Acids Res 41:7635–7655]. We report here the discovery and characterization of two bases, 5-(2-aminoethoxy)methyluridine (5-NeOmdU) and 5-(2-aminoethyl)uridine (5-NedU), in the virion DNA of ViI and M6 phages, respectively. Furthermore, we show that recombinant expression of five gene products encoded by phage ViI is sufficient to reconstitute the formation of 5-NeOmdU in vitro. These findings point to an unexplored diversity of DNA modifications and the underlying biochemistry of their formation.
The tailed viruses of bacteria (bacteriophages or phages) contain the greatest variety of known biologically derived deoxyribonucleotide modifications (1) compared with any group of cellular organisms. In addition to methylated bases (e.g., N6-methyladenosine, N4-methylcytosine, and 5-methylcytosine) commonly found in cellular organisms, bacteriophages can also contain modifications ranging in size from an amino group appended to the N2 position of adenosine (N2-aminoadenine) as found in the Synechocystis phage S-2L (2) to complex hypermodifications, such as glucosyl-phosphoglucuronolactone-dihydroxypentauracil, found in the Bacillus phage SP-15 (3, 4)—this latter modification is predicted to append a 488-Da side chain to the thymidine nucleotide.
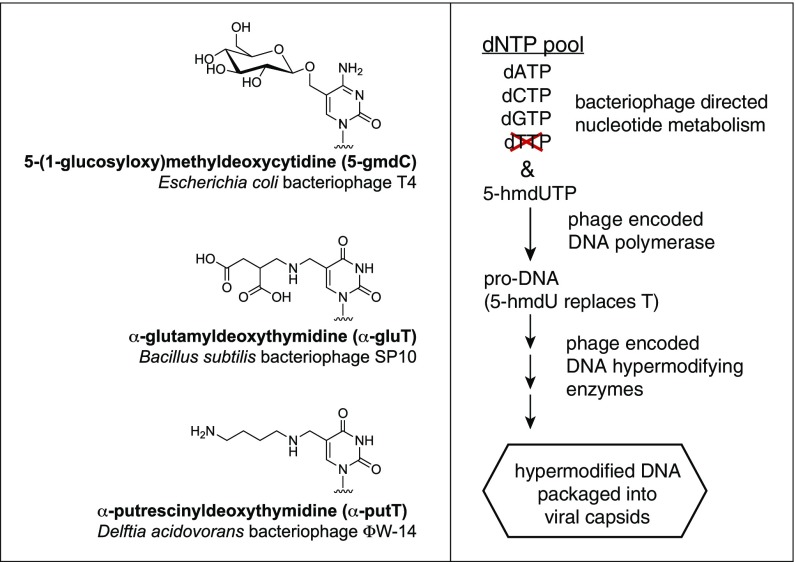
Details of the genetic and biochemical basis for DNA hypermodification have been determined in some bacteriophages, including the Escherichia phage T4, the Delftia phage ΦW-14, and the Bacillus phage SP10 (reviewed in ref. 1), with hypermodifications that are represented in Fig. 1. The phages ΦW-14 and SP10 hypermodify their DNA bases through a combination of pre- and postreplicative mechanisms (schematically represented in Fig. 1) (1). These two phages synthesize 5-hmdUMP from dUMP using a thymidylate synthase homolog—dUMP hydroxymethyltransferase (5); 5-hmdUMP is converted to 5-hmdUTP and incorporated into newly replicated phage DNA by a phage-encoded DNA polymerase. The 5-hmdU bases are then converted in situ to either thymidine or the hypermodified base in a sequence-specific fashion (6, 7). How 5-hmdU is converted to thymidine during phage replication is still unknown. The hypermodification reaction proceeds through a two-step modification of 5-hmdU on the DNA polymer coincident with or immediately after DNA replication (7, 8). A phage-encoded kinase (or pyrophosphotransferase) modifies 5-hmdU at the hydroxymethyl moiety to yield 5-((O-pyrophosphoryl)hydroxymethyl)uridine (5-PPmdU) (7, 8). The pyrophosphorylation of 5-hmdU could activate the 5-methylene carbon for nucleophilic substitution, with the pyrophosphate serving as a leaving group.
Fig. 1.
Examples of phage hypermodified pyrimidines and the generalized DNA thymidine (T) hypermodification pathway of phages SP10 and ΦW-14.
Previous work by Aravind and coworkers (9) predicted an enzyme family responsible for the formation of 5-PPmdU from 5-hmdU. In that work, ORFs proposed to encode kinases synthesizing 5-PPmdU were identified in ΦW-14, SP10, the Salmonella phage ViI, and the Pseudomonas phage M6 in addition to other bacteriophage genomes. For the purposes of discussion in this work, we will refer to this enzyme as 5-hmdU DNA kinase (5-HMUDK). We reasoned that the bacteriophages M6, ViI, and others encoding 5-HMUDK homologs should contain thymidine hypermodifications that would be either identical or similar to those found in ΦW-14 and SP10. In the study reported here, we describe the discovery of two thymidine hypermodifications, 5-(2-aminoethyl)uridine (5-NedU) and 5-(2-aminoethoxy)methyluridine (5-NeOmdU), in virion DNA from M6 and ViI, respectively. The chemical nature of these modifications is identified and confirmed using liquid chromatography-tandem MS (LC-MS/MS), NMR, chemical synthesis, and amine-reactive labeling. Furthermore, we reconstitute the biosynthesis of 5-NeOmdU in vitro using recombinant Escherichia coli extracts expressing candidate DNA hypermodification genes identified here and by Aravind and coworkers (9).
Results
Distinct Groups of Phage Encode a Putative 5-HMUDK.
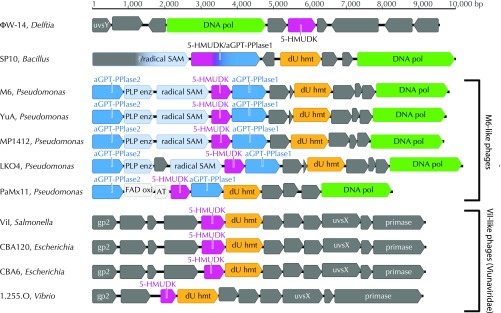
Bacteriophages ΦW-14 and SP10 encode a putative 5-HMUDK, first predicted by Aravind and coworkers (9), to participate in the biosynthesis of hypermodified thymidine derivatives. We hypothesized that other 5-HMUDK–encoding phages would similarly contain thymidine hypermodifications in their DNAs. In an effort to characterize the extent and diversity of the bacteriophage genomes encoding this kinase, homologs of SP10 and ΦW-14 5-HMUDKs were identified using BLAST (10). Fully sequenced bacteriophage genome sequences encoding a putative 5-HMUDK were collected from GenBank (11) (Table 1), and the regions surrounding the kinase gene were visualized as genomic maps. Notably, many genomes showed a high degree of conservation not only in their overall gene content but in their gene organization as well. This synteny is illustrated in Fig. 2 through subgenomic maps detailing the regions surrounding the 5-HMUDK gene of a subset of the phage genomes listed in Table 1.
Table 1.
Selected bacteriophages encoding putative 5-HMUDK
| Phage | Host | Accession no. | Morphotype | Source | Location | Ref. |
| ΦW-14 | Delftia acidovorans | NC_013697 | Myoviridae | Sewage | Vancouver, Canada | 12 |
| SP10 | Bacillus subtilis W23 | AB605730 | Myoviridae | Soil | Unpublished | 13 |
| Vi1-like | ||||||
| SJ3 | Salmonella enterica serovar Typhimurium g4232 | NC_024122 | Myoviridae | Wastewater | Indiana, United States | 14 |
| Marshall | S. enterica serovar Enteritidis | NC_022772 | Myoviridae | Sewage | Texas, United States | 15 |
| 0507-KN2-1 | Klebsiella pneumoniae Ca0507 (KN2) | NC_022343 | Myoviridae | Sewage | Taipei City, Taiwan | 16 |
| MAM1 | Serratia plymuthica A153 | NC_020083 | Myoviridae | Sewage | Cambridge, United Kingdom | 17 |
| LIMEstone | Dickeya solani | NC_019925 | Myoviridae | Crop soils | Merelbeke, Belgium | 18 |
| SKML-39 | S. enterica | NC_019910 | Myoviridae | Estuary | Baltimore, United States | 69 |
| ΦSH19 | S. typhimurium U288 | NC_019530 | Myoviridae | Sewage | United Kingdom | 19 |
| PhaxI | E. coli O157:H7 | NC_019452 | Myoviridae | Sewage | Tehran, Iran | 20 |
| CBA120 | E. coli O157:H7 (NCTC 12900) | NC_016570 | Myoviridae | Cattle feces | Southwest United States | 21 |
| SFP10 | E. coli O157:H7 | NC_016073 | Myoviridae | Slurry | Seoul Forest, South Korea | 22 |
| Vi01 | S. enterica serovar Typhi | NC_015296 | Myoviridae | Human feces | Canada | 23 |
| ΦSboM-AG3 | Shigella boydii | NC_013693 | Myoviridae | Wastewater | Guelph, Canada | 24 |
| SJ2 | Salmonella serovar Enteritidis C721 | KJ174317 | Myoviridae | Chicken egg | Guelph, Canada | 25 |
| Maynard | S. enterica serovar Typhimurium | KF669654 | Myoviridae | Sewage | Texas, United States | 26 |
| ECML-4 | E. coli O157:H7 | JX128257 | Myoviridae | Harbor water | Baltimore, United States | 70 |
| 1.255.O | V. splendidus 10N.286.45.F1 | MG592621 | Myoviridae | Seawater | Massachusetts, United States | 27 |
| CBA6 | E. coli O157:H7 (NCTC 12900) | KM389296 | Myoviridae | Cattle feces | Southwest, United States | 21 |
| M6-like | ||||||
| M6 | P. aeruginosa M6,9 | NC_007809 | Siphoviridae | Unknown | Unknown | 28 |
| YuA | P. aeruginosa PAO1 | NC_010116 | Siphoviridae | Pond water | Moscow, Russia | 29 |
| MP1412 | P. aeruginosa PAO1 | NC_018282 | Siphoviridae | Sewage water | South Korea | 30 |
| PAE1 | P. aeruginosa strain PAO9505 | NC_028980 | Siphoviridae | Sewage water | Bendigo, Australia | 31 |
| LKO4 | P. aeruginosa | KC758116 | Siphoviridae | Unpublished | Greece | |
| AN14 | Pseudomonas | KX198613 | Siphoviridae | Freshwater | Lake Baikal, Siberia | |
| Ab18 | P. aeruginosa PAO1 | NC_026594 | Siphoviridae | Sewage | Abidjan, Côte d’Ivoire | 32 |
| PaMx11 | P. aeruginosa PAO1 | NC_028770 | Siphoviridae | Wastewater | Central Mexico | 33 |
Fig. 2.
Genomic maps of the regions surrounding the 5-HMUDK gene of bacteriophages listed in Table 1. GenBank accession numbers as well as the name and host of the bacteriophages are indicated. Notably, 5-HMUDK coassociates with dUMP hydroxymethyltransferase (dU hmt), a member of the thymidylate synthase superfamily.
From this analysis, four distinct groups of phages emerged. At the time that this analysis was performed, the hypermodified phages SP10 and ΦW-14 were essentially unique (i.e., they do not share syntenic regions of nucleotide sequence homology with any other phage encoding a putative 5-HMUDK). Of two other groups of phages encoding 5-HMUDK, one is composed exclusively of phages belonging to the Viunaviridae genus of dsDNA bacteriophages (34, 35). This group is defined by relatedness to the Salmonella phage ViI, a phage originally isolated in 1936 (23) and used by others in phage typing schemes for characterizing Salmonella serotypes (36). The members of the other 5-HMUDK–encoding phage group are all related in sequence and gene organization to the Pseudomonas phage M6 (28, 37), a siphovirus (38, 39) also originally isolated for use in phage typing schemes. We chose Salmonella phage ViI and Pseudomonas phage M6 (available from the ATCC) (Materials and Methods) as representatives of each of these groups for our studies.
Phages ViI and M6 Have Noncanonical Bases.
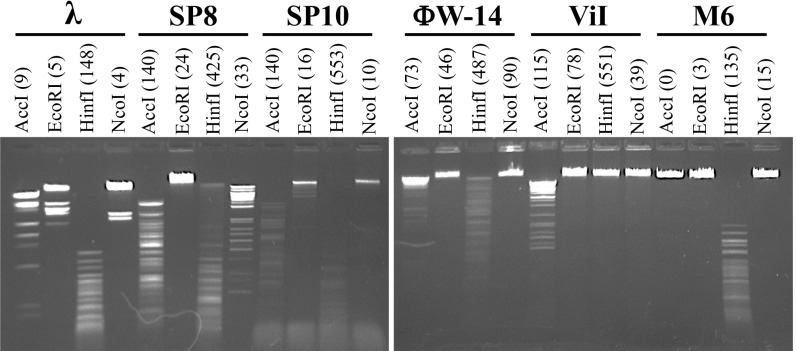
Bacteriophage hypermodified DNAs are often resistant to a range of restriction endonucleases in vitro (40). As seen in Fig. 3, genomic DNA extracted from Salmonella phage ViI and Pseudomonas phage M6 is resistant to cleavage by certain classical type II restriction enzymes. For example, NcoI does not cleave DNA from phages ViI and M6, although it can cleave canonical DNA from bacteriophage λ as well as DNA from the Bacillus phage SP8, the latter containing 5-hmdU fully substituting for thymidine (41). Additionally, NcoI does not cleave hypermodified DNAs of Bacillus phage SP10 and Delftia phage ΦW-14. DNAs of ViI and M6 are resistant to cleavage by EcoRI, although this enzyme is also inhibited by 5-hmdU. Although EcoRI is sensitive to CpG methylation, NcoI is not inhibited by dam, dcm, or cytidine–guanosine sequence dinucleotide (CpG) methylation, further suggesting the presence of noncanonical nucleotides in M6 and ViI.
Fig. 3.
Restriction digests of modified and unmodified bacteriophage genomic DNAs. Genomic DNA extracted from the indicated bacteriophages was incubated with restriction enzymes AccI, EcoRI, HinfI, and NdeI. The predicted number of cut sites in each bacteriophage sequence is shown in parentheses next to the given enzyme; λ contains canonical DNA bases only. Phage SP8 DNA contains 5-hmdU, replacing thymidine. Phages SP10 and ΦW-14 contain hypermodified thymidines.
Interestingly, we find that three RNA guided DNA endonucleases of CRISPR-Cas systems each can cleave genomic DNA from phage ViI to varying degrees. Four of four single guide RNA (sgRNA)-targeted sequences of ViI are specifically cleaved by Streptococcus pyogenes Cas9, two of four sites are cleaved by Staphylococcus aureus Cas9, and at least two of four sites are cleaved by Lachnospiraceae bacterium Cas12a (also known as Cpf1) (SI Appendix, Fig. S1). We were not able to determine if the ViI sequences targeted for cleavage contained hypermodified bases or if any bases of the protospacer adjacent motif (PAM) sequences in ViI DNA were hypermodified.
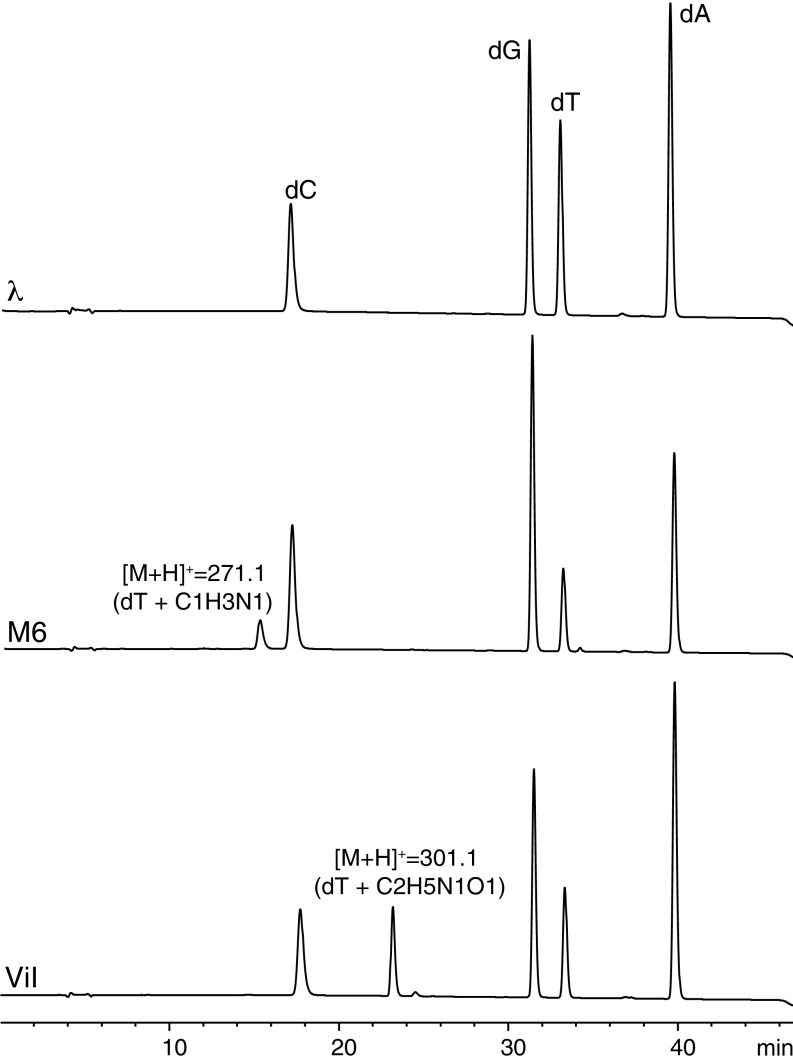
To determine if the restriction enzyme-inhibiting features on ViI and M6 DNAs indeed correspond to noncanonical bases, we performed LC-MS analysis of the nucleoside content obtained from enzymatic digestion of each phage genomic DNA. HPLC traces from each of these samples are shown in Fig. 4. In addition to the peaks corresponding to the canonical nucleosides 2′-deoxyadenosine, 2′-deoxyguanosine, 2′-deoxycytidine, and thymidine, the phages ViI and M6 each have a fifth peak. The retention time (23.2 min) and the mass (301 Da) of the fifth nucleoside peak of ViI are identical to those found for the ViI-like phages 1.255.O, CBA6, and CBA120 (SI Appendix, Fig. S1). The 301-Da nucleoside replaces ∼40% of thymidines in ViI and ViI-like DNA (SI Appendix, Table S1). The retention time and mass of the fifth nucleoside peak of phage M6 are 15.1 min and 271 Da, respectively. The 271-Da nucleoside replaces ∼30% of thymidines in M6 DNA (SI Appendix, Table S1).
Fig. 4.
HPLC traces and MS analysis of bacteriophage M6 and ViI nucleosides. The trace in Top was obtained from bacteriophage λ to show the retention of canonical nucleosides. M6 and ViI show a fifth major peak corresponding to the hypermodified base. The protonated molecular ion detected for each hypermodified base is indicated as well as a hypothetical combination of atoms to account for the observed masses. dA, 2′-deoxyadenosine; dG, 2′-deoxyguanosine; dC, 2′-deoxycytidine; dT, thymidine.
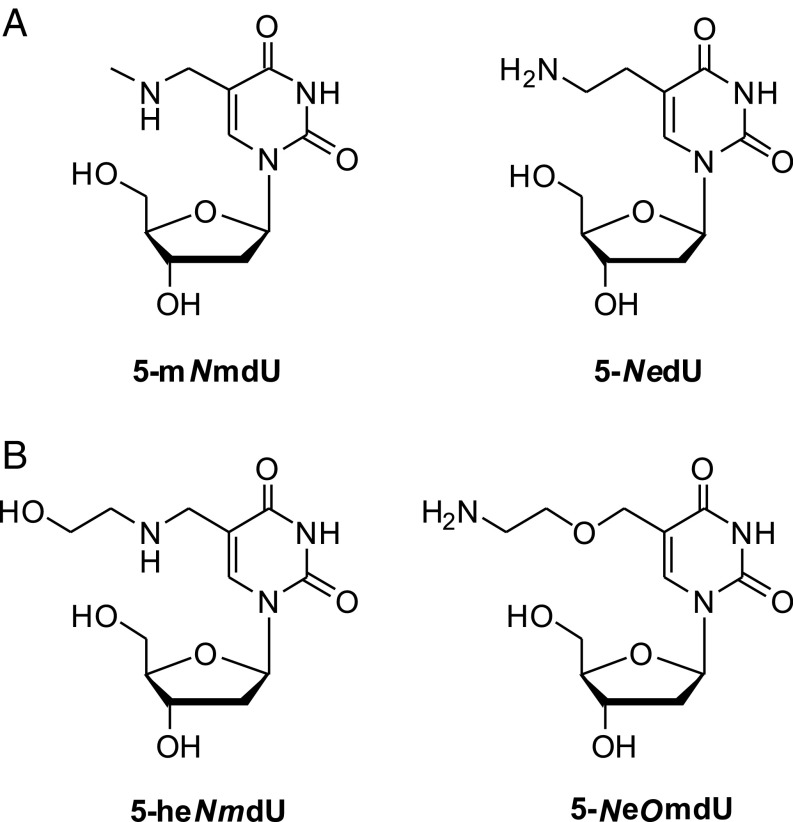
The mass difference between the ViI-modified base and thymidine corresponds to two additional carbons, five hydrogens, one nitrogen, and one oxygen. The difference between the M6-modified base and thymidine is one extra carbon, three hydrogens, and one nitrogen. Since the DNA of ΦW-14 and SP-10 contain thymidines hypermodified with putrescinyl and glutamyl moieties, respectively, via an N-C linkage (7, 42), we initially hypothesized that ViI and M6 would also feature N-C–bonded thymidine adducts. In the case of phage ViI, the adduct could originate from the appendage of a group of formula C2H5NO, such as ethanolamine, to give 5-((2-hydroxyethyl)amino)methyluridine (5-heNmdU). In the case of phage M6, we envisioned a modified base comprising a methylamine substitution to form 5-(methylamino)methyluridine (5-mNmdU). The proposed structures of these nucleosides are shown in Fig. 5.
Fig. 5.
Proposed structures of phage M6 (A) and ViI (B) modifications. 5-NedU and 5-NeOmdU are shown to be the actual modifications in this work.
To test whether these hypothetical structures indeed correspond to the native phage modifications, we chemically synthesized 5-mNmdU and 5-heNmdU according to a method adapted from Saavedra (43). Briefly, starting with commercially available 2′-deoxy-5-formyluridine, 5-mNmdU and 5-heNmdU were obtained by reductive alkylation with methyl amine or 2-ethanolamine, respectively, in the presence of NaBH4 (SI Appendix, Fig. S3). These compounds were purified by reverse-phase preparative HPLC and characterized by NMR and high-resolution MS (HRMS).
To our surprise, the synthetic and naturally modified nucleosides of both M6 and ViI, other than having identical masses, exhibited markedly different HPLC retention times. Peaks of synthetic 5-mNmdU and 5-heNmdU were detected at 12.0 and 12.8 min, respectively, while those of M6 and ViI were observed at 15.1 and 23.2 min. Per our hypothesis, the hypermodification of phages encoding a 5-HMUDK are derived from further elaboration of the hydroxyl moiety of 5-hmdU in DNA. The fact that our predicted structures did not match the ones obtained from M6 and ViI DNA digests suggested that these phage families may follow a different substitution pattern than the N-C–linked thymidine modifications encountered for phages ΦW-14 and SP-10. Hence, we presumed that the M6 modification could originate from an inverse relationship between the incoming C and N atoms as shown in Fig. 5A. The resulting 5-NedU would feature a terminal primary amine and a C-C linkage between the hypermodification and the thymine base. In the case of the ViI, the hypermodification presenting a terminal amine but connected to the thymine via an ether linkage to give 5-NeOmdU seemed the most plausible among several isomeric structures (Fig. 5B). For both 5-NedU (M6) and 5-NeOmdU (ViI), the proposed primary amino group could project into the major groove of DNA and interact favorably with the phosphate backbone.
Fragmentation Indicates a C-C Modification in Phage M6.
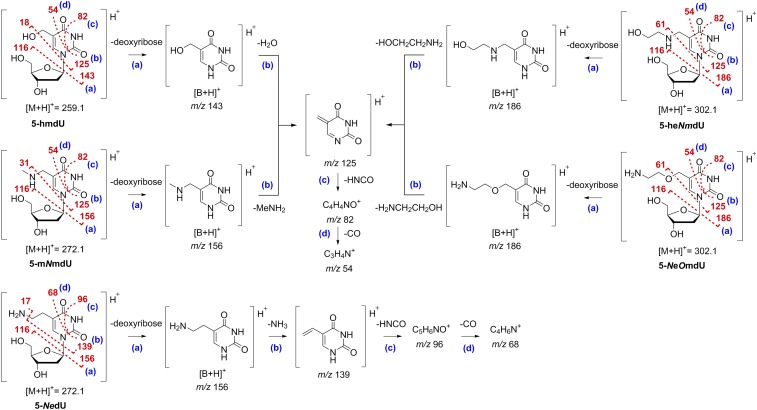
To further infer the atom connectivity of the hypermodified M6 and ViI nucleosides, we then performed a series of collision-induced dissociation (CID) MS/MS experiments. Electrospray ionization (ESI)-MS/MS spectra of 5-hmdU, 5-mNmdU, and the unknown hypermodified nucleoside from phage M6 are shown in SI Appendix, Figs. S4–S6. A generalized fragmentation pathway is shown in Fig. 6. Low-energy CID spectra (SI Appendix, Figs. S4, Upper, S5, row 1, and S6, row 1) show the formation of the [M + H]+ ions of 5-hmdU, 5-mNmdU, and the unknown M6 nucleoside at m/z 259, 272, and 272, respectively. The products corresponding to the cleavage of the glycosidic bond with loss of neutral 2′-deoxyribose and yielding protonated nucleobase [B + H]+ ions were detected at m/z 143, 156, and 156, respectively. Minor peaks resulting from ribose dehydration of [M + H]+ ions (e.g., at m/z 241, 213, and 205 for 5-hmdU and at m/z 223 and 205 for 5-mNmdU) as well as from the cleavage of the glycosidic bond with loss of the protonated 2′-deoxyribose ion (m/z 117) and its further dehydration ions (m/z 99 and 81) were also observed. The loss of neutral 2′-deoxyribose and, in the same vein, the presence of protonated 2′-deoxyribose ion at m/z 117 in the spectra provided conclusive evidence that the M6 modification was present at the nucleobase and not present at the sugar moiety. High-energy CID spectra (SI Appendix, Figs. S4, Lower, S5, row 4, and S6, row 4) indicated losses of water (5-hmdU) or methylamine (5-mNmdU) from the protonated nucleobase [B + H]+ ions to give origin to a common fragment ion at m/z 125. Subsequent losses of HNCO and CO produced ions at m/z 82 and 54, respectively, which are consistent with well-documented fragmentations for uracil derivatives (44–46).
Fig. 6.
CID pathways of protonated 5-hmdU, 5-mNmdU, 5-NedU, 5-heNmdU, and 5-NeOmdU. Proposed CID pathways are based on well-documented fragmentations for uracil derivatives.
In the case of the M6 modification, MS/MS spectra displayed fragment ions at m/z 156, 139, 96, and 68, which could be attributed similarly to the loss of 2′-deoxyribose to form [B + H]+ followed by losses of ammonia [B+H-NH3]+, HNCO [B+H-NH3-HNCO]+, and CO [B+H-NH3-HNCO-CO]+, respectively. Comparative analysis suggests the presence of an additional CH2 group in all main fragments, which is in agreement with the formation of a styrenoid cation (Fig. 6) (47), and supports the occurrence of a C-C–substituted thymine. It is also important to note that the similarity of the UV spectra (SI Appendix, Table S2) of the unknown M6 modification (λmax 266 nm) to that of thymine (λmax 265 nm) rather than isothymine (6-methyluracil, λmax 261 nm) (47) further strengthens our presumption that the uridine substitution is, in fact, at the five position. Finally, since the loss of NH3 directly from the uracil ring has been reported to be prohibited in 5-hmdU and related structures (46), the facile loss of ammonia observed for the unknown M6 nucleoside substantiated the presence of a terminal primary amine. All of the results taken together led us to conclude that the hypermodification found in phage M6 is 5-NedU.
Next, we compared the CID mass spectra of the unknown ViI modification with those of the synthetic 5-heNmdU. For both nucleosides, low-energy CID (SI Appendix, Figs. S7, row 1 and S8, row 1) showed the formation of two abundant ions at m/z 302 and 186, which correspond to [M + H]+ and [B + H]+, respectively. Additional fragmentation of protonated 5-heNmdU resulted in loss of ethanolamine to give rise to the fragment ion [B+H-HOCH2CH2NH2]+ at m/z 125. The peak corresponding to the protonated form of ethanolamine was also found at m/z 62. As was the case with 5-hmdU and 5-mNmdU, loss of HNCO via retro Diels–Alder followed by successive loss of CO was the major pathway of dissociation from the common ion at m/z 125, and it produced the fragment ions m/z 82 and 54 (Fig. 6 and SI Appendix, Figs. S7 and S8). In addition to [M + H]+ and [B + H]+, the fragmentation of the protonated unknown ViI nucleoside generated a series of fragment ions at m/z 125, 82, and 54 that paralleled those of 5-heNmdU. These results led us to surmise that 5-heNmdU and the unknown ViI modification shared the same uridine backbone but had an inverse arrangement of the heteroatom substituents at the five position, such as the one found in 5-NeOmdU. Since we could not obtain any further conclusive structural information from these fragmentation studies, we decided to isolate the unknown ViI nucleoside and perform NMR analysis in attempt to unambiguously elucidate its structure.
NMR Reveals an Ethoxy Linkage in ViI Modification.
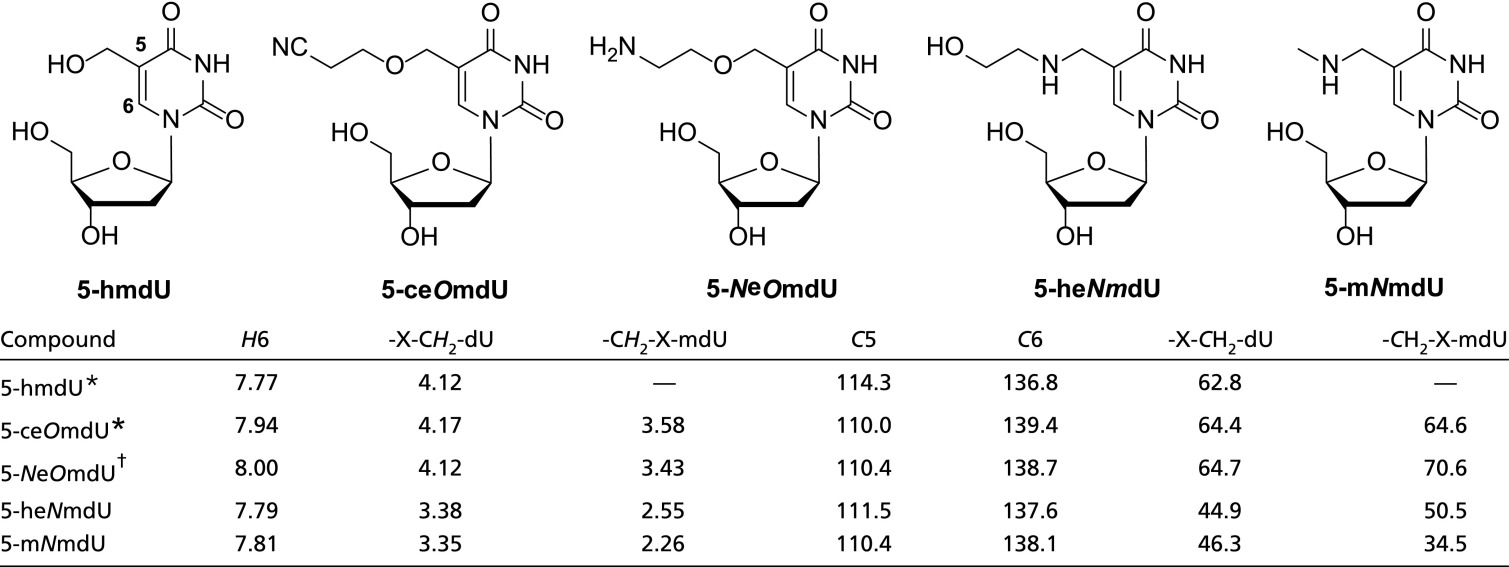
To obtain sufficient amounts of the unknown nucleoside for NMR analysis, 18 L of early log-phase host Salmonella typhi was infected with freshly prepared plaques of ViI at about one plaque per 250 mL. The resultant phage particles were purified, and their genomic DNA was extracted as described in Materials and Methods. Approximately 0.4 g of DNA was obtained and subsequently processed in four 0.1-g batches. DNA was extensively dialyzed against water, and then, 0.1 g of dialyzed genomic DNA was digested to free nucleosides. After HPLC purification using a gradient of ammonium acetate buffer, pH 4.5, and methanol and pooling the desired fractions, ∼9 mg of the unknown nucleoside was isolated from the total starting material. This nucleoside was subjected to 1H and 13C NMR analyses. We compared the NMR spectra of the unknown ViI nucleoside with those of the synthetic 5-heNmdU and 5-mNmdU as well as with those of the known compounds 5-hmdU and 5-(2-cyanoethoxy)methyluridine (5-ceOmdU) (48).
As depicted in Table 2, there is a strong correlation between the NMR chemical shifts of 5-ceOmdU and the proposed ViI nucleoside 5-NeOmdU and between the chemical shifts of 5-heNmdU and 5-mNmdU. The presence of an ethoxy modification (X = O) off the 5-methyl group of the thymine ring in 5-ceOmdU and 5-NeOmdU was evidenced by the downfield shift of the methylene hydrogens immediately adjacent to the oxygen atom [4.17 and 4.12 ppm for -OCH2-dU and 3.58 and 3.43 ppm for -CH2O-mdU, respectively relative to those of the compounds carrying an ethylamine modification (X = N)]. The cognate signals of 5-heNmdU and 5-mNmdU were detected at 3.38 and 3.35 ppm (-NCH2-dU) and 2.55 and 2.26 ppm (-CH2N-mdU), respectively (Table 2) (48). A small but consistent downfield shift was also observed for the thymine H6 of 5-ceOmdU (7.94 ppm) or 5-NeOmdU (8.00 ppm) relative to that of 5-heNmdU (7.79 ppm) or 5-mNmdU (7.81 ppm). All other 1H NMR signals from these four compounds were essentially identical (SI Appendix, Fig. S9).
Table 2.
Selected 1H and 13C NMR chemical shift data of 5-substituted uridines (in parts per million)
 |
From Hansen et al. (48).
Proposed structure for the unknown ViI nucleoside (this work). DMSO-d6 was used as internal standard.
A very similar outcome was revealed by 13C NMR analysis (Table 2). The ethoxy modification in 5-ceOmdU and 5-NeOmdU caused an expected downfield shift of signals in the vicinity of the oxygen atom [64.4 and 64.7 ppm (OCH2-dU) and 64.6 and 70.6 ppm (-CH2O-mdU) relative to the corresponding signals in the ethylamine-modified 5-heNmdU and 5-mNmdU, 44.9 and 46.3 ppm (-NCH2-dU) and 50.5 and 34.5 ppm (-CH2N-mdU), respectively] (Table 2). Again, a smaller but significant downfield shift was obtained for the thymine C6 of 5-ceOmdU (139.4 ppm) or 5-NeOmdU (138.7 ppm) relative to that of 5-heNmdU (137.6 ppm) or 5-mNmdU (138.1 ppm). No significant difference was observed for thymine C5 or any other 13C NMR signal from these four compounds (SI Appendix, Fig. S10). Based on these results, we, therefore, confirmed the identity of the unknown ViI nucleoside as 5-NeOmdU. It is worth noting that the reported 1H and 13C NMR signals for 5-hmdU [in particular, HOCH2-dU (4.12 ppm) and HOCH2-dU (62.8 ppm)] also corroborated well with the presence of an oxygen immediately after the methyl group of the thymine nucleobase. In support of the determination of the ViI modification as 5-NeOmdU (which should contain a terminal primary amino group), we find that ViI genomic DNA can be specifically labeled with the amine reactive dye 3-(4carboxybenzoyl)quinoline-2-carboxaldehyde (SI Appendix, Fig. S11).
Phage Genes Unique to Thymidine Hypermodifying Phages.
The finding that, like SP10 and ΦW-14, the ViI- and M6-like bacteriophages hypermodify their thymidines affords the possibility to narrow the search space for genes encoding thymidine hypermodification functions according to the following logic: only genes found within some or all of the hypermodifying phage genomes but not in genomes of phages containing nonhypermodified thymidines (λ, T4, and Bacillus phage SPO1) should be directly involved in the formation of hypermodified thymidines. Through pairwise BLAST comparisons and manual curation of lists of annotated reading frames from the thymidine hypermodifying phages M6, SP10, ViI, and ΦW-14, we found four classes of genes that appear to be unique to the thymidine hypermodifying phages (Table 3). These genes are predicted to encode the following products: a putative 5-HMUDK, a pyridoxal-phosphate (PLP)–dependent enzyme, a predicted radical S-adenosyl-l-methionine (SAM) enzyme, and an enzyme predicted to have a Helix-hairpin-Helix DNA base glycosylase fold by Phyre2 (49). These findings agree with observations and predictions previously made by Aravind and coworkers (9). The DNA base glycosylase-like gene product has been named by Aravind and coworkers (9) an “alpha-glutamyl/putrescinyl thymidine pyrophosphorylase” (aG/PT-PPlase) after its predicted activity toward 5-PPmdU during DNA hypermodification. Within the genomes that we examined (SP10, ΦW-14, ViI, M6, λ, T4, and Bacillus phage SPO1), the aG/PT-PPlase appeared to be restricted to the hypermodifying phages; however, homologs are also found within cellular organisms and viruses (9).
Table 3.
Genes unique to thymidine hypermodifying bacteriophages
| Predicted | ΦW-14 | SP10 | ViI | M6 |
| Kinase | gp37 (8683983) | gp186N* (14007383) | gp67 (10351655) | gp54 (5237077) |
| Kinase2 | N/d | gp218 (14007415) | gp243 (10351581) | N/d |
| PLP enzyme | N/d | N/d | gp226 (10351570) | gp52 (5237094) |
| Radical SAM | N/d | gp185 (14007382) | N/d | gp53 (5237052) |
| aG/PT-PPlase1 | gp72 (8684019) | gp186C* (14007383) | gp160 (10351543) | gp51 (5237089) |
| aG/PT-PPlase2 | gp109 (8684057) | N/d | gp247 (10351584) | gp55 (5237058) |
A National Center for Biotechnology Information Gene ID number is indicated in parentheses after the gene product number. N/d, none detected.
Domain fusion.
The locations of these putative hypermodification genes within their respective genomes are schematically shown as colored boxes in Fig. 2 and SI Appendix, Fig. S12. The M6-like family of phages shows the tightest clustering of genes predicted to function in hypermodification. Of this grouping, the Pseudomonas phages PAO1_Ab18 and PaMx11 differ in having predicted flavin adenine dinucleotide (FAD) and acetyl transferase genes instead of PLP-dependent and radical SAM enzyme-encoding genes at the orthologous locus (Fig. 2). Notably, the predicted hypermodification biosynthesis genes are often found near genes for dUMP hydroxymethyl transferase and phage-encoded DNA polymerase (indicated in yellow and green, respectively, in Fig. 2 and SI Appendix, Fig. S12).
Reconstitution of Base Hypermodification from Recombinant E. coli Lysates.
While working toward the in vitro reconstitution thymidine hypermodification, we first attempted to clone an apparent gene cluster of phage M6 containing five genes predicted to function in base hypermodification (Fig. 2 and column 5 in Table 3). However, we were unable to clone the entire operon as a single construct, possibly due to toxicity of one or more gene products encoded within the operon. However, we successfully cloned ViI genes encoding gene product 67 (gp67), gp160, gp226, gp243, and gp247 (Table 3) into plasmids for expression in E. coli under control of T7-inducible promoters. After confirming protein expression, individual lysates from cultures expressing each of these proteins were mixed together and incubated with biotinylated restriction fragments of genomic DNA from the Bacillus phage SP8, which contains 5-hmdU fully substituting for thymidine. After incubation, the biotinylated DNA was recovered using streptavidin-coated magnetic beads, washed in buffer, and subsequently digested to individual nucleosides for LC-MS analyses.
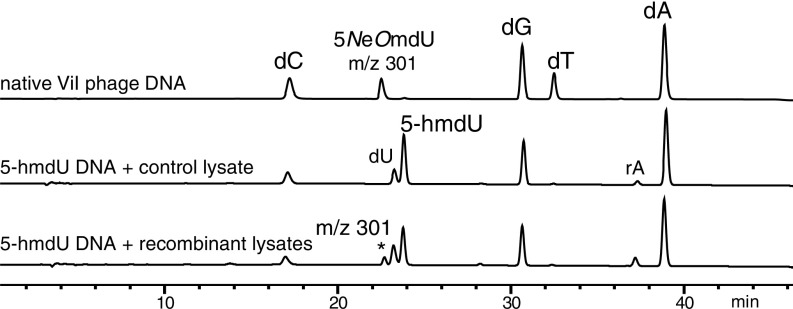
Fig. 7 shows HPLC traces from nucleoside digests of SP8 substrate DNA treated with nonrecombinant E. coli lysate (negative control), recombinant lysate-treated substrate DNA, and the native hypermodified DNA. Both traces obtained from the lysate-treated DNA contain a peak corresponding to 2′-deoxyuracil produced by background cytidine deaminase activity of the E. coli host lysate. Noticeably, substrate DNA treated with a mixture of the five lysates containing expressed gp67, gp160, gp226, gp243, and gp247 (Table 3) contained an additional peak of identical retention time and mass as 5-aminoethoxymethyl-2′-deoxyuridine found in native phage ViI genomic DNA. Thus, this combination of gene products, potentially together with unknown cytoplasmic cofactors, is sufficient to produce this hypermodification.
Fig. 7.
In vitro reconstitution of 5-NeOmdU from recombinant bacterial lysates. Substrate DNA containing 5-hmdU incubated in mixed lysates from E. coli expressing ViI gp67, gp160, gp226, gp243, and gp247 contains a nucleotide product (301; denoted by the asterisk) of identical mass and retention time as the native modification of ViI. dA, 2’-deoxyadenosine; dG, 2’-deoxyguanosine; dC, 2’-deoxycytidine; dT, thymidine; rA, ribo adenosine.
Discussion
The hypermodified thymidines described in this work imply an even wider variety of nucleotide modifications synthesized by bacteriophages and cells. Bacteriophage ViI and its closely related phages CBA6, CBA120, and 1.255.O contain a C-O bond linking the aminoethanoyl moiety to the 5-methyl group of thymidine (5-NeOmdU). The high degree of sequence similarity and synteny of these phages to other members of the ViI-like group listed in Table 1 and illustrated in Fig. 2 indicate that they likely contain 5-NeOmdU as well. The M6-modified base contains an aminomethyl group attached via a C-C linkage to the 5-methyl group of thymidine (5-NedU). Based on our findings, we predict here that Pseudomonas phages YuA, MP1412, PAE1, LKO4, and AN14 similarly contain 5-NedU. It should be noted that, within the M6 family of Pseudomonas phages discussed here, PAO1_Ab18 and PaMx11 have a predicted FAD-dependent oxidoreductase and an acetyltransferase gene replacing the predicted PLP-dependent enzyme and radical SAM enzyme-encoding genes found at the same location within the genomes of the other M6-like family members listed in Table 1 and diagrammed in Fig. 2. We hypothesize that these two phages contain hypermodifications that differ from the 5-NedU of phage M6.
Each of the four phage groups described here encodes a putative 5-HMUDK implicated in the hypermodification process (9). Despite the lack of similarity between SP10 and ΦW-14 gene arrangements, both of their hypermodifications are composed of thymidines modified via an N-C linkage off the 5-methyl group (7, 42). Although the attached glutamyl and putrescinyl moieties are different in their structure, the phages nonetheless use the same intermediate in the formation of the hypermodification (i.e., a pyrophosphorylated 5-hmdU) (7, 8). The occurrence of a 5-PPmdU intermediate would suggest a mechanism in which the hypermodifications are formed via displacement at a pyrophosphate-activated 5-hmdU. Given that 5-NeOdU contains an ether bond, it is tempting to imagine cytoplasmic metabolites bearing free amine or hydroxyl groups as donors for nucleophilic substitution at pyrophosphorylated 5-hmdU for an even greater variety of possible modifications. However, phage hypermodifications are not limited to N-C or O-C bonds. The 5-NedU modification of M6 contains a C-C bond. This hypermodification is more difficult to rationalize via a simple nucleophilic substitution. It remains to be determined if 5-NedU forms through a charged or radical intermediate, such as might be suggested by the presence of a radical SAM enzyme-encoding gene in the M6-like phages. Collectively, the modifications observed to date in 5-HMUDK–encoding phages are now known to encompass N-C, O-C, and C-C hypermodified pyrimidines.
We show here that five recombinantly expressed genes (ViI gp67, gp160, gp226, gp243, and gp247) are sufficient to reconstitute 5-NeOmdU synthesis in vitro using a crude lysate system. It is not yet known which of these five are necessary. Of the five genes, two pairs are paralogs (gp67 and gp243 are predicted 5-HMUDKs, and gp160 and gp247 are predicted to have a DNA glycosylase fold) (Table 3). This redundancy suggests that three classes of enzymatic activity are minimally necessary for thymidine hypermodification. The metabolic origin of the additional atoms constituting the ViI hypermodifying moiety has not yet been determined, although given that we can reconstitute 5-NeOmdU in lysates, it is likely a metabolite normally present in cytoplasm of E. coli. For ΦW-14, the putrescine moiety has been shown to derive from ornithine (50). Glutamyl thymidine is likely derived from glutamate, which is the most abundant solute in bacterial cells (51). Less obvious is how 5-NeOmdU and 5-NedU are derived. To the best of our knowledge, ethanolamine and methylamine are not present at above-micromolar levels in bacterial cells (51) and therefore, are unlikely to be the direct precursors to the hypermodifying moieties. Although PLP-dependent enzymes and radical SAM enzymes encompass very diverse functionalities, they are often found in amino acid metabolic pathways. The known modifications of ΦW-14 and SP10 utilize components of amino acid metabolism. Together with enzymes having similarity to DNA glycosylases, perhaps these enzymes utilize amino acids as precursors. For example, an ethanolamine group could be derived from the decarboxylation of serine, and a methylamine could be derived from the decarboxylation of glycine. These hypotheses await experimental testing.
An obvious possible function of thymidine hypermodification is to counteract bacterial immunity systems. For example, bacteriophage T4 contains glucosylated hydroxymethylcytidines that allow T4 DNA to resist cleavage by a variety of restriction endonucleases both in vitro (40) and in vivo (52). Presumably, the presence of glucose in the major groove of DNA sterically blocks either the recognition of DNA by endonucleases or the mechanism of cleavage. As seen in Fig. 3, thymidine hypermodifications in ViI and M6 also inhibit cleavage by certain restriction endonucleases. Resistance to host restriction would be expected to confer a fitness advantage to phages encoding thymidine hypermodification machinery, and the global distribution of phages encoding thymidine hypermodifications (Table 1) suggests that this is the case.
Less clear is the effect that the thymidine hypermodification might have on CRISPR-based adaptive immunity systems. We observed cleavage of ViI DNA by each of three Cas RNA guided DNA endonucleases in vitro (SI Appendix, Fig. S1). Engineered Cas9 systems in E. coli have been shown to restrict glucosylated hydroxymethylcytidine of phage T4 (53) in vivo, but under slightly different experimental conditions, T4 could bypass Cas9 cleavage (54). Thymidine hypermodifications could potentially interfere with at least three aspects of Cas endonuclease function: contacts between Cas endonuclease and DNA backbone during R-loop formation, pairing between the sgRNA and target sequence on DNA, and Cas-endonuclease–DNA interactions at the PAM site. Although the exact sequence motifs of thymidine hypermodifications are not yet known, the length of the target sequences (∼20 nt) and the fact that at least 8 of 12 sites targeted in our experiment are nonetheless cleaved (SI Appendix, Fig. S1) suggest that hypermodification does not interfere with Cas endonuclease binding and R-loop formation. However, it is not known if a hypermodification occurs within any of the PAM sequences of the DNA sites that we targeted. Therefore, we hypothesize that, if any inhibition of Cas endonuclease function by thymidine hypermodification was to occur, it would be mediated by steric interference between modified PAM sequences and the Cas endonuclease.
Bacteria are in a constant arms race with their viral predators (55), and the existence of the 5-NeOmdU and 5-NedU modifications points to the possibility of host mechanisms targeting these modifications. Certain strains of E. coli, such as CT596 and UT189, encode a hypermodification-dependent restriction endonuclease called GmrSD (56, 57). GmrSD specifically recognizes and cleaves glucosylated 5-hmdC of bacteriophage T4 but leaves canonical cytidine DNA intact. GmrSD likely contains distinct cleavage and reader domains that specifically recognize the modified base and hydrolyze the phosphodiester backbone, respectively (56). Some bacteria encode up to five GmrSD homologs, suggesting that paralogous members of this family may recognize different types of hypermodified pyrimidines (56), including the kinds described in this work.
In addition to potential roles in virus–host interactions, the thymidine hypermodifications may have structural effects on DNA stability of both the free polymer and that of DNA tightly packaged in the phage capsid. Putrescinyl thymidine, 5-NeOmdU, and 5-NedU each terminate in a solvent-exposed positively charged amino group likely located in the major groove. This positive charge might serve the function of countering the negative charge of the phosphodiester backbone to overcome repulsion and facilitate packaging of DNA in the virion capsid, a stabilizing function ordinarily provided by divalent metal cations, such as Mg2+ and Ca2+. Indeed, although the genome of ΦW-14 is ∼7% shorter than T4, it is packaged into a 28% smaller volume (58). Additionally, the Tm of DNA from ΦW-14 is 5 °C to 9 °C higher than expected based on percentage of guanosine and cytidine (GC) content (42). The hypermodifications of ViI and M6 might have similar physicochemical effects on their capsid-packaged genomic DNAs, potentially impacting phage viability under different environmental conditions.
Among the set of genes identified in Table 3, the putative kinase (5-HMUDK) and the DNA glycosylase-like genes are the only genes absolutely shared between SP10, ΦW-14, ViI, and M6. Homologs to these DNA glycosylase-like genes are found not only among bacteriophages but in eubacterial genomes as well (SI Appendix, Fig. S12). Recently, through a combination of bioinformatic analyses and chemical methods, a diverse class of naturally occurring queuosine-like 2′-deoxy-7-deazaguanosine–derived DNA modifications was discovered in the DNA of the E. coli phage 9g as well as in pathogenic enteric bacteria (59). These guanosine hypermodifications were shown to inhibit cleavage of phage 9g DNA by restriction endonucleases in vitro (60), and a genomic island including 2′-deoxy-7-deazaguanosine biosynthetic genes was shown to be a barrier to transformation of Salmonella by unmodified DNA in vivo (59), showing that viruses and cells can co-opt the same complex DNA modifications, each for their own survival. We observe homologs of ViI gp160 and gp247 distributed among viruses, basidiomycete fungi, and diverse eubacteria, including soil-dwelling actinomycetes and medically relevant enteric gamma-proteobacteria (SI Appendix, Fig. S13), suggesting a much larger, unexplored diversity of nucleotide modifications and biological roles. The approaches described here show that thymidine hypermodifications are experimentally tractable, and additional investigation of their biosynthesis may reveal nucleic acid biochemistries, uncover biological functions, and lead to innovative experimental tools for nucleic acid manipulation.
Materials and Methods
Chemicals, Media, Enzymes, and Microbes.
Unless otherwise specified, all chemicals were obtained from Sigma-Aldrich and used without additional purification. Media and media components were sourced from Beckton Dickinson-Difco. All enzymes used in this study were obtained from New England Biolabs (NEB). The following strains were purchased from the American Type Culture Collection (ATCC): bacteriophage ViI (ATCC 27870-B1), the ViI host Salmonella enterica ssp. enterica serovar Typhi (ATCC 27870), bacteriophage M6 (ATCC BAA-31-B1), and the M6 host Pseudomonas aeruginosa (ATCC BAA-31). Vibrio phage 1.255.O._10N.286.45.F1 (hereafter referred to as 1.255.O) and its host, Vibrio splendidus 10N.286.45.F1, were isolated from littoral zone seawater at Canoe Cove (Nahant, MA) on October 13, 2010 (27). Bacteriophages CBA6 and CBA120 as well as their host E. coli Serotype O157:H7 National Collection of Type Cultures (NCTC) strain 12900 (also known as ATCC 700728) were provided by Elizabeth Kutter, Evergreen State College, Olympia, WA.
Growth and Purification of Bacteriophages.
Escherichia and Salmonella were grown in Rich Broth composed of 10 g tryptone, 5 g NaCl, and 5 g yeast extract per 1 L. Pseudomonas was grown on trypticase soy broth (61). Vibrio was cultured in Zobell Marine Broth 2216 (61). For solid medium, bactoagar was added at 1.5% to the liquid medium base before autoclaving. Large-scale bacteriophage lysates were prepared as described previously (62), purified by CsCl density ultracentrifugation, and purified according to previous methods (63). Additional details of phage purification and preparation of phage genomic DNA are given in SI Appendix.
Isolation and Characterization of Bacteriophage-Modified Nucleosides.
Phage ViI.
Dialyzed phage ViI DNA (100 mg) was digested at 0.1 mg/mL final concentration to nucleosides by treatment overnight at 37 °C with the Nucleoside Digestion Mix in 1 mL of 1× Nucleoside Digestion Mix buffer (NEB). After passage through a 3-kDa molecular mass cutoff Vivaspin 20 spin column (GE Healthcare Bio-Sciences) to remove proteins, the flow through was collected and passed through a 0.22-µm membrane (Corning) to remove any particles before HPLC purification. The filtrate was lyophilized to dryness and then redissolved in water. The phage ViI hypermodified nucleoside was purified by reversed-phase preparative HPLC on a Waters Atlantis T3 (Waters Corp.) semipreparative column (19 × 250 mm, 5 µm) as described in the SI Appendix. A white powder was obtained after lyophilization. HRMS m/z calculated for C12H19N3O6 [M + H]+, 302.1347; observed, 302.1366. 1H NMR (500 MHz, DMSO-d6) δ 8.00 (s, 1H), 6.16 (t, J = 6.7 Hz, 1H), 4.25 (q, J = 4.1 Hz, 1H), 4.18–4.04 (m, 2H), 3.78 (q, J = 3.3 Hz, 1H), 3.60 (dd, J = 11.9, 3.5 Hz, 1H), 3.55 (dd, J = 11.8, 3.5 Hz, 1H), 3.43 (t, J = 5.1 Hz, 2H), 2.74 (s, 2H), 2.10 (dd, J = 6.8, 4.6 Hz, 2H). 13C NMR (126 MHz, DMSO-d6) δ 162.7, 150.3, 138.7, 110.4, 87.5, 84.2, 70.6, 70.3, 64.7, 61.1, 40.5.
Phages 1.255.O, CBA6, CBA120, and M6.
Five micrograms of virion DNA from each of the phages 1.255.O, CBA6, CBA120, and M6 DNA were digested to nucleosides at 0.1–0.2 μg/μL final concentration by treatment overnight at 37 °C with 5 μL of the Nucleoside Digestion Mix in 1× Nucleoside Digestion Mix Reaction Buffer (NEB). The resulting DNA nucleoside mixtures were directly analyzed by reversed-phase LC/MS without additional purification. Phage M6 nucleosides were also analyzed by fragmentation-based LC-MS/MS. Phages 1.255.O, CBA6, and CBA120: MS m/z C12H19N3O6 [M + H]+, 302.1; observed, 302.1. Phage M6: MS m/z C11H17N3O5 [M + H]+, 272.1; observed, 272.1.
Chemical Synthesis of Nucleoside Standards.
5-mNmdU and 5-heNmdU were synthesized following a method adapted from Saavedra (43). Additional details of their synthesis are given in SI Appendix.
LC-MS/MS and Fragmentation Analysis.
LC-MS/MS was performed on an Agilent 1290 ultrahigh performance liquid chromatograph equipped with a G4212A diode array detector and a 6490A triple quadrupole mass detector operating in the positive ESI mode. Ultra-HPLC was performed on a Waters XSelect HSS T3 XP column (2.1 × 100 mm, 2.5-µm particle size) at a flow rate of 0.6 mL/min with a binary gradient from 1% solvent A (10 mM ammonium formate, pH 4.4) to 100% B (methanol). Absorbance was monitored at 260 nm. MS/MS fragmentation spectra were recorded in the positive product–ion mode with the following parameters: gas temperature 200 °C, gas flow 14 L/min, nebulizer 45 psi, sheath gas temperature 350 °C, sheath gas flow 11 L/min, capillary voltage 2 kV, nozzle voltage 1.5 kV, fragmentor voltage 380 V, and collision energy 5–65 V.
Computational Methods.
Homology searches were performed using web- and local application-based implementations of BLAST and position-specific iterated BLAST (10, 64). Multiple sequence alignments were generated using MAFFT (65) with the JTT100 scoring matrix (66). Phylogenetic trees were generated using PHYML with the LG substitution model (67). Most bioinformatic operations described were performed within the Geneious software package (68). Protein function and fold were predicted using the Phyre2 structure prediction server (49).
Preparation and Assay of Hypermodification Reactions.
SP8 genomic DNA was digested to completion with restriction endonuclease HpyCH4IV according to the manufacturer's guidelines. After heat inactivation at 80 °C for 20 min, the digest was purified using a Monarch nucleic acid purification kit (NEB). Fragmented SP8 genomic DNA at a final concentration of 200 ng/µL was biotinlylated by incubating 1 U/µL of Taq DNA polymerase containing 50 µM biotin-16-dCTP at 68 °C for 3 h in 1× ThermoPol buffer (NEB). After cooling the reaction to room temperature, the biotinylated DNA was purified using a QIAquick nucleotide removal kit (Qiagen).
Codon-optimized phage ViI candidate hypermodification genes were synthesized and cloned into pET28b vectors by Genscript’s custom gene synthesis services (Genscript). These plasmids were used to transform E. coli strain NEB T7 Express. Proteins were expressed in 5 mL LB medium inoculated 1:50 from an overnight starting culture. Cultures were incubated at 37 °C with agitation until they reached an OD600 ∼ 0.6 and cooled to room temperature before isopropyl β-d-1-thiolgalactopyranoside was added at a final concentration of 0.1 mM. Induced cultures were then incubated overnight at 16 °C with shaking. Cells were harvested by centrifugation and suspended in lysis buffer (10 mM Tris⋅HCl, pH 8, 100 mM NaCl, 10 mM KCl, 1 mM PMSF). Cell disruption was carried out using a Q500 micro tip sonicator (Qsonica) for 2 min with a 33% duty cycle and 30% power. The resulting lysate was clarified by centrifugation at 21,000 × g at 4 °C for 15 min, and the supernatant was collected. Expression of each gene product candidate was confirmed by SDS/PAGE analysis. Aliquots of lysates were frozen in liquid nitrogen and stored at −80 °C until use.
Biotinylated SP8 genomic DNA fragment (4 µg) was added into a lysate mixture containing ∼20 µg total protein in ATP (1 mM), RNaseA (50 µg/µL), and reaction buffer (25 mM Tris⋅HCl, pH 7.5, 5 mM β-mercaptoethanol, 5 mM MgCl2, 25 mM KCl). The combined DNA lysate mixture was incubated at 37 °C for 1 h and then quenched by addition of an equal volume of quenching/bead binding buffer (40 mM Tris⋅HCl, pH 7.6, 200 mM NaCl, 2 mM DTT, 15% PEG 6000, 20 mM EDTA). Streptavidin-coated magnetic beads (NEB) were used to capture and separate the biotinylated SP8 DNA from endogenous nucleic acids. The beads (30 µL slurry) were conditioned by washing in 20 mM Tris⋅HCl, pH 7.5, 0.5 M NaCl, and 1 mM EDTA. Beads were added to the DNA lysate quenching buffer and incubated at ambient temperature for 10 min. The DNA-bound beads were captured by a magnet, and the supernatant was discarded. After two wash cycles, the DNA-bound beads were suspended in 43 µL of deionized water. To release the nucleosides for analysis, 5 µL of Nucleoside Digestion Mix Buffer (50 mM NaOAc, pH 5.4, 1 mM ZnCl2; NEB) and 2 µL of Nucleoside Digestion Mix were added to the bead suspension and incubated at 37 °C for 2 h. The beads were removed with a magnet, and the supernatant was subjected to LC/MS analysis.
Supplementary Material
Acknowledgments
We thank Prof. Betty Kutter (Evergreen State College) for sharing microbial strains. Dr. Lana Saleh (NEB) provided a critical reading of the manuscript throughout its preparation. Y.-J.L., N.D., S.E.W., S.M., M.E.F., C.G., I.R.C., and P.R.W. acknowledge guidance from Christopher Noren, William Jack, and Richard J. Roberts and financial support from NEB.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714812115/-/DCSupplemental.
References
- 1.Weigele P, Raleigh EA. Biosynthesis and function of modified bases in bacteria and their viruses. Chem Rev. 2016;116:12655–12687. doi: 10.1021/acs.chemrev.6b00114. [DOI] [PubMed] [Google Scholar]
- 2.Kirnos MD, Khudyakov IY, Alexandrushkina NI, Vanyushin BF. 2-aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature. 1977;270:369–370. doi: 10.1038/270369a0. [DOI] [PubMed] [Google Scholar]
- 3.Walker MS, Mandel M. Biosynthesis of 5-(4‘5’-dihydroxypentyl) uracil as a nucleoside triphosphate in bacteriophage SP15-infected Bacillus subtilis. J Virol. 1978;25:500–509. doi: 10.1128/jvi.25.2.500-509.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehrlich M, Ehrlich KC. A novel, highly modified, bacteriophage DNA in which thymine is partly replaced by a phosphoglucuronate moiety covalently bound to 5-(4′,5′-dihydroxypentyl)uracil. J Biol Chem. 1981;256:9966–9972. [PubMed] [Google Scholar]
- 5.Neuhard J, Maltman KL, Warren RA. Bacteriophage phi W-14-infected Pseudomonas acidovorans synthesizes hydroxymethyldeoxyuridine triphosphate. J Virol. 1980;34:347–353. doi: 10.1128/jvi.34.2.347-353.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren RA. Ordered distribution of α-putrescinylthymine in the DNA of bacteriophage ΦW-14. Curr Microbiol. 1981;6:185–188. [Google Scholar]
- 7.Witmer H. Synthesis of deoxythymidylate and the unusual deoxynucleotide in mature DNA of Bacillus subtilis bacteriophage SP10 occurs by postreplicational modification of 5-hydroxymethyldeoxyuridylate. J Virol. 1981;39:536–547. doi: 10.1128/jvi.39.2.536-547.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maltman KL, Neuhard J, Warren RA. 5-[(Hydroxymethyl)-O-pyrophosphoryl]uracil, an intermediate in the biosynthesis of alpha-putrescinylthymine in deoxyribonucleic acid of bacteriophage phi W-14. Biochemistry. 1981;20:3586–3591. doi: 10.1021/bi00515a043. [DOI] [PubMed] [Google Scholar]
- 9.Iyer LM, Zhang D, Burroughs AM, Aravind L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 2013;41:7635–7655. doi: 10.1093/nar/gkt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Benson DA, et al. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kropinski AMB. 1965. Isolation and characterization of a bacteriophage active against pseudomonas acidovorans. MSc thesis (University of British Columbia, Vancouver, Canada)
- 13.Thorne CB. Transduction in Bacillus subtilis. J Bacteriol. 1962;83:106–111. doi: 10.1128/jb.83.1.106-111.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wall SK, Zhang J, Rostagno MH, Ebner PD. Phage therapy to reduce preprocessing Salmonella infections in market-weight swine. Appl Environ Microbiol. 2010;76:48–53. doi: 10.1128/AEM.00785-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luna AJ, Wood TL, Chamakura KR, Kuty Everett GF. Complete genome of Salmonella enterica serovar enteritidis myophage Marshall. Genome Announc. 2013;1:e00867-13. doi: 10.1128/genomeA.00867-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu C-R, Lin T-L, Pan Y-J, Hsieh P-F, Wang J-T. Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PLoS One. 2013;8:e70092. doi: 10.1371/journal.pone.0070092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matilla MA, Salmond GPC. Complete genome sequence of Serratia plymuthica bacteriophage ΦMAM1. J Virol. 2012;86:13872–13873. doi: 10.1128/JVI.02702-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adriaenssens EM, et al. T4-related bacteriophage LIMEstone isolates for the control of soft rot on potato caused by ‘Dickeya solani’. PLoS One. 2012;7:e33227. doi: 10.1371/journal.pone.0033227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooton SPT, Atterbury RJ, Connerton IF. Application of a bacteriophage cocktail to reduce Salmonella Typhimurium U288 contamination on pig skin. Int J Food Microbiol. 2011;151:157–163. doi: 10.1016/j.ijfoodmicro.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 20.Shahrbabak SS, et al. Isolation, characterization and complete genome sequence of PhaxI: A phage of Escherichia coli O157: H7. Microbiology. 2013;159:1629–1638. doi: 10.1099/mic.0.063776-0. [DOI] [PubMed] [Google Scholar]
- 21.Oot RA, et al. Prevalence of Escherichia coli O157 and O157:H7-infecting bacteriophages in feedlot cattle feces. Lett Appl Microbiol. 2007;45:445–453. doi: 10.1111/j.1472-765X.2007.02211.x. [DOI] [PubMed] [Google Scholar]
- 22.Park M, et al. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl Environ Microbiol. 2012;78:58–69. doi: 10.1128/AEM.06231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craigie J, Brandon KF. Bacteriophage specific for the O‐resistant V form of B. typhosus. J Pathol. 1936;43:233–248. [Google Scholar]
- 24.Anany H, et al. A Shigella boydii bacteriophage which resembles Salmonella phage ViI. Virol J. 2011;8:242. doi: 10.1186/1743-422X-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Favrin SJ, Jassim SA, Griffiths MW. Development and optimization of a novel immunomagnetic separation- bacteriophage assay for detection of Salmonella enterica serovar enteritidis in broth. Appl Environ Microbiol. 2001;67:217–224. doi: 10.1128/AEM.67.1.217-224.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatsch CO, Wood TL, Chamakura KR, Kuty Everett GF. Complete genome of Salmonella enterica serovar Typhimurium myophage Maynard. Genome Announc. 2013;1:e00866-13. doi: 10.1128/genomeA.00866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kauffman AKM. 2014. Demographics of lytic viral infection of coastal ocean vibrio. PhD dissertation (Massachusetts Institute of Technology, Cambridge, MA)
- 28.Sjöberg L, Lindberg AA. Phage typing of Pseudomonas aeruginosa. Acta Pathol Microbiol Scand. 1968;74:61–68. doi: 10.1111/j.1699-0463.1968.tb03455.x. [DOI] [PubMed] [Google Scholar]
- 29.Ceyssens PJ, et al. The genome and structural proteome of YuA, a new Pseudomonas aeruginosa phage resembling M6. J Bacteriol. 2008;190:1429–1435. doi: 10.1128/JB.01441-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae HW, Chung IY, Sim N, Cho YH. Complete genome sequence of Pseudomonas aeruginosa siphophage MP1412. J Virol. 2012;86:9537. doi: 10.1128/JVI.01403-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dyson ZA, Seviour RJ, Tucci J, Petrovski S. Genome sequences of Pseudomonas oryzihabitansPhage POR1 and Pseudomonas aeruginosaPhage PAE1. Genome Announc. 2016;4:e01515-15. doi: 10.1128/genomeA.01515-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Essoh C, et al. Investigation of a large collection of Pseudomonas aeruginosa bacteriophages collected from a single environmental source in Abidjan, Côte d’Ivoire. PLoS One. 2015;10:e0130548. doi: 10.1371/journal.pone.0130548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sepúlveda-Robles O, Kameyama L, Guarneros G. High diversity and novel species of Pseudomonas aeruginosa bacteriophages. Appl Environ Microbiol. 2012;78:4510–4515. doi: 10.1128/AEM.00065-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickard D, et al. A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica serovar Typhi. J Bacteriol. 2010;192:5746–5754. doi: 10.1128/JB.00659-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adriaenssens EM, et al. A suggested new bacteriophage genus: “Viunalikevirus.”. Arch Virol. 2012;157:2035–2046. doi: 10.1007/s00705-012-1360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felix A, Callow BR. Typing of paratyphoid B bacilli by Vi bacteriophage. BMJ. 1943;2:127–130. doi: 10.1136/bmj.2.4308.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwan T, Liu J, Dubow M, Gros P, Pelletier J. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J Bacteriol. 2006;188:1184–1187. doi: 10.1128/JB.188.3.1184-1187.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradley DE, Pitt TL. Pilus-dependence of four Pseudomonas aeruginosa bacteriophages with non-contractile tails. J Gen Virol. 1974;24:1–15. doi: 10.1099/0022-1317-24-1-1. [DOI] [PubMed] [Google Scholar]
- 39.Ackermann HW, Cartier C, Slopek S, Vieu JF. Morphology of Pseudomonas aeruginosa typing phages of the Lindberg set. Ann Inst Pasteur Virol. 1988;139:389–404. doi: 10.1016/s0769-2617(88)80075-3. [DOI] [PubMed] [Google Scholar]
- 40.Huang LH, Farnet CM, Ehrlich KC, Ehrlich M. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 1982;10:1579–1591. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kallen RG, Simon M, Marmur J. The new occurrence of a new pyrimidine base replacing thymine in a bacteriophage DNA:5-hydroxymethyl uracil. J Mol Biol. 1962;5:248–250. doi: 10.1016/s0022-2836(62)80087-4. [DOI] [PubMed] [Google Scholar]
- 42.Kropinski AM, Bose RJ, Warren RA. 5-(4-Aminobutylaminomethyl)uracil, an unusual pyrimidine from the deoxyribonucleic acid of bacteriophage phiW-14. Biochemistry. 1973;12:151–157. doi: 10.1021/bi00725a025. [DOI] [PubMed] [Google Scholar]
- 43.Saavedra JE. Reductive alkylation of. beta.-alkanolamines with carbonyl compounds and sodium borohydride. J Org Chem. 1985;50:2271–2273. [Google Scholar]
- 44.Nelson CC, McCloskey JA. Collision-induced dissociation of uracil and its derivatives. J Am Soc Mass Spectrom. 1994;5:339–349. doi: 10.1016/1044-0305(94)85049-6. [DOI] [PubMed] [Google Scholar]
- 45.Dudley E. Analysis of urinary modified nucleosides by mass spectrometry. In: Banoub JH, editor. Mass Spectrometry of Nucleosides and Nucleic Acids. Limbach; Pittsburgh: 2010. pp. 163–194. [Google Scholar]
- 46.Cao H, Wang Y. Collisionally activated dissociation of protonated 2′-deoxycytidine, 2′-deoxyuridine, and their oxidatively damaged derivatives. J Am Soc Mass Spectrom. 2006;17:1335–1341. doi: 10.1016/j.jasms.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 47.Hayashi H, Nakanishi K, Brandon C, Marmur J. Structure and synthesis of dihydroxypentyluracil from bacteriophage SP-15 deoxyribonucleic acid. J Am Chem Soc. 1973;95:8749–8757. doi: 10.1021/ja00807a041. [DOI] [PubMed] [Google Scholar]
- 48.Hansen AS, Thalhammer A, El-Sagheer AH, Brown T, Schofield CJ. Improved synthesis of 5-hydroxymethyl-2′-deoxycytidine phosphoramidite using a 2′-deoxyuridine to 2′-deoxycytidine conversion without temporary protecting groups. Bioorg Med Chem Lett. 2011;21:1181–1184. doi: 10.1016/j.bmcl.2010.12.098. [DOI] [PubMed] [Google Scholar]
- 49.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maltman KL, Neuhard J, Lewis HA, Warren RA. Synthesis of thymine and alpha-putrescinylthymine in bacteriophage phi W-14-infected Pseudomonas acidovorans. J Virol. 1980;34:354–359. doi: 10.1128/jvi.34.2.354-359.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Revel HR, Luria SE. DNA-glucosylation in T-even phage: Genetic determination and role in phagehost interaction. Annu Rev Genet. 1970;4:177–192. doi: 10.1146/annurev.ge.04.120170.001141. [DOI] [PubMed] [Google Scholar]
- 53.Yaung SJ, Esvelt KM, Church GM. CRISPR/Cas9-mediated phage resistance is not impeded by the DNA modifications of phage T4. PLoS One. 2014;9:e98811. doi: 10.1371/journal.pone.0098811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bryson AL, et al. Covalent modification of bacteriophage T4 DNA inhibits CRISPR-Cas9. mBio. 2015;6:e00648-15. doi: 10.1128/mBio.00648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 56.Machnicka MA, Kaminska KH, Dunin-Horkawicz S, Bujnicki JM. Phylogenomics and sequence-structure-function relationships in the GmrSD family of Type IV restriction enzymes. BMC Bioinformatics. 2015;16:336. doi: 10.1186/s12859-015-0773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bair CLC, Black LWL. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J Mol Biol. 2007;366:768–778. doi: 10.1016/j.jmb.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scraba DG, Bradley RD, Leyritz-Wills M, Warren RA. Bacteriophage phi W-14: The contribution of covalently bound putrescine to DNA packing in the phage head. Virology. 1983;124:152–160. doi: 10.1016/0042-6822(83)90298-2. [DOI] [PubMed] [Google Scholar]
- 59.Thiaville JJ, et al. Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc Natl Acad Sci USA. 2016;113:E1452–E1459. doi: 10.1073/pnas.1518570113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai R, Corrêa IR, Xu MY, Xu S-Y. Restriction and modification of deoxyarchaeosine (dG+)-containing phage 9 g DNA. Sci Rep. 2017;7:8348. doi: 10.1038/s41598-017-08864-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Atlas RM. Handbook of Microbiological Media. 4th Ed CRC; Boca Raton, FL: 1993. [Google Scholar]
- 62.Lech K. Growing lambda-derived vectors. Curr Protoc Mol Biol. 2001;1:1.12. doi: 10.1002/0471142727.mb0112s13. [DOI] [PubMed] [Google Scholar]
- 63.Lech K, Reddy KJ, Sherman LA. Preparing lambda DNA from phage lysates. Curr Protoc Mol Biol. 2001;1:1.13. doi: 10.1002/0471142727.mb0113s10. [DOI] [PubMed] [Google Scholar]
- 64.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 67.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 68.Kearse M, et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pasternack G, Sulakvelidze A. 2014. US Patent 8,685,696 B2.
- 70.Sulakvelidze A, Pasternack GR. 2009. US Patent 7,635,584 B2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.