Significance
How species differ from each other is a key question in biology. However, genetic mapping between species often fails because of sterile hybrid crosses. Here, we have developed a technique called in vitro recombination to circumvent breeding. We induced genetic reshuffling through mitotic recombination with the drug ML216 and mapped trait variations in a dish. Starting with hybrid embryonic stem cells between the Mus musculus laboratory mouse and Mus spretus spanning ∼1.5 million years of divergence, we show that it is possible to map the gene responsible for differential resistance to the drug tioguanine in as few as 21 days. Our technique opens up experimental avenues in genetic mapping of various traits and diseases across mouse species.
Keywords: interspecific hybrids, mitotic recombination, QTL mapping, genetics, evolution
Abstract
Discovering the genetic changes underlying species differences is a central goal in evolutionary genetics. However, hybrid crosses between species in mammals often suffer from hybrid sterility, greatly complicating genetic mapping of trait variation across species. Here, we describe a simple, robust, and transgene-free technique to generate “in vitro crosses” in hybrid mouse embryonic stem (ES) cells by inducing random mitotic cross-overs with the drug ML216, which inhibits the DNA helicase Bloom syndrome (BLM). Starting with an interspecific F1 hybrid ES cell line between the Mus musculus laboratory mouse and Mus spretus (∼1.5 million years of divergence), we mapped the genetic basis of drug resistance to the antimetabolite tioguanine to a single region containing hypoxanthine–guanine phosphoribosyltransferase (Hprt) in as few as 21 d through “flow mapping” by coupling in vitro crosses with fluorescence-activated cell sorting (FACS). We also show how our platform can enable direct study of developmental variation by rederiving embryos with contribution from the recombinant ES cell lines. We demonstrate how in vitro crosses can overcome major bottlenecks in mouse complex trait genetics and address fundamental questions in evolutionary biology that are otherwise intractable through traditional breeding due to high cost, small litter sizes, and/or hybrid sterility. In doing so, we describe an experimental platform toward studying evolutionary systems biology in mouse and potentially in human and other mammals, including cross-species hybrids.
Discovering the genetic changes underlying the phenotypic differences between species is a central goal in evolutionary genetics (1). However, hybrid crosses between even recently diverged species in animals often suffer from hybrid sterility (1, 2), greatly complicating genetic mapping of trait variation, especially in mammals. On the other hand, within-species genetic mapping has been tremendously successful in linking genetic polymorphisms to trait variations in innumerable organisms since the early 20th century (3–5). Genetic mapping typically involves breeding and analyzing mapping panels to identify genetic loci controlling trait variations, or quantitative trait loci (QTL). The ability to disentangle linked genetic associations determines mapping resolution and mainly depends on the number of meiotic cross-overs. Accordingly, researchers are driven to create ever-larger mapping populations and/or accumulating recombination over at least two, often many generations to achieve gene-level mapping resolutions (6–8). In the mouse, genetic studies are complicated by the relatively long generation times and small litter sizes, which often decline with increased inbreeding. Consequently, compared with other classical model organisms like yeast, worms, and Arabidopsis, genetic mapping in the mouse requires far greater resources (6–8), such that relatively few traits have been mapped to the gene level (but see landmark studies identifying Tlr4 and Prdm9) (9, 10). This challenge was particularly acute for crosses at or beyond the species level, where hybrid sterility often makes it impossible to generate a panel (if hybrids are viable in the first place). Nonetheless, the potential to reveal unique biology occurring at the species boundaries in mammalian evolution makes such panels worthy attempts (11–15), even allowing for lower mapping resolution. This is because evolutionary changes in trait architecture (including at the cellular or tissue level) can reveal much about the underlying evolutionary process. Should genetic exchange in hybrid animal genomes become feasible, direct genetic mapping of species differences would become routinely possible.
We set out to establish a universal method that allows genetic mapping in mammals without breeding, even across divergent species with viable but otherwise sterile hybrids. We opted to use mouse embryonic stem (ES) cells to take advantage of the full spectrum of genetic manipulations available in tissue culture. A minimal system will have the two following features: an ability to induce on-demand extensive genetic exchange; and genetic (and trait) variation such as those found in F1 hybrid ES cells, ideally between species.
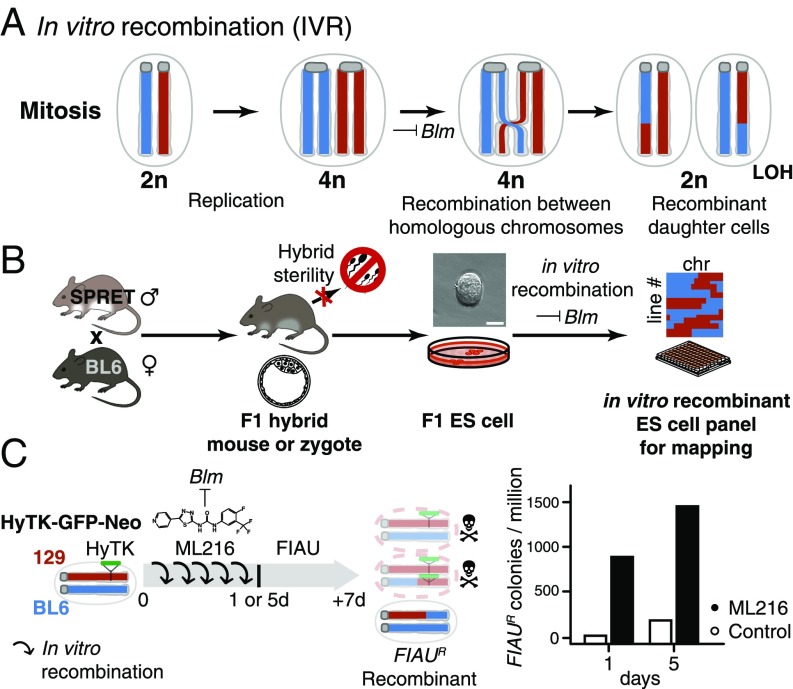
Intriguingly, the technique to create genetic variation through recombination has been in broad use in the mouse genetics community, albeit never explicitly in F1 hybrid ES cells with the goal of genetic mapping. In 2004, two independent groups showed that recessive, biallelic mutants could be reliably recovered in mouse ES cells without breeding by suppressing the DNA helicase Bloom syndrome (Blm; Fig. 1A) (16, 17). Yusa and coworkers (18) showed that these recessive phenotypes were revealed through mitotic recombination between homologous chromosomes. We reasoned that the same mechanism could be leveraged to generate genome-wide random mitotic recombination. This mechanism enabled the creation of panels of arbitrary size carrying recombinant genomes, while avoiding the limitations of hybrid sterility or inbreeding depression (Fig. 1B).
Fig. 1.
IVR via Blm helicase suppression. (A) Blm encodes a helicase normally active during mitosis. Loss of Blm activity leads to increased improper sister chromatid exchange as well as recombination between homologous chromosomes. Mitotic recombination can give rise to recombinant diploid daughter cells with LOH between the breakpoint and the telomeres. (B) IVR allowed the circumvention of hybrid sterility in crosses between the laboratory mouse, e.g., BL6, and a murine sister species SPRET. (BL6 × SPRET)F1 hybrid mice were viable and allowed derivation of F1 ES cells despite male sterility (25). Applying IVR to F1 ES cells allowed rapid and efficient generation of recombinant ES cell panels for genetic mapping. (Scale bar: 50 µm.) chr, chromosome. (C) Efficiency of IVR was estimated by colony survival assay. We estimated the recombination rate between homologous chromosomes with cells hemizygous for a dominant selectable marker (HyTK; green). We induced IVR by adding a small-molecule BLM inhibitor, ML216 (19), to the culturing medium for 1 or 5 d. Under FIAU negative selection, cells having undergone mitotic recombination to become homozygous for the wild-type BL6 alleles (blue) survived, while nonrecombined cells or recombinant cells retaining the HyTK transgene metabolized FIAU, resulting in cell death due to misincorporation of toxic nucleotide analogs (top and middle cells with red chromosomes). Under ML216 treatment (25 µM), IVR rate was estimated to be 2.9 × 10−4 per cell per generation, yielding 800–1,500 FIAU-resistant colonies per million following treatment.
Results
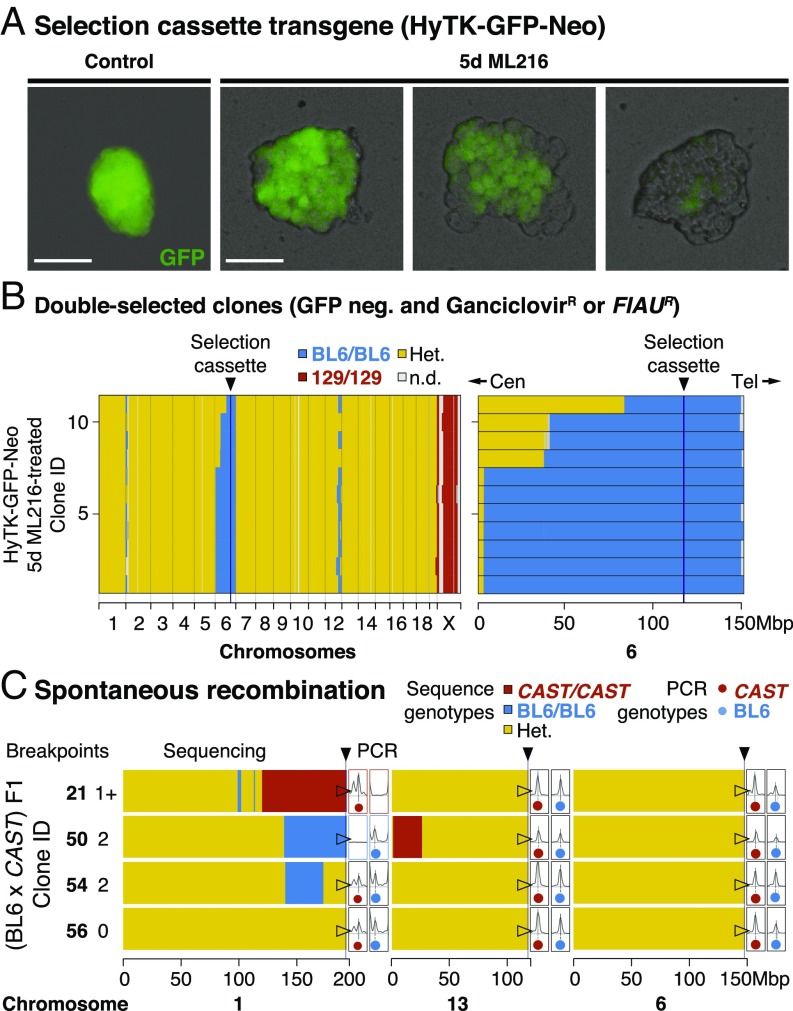
To test if BLM inhibition could lead to elevated homologous recombination rates in mitosis, we inhibited BLM in a number of mouse ES cell lines using a recently discovered small-molecule inhibitor, ML216 (Fig. 1C) (19). As a first test, we started with F1 ES cells between the laboratory mouse strains C57BL/6J (“BL6” in short) and 129 that carried a targeted transgene as a hemizygous allele at the ROSA26 locus on distal chromosome 6. We estimated homologous recombination by counting colony survival under fialuridine (FIAU) treatment, which selected against the transgene consisting of hygromycin phosphotransferase–thymidine kinase (HyTK) and a green fluorescent protein (GFP; Fig. S1). We found that BLM inhibition led to highly elevated rates of homologous recombination, as revealed by increased numbers of FIAU-resistant colonies [Fig. 1C; in vitro recombination (IVR) rate: 2.9 × 10−4 per cell per generation] and the appearance of mosaic GFP expression within a colony (Fig. 2 A, Right). This is consistent with reported rates under Blm suppression or disruption (targeted tetracycline inhibition or knockout alleles: 2.3 × 10−4 to 4.2 × 10−4 vs. wild-type rates between 8.5 × 10−6 and 2.3 × 10−5) (16, 17). The small-molecule BLM inhibitor ML216 offers unique experimental advantages, because its application is simple, rapid, and reversible, eliminating the use of transgenes against Blm (16, 17) or repeated transfections of small interfering RNA to achieve continued suppression of Blm. Importantly, elevated homologous recombination under BLM inhibition was not associated with increased aneuploidy (n = 154 metaphase spreads; Mann–Whitney U test, W = 1,871, h1 > 0, nonsignificant; Fig. S2A). Furthermore, ML216-treated ES cells retained robust expression of NANOG, a key stemness marker (Fig. S3B).
Fig. 2.
Widespread IVR across a range of evolutionary divergence. (A) Selection cassette transgene (HyTK-GFP-Neo). ES cell colonies displayed mosaic GFP expression within a colony when cultured with ML216, but not under control conditions, consistent with homologous recombination and loss of GFP through IVR. Recombination between homologous chromosomes could result in daughter cells with two wild-type (BL6 allele; dark) or transgenic copies (129 allele; bright). Early recombination events followed by random cell loss during clonal expansion could produce completely dark colonies. (Scale bars: 100 µm.) (B) Double-selected clones. After expansion under negative selection against the transgene (both ganciclovir and FIAU kill cells expressing HyTK), 11 ganciclovir-resistant and GFP-negative colonies were whole-genome sequenced. Selection favored loss of transgene (homozygous BL6/BL6 genotypes) at distal chromosome 6. In contrast to normal meiotic recombination (averaging one or more cross-overs per chromosome pair), mitotic recombination typically affected only a single chromosome pair: Much of the genome remained heterozygous (Het.; yellow), with the exception of the transgene-carrying chromosome 6 (mostly BL6/BL6; blue), the single 129 chromosome X (male; 129; red), and at tips of certain chromosomes (e.g., chromosomes 1 and 12). Mitotic recombination events converted genotypes telomeric to the breakpoint toward homozygosity (LOH; yellow to blue). Cen, centromere; n.d., not determined; Tel, telomere. (C) Spontaneous recombination. IVR also occurred in cells carrying divergent genomes with no transgenes. (BL6 × CAST) F1 hybrid ES cells were treated with ML216 and screened by PCR genotyping at diagnostic telomeric markers. Selected clones (two recombinant and control clones each) were whole-genome sequenced, showing recombination events toward both homozygous genotypes, consistent with PCR genotype screening results (total breakpoints per clone ranged from zero to two). Additional recombination events were also recovered, even though the chromosome 1 telomeric marker remained heterozygous (clone 54). These clones also carried nonrecombined chromosomes (e.g., chromosome 6; fully heterozygous; yellow).
To determine the frequency and distribution of mitotic cross-overs under ML216-mediated BLM inhibition, we sequenced and compared the genomes of 11 clones that survived ganciclovir selection (a FIAU alternative; Fig. 2B). We found that the transgene had been lost in all sequenced clones (n = 11 of 11 vs. 9 of 826; Fisher exact test, P < 2.2 × 10−16). Instead, large chromosome segments had undergone mitotic cross-over, removing the transgene in the process. Notably, cross-over breakpoints were recovered not at the transgene itself, but up to many megabases away (all 11 cross-overs were centromeric to chromosome 6; 113 Mbp; Fig. 2B; note that shared breakpoints here may correspond to related clones). This was consistent with the negative selection, because chromosome segments telomeric to the cassette should have no effect on selection. We also treated the F1 hybrid ES cell line (BL6 × CAST) F1 (20) derived from BL6 and Mus castaneus (CAST/EiJ, abbreviated to CAST; diverged ∼0.5 million years ago, or 7.9 SNPs per kbp; compare 1.7 SNPs per kbp between BL6 and 129) with ML216 but otherwise grown without selection. We screened 136 randomly picked ML216-treated clones for loss of heterozygosity (LOH) recombinants and recovered recombinants in both BL6/BL6 and CAST/CAST directions on chromosome 1. Sequencing of representative clones revealed conversion from F1 heterozygous genotypes toward both homozygous genotypes at the telomeres (Fig. 2C, clones 21 and 50; note also additional recombination on chromosome 13). In contrast, control nonrecombinant clones retained heterozygosity at the telomeres (clones 54 and 56). However, even here we discovered a single clone carrying additional internal recombinants on chromosome 1 (Fig. 2C). In both experiments, we noted that cross-overs created by mitotic recombination usually occurred only on one or few chromosomes at a time (Fig. 2 B and C and Fig. S4) (18, 21), unlike in meiosis with typically one cross-over per chromosome arm. Together, the data show that BLM inhibition efficiently generated IVR across wide evolutionary distance, and IVR ES cell panels may constitute genetically distinct lineages suitable for genetic mapping.
Our experiments to determine IVR rate demonstrated that the collective location of recombination breakpoints could reveal the position of the selectable transgene (HyTK or GFP), but under mitotic recombination, the critical interval was defined primarily on the centromeric side due to telomeric LOH. To further illustrate the potential of this approach, we used IVR to map naturally occurring variations. One classical polymorphism is the 25- to 75-fold increased activity of the Mus spretus “a” allele of hypoxanthine–guanine phosphoribosyltransferase (Hprta) compared with the laboratory mouse Hprtb allele (22). Importantly, HPRT metabolizes the antimetabolite tioguanine (6-TG) and causes cytotoxicity. It should be noted that, besides the known Hprt allozyme polymorphism, 6-TG susceptibility itself has not been mapped genetically within or between mouse species. Here, we expected ES cells carrying Hprta to be highly susceptible to 6-TG treatment, whereas Hprtb/b or Hprt−/− ES cells should survive far higher 6-TG concentrations (Fig. S5). We set out to map the QTL for differential 6-TG susceptibility using a bulk segregant assay simply by comparing allele frequencies across the genome between pools of 6-TG–susceptible and –resistant ES cells.
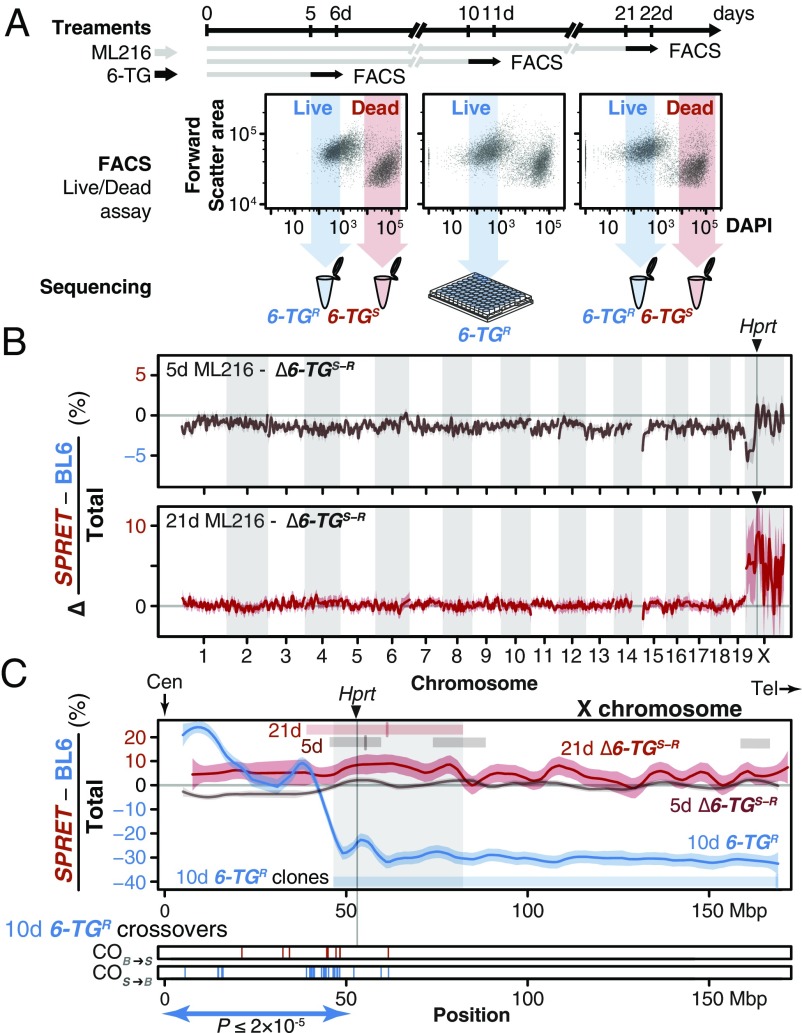
We first confirmed the absence of chromosome-scale rearrangements between the parental strains that could preclude mapping using the de novo assembled genomes of the parental strains made available by the Wellcome Trust Sanger Institute (BL6 and SPRET/EiJ, abbreviated to SPRET here; ∼1.5 million years of divergence or 16.0 SNPs per kbp) (23, 24). We generated IVR panels by treating a female (BL6 × SPRET)F1 hybrid ES cell line (“S18”) (25) with ML216 over 5, 10, and 21 d (Fig. 3A). The use of a female ES cell line, which carried two active X chromosomes before the onset of X inactivation during differentiation (26), allowed direct selection on the alternative Hprta and Hprtb alleles. After confirming biallelic Hprt expression in S18 cells using quantitative PCR, we treated control and IVR S18 cells with 6-TG and determined cell viability via a 4′,6-diamidino-2-phenylindole (DAPI) exclusion assay. Damaged cells with ruptured membrane exhibited rapid uptake of DAPI, a feature unaffected by ML216 treatment, and were distinguishable by fluorescence-activated cell sorting (FACS; “live” proportions under ML216 treatment vs. live proportions under 6-TG treatment, n = 5 paired treatments; Kruskal–Wallis test, χ2 = 13.17, df = 1, P < 0.0003; Fig. 3A and Fig. S6). We separately recovered and sequenced each “resistant” (6-TGR) and “susceptible” (6-TGS) pool (Fig. 3A). Under both 5- and 21-d ML216 treatment, a large skew toward excess SPRET coverage was observed on chromosome X in the 6-TGS relative to the 6-TGR pool (Fig. 3 A and B; differential SPRET bias as a fraction of all reads in adjacent megabase windows between the two pools, with raw SPRET bias ranging from 1 for SPRET-only to −1 for BL6-only). This was in stark contrast to the genomic background, which showed little bias for either SPRET or BL6 contributions (Fig. 3 A and B, also Fig. S6A). The region with the greatest SPRET bias was found on chromosome X near Hprt (Fig. 3C; common region found in both 5- and 21-d ML216 treatments between 49 and 80 Mbp; also see chromosome X in Fig. S6A and discussion in SI Text). Here, our forward genetic mapping for 6-TG susceptibility clearly identified a single locus, suggesting that 6-TG susceptibility depended only on Hprt genotypes. To confirm the role of Hprt in mediating differential 6-TG susceptibility beyond bulk sequencing, we also sequenced 46 individual 6-TGR IVR clones after 10-d ML216 treatment to determine their recombination breakpoints (Fig. 3C). Echoing the skewed cross-over patterns centromeric to the HyTK selection cassette (Fig. 2B), we observed more SPRET-to-BL6 than BL6-to-SPRET centromeric recombinants (n = 35 vs. 8, P ≤ 2 × 10−5, exact binomial test, h1 ≥ h0) and also ruled out aneuploidy or deletion of Hprta as major contributors to 6-TG resistance. We note, however, that despite the strongly skewed ratio of 27 BL6/BL6 homozygous clones at the Hprt locus, of 46 total recovered clones, we still observed 9 heterozygotes and 10 SPRET/SPRET homozygous clones (BL6/BL6 at the Hprt locus: 58.6%; χ2 test using observed allele counts, χ2 = 13.17, df = 2, P ≤ 0.002). Close examination of our flow cytometry data suggests that the original DAPI-only FACS gating may not have been sufficiently exclusive (as opposed to gating using additional channels; Fig. 3A and Fig. S5, Right). Other alternative explanations may be a quantitative, rather than absolute allelic difference in susceptibility to 25 µM 6-TG treatment (Fig. S5) or other mutation(s) at Hprt or elsewhere leading to 6-TG resistance. We concluded that we were able to perform forward genetic mapping using IVR and recover a single region containing Hprt as the most likely gene underlying 6-TG susceptibility differences between BL6 and SPRET.
Fig. 3.
In vitro genetic mapping of variation in 6-TG susceptibility between divergent species. (A) A female ES cell line S18 derived from a M. spretus and BL6 F1 interspecific hybrid was treated with ML216 (25 µM) and subjected to the antimetabolite 6-TG for 1 d before FACS. ES cells were evaluated for viability based on DAPI exclusion. Resistant and susceptible (6-TGR and -TGS) subpopulations were gated conservatively (shaded arrows) and pooled for sequencing. Individual clones from the 10-d ML216 treatment were cultured and whole-genome sequenced. (B) An excess of SPRET contribution on chromosome X between the 6-TGR and -TGS pools suggested that a single locus conferred 6-TG susceptibility. Allele counts were shown as the difference in SPRET bias between the 6-TGs and the 6-TGR samples (as an internal ML216 treatment control) after 5-d (brown) and 21-d (red) ML216 treatment (mean differential SPRET bias ± SEM in megabase windows). In both cases, the genome-wide peak window contains the Hprt gene with the SPRET allele showing significantly increased susceptibility. (C) Detailed view of chromosome X, showing differential SPRET bias for the 5-d (brown) and 21-d (red) ML216 treatment as described above. In addition, 6-TGR clones following 10-d ML216 treatment were sequenced to determine recombination breakpoints. In contrast to the differential bias toward SPRET observed in the susceptible 5- and 21-d ML216 samples, the raw SPRET bias in the solitary 6-TG–resistant sample showed an opposite skew toward BL6 in the 10-d ML216-treatment (blue). Regions deviating from 0 (thus showing bias) after local smoothing in each sample are shown as bars with mark showing maximum skew. Together, they define a common region (shaded) on chromosome X containing Hprt. Cross-overs in individual 6-TGR clones (10-d ML216 treatment) recombined significantly more likely in the SPRET-to-BL6 direction (S > B = 37; B > S = 5; P ≤ 2 × 10−5) between the centromere (Cen) and Hprt, consistent with strong selection favoring the BL6 Hprtb allele. In contrast, only three additional cross-overs were detected telomeric to Hprt. Tel, telomere.
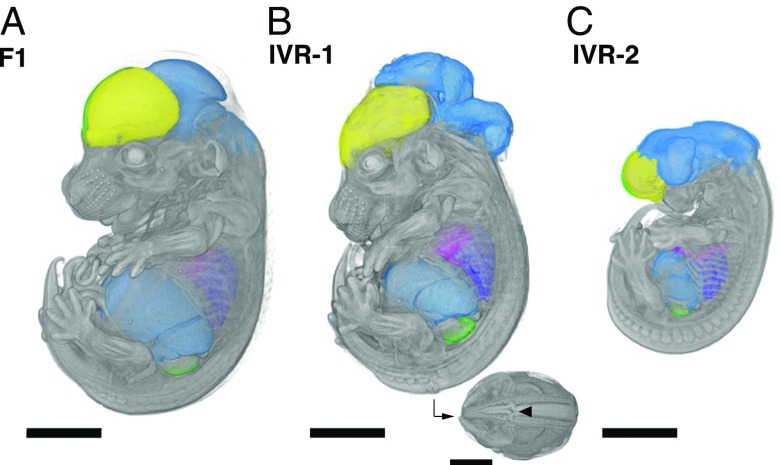
The ability to easily circumvent hybrid sterility in evolutionarily divergent murine species led us to ask what developmental phenotypes may arise from such otherwise inaccessible genetic configurations (M. spretus—laboratory mouse hybrid males are sterile, following Haldane’s rule; back-crosses using female hybrids are possible but extremely challenging) (15). Assaying developmental phenotypes from evolutionarily divergent hybrid ES cells is nontrivial, because hybrid sterility makes conventional rederivation impossible due to a block in germ-line transmission in the chimera generation. Instead, we directly produced embryos via laser-assisted morula injection, a technique widely used for direct phenotyping of fully ES cell-derived founder mice (27), using two karyotypically normal but genetically distinct IVR ES cell lines along with control, nonrecombined S18 cells (IVR 1 and 2; Figs. S2 and S7 and Movies S1–S3). We succeeded in obtaining multiple embryos per line at embryonic day (E) 14.5 of development (n = 36; 24 from IVR lines vs. n = 9 untreated S18 line). Using high-resolution microcomputer tomography (microCT), we observed that the embryos from the untreated clones showed uniformly normal development, whereas embryos from both IVR lines showed both normal development and dramatic craniofacial and neural tube closure defects (two abnormal embryos of four scanned embryos in IVR line 1; two of seven in line 2; and zero of six from the original S18 line; Fig. 4, Fig. S8, and Movies S1–S3). Neural tube and craniofacial defects are among the most common developmental defects due to the complex coordination of cell migration and cell–cell communications, which may be impaired due to novel genetic combinations in the IVR lines, or incompatibility between the host and ES cell-derived cells in a chimera (Fig. S7). Besides major developmental defects, we also made 3D measurements from various organs, including subregions of the brain, heart, and liver, in individual embryos from each ES cell line (Fig. S8). This approach illustrated the feasibility of generating recombinant mice from sterile mouse crosses, which may make it possible to map the genetic basis of evolutionary developmental variation between species.
Fig. 4.
Accessing developmental phenotypes in recombinants between evolutionarily divergent species. Embryos at midgestation (14.5 d after fertilization) were derived from nonrecombinant F1 S18 ES cells (A) and IVR lines 1 (B) and 2 (C; see Methods for details). Embryos were dissected, contrast-stained, and scanned by using X-ray microCT at 9.4-µm resolution. The high scanning resolution allowed identification and precise measurements of individual organs (colorized here). Major developmental craniofacial and neural tube closure defects were observed in the IVR lines (B; caudal view with arrowhead indicates neural tube lesion). (Scale bars: 200 µm.)
Discussion
A central goal of evolutionary genetics is to identify how mutations arose during evolution and influenced phenotypes. For many organisms, a major barrier has been the inability to reliably generate diverse and large mapping panels of sufficient evolutionary diversity. Here, we describe a simple and robust method to make “in vitro crosses,” resulting in panels with inter-cross-like homozygous genotypes from otherwise sterile interspecific hybrid crosses. Being able to bring forth genetic diversity in a Petri dish creates the unique opportunity to conduct mouse genetic mapping at unprecedented speeds with “flow mapping” [similar to “X-QTL” in yeast (28)] or arbitrarily large panels unmatched by most other model organisms, except possibly yeast (21, 28). As renewable stem cells, IVR panels can be expanded, archived, and shared, offering a cellular resource with many of the advantages sought from traditional community resources such as recombinant inbred (RI) line panels. Furthermore, we have shown that our IVR method works in a broad range of ES cells. With millions of potentially recombinant (thus genetically distinct) ES cells in a Petri dish, we demonstrated how IVR enabled mapping of QTLs for drug resistance in as few as 6 d (with an estimated total of five doublings over 5 d). Putting this in context, such an experiment using traditional mouse crosses would have taken 450 d, based on the typical mouse generation time of 90 d, assuming that hybrid sterility could be overcome and allowing for selfing.
We see IVR as a complementary extension to classical organismal genetic mapping panels. In the mouse, the largest organismal RI panel BXD contains ∼160 lines (with most published work based on the ∼35 original BXD strains) (29), and attempts in generating panels incorporating greater divergences encountered enormous challenges (30). Nevertheless, mouse RI resources represented some of the most powerful tools available to dissect system genetics in the mouse, the prime biomedical model organism (31). Seen in this light, IVR represents an alternative approach that could greatly extend the available renewable resources, not least because the genotype combinations between divergent species are hitherto impossible to obtain in the first place.
Recently, Sadhu et al. (21) also achieved a major advance in genetic mapping using CRISPR/Cas9-mediated mitotic recombination in yeast. In contrast to CRISPR targeting, our transgene-free approach offers the simplicity of inducing genome-wide recombinants by the simple addition of a single inexpensive small molecule to the tissue culture medium. Going forward, we envision a combined, complementary approach to IVR: using BLM inhibition for mapping panel generations and efficient QTL identification, then switching to targeted transgene-based screening or CRISPR/Cas9-based IVR for fine-scale mapping.
In addition to the traits we have investigated, M. spretus and the Mus musculus laboratory mouse differ in a number of distinct traits, such as longevity and telomere lengths (32), cancer and inflammation resistance (33, 34), and metabolism (35). Many of these traits have tissue or cellular models suitable for IVR mapping panels or flow mapping through fluorescent detection of specific proteins or metabolites. Future experiments may also probe even greater evolutionary divergence: Early work has shown that F1 hybrids spanning as much as 6 million years between M. musculus and Mus caroli was viable (36). Given active development in single-cell genomics and disease modeling from patient-specific induced pluripotent stem cells, including organoids or organ-on-a-chip microfluidics systems, we anticipate that the in vitro recombinant platform can be broadly applied to accelerate the identification of the genetic basis of many traits and diseases.
Materials and Methods
Animal Care and Use.
All animal experiments have been approved by the applicable animal welfare ethics committees: Faculty of Sciences, Ghent University (reference no. 06/022); and Landesdirektion Sachsen, Germany, permit 24-9168.11-9/2012-5.
Cell Culture.
Unless otherwise stated, ES cell lines were cultured on SNL76/7-4 feeder cells in attachment factor (AF)-coated plates under 2i medium, supplemented with leukemia inhibitory factor (LIF) (see SI Methods for details).
BLM Inhibition Using ML216.
BLM inhibition was performed by using 25 µM ML216 (Sigma-Aldrich) in 2i/LIF medium on inactivated feeders.
Generation of HyTK-EGFP-Neo Cell Line.
Starting with G4 ROSALUC B12 ES cells (37), we replaced the cassette at the ROSA26 locus with a cassette carrying two selectable markers, HyTK and enhanced GFP (EGFP; selectable in fluorescence-assisted cell sorting; Fig. S1; see SI Methods for details). Successful replacement of the cassette was confirmed through selection by using geneticin (G148; ThermoFisher Scientific) and genotyping.
Colony Survival Assay.
HyTK-EGFP-Neo cells were seeded at a density of 5 × 105 per 10 cm AF per feeder plate, followed by 25 µM ML216 treatment for 1 or 5 d. Before the start of negative selection, cells were replated at 2 × 105 per 10 cm AF per feeder plate, and FIAU (0.2 µM; Sigma-Aldrich) or ganciclovir (10 µM; Sigma-Aldrich) selection was applied for 5 d. Colonies were stained with the alkaline phosphatase kit (EMD Millipore), photographed, and counted to determine survival rates. The entire procedure was repeated multiple times, and the survival colony counts were averaged to determine IVR rate.
Screening for Spontaneous Recombinant ES Cell Colonies.
Cells were treated with ML216 for 2 d at a concentration of 5 µM and then for 3 d at a concentration of 25 µM. Cells were then replated and cultured for 5 d in 2i/LIF without ML216. A total of 189 colonies were randomly picked (without selection), of which 136 were screened with multiplexed genotyping.
Multiplexed Genotyping for Detection of LOH.
Diagnostic markers between BL6, CAST, and SPRET strains at tips of each chromosome were designed to track the presence of each allele. The markers were amplified with fluorescence-tagged primers as proposed in ref. 38 (see SI Methods and Table S1 for details). The PCRs were pooled at equimolar proportions and analyzed with a 3730xl DNA Analyzer capillary sequencer (ThermoFisher Scientific) and scored for conversion from heterozygous into homozygous genotypes (LOH) at the tips of each chromosome.
The 6-TG Treatment and DAPI Exclusion Assay.
Before the main experiments, killing curves for 6-TG (Sigma-Aldrich) was performed by using a WST-1 assay (Roche) according to the manufacturer’s instructions (Figs. S3A and S5, Left). For the main experiments, the S18 ES cell line was cultured for 5, 10, or 21 d with 25 µM ML216. Following the designated ML216 treatment, the cells were replated and treated with 25 µM 6-TG in 2i/LIF. We determined “live/dead” cell viability by using DAPI staining (1 µg/mL; Sigma-Aldrich) after 1 d of 25 µM 6-TG treatment.
FACS.
Flow cytometry was performed by using an Aria II Cell Sorter (Becton Dickinson GmbH). We defined the 6-TGR and -TGS populations by using the DAPI exclusion assay in reference ES cell populations. In sorting experiments, ML216-treated or control 6-TGR and -TGS population were recovered for sequencing. For quantification, we performed post hoc analysis using the R Bioconductor package flowCore (39), principally by clustering using the forward scatter area and DAPI/Pacific Blue-A channels into live and dead clusters using mclust (Version 5.2) (40, 41) in 6-TG–treated experiments, considering ML216-treated and controls separately (Fig. S5).
Sequencing and Analysis Pipeline.
Sequencing libraries for high-throughput sequencing were generated by using the Nextera DNA Library Prep Kit (Illumina, Inc.) according to manufacturer’s recommendations or equivalent purified Tn5 transposase as described in ref. 42. Each sample (FACS-sorted clones, single colonies or pooled cells) was barcoded through PCR extension by using an i7-index primer (N701–N763) and the N501 i5-index primer. Pooled libraries were sequenced by a HiSeq 3000 (Illumina) and analyzed by using a custom pipeline (see SI Methods for details). We performed genotyping based on allelic coverage per megabase using known informative variants between the BL6, CAST, and SPRET strains (Mouse Genomes Project Version 3 dbSNP Version 137 release) (24). Scripts are available at: https://github.com/evolgenomics/HybridMiX.
Laser-Assisted Morula Injection.
ES cell-derived embryos were obtained through injection into eight-cell-stage embryos (morulae) as described in ref. 27. The introduction of excess ES cells was expected to lead to embryos with fully ES cell contributions. At 14 d after the injection and subsequent embryo transfer into surrogates (approximating developmental stage E14.5), the gestation was terminated, and embryos were dissected and fixed with 4% paraformaldehyde for microCT scanning. Due to sample preparation, genotyping of scanned embryos was not performed. Instead, control (unscanned) embryos were dissociated and genotyped at diagnostic loci, confirming ES cell contributions from the respective cell lines.
microCT.
Soft-tissue X-ray contrast staining was done via 4-d perfusion in 25% Lugol’s or iodine potassium iodide solution. Then the embryos were rehydrated, mounted in 1% low-melting agarose, and scanned with a Skyscan 1173 instrument (Bruker Corporation) at 9.96-μm resolution (0.5-mm aluminum filter, energy at 70 kV and 110 µA). Image analysis, segmentation, and visualizations were performed by using Amira (Version 6.2.0; FEI) with the XImagePAQ extension 6.2.
Supplementary Material
Acknowledgments
We thank Felicity Jones for experimental design, helpful discussion and input, and improving the manuscript; Caroline Schmid for animal husbandry; Sebastian Kick for microCT scanning; the Y.F.C. and Jones laboratory members for support, scientific input, and improving the manuscript; Christa Lanz and Ilja Bezrukov for assistance with high-throughput sequencing and associated data processing; Andre Noll for high-performance computing support; and Cornelia Grimmel and Stella Autenrieth for technical assistance with FACS. Cell sorting, flow cytometry sample acquisition, and/or data analysis was done on shared instruments of the Flow Cytometry Core Facility Tübingen. RV-L3-HyTK-2L and pCAG-Flpo were gifts from Geoff Wahl and Massimo Scanziani (Addgene plasmids 11684 and 60662). The G4 ROSALUC ES cell line was a gift from Jody J. Haigh. We thank Hua Tang, David M. Kingsley, Karsten Borgwardt, and Detlef Weigel for input and discussion on experimental design. We thank our reviewers for their thoughtful input, which has greatly improved the manuscript. J.P.L.C. is supported by the International Max Planck Research School “From Molecules to Organisms.” P.M. was supported by the Fulbright US Student Program. T.H. and C.L. are supported by Ghent University. Y.F.C. is supported by the Max Planck Society and European Research Council Starting Grant 639096 “HybridMiX.”
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. PRJNA390071). Scripts are available at https://github.com/evolgenomics/HybridMiX.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717474115/-/DCSupplemental.
References
- 1.Darwin C. On the Origin of Species by Means of Natural Selection. John Murray; London: 1859. [Google Scholar]
- 2.Dejager L, Libert C, Montagutelli X. Thirty years of Mus spretus: A promising future. Trends Genet. 2009;25:234–241. doi: 10.1016/j.tig.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackay TFC, Stone EA, Ayroles JF. The genetics of quantitative traits: Challenges and prospects. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- 5.Allen Orr H. The genetics of species differences. Trends Ecol Evol. 2001;16:343–350. doi: 10.1016/s0169-5347(01)02167-x. [DOI] [PubMed] [Google Scholar]
- 6.Churchill GA, et al. Complex Trait Consortium The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 7.Nicod J, et al. Genome-wide association of multiple complex traits in outbred mice by ultra-low-coverage sequencing. Nat Genet. 2016;48:912–918. doi: 10.1038/ng.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker CC, et al. Genome-wide association study of behavioral, physiological and gene expression traits in outbred CFW mice. Nat Genet. 2016;48:919–926. doi: 10.1038/ng.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 10.Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 11.Turner LM, White MA, Tautz D, Payseur BA. Genomic networks of hybrid sterility. PLoS Genet. 2014;10:e1004162. doi: 10.1371/journal.pgen.1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White MA, Ikeda A, Payseur BA. A pronounced evolutionary shift of the pseudoautosomal region boundary in house mice. Mamm Genome. 2012;23:454–466. doi: 10.1007/s00335-012-9403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White MA, Stubbings M, Dumont BL, Payseur BA. Genetics and evolution of hybrid male sterility in house mice. Genetics. 2012;191:917–934. doi: 10.1534/genetics.112.140251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forejt J. Hybrid sterility in the mouse. Trends Genet. 1996;12:412–417. doi: 10.1016/0168-9525(96)10040-8. [DOI] [PubMed] [Google Scholar]
- 15.Burgio G, et al. Interspecific recombinant congenic strains between C57BL/6 and mice of the Mus spretus species: A powerful tool to dissect genetic control of complex traits. Genetics. 2007;177:2321–2333. doi: 10.1534/genetics.107.078006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- 17.Yusa K, et al. Genome-wide phenotype analysis in ES cells by regulated disruption of Bloom’s syndrome gene. Nature. 2004;429:896–899. doi: 10.1038/nature02646. [DOI] [PubMed] [Google Scholar]
- 18.Yamanishi A, et al. Enhancement of microhomology-mediated genomic rearrangements by transient loss of mouse Bloom syndrome helicase. Genome Res. 2013;23:1462–1473. doi: 10.1101/gr.152744.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen GH, et al. A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem Biol. 2013;20:55–62. doi: 10.1016/j.chembiol.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barakat TS, Rentmeester E, Sleutels F, Grootegoed JA, Gribnau J. Precise BAC targeting of genetically polymorphic mouse ES cells. Nucleic Acids Res. 2011;39:e121. doi: 10.1093/nar/gkr550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadhu MJ, Bloom JS, Day L, Kruglyak L. CRISPR-directed mitotic recombination enables genetic mapping without crosses. Science. 2016;352:1113–1116. doi: 10.1126/science.aaf5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson GG, Chapman VM. Altered turnover of hypoxanthine phosphoribosyltransferase in erythroid cells of mice expressing Hprt a and Hprt b alleles. Genetics. 1987;116:313–320. doi: 10.1093/genetics/116.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mouse Genomes Project, Wellcome Trust Sanger Institute 2017 Data from Genome Evaluation Browser, SPRET_EiJ. Available at mice-geval.sanger.ac.uk/SPRET_EiJ_R20150909/Info/Index. Accessed January 28, 2017.
- 24.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochepied T, et al. Breaking the species barrier: Derivation of germline-competent embryonic stem cells from Mus spretus x C57BL/6 hybrids. Stem Cells. 2004;22:441–447. doi: 10.1634/stemcells.22-4-441. [DOI] [PubMed] [Google Scholar]
- 26.Barakat TS, Gribnau J. X chromosome inactivation and embryonic stem cells. Adv Exp Med Biol. 2010;695:132–154. doi: 10.1007/978-1-4419-7037-4_10. [DOI] [PubMed] [Google Scholar]
- 27.Poueymirou WT, et al. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat Biotechnol. 2007;25:91–99. doi: 10.1038/nbt1263. [DOI] [PubMed] [Google Scholar]
- 28.Ehrenreich IM, et al. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464:1039–1042. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams RW, Williams EG. Resources for systems genetics. Methods Mol Biol. 2017;1488:3–29. doi: 10.1007/978-1-4939-6427-7_1. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Cross Consortium The genome architecture of the collaborative cross mouse genetic reference population. Genetics. 2012;190:389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andreux PA, et al. Systems genetics of metabolism: The use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150:1287–1299. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding H, et al. Regulation of murine telomere length by Rtel: An essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 33.Mahieu T, et al. The wild-derived inbred mouse strain SPRET/Ei is resistant to LPS and defective in IFN-β production. Proc Natl Acad Sci USA. 2006;103:2292–2297. doi: 10.1073/pnas.0510874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puimège L, et al. Glucocorticoid-induced microRNA-511 protects against TNF by down-regulating TNFR1. EMBO Mol Med. 2015;7:1004–1017. doi: 10.15252/emmm.201405010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song Y, et al. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Curr Biol. 2011;21:1296–1301. doi: 10.1016/j.cub.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West JD, Frels WI, Papaioannou VE, Karr JP, Chapman VM. Development of interspecific hybrids of Mus. J Embryol Exp Morphol. 1977;41:233–243. [PubMed] [Google Scholar]
- 37.Haenebalcke L, et al. Efficient ROSA26-based conditional and/or inducible transgenesis using RMCE-compatible F1 hybrid mouse embryonic stem cells. Stem Cell Rev. 2013;9:774–785. doi: 10.1007/s12015-013-9458-z. [DOI] [PubMed] [Google Scholar]
- 38.Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- 39.Hahne F, et al. flowCore: A bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc. 2002;97:611–631. [Google Scholar]
- 41.Fraley C. 2012. mclust Version 4 for R: Normal Mixture Modeling for Model-Based Clustering, Classification, and Density Estimation (University of Washington, Seattle)
- 42.Picelli S, et al. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014;24:2033–2040. doi: 10.1101/gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.