Significance
A substantial number of known genetic disorders have their origin in mutations that cause misfolding or dysfunction of G protein-coupled receptors (GPCRs). Pharmacological chaperones can rescue such mutant receptors from the endoplasmic reticulum by stabilizing protein conformations that support trafficking into the target membrane. Rhodopsin-mediated retinitis pigmentosa is a misfolding disease that might be targeted by PCs. Here we present a structure-based drug design approach to identify nonretinal compounds that bind and stabilize the receptor. Surprisingly, selected hits induce a previously unknown conformation of the seven-transmembrane helix bundle. Our study thus provides a remarkable example for compound class discovery and for the adaptability of GPCRs to chemically diverse ligands.
Keywords: structural biology, drug discovery, chemical biology, rare diseases, ophthalmology
Abstract
In the degenerative eye disease retinitis pigmentosa (RP), protein misfolding leads to fatal consequences for cell metabolism and rod and cone cell survival. To stop disease progression, a therapeutic approach focuses on stabilizing inherited protein mutants of the G protein-coupled receptor (GPCR) rhodopsin using pharmacological chaperones (PC) that improve receptor folding and trafficking. In this study, we discovered stabilizing nonretinal small molecules by virtual and thermofluor screening and determined the crystal structure of pharmacologically stabilized opsin at 2.4 Å resolution using one of the stabilizing hits (S-RS1). Chemical modification of S-RS1 and further structural analysis revealed the core binding motif of this class of rhodopsin stabilizers bound at the orthosteric binding site. Furthermore, previously unobserved conformational changes are visible at the intradiscal side of the seven-transmembrane helix bundle. A hallmark of this conformation is an open channel connecting the ligand binding site with the membrane and the intradiscal lumen of rod outer segments. Sufficient in size, the passage permits the exchange of hydrophobic ligands such as retinal. The results broaden our understanding of rhodopsin’s conformational flexibility and enable therapeutic drug intervention against rhodopsin-related retinitis pigmentosa.
Retinitis pigmentosa (RP) unites a class of hereditary diseases causing the progressive degeneration of the photoreceptor system (1) and visual impairment in 1.5 million patients worldwide (2). Predominantly affected are rod cells in the retina of the eye that aggregate and progressively affect cone cell viability (3). One reason is the inherited RP mutation P23H in rhodopsin discovered nearly 30 y ago as one of the first examples for a disease-related G protein-coupled receptor (GPCR) mutation (4). Over 100 more followed over the years, sparking the idea to treat this common cause of age-related blindness by directly targeting the mutated photoreceptor opsin (5, 6). In contrast to most GPCR drugs that modulate signaling, drugs acting on opsin mutants have to intervene early in protein synthesis and folding to compensate for the observed decrease in protein stability (7) and for the fatal consequences for cell metabolism and rod and cone cell survival (3).
A pharmacological chaperone (PC) treatment of rhodopsin-mediated RP can stabilize and increase opsin resistance to facilitate correct transport to the disk membranes of the rod outer segment (ROS). This is in analogy to the effect of known orthosteric and allosteric ligands that promote expression and correct protein folding and trafficking for a number of GPCRs (8, 9). In a mouse model of failed puberty, PC therapy could restore the fertility of mice by correctly routing the mutated gonadotropin-releasing hormone receptor to the plasma membrane (10). Notably, a very small subset of organic molecules was able to address most of the mutants causing the disease phenotype (10). Similarly, the endogenous ligand 11-cis retinal assists folding and integration of opsin RP mutants in vitro and in vivo (7, 11–14). However, the toxicity of 11-cis retinal and its regulated transport to and from the ROS limits its potential as pharmacological chaperone (15, 16). Here, we screened for nonretinoid drug-like rhodopsin stabilizers and studied their molecular interactions with the photoreceptor protein by X-ray crystallography. Comparing our pharmacologically stabilized structures of opsin to existing active- and inactive-state rhodopsin structures, we identify distinctive conformational rearrangements and an open ligand channel to the intradiscal side.
Results and Discussion
Nonretinal Opsin-Stabilizing Compounds.
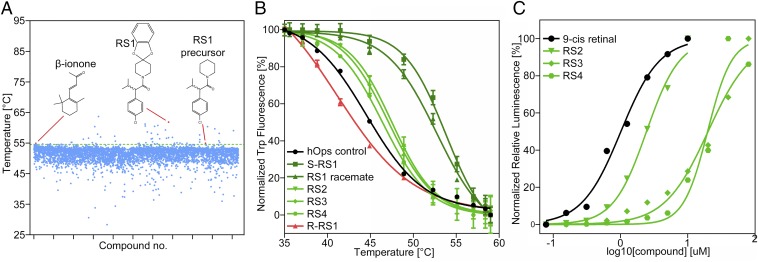
In congenital stationary night blindness, the mild and nonprogressive phenotype of RP, mutations lead to increased basal activity by selectively stabilizing the orthosteric binding site (17, 18). In contrast, severe RP mutants have a destabilizing effect on rhodopsin protein (5, 6). To counterbalance, pharmacological rescue aims at intervening early in protein genesis to improve folding and trafficking in vivo (14) and in vitro (7, 11–13), as demonstrated so far only for retinoid-like molecules (16). Binding and stabilization of mutated receptor protein initiates the pharmacological rescue after compounds enter the cell to prevent the protein from accumulation at the endoplasmic reticulum (12). Here, we screened for nonretinal molecules that bind and stabilize purified human opsin to investigate their molecular modes of binding. We assembled a focused library using virtual screening of Roche’s compound collection. Selection methods were based on ligand similarity and structural fit to ground-state rhodopsin [Protein Data Bank (PDB) ID: 1GZM] (19) or light-activated rhodopsin (PDB ID: 4A4M) (20). Focused screening of this library followed by hit expansion of the most promising hits was performed using a thermofluor assay and purified, functionally neutral, stabilized human opsin N2C/N282C. Within the screened library about 0.5% of the 5,186 compounds had a stabilizing effect of more than two SDs above the DMSO control (corresponding to 2.6 °C) with the strongest hit stabilizing up to 9 °C (Fig. 1A). Notably, β-ionone is a known binder of opsin (21) and was unintentionally included in the focused library and thus acted as a blind control hit above the defined threshold. Compounds were further validated by thermal shift fluorescence-detection size-exclusion chromatography (FSEC-TS) (Fig. 1B), yielding the hits RS1-4 for further investigation.
Fig. 1.
Focused screening for opsin-stabilizing compounds. (A) Single-point thermofluor screening at 47 μM compound and 1.4 μM protein concentration. Hits are above the threshold of 54.6 °C (green line). (B) FSEC-TS hit validation at 0.26 µM human opsin N2C N282C in the absence and presence of 10 µM compound concentration (triplicate). Stabilization by the S but not the R enantiomer of RS1 suggests stereoselective binding to opsin. (C) Pharmacotrafficking assay demonstrating cell surface trafficking of disease-mutated opsin P23H at EC50 values (duplicate) of 1.0 µM (9-cis retinal), 2.4 µM (RS2), 16.7 µM (RS4), and 20 µM (RS3).
To clarify that opsin-stabilizing compounds effectively outcompete 11-cis retinal within the binding pocket once the rescued mutant protein has been targeted to the rod outer membrane, we measured the regeneration of rhodopsin in the presence of RS1-4 by UV/VIS spectroscopy (SI Appendix, Fig. S1). Expectedly, compounds did not interfere with the formation of a stable rod cell pigment between opsin protein and the covalent, irreversible binder 11-cis retinal.
To explore if the molecules derived from a thermal stabilization screening are generally able to rescue mutated opsin in a cellular context, we employed a commercially available cellular pharmacotrafficking assay based on the RP-causing mutation P23H (22). We measured compound-dependent rescue and increased opsin transport to the plasma membrane for RS2-4 but not for RS1 as it likely lacks cell-penetrating features (Fig. 1C).
Promiscuous Binding Pocket Similar to Other Class-A GPCRs.
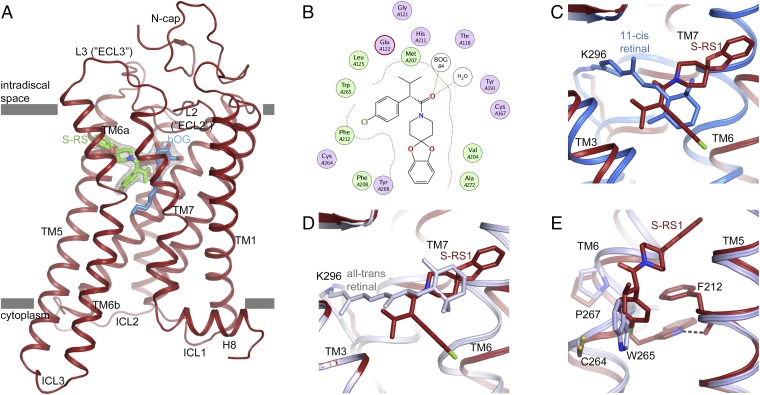
To generate a molecular understanding for opsin stabilization, we crystallized bovine opsin and soaked crystals with the selected hits including S-RS1, R-RS1, racemate R/S-RS1, and RS2-4. As opposed to RS2-4, we were able to determine the crystal structure of the S-RS1–opsin complex at 2.4 Å resolution (SI Appendix, Table S1). Molecular replacement and structural refinement resulted in well-resolved electron density for the receptor, the rhodopsin stabilizer S-RS1, and neighboring β-octyl gluocoside (OG) molecules (Fig. 2A). S-RS1 binds within the orthosteric binding pocket of class-A GPCRs that is located between transmembrane helices (TM) 3, 5, 6, and 7 at the level of the outer-membrane leaflet (SI Appendix, Fig. S2). Compared with dark-state rhodopsin with bound 11-cis retinal, the bottom of the binding pocket is shifted by 3 Å toward the cytoplasmic site with S-RS1 penetrating rather deep into the seven-transmembrane helix bundle, similar to doxepin in the histamine 1 receptor (SI Appendix, Fig. S3) (23).
Fig. 2.
Opsin stabilization by S-RS1. (A) Side view of the S-RS1–opsin complex in cartoon representation. S-RS1 and a neighboring β-octyl glucoside shown in green and blue sticks are located at the orthosteric GPCR binding pocket. Experimental electron density (Fo-Fc) maps (gray mesh) for the ligands contoured at 3 σ. (B) MOE-calculated protein–ligand plot identifying mainly hydrophobic amino acids and two hydrogen bonds (dotted lines) that contribute to ligand binding. Hydrophilic residues are purple, blue rings indicate basic groups, red rings indicate acidic groups, and hydrophobic residues are green. (C and D) Superposition of the S-RS1–opsin complex with (C) dark-state (blue; PDB ID: 1GZM) and (D) meta-II active-state rhodopsin (blue-white; PDB ID: 3PQR) highlighting the overlap of the functional groups of the ligand with retinal’s β-ionone ring in two different conformations. (E) S-RS1–induced outward shift of Trp2656.48 within the highly conserved CWxP motif. The initial χ angle of −81° in opsin (light blue; PDB ID: 4J4Q) changes to −174° in the S-RS1 complex.
Receptor–ligand interactions are mostly hydrophobic with 15 opsin residues involved in ligand binding. The majority is also part of the native retinal binding pocket (Fig. 2B and SI Appendix, Table S2) with Lys1253.40, Cys1674.56, Val2045.39, and Cys2646.47 being the only residues exclusive for S-RS1 binding. Interestingly, S-RS1 does not reach out for Lys2967.43, the key residue for covalent retinal binding to opsin. Instead, space is cleared for one of four OG detergent molecules identified in the structure (Fig. 2A and SI Appendix, Fig. S4). The head group of this central OG molecule, but not its lipophilic tail, is in a similar position to that of OG identified previously in the crystal structure of opsin in the active conformation (SI Appendix, Fig. S4A) (24). Since OG was only present in the crystallization buffer but was absent during drug screening and characterization, S-RS1 binding and stabilization of opsin must be independent from OG molecules. Possibly, other more hydrophobic molecules including lipids can penetrate the receptor also in the absence of S-RS1. The positions of OG molecules inside and outside of the receptor may thus be relevant for the recently proposed lipid scramblase function of opsin (SI Appendix, Fig. S4B) (25). More importantly, it is clear from our data that opsin can accommodate a range of chemically different ligands within its binding pocket, just like other GPCRs with a larger variety of natural ligands.
From Structural Hit to Chemical Design and S-RS1 Derivative Structures.
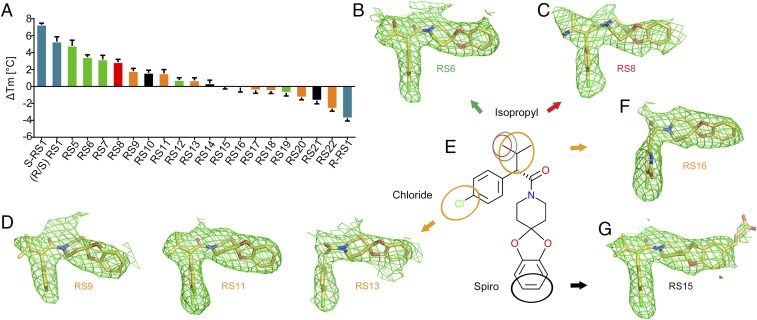
To gain deeper insight into the binding mode of the rhodopsin stabilizer S-RS1, we performed a limited chemical modification analysis in combination with thermofluor analysis and structure determination (Fig. 3 A–G and SI Appendix, Table S3). Seven opsin structures in complex with S-RS1 derivatives were determined between 2.5 and 3.1 Å resolution with indistinguishable overall protein conformations or side-chain orientations. Overall, we observed a correlation between the stabilizing effect for a particular compound and the success rate for its cocrystallization and structure determination (SI Appendix, Fig. S5).
Fig. 3.
Crystal structures and thermofluor analysis of S-RS1 derivatives. (A) Experimentally determined ΔTm values for RS1 stereoisomers (blue) and derivatives grouped by alterations at the isopropyl and chlorine (orange), isopropyl (green, red), or spiro function (black). (B–G) Side view of the ligand binding site of opsin (model not shown) in complex with compounds in stick representation. The compound color code reflects each group for limited chemical modification. Experimental electron density maps (Fo-Fc) in green mesh are contoured at 3 σ. (E) Chemical structure of S-RS1.
Of particular interest is the close interaction between the isopropyl group in position C21 of S-RS1 and opsins carboxyl group of Glu1223.37 in TM3. This location, together with the adjacent Gly1213.36, plays a role in retinal selectivity (26), and S-, M-, and L-cone opsins feature distinct amino acids at this position. Opsin exhibits stereoselectivity for the S enantiomer of the chiral compound S-RS1 likely due to steric hindrances for the R enantiomer. Structure refinement using the R enantiomer R-RS1 resulted in structural clashes. Dose–response thermofluor analysis of both S and R configurations of RS1 revealed that no stabilization could be measured for the R enantiomer (Fig. 1B and SI Appendix, Fig. S6). Modification of S-RS1 with an amino group at position C24 or a hydroxyl group at position C25 was neutral for stabilization yet resulted in crystal structures and modified hydrophilic groups in H-bonding distance to Glu1223.37. Modifications impacting position C21 prevented cocrystallization.
The chlorine of the S-RS1 phenyl chlorine ring forms interactions with two crucial binding pocket residues, namely, the sulfhydryl group of Cys2642.47 and several atoms of Trp2652.48. Removal of the chlorine as well as addition of an extra chlorine in position C23 diminished stabilization although we observed only minimal changes in the binding mode. Larger modification at these positions and of the ring size abolished binding completely. This renders the region critical for binding similar to the position C21 isopropyl group limiting S-RS1 modification for improved solubility and permeability.
We speculated about the spiro ring modification of S-RS1 to improve solubility and affinity, since an initial hit of thermofluor screening and precursor of S-RS1 did not contain this ring function (Fig. 1A). To probe the large cavity, we added a carboxyl group connected with a linker to C16 of the S-RS1 spiro ring resulting in additional interactions with Tyr191ECL2 and Met2887.35, while a rotamer change buries Phe2766.59 into the hydrophobic environment surrounding the receptor. Both thermofluor and FSEC-TS analysis revealed maximal stabilization among the S-RS1 derivatives. This side of the binding pocket may thus be a location to further probe the chemical space and improve on the initial rhodopsin stabilizers. The methods we used in our approach, including virtual screening, thermal shift analysis, retinal competition, cellular trafficking, and structural analysis, are very robust and suggest good applicability for drug discovery. Furthermore, the results indicate that rhodopsin is a tractable target for structure-based drug design and that the early compounds described in this study can be further optimized.
A Ligand Channel Toward the Extracellular Side.
Ligand-induced conformational changes are a hallmark of GPCR activation. In rhodopsin, translocation of retinal’s β-ionone ring is a critical step during receptor activation. It allows TM6 to rotate into the active conformation and to provide access to the G protein binding site (27–29). Interestingly, structure comparison reveals that the positions of all stabilizing S-RS1–based compounds simultaneously overlap with the positions of retinal’s β-ionone ring in the dark and active meta-II states. In other words, the angular shape of S-RS1 stabilizes opsin simultaneously at locations of the orthosteric binding pocket known for the agonist all-trans retinal or the inverse agonist 11-cis retinal. The chlorinated phenyl ring is located at the position of the β-ionone ring in the dark state, and the spiro group overlaps with the position in the meta-II conformation (Fig. 2 C and D and SI Appendix, Fig. S7). Occupation of these positions by the compound forces Trp2656.48 of the highly conserved CWxP sequence motif found in class-A GPCRs to rotate into a new rotamer position outside the 7TM bundle (Fig. 2E). The outward position is maintained by a hydrogen bond with the backbone carbonyl of Phe2125.47 and π-stacking with the phenyl group of Phe2125.47 in TM5. This dislocation of Trp2656.48 toward a more polar environment was not reported in previous crystal structures of rhodopsin or other class-A GPCRs, but is in good agreement with predictions from linear dichroism (30) and UV/VIS spectroscopy (31–33). Importantly, investigation of rhodopsin decay kinetics by NMR spectroscopy with 15N-labeled tryptophan further supports the view of a position of Trp2656.48 during rhodopsin decay that is different from both positions in the dark and meta-II states (34).
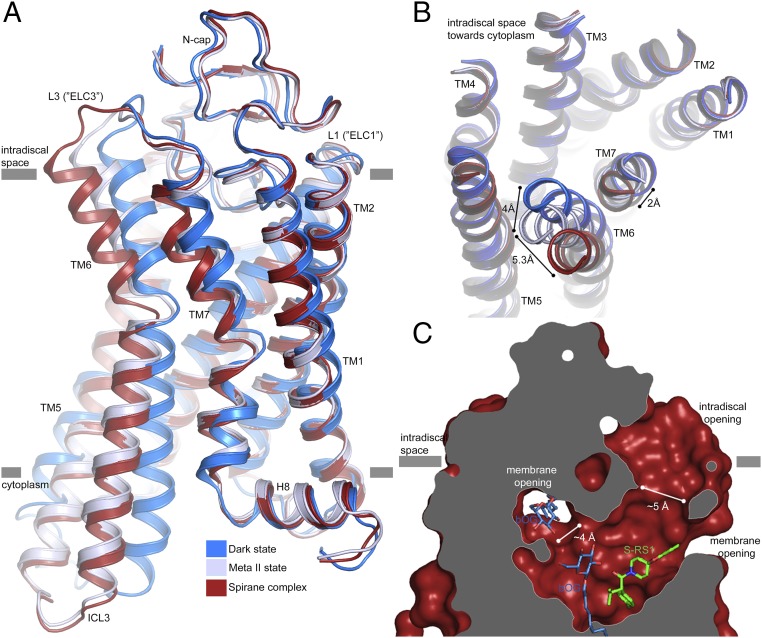
On the intradiscal side, repositioning of Trp2652.48 is accompanied by a significant rearrangement of the seven transmembrane helix bundle (Fig. 4A). In particular, TM6 shifts by 4 and 5.3 Å in comparison with the meta-II and dark states, respectively (Fig. 4B). Smaller adjustments are visible for TM7 as well as for intradiscal loops L2 and L3 that correspond to the extracellular loops ECL2 and ECL3 of other GPCRs. This conformational reorganization creates a large opening, defined by residues of TM5, TM6, L2, and L3 that connect the orthosteric binding site with the lipid bilayer and the lumen of the rod outer segment disk (Fig. 4C and SI Appendix, Fig. S8). Residues exposed at the extracellular boundary are primarily hydrophobic and highly conserved (Phe2085.43, Phe2125.47, Tyr2686.51, Val2045.39, Phe2766.59, Ala2726.55, Phe283, Tyr191, Tyr192, and Met2887.35) (SI Appendix, Fig. S9). We also observe a second cavity located between TM1 and TM7 pointing toward the cytosolic leaflet of the membrane. The boundary is primarily lined by hydrophobic residues of low conservation (Phe2937.40, Ile2867.33, Met391.34, Leu401.35, and Met183) similar to the ones reported earlier (SI Appendix, Fig. S9) (35). This second cavity is much smaller but large enough in size to harbor a detergent molecule whose head group is buried inside, the alkyl chain pointing toward the membrane (SI Appendix, Fig. S4).
Fig. 4.
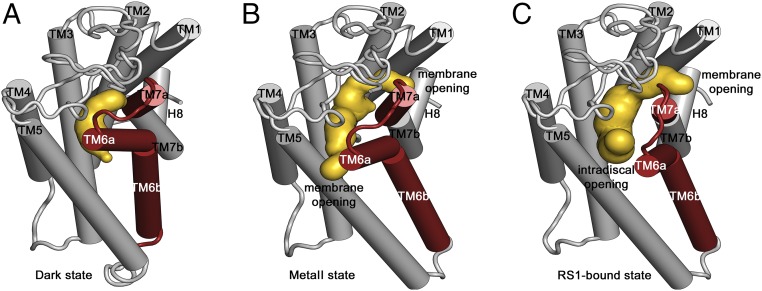
A ligand channel in S-RS1–stabilized rhodopsin. (A) Side view and (B) top view onto the superposition of the S-RS1–bound conformation (red) of opsin-, dark- (PDB ID: 1GZM, blue), and meta-II–state rhodopsin (PDB ID: 3PQR, blue-white) visualizing the helical movements in TM6 and TM7 at the intradiscal side of the membrane. The complex has a higher structural similarity to meta-II (rmsd = 0.4 Å) than dark-state rhodopsin (rmsd = 1.6 Å). (C) Ligand channel in surface representation sliced perpendicular to the membrane highlighting an intradiscal opening formed between L2 (“ECL2”), TM5, and TM6a as well as a membrane opening, large enough to permit the exchange of hydrophobic ligands including retinal.
In the closely related β-adrenergic receptors, a channel through the extracellular domain allows entry and exit of the diffusible ligands needed for activation (36, 37). In the case of dark-state rhodopsin, this channel is not observed (Fig. 5A). On the other hand, retinal was proposed to enter and exit to the lipidic membrane through two openings located between TMs 1 and 7 and TMs 5 and 6 in the near-identical active opsin and meta-II conformations (Fig. 5B) (20, 35), yet an extensive mutational analysis of this channel remained inconclusive (38). Recent studies established that retinals bind selectively to different conformations with 11-cis retinal entering through a putative new open conformation not seen in previous crystal structures (39, 40). The diameter of the intradiscal channel in our structure is between 4.8 Å and 6.8 Å in diameter, i.e., wide enough to allow passage of retinals (Fig. 5C), and is in good agreement with mutational data for binding and release of retinal (38). It is thus tempting to speculate that the surprisingly open conformation of the S-RS1–opsin complex (Fig. 5C) may represent a physiologically relevant transition state by which retinals can enter or leave the orthosteric binding site.
Fig. 5.
Ligand channel comparison of dark, meta-II, and S-RS1–bound opsin. Cartoon representation of the receptor depicting unaltered transmembrane helix regions (gray) and region of ligand-induced conformational changes (red). The caver analysis reveals the void pathways of the receptor in yellow of (A) dark-state rhodopsin bound to 11-cis retinal featuring an occluded pocket, (B) light-activated meta-II rhodopsin with bound all-trans retinal showing two small openings toward the membrane, and (C) S-RS1–bound opsin identifying a ligand channel toward the membrane and intradiscal space suggesting a potential route for retinal exchange and ligand-induced receptor breathing.
Conclusion
One of the greatest challenges to make use of structure–activity relationships in GPCRs is their conformational flexibility. Atomic resolution information provides a practical entry point for rational drug design. Pharmacologically stabilized opsin is a reminder of how unnatural ligands can stabilize previously unresolved protein conformations, even in a tightly regulated GPCR such as rhodopsin.
Our study clearly demonstrates that rhodopsin is a pharmacological target similar to other GPCRs and remains a valuable system to study GPCRs. The S-RS1 compound and derivatives are particularly interesting because their angular shape simultaneously probes the orthosteric binding pocket similar to how the agonistic or inverse agonistic endogenous ligand do individually. A next generation of pharmacological chaperones will thus not only stabilize a particular conformation but will likely support different stages of the folding pathway providing a valuable framework for the pharmacological rescue of rhodopsin-mediated retinitis pigmentosa. Reversible binding will further allow 11-cis retinal to form a stable pigment once stabilized opsin reaches the rod outer segment membrane.
Materials and Methods
In Silico Selection of Focused Library.
Three different types of 2D ligand- and 3D shape-based similarity calculations using all-trans retinal and three published retinoid opsin binders (41, 42) as references, as well as two structure-based methods, namely, pharmacophore searching and docking, were used to compile the focused library. The crystal structure of rhodopsin with bound all-trans retinal [PDB ID: 4A4M (20)] was used as a template for a pharmacophore model performed in the software program MOE (Molecular Operating Environment software package; available at https://www.chemcomp.com/MOE-Molecular_Operating_Environment.htm) including five hydrophobic features covering the β-ionone ring and the region close to the ligand head group, as well as excluded volume features for the surrounding protein except for the flexible, retinal-binding Lys2967.43. Additional pharmacophore searches were performed with models mimicking a potential interaction with Lys2967.43. Molecular docking was performed with an additional hydrogen-bonding constraint between any ligand atom and NZ of Lys2967.43 to exclude covalent binders.
Protein Production, Crystal Soaking, and Structure Determination.
A detailed description of the cloning, expression, purification, and crystallization process of human opsin N2C/N282C and bovine opsin N2C/D282C can be found here (29, 43, 44). Crystal soaks using 3.75–5 mM compound concentrations were incubated for 24 h before flash-cooling of crystals with liquid nitrogen. Statistics of structure determination are listed in SI Appendix, Table S1.
Automated Thermofluor Assay.
A published assay (45) based on the N-4-(7-diethylamino-4-metyl-3-coumarinyl)phenylmalemeide (cpm) dye was adapted and automated to screen for stabilizing compounds. Melting curves were obtained applying a temperature gradient from 25 to 95 °C and a heating rate of 0.25 °C/s. All liquid-handling steps were performed using a Bravo Automated Liquid handling platform and a 96LT head. Melting temperatures (TM) were calculated and analyzed by automated scripts based on a fit to the Boltzmann equation.
Fluorescence-Detection Size-Exclusion Chromatography Thermal Shift Assay.
The method was used to validate the compound-induced stabilization of human opsin N2C/N282C (46). Melting temperatures were determined in duplicate by incubation of 0.26 µM human opsin N2C N282C with and without compounds at 10 µM concentration (10 mM stock solution in DMSO) for 10 min in a Prime Thermal cycler at 12 different temperatures between 35 and 60 °C in an Eppendorf twin.tec PCR plate in a buffer containing 10 mM Hepes (HCl) pH 7.4, 200 mM NaCl, 0.125% (wt/vol) n-decyl-β-D-maltopyranoside. Nonaggregated protein was quantified using tryptophan fluorescence after injection of 4 µL protein solution and separation on an HPLC system (γexcitation of 280 nm, γemission of 350 nM). Melting temperatures (TM) were determined using the Boltzmann equation fit in GraphPad Prism including normalization defining 0% as the smallest and 100% as the largest identified value within the tryptophan fluorescence dataset.
Pharmacotrafficking Complementation Assay.
Commercial rhodopsin P23H stable cell lines (ID 93–1077C3; DiscoverRx) were used to express a C-terminal fusion of mouse P23H opsin with the small β-galactosidase subunit and the fusion of the large subunit of β-galactosidase and the pleckstrin homology domain of phospholipase C-δ as described (22). For measurements, the cell medium was exchanged to reaction reagent, and ∼5,000 cells per reaction were transferred to a 384-well plate. Detection of cell surface expression was performed with PathHunter Detection Kit reagent in a luminescence plate reader. Measurements were evaluated with GraphPad Prism using the normalized dose–response curve with the lowest value being defined as 0% and the highest value as 100% in the compound dataset. All experiments were done under red-light conditions due to the instability of the control 9-cis retinal to normal light.
Compound Production and Characterization.
S- and R-RS1, the racemate, and additional compounds were synthesized at Chembiotek and purified. All reactions were monitored by TLC, and the reaction construct was confirmed by NMR. The purity of final compounds as measured by liquid chromatography coupled to mass spectrometry was at least >95%. The two enantiomers of RS1 were separated on a Reprosil Chiral NR column. The identity of compounds S-RS1 and R-RS1 was further confirmed by small-molecule crystallography (SI Appendix, Table S4).
Supplementary Material
Acknowledgments
We are grateful to Ralf Thoma, Guido Hartmann, Andreas Kuglstatter, and Sascha Fauser for fruitful discussions. We would like to express our gratitude to Andreas Topp for his expertise and help for assay development and screening. We thank Marcello Foggetta, Martin Siegrist, Agnes Baronina, and Christian Miscenic for technical assistance for cell line and biomass production. We thank Anke Eisenmann for programming automated data evaluation software as well as Rene Rietmann, Tom Kissling, Alexandre Zimmermann, Martin Brunner, Daniel Zimmerli, Inken Plitzko, Christian Bartelmus, Eric Kuzsnir, and Arne Rufer for help relating to compounds and screening. We also thank Jörg Benz for advice and expertise during crystallization as well as Expose GmbH and the team at the beam line X10SA at Swiss Light Source. D.M. was supported by the Roche Postdoctoral Fellowship (RPF ID: 298) and a European Molecular Biology Organization long-term fellowship (ALTF-455). We further acknowledge the Swiss National Science Foundation for Grants 310030_153145 (to G.F.X.S.) and 31003A_159558 (to J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org [PDB ID codes 6FK6 (S-RS01), 6FK7 (RS06), 6FK8 (RS08), 6FK9 (RS09), 6FKA (RS11), 6FKB (RS13), 6FKC (RS15) and 6FKD (RS16)]. Additional data related to this publication are available at the Cambridge Structural Database, https://www.ccdc.cam.ac.uk/solutions/csd-system/components/csd/ [CCDC codes 1819787 (R-RS1) and 1819788 (S-RS1)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718084115/-/DCSupplemental.
References
- 1.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 2.Ferrari S, et al. Retinitis pigmentosa: genes and disease mechanisms. Curr Genomics. 2011;12:238–249. doi: 10.2174/138920211795860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Punzo C, Kornacker K, Cepko CL. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci. 2009;12:44–52. doi: 10.1038/nn.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dryja TP, et al. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- 5.Sung CH, Schneider BG, Agarwal N, Papermaster DS, Nathans J. Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci USA. 1991;88:8840–8844. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krebs MP, et al. Molecular mechanisms of rhodopsin retinitis pigmentosa and the efficacy of pharmacological rescue. J Mol Biol. 2010;395:1063–1078. doi: 10.1016/j.jmb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Mendes HF, Cheetham ME. Pharmacological manipulation of gain-of-function and dominant-negative mechanisms in rhodopsin retinitis pigmentosa. Hum Mol Genet. 2008;17:3043–3054. doi: 10.1093/hmg/ddn202. [DOI] [PubMed] [Google Scholar]
- 8.Tao YX, Conn PM. Chaperoning G protein-coupled receptors: from cell biology to therapeutics. Endocr Rev. 2014;35:602–647. doi: 10.1210/er.2013-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Generoso SF, et al. Pharmacological folding chaperones act as allosteric ligands of Frizzled4. Nat Chem Biol. 2015;11:280–286. doi: 10.1038/nchembio.1770. [DOI] [PubMed] [Google Scholar]
- 10.Janovick JA, et al. Restoration of testis function in hypogonadotropic hypogonadal mice harboring a misfolded GnRHR mutant by pharmacoperone drug therapy. Proc Natl Acad Sci USA. 2013;110:21030–21035. doi: 10.1073/pnas.1315194110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noorwez SM, et al. Retinoids assist the cellular folding of the autosomal dominant retinitis pigmentosa opsin mutant P23H. J Biol Chem. 2004;279:16278–16284. doi: 10.1074/jbc.M312101200. [DOI] [PubMed] [Google Scholar]
- 12.Noorwez SM, et al. Pharmacological chaperone-mediated in vivo folding and stabilization of the P23H-opsin mutant associated with autosomal dominant retinitis pigmentosa. J Biol Chem. 2003;278:14442–14450. doi: 10.1074/jbc.M300087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohgane K, Dodo K, Hashimoto Y. Retinobenzaldehydes as proper-trafficking inducers of folding-defective P23H rhodopsin mutant responsible for retinitis pigmentosa. Bioorg Med Chem. 2010;18:7022–7028. doi: 10.1016/j.bmc.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Li T, et al. Effect of vitamin A supplementation on rhodopsin mutants threonine-17–> methionine and proline-347–> serine in transgenic mice and in cell cultures. Proc Natl Acad Sci USA. 1998;95:11933–11938. doi: 10.1073/pnas.95.20.11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saari JC. Vitamin A metabolism in rod and cone visual cycles. Annu Rev Nutr. 2012;32:125–145. doi: 10.1146/annurev-nutr-071811-150748. [DOI] [PubMed] [Google Scholar]
- 16.Noorwez SM, Ostrov DA, McDowell JH, Krebs MP, Kaushal S. A high-throughput screening method for small-molecule pharmacologic chaperones of misfolded rhodopsin. Invest Ophthalmol Vis Sci. 2008;49:3224–3230. doi: 10.1167/iovs.07-1539. [DOI] [PubMed] [Google Scholar]
- 17.Singhal A, et al. Insights into congenital stationary night blindness based on the structure of G90D rhodopsin. EMBO Rep. 2013;14:520–526. doi: 10.1038/embor.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singhal A, et al. Structural role of the T94I rhodopsin mutation in congenital stationary night blindness. EMBO Rep. 2016;17:1431–1440. doi: 10.15252/embr.201642671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 20.Deupi X, et al. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci USA. 2012;109:119–124. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumoto H, Yoshizawa T. Existence of a beta-ionone ring-binding site in the rhodopsin molecule. Nature. 1975;258:523–526. doi: 10.1038/258523a0. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, et al. A high-throughput drug screening strategy for detecting rhodopsin P23H mutant rescue and degradation. Invest Ophthalmol Vis Sci. 2015;56:2553–2567. doi: 10.1167/iovs.14-16298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimamura T, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JH, et al. Opsin, a structural model for olfactory receptors? Angew Chem Int Ed Engl. 2013;52:11021–11024. doi: 10.1002/anie.201302374. [DOI] [PubMed] [Google Scholar]
- 25.Goren MA, et al. Constitutive phospholipid scramblase activity of a G protein-coupled receptor. Nat Commun. 2014;5:5115. doi: 10.1038/ncomms6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han M, Lin SW, Smith SO, Sakmar TP. The effects of amino acid replacements of glycine 121 on transmembrane helix 3 of rhodopsin. J Biol Chem. 1996;271:32330–32336. doi: 10.1074/jbc.271.50.32330. [DOI] [PubMed] [Google Scholar]
- 27.Deupi X, Standfuss J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr Opin Struct Biol. 2011;21:541–551. doi: 10.1016/j.sbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 29.Standfuss J, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chabre M, Breton J. Orientation of aromatic residues in rhodopsin. Rotation of one tryptophan upon the meta I to meta II transition afer illumination. Photochem Photobiol. 1979;30:295–299. doi: 10.1111/j.1751-1097.1979.tb07150.x. [DOI] [PubMed] [Google Scholar]
- 31.Rafferty CN, Muellenberg CG, Shichi H. Tryptophan in bovine rhodopsin: its content, spectral properties and environment. Biochemistry. 1980;19:2145–2151. doi: 10.1021/bi00551a023. [DOI] [PubMed] [Google Scholar]
- 32.Lin SW, Sakmar TP. Specific tryptophan UV-absorbance changes are probes of the transition of rhodopsin to its active state. Biochemistry. 1996;35:11149–11159. doi: 10.1021/bi960858u. [DOI] [PubMed] [Google Scholar]
- 33.Kochendoerfer GG, Kaminaka S, Mathies RA. Ultraviolet resonance Raman examination of the light-induced protein structural changes in rhodopsin activation. Biochemistry. 1997;36:13153–13159. doi: 10.1021/bi971541c. [DOI] [PubMed] [Google Scholar]
- 34.Stehle J, et al. Characterization of the simultaneous decay kinetics of metarhodopsin states II and III in rhodopsin by solution-state NMR spectroscopy. Angew Chem Int Ed Engl. 2014;53:2078–2084. doi: 10.1002/anie.201309581. [DOI] [PubMed] [Google Scholar]
- 35.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 36.Wang T, Duan Y. Ligand entry and exit pathways in the beta2-adrenergic receptor. J Mol Biol. 2009;392:1102–1115. doi: 10.1016/j.jmb.2009.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.González A, Perez-Acle T, Pardo L, Deupi X. Molecular basis of ligand dissociation in β-adrenergic receptors. PLoS One. 2011;6:e23815. doi: 10.1371/journal.pone.0023815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piechnick R, et al. Effect of channel mutations on the uptake and release of the retinal ligand in opsin. Proc Natl Acad Sci USA. 2012;109:5247–5252. doi: 10.1073/pnas.1117268109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schafer CT, Farrens DL. Conformational selection and equilibrium governs the ability of retinals to bind opsin. J Biol Chem. 2015;290:4304–4318. doi: 10.1074/jbc.M114.603134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schafer CT, Fay JF, Janz JM, Farrens DL. Decay of an active GPCR: Conformational dynamics govern agonist rebinding and persistence of an active, yet empty, receptor state. Proc Natl Acad Sci USA. 2016;113:11961–11966. doi: 10.1073/pnas.1606347113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garvey DS. 2012. US Patent Appl WO2012174064A1.
- 42.Garvey DS, Larosa GJ, Greenwood JR, Frye LL. 2011. US Patent Appl WO2011155983A1.
- 43.Mattle D, Singhal A, Schmid G, Dawson R, Standfuss J. Mammalian expression, purification, and crystallization of rhodopsin variants. Methods Mol Biol. 2015;1271:39–54. doi: 10.1007/978-1-4939-2330-4_3. [DOI] [PubMed] [Google Scholar]
- 44.Xie G, et al. Preparation of an activated rhodopsin/transducin complex using a constitutively active mutant of rhodopsin. Biochemistry. 2011;50:10399–10407. doi: 10.1021/bi201126r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexandrov AI, Mileni M, Chien EYT, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Hattori M, Hibbs RE, Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure. 2012;20:1293–1299. doi: 10.1016/j.str.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.