Significance
Single-base substitutions are capable of producing transformative phenotypic changes. While methods to classify such mutations are well established, it is difficult to modulate or preclude their occurrence in a direct and efficacious manner. In this study, we refine the specificity of the CRISPR-Cas9 system and present a general framework for proactively preventing the occurrence of point mutations. This “mutation prevention system” is a broadly useful tool for the study and control of DNA substitutions, particularly in contexts where an associated phenotype or evolutionary pathway is undesirable.
Keywords: Cas9, tuned gRNA, tgRNA, mutation prevention, synthetic biology
Abstract
Here, we present a generalized method of guide RNA “tuning” that enables Cas9 to discriminate between two target sites that differ by a single-nucleotide polymorphism. We employ our methodology to generate an in vivo mutation prevention system in which Cas9 actively restricts the occurrence of undesired gain-of-function mutations within a population of engineered organisms. We further demonstrate that the system is scalable to a multitude of targets and that the general tuning and prevention concepts are portable across engineered Cas9 variants and Cas9 orthologs. Finally, we show that the mutation prevention system maintains robust activity even when placed within the complex environment of the mouse gastrointestinal tract.
There are few biological perturbations that rival point mutations when it comes to the power to affect phenotypic change. Single-base substitutions endow pathogens with resistance to antibiotics and rogue cells with oncogenic potential (1, 2). Despite decades of research into the causes and effects of point mutations, no tool exists to directly prevent their occurrence. At best, we can track when mutations occur and utilize them as prognostic factors to predict emergent properties or chemotherapeutic outcome (3, 4). Here we describe an in vivo “mutation prevention” system that can prevent the occurrence of targeted point mutations with little to no latent toxicity to the host organism.
The system is predicated on Streptococcus pyogenes Cas9 (hereon referred to as Cas9) and its orthologs: endonucleases directed to a target locus via hybridization between an associated guide RNA (gRNA) and a target site near a required protospacer adjacent motif (PAM) (5–8). While incredibly plastic, Cas9 suffers from difficult-to-predict nonspecific activity, which can be tolerant to multiple mismatches between the gRNA and the inappropriately bound locus (9–20). Several groups have presented approaches that confer greater specificity to Cas9 (in some cases demonstrating single-nucleotide specificity) (13, 21–29). These techniques, including those involving engineered Cas9 proteins, can still fall short in applications where near-absolute single-nucleotide discrimination is required, necessitating the introduction of alternative approaches. To endow Cas9 with the ability to discriminate between single-nucleotide polymorphisms (SNPs), we developed a screening methodology that confers single-nucleotide specificity through the selection of a tuned guide RNA (tgRNA).
Results
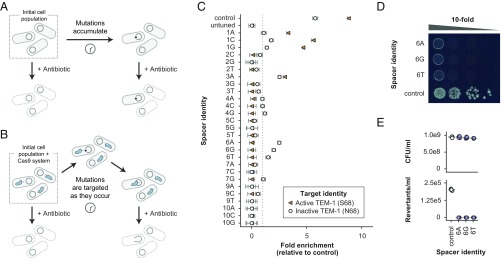
To demonstrate the feasibility of preventing the emergence of point mutations with a Cas9/tgRNA system, we generated a strain of Escherichia coli deficient in mismatch repair (MG1655-mutS::kan), to increase mutation rates, and harboring a plasmid encoding a catalytically inactive version of the TEM-1 β-lactamase (TEM-1-S68N) (30, 31). The active version of TEM-1 confers resistance to β-lactam antibiotics, such as ampicillin (32). Under typical growth conditions, errors in DNA replication and repair lead to the accumulation of mutations over time. Some of these mutations result in the reversion of the inactive N68 allele to its catalytically active form, S68. The number of revertants within the population is quantified by plating overnight cultures to solid media containing ampicillin (Fig. 1A).
Fig. 1.
Mutation prevention system overview and performance. (A) Mutations conferring antibiotic resistance occur stochastically over time in native bacterial populations. When antibiotic pressure is applied to the population, cells with resistance-conferring mutations survive, while wild-type cells die. (B) Cells with the mutation prevention system grow and divide normally. When a targeted resistance-causing mutation occurs it is cut by Cas9, causing its loss from the population. (C) Screening for tgRNAs designed to prevent the reversion of inactive TEM-1 to its catalytically active form (conferring ampicillin resistance). In this assay, properly tuned gRNAs are expected to be enriched in the presence of the inactive target (N68) and depleted in the presence of the active target (S68). The y axis indicates the additional tuning mismatch that is inserted into each respective gRNA. The dotted line represents a relative fold enrichment of 1, above which library members are considered enriched and below which they are considered depleted; n = 3 independent biological replicates. (D) Representative spot assay demonstrating the performance of mutation prevention systems containing the three most discriminatory tgRNAs (as indicated by the screening process). Each spot represents a 10-fold serial dilution. (E) The most discriminatory tgRNAs consistently prevent mutations to baseline levels, while not affecting the overall number of colony-forming units present within the culture under nonselective conditions; n = 5 independent biological replicates. All reversion rates are significant relative to the control (P < 0.01); all cfu/mL counts are not significant relative to the control (P > 0.01). For all experiments the control gRNA represents a guide that targets a sequence not present within the E. coli genome. All error bars represent SEM.
We postulated that we could prevent these reversions by introducing a Cas9 system tuned to cut the active TEM-1 variant while exhibiting undetectable activity against the inactive allele. In practice, such a system would benignly persist within cells until the occurrence of the reversion. Cas9 would then cut at the active allele, causing irreparable genetic damage and a decrease in the overall number of ampicillin-resistant cells (Fig. 1B). By design, the inactive form of TEM-1 differs from the active form by a single-nucleotide substitution (203G > A). Our initial, naive system included a gRNA with full complementarity to the active S68 allele (203G) and thus only differed from the inactive N68 allele by a single nucleotide. As expected, this approach performed poorly, as the Cas9/gRNA system cut both the active and inactive forms efficiently. Consequently, the system exhibited high toxicity even in the absence of the reversion (data not shown). Furthermore, we found that even the high-fidelity eCas9 and Cas9-HF1 variants failed to confer single-nucleotide discrimination with the naive one-off gRNA, as indicated by a sharp decrease in viability for cells carrying the system (25, 26) (SI Appendix, Table S1).
Previous work examining the factors that influence Cas9 behavior has shown that its activity is easily modulated by the introduction of mismatches within the gRNA that prevent full complementarity between it and the target site (5, 10, 11). We posited that, for a given pair of targets that differ by a single base, there may exist a set of mismatches within the gRNA that would eliminate activity on one of the variants while maintaining robust activity on the other. To test our hypothesis, we screened a library of gRNAs that each differed from the active TEM-1 allele (S68) by a single mismatch and from the inactive TEM-1 allele (N68) by two mismatches (SI Appendix, Fig. S1). We focused our mutational analysis on the region within the gRNA that interacts with the bases proximal to the PAM at the target locus, as these residues have been shown to be most critical for Cas9 activity (5, 9–11).
Our screen was designed such that an ideal tgRNA candidate was expected to be markedly depleted when in the presence of the functional S68 allele (signifying that it cut the undesired TEM-1 variant that we wish to prevent) and strongly enriched when tested against the nonfunctional N68 allele (suggesting that it did not cut the desired TEM-1 variant we wish to maintain). Upon performing the screen, we identified several promising tgRNA candidates that were subjected to further characterization (Fig. 1C).
Individual testing of these select library members revealed that the fold depletion we observe within the screen strongly correlates with their respective activities in isolation (SI Appendix, Fig. S2). This validates the capability of our library approach to accurately identify the most discriminatory gRNAs. Based on the results of our screen and subsequent validation, we selected the three candidate tgRNAs with the greatest discriminatory power, 6A, 6G, and 6T, for additional analysis. We tested their ability to prevent emergence of the active TEM-1 allele from within a population of cells containing only the inactive TEM-1 variant. As predicted, all three tgRNAs were able to prevent reversion to the active TEM-1 allele by several orders of magnitude over cells with a nonfunctional control gRNA, while also exhibiting minimal levels of baseline toxicity (Fig. 1 D and E).
Because SP-Cas9 represents one of thousands of known Cas9 proteins, each with differences in efficiency, specificity, and PAM requirements, we sought to verify that our approach was generalizable to two commonly used Cas9 orthologs: NM-Cas9 and ST1-Cas9 (33, 34). We leveraged the same TEM-1 reversion assay and gRNA screening technique to identify tgRNAs with high discriminatory power between the active and inactive targets. Each screen yielded promising tgRNA candidates that proved to be highly effective at preventing the TEM-1 reversion mutation with either NM-Cas9 or ST1-Cas9, respectively (SI Appendix, Fig. S3).
Having demonstrated the efficacy of the mutation prevention concept with our exogenous TEM-1 model, we sought to apply the same approach to prevent endogenous mutations. Toward this goal, we set out to prevent several common mutations within E. coli that confer resistance to two clinically relevant antibiotics: streptomycin and rifampicin (35).
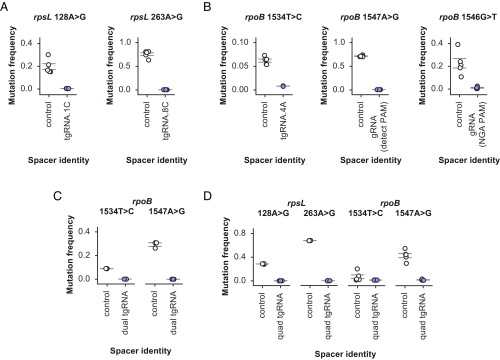
Streptomycin is an aminoglycoside antibiotic that inhibits protein synthesis by binding to bacterial ribosomes (36). There are well-characterized ribosomal protein mutations that confer resistance to streptomycin, including commonly observed substitutions in rpsL (37). We designed mutation prevention systems for two of the highest-frequency resistance-bearing mutations within rpsL: 128A > G and 263A > G. Our screening process yielded two effective tgRNAs, both of which prevented the occurrence of their targeted mutations to below the detection limits of our next-generation sequencing assay, while causing no apparent toxicity to nonmutant cells (Fig. 2A and SI Appendix, Table S2).
Fig. 2.
Endogenous mutation prevention within E. coli. (A) Frequency of rpsL 128A > G and rpsL 263A > G alleles within populations of streptomycin-resistant cells containing either tgRNA rpsL.128A > G.1C or tgRNA rpsL.263A > G.8C, respectively, compared with cells with a control gRNA. (B) Frequency of rpoB 1534T > C, rpoB 1547A > G and rpoB 1546G > T mutations within populations of rifampicin-resistant cells. For rpoB 1534T > C, a tgRNA containing an additional mismatch at the −4A position was employed to prevent the occurrence of the mutation. With rpoB.1547A > G, the mutation generates a PAM that induces Cas9 targeting. Because the rpoB.1546G > T mutation is not near a conventional Cas9 PAM, the Cas9-VQR variant with an alternative PAM requirement was employed along with an appropriate gRNA to prevent the mutation. (C) Simultaneous multiplex mutation prevention was performed against both rpoB 1534T > C and 1547A > G mutations in the same population of rifampicin-resistant cells by simultaneously expressing a pair of tgRNAs (denoted as dual tgRNA) along with the necessary Cas9 machinery. (D) Similar to C except a total of four mutations (rpsL 128A > G and 263A > G, rpoB 1534T > C and 1547A > G) were simultaneously prevented by using an array of four previously validated tgRNAs (denoted “quad tgRNA”) along with Cas9 protein. For all experiments the control gRNA represents a guide that targets a sequence not present within the E. coli genome. All error bars represent SEM, n = 5 independent biological replicates for all experiments, except for C (n = 3). Experimental means are significant relative to control means (P < 0.01) with the exception of rpob0.1534T > C.4A in D (P = 0.14).
Rifampicin is a widely used antibiotic that inhibits RNA polymerase function, with resistance arising from several well-documented mutations within the rpoB gene (38, 39). To further demonstrate the plasticity of our approach, we targeted a series of the prominent rifampicin-resistance mutations and generated unique tgRNAs against each using three disparate design principles. To prevent the rpoB 1534T > C mutation, we screened a series of gRNAs and identified a canonical tgRNA, bearing additional mutations, that was highly efficient (Fig. 2B). Targeting the rpoB 1547A > G mutation was simplified by the fact that the mutation generates a PAM that is not present within the wild-type rpoB gene. A gRNA designed to bind in the presence of the generated PAM conferred marked mutation prevention (Fig. 2B). Finally, to prevent the rpoB 1546G > T mutation, we employed the Cas9-VQR variant with altered PAM specificity (40). This was necessary given the lack of a canonical PAM sequence near the mutation site. Cas9-VQR, in the presence of an appropriate gRNA, led to efficient mutation prevention (Fig. 2B).
Having demonstrated both the portability and flexibility of our mutation prevention concept, we next assessed its scalability by targeting multiple mutations within rpoB with a single system. There was no obvious decrease in efficiency upon multiplexing, with both rpoB 1534T > C and 1547A > G mutations being simultaneously prevented within the engineered strain (Fig. 2C). To further probe the limits of our multimutation prevention system, we sought to simultaneously inhibit all of the previously targeted endogenous mutations (except rpoB 1546G > T, which requires the Cas9-VQR variant) within a single strain. Cells carrying a full complement of four tgRNAs exhibited minimal toxicity compared with cells carrying nontargeting control gRNAs and retained robust mutation prevention against all targeted loci (Fig. 2D and SI Appendix, Table S2).
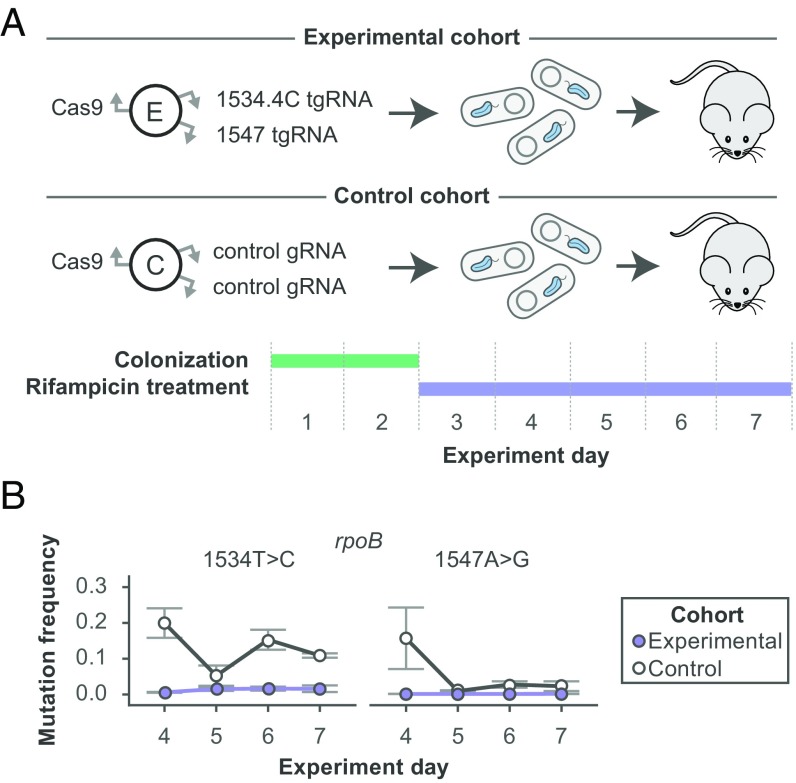
We next wanted to assess the robustness of our system by preventing point mutations from emerging within a complex in vivo environment. Toward this goal we inoculated gnotobiotic mice with one of two strains containing either a pair of nontargeted gRNAs or a pair of tgRNAs directed to prevent mutations that confer rifampicin resistance within E. coli. Two days after colonization, mice were provided rifampicin within their drinking water and the spectrum of rifampicin resistance mutations was analyzed in bacterial cells recovered from fecal samples collected on days 4–7, postcolonization (Fig. 3A). We observed minimal frequencies of the targeted point mutations within the engineered strain, relative to the much higher frequencies within the control, across all days (Fig. 3B).
Fig. 3.
Longitudinal mutation prevention in a complex in vivo environment. (A) Two cohorts of mice were colonized with either an experimental E. coli strain encoding the dual rpoB 1534T > C/1547A > G mutation prevention system or a control E. coli strain encoding a system with two gRNAs that do not cut anywhere in its genome. After a 2-d initial colonization, mice were provided water containing the antibiotic rifampicin for the remaining 5 d of the experiment. (B) The mutation prevention system successfully prevented the occurrence of the two targeted mutations for the duration of the rifampicin treatment. All error bars represent SEM, n = 2 independent biological replicates for control cohort, n = 3 independent biological replicates for experimental cohort.
Discussion
It is expected that the mutator strain of E. coli utilized within all of our experiments would commensurately increase the frequency of mutations that disable the mutation prevention system itself (3, 30). Given that the system is likely breaking at an appreciable rate, the fact that we do not see obvious degradation in its performance over time likely indicates that the system places a negligible fitness burden on the host organism. If the system were to place a significant fitness burden on its host, one would expect individuals with attenuated or broken systems to rapidly overtake the population, thus facilitating escape from its selection. It is thanks to this minimal fitness burden that escape is an extremely rare event. This is due to the fact that during any given point in time there is only a small population of cells within the total population that have inactivated the system and it is within this rare population that a second targeted gain-of-function mutation must occur. This “two-hit” requirement effectively makes escape from our mutation prevention strategy an extremely rare event and explains our efficacy at preventing undesired mutations. The robust performance of the mutation prevention system within the mouse gastrointestinal tract is particularly notable given its tolerance of multiday rifampicin selection, the absence of the active selection for the episomal plasmid encoding our system, and the fact that these experiments were performed within a highly mutagenic mismatch repair deficient background of E. coli.
Although our mutation prevention strategy requires antecedent knowledge of the target mutation(s), we note that there is already a plethora of known gain-of-function mutations that endow living cells with undesired phenotypes. In these cases, users may want to prevent a subset of these mutations to gain deeper insight into the existence and effects of other rarer alleles. In addition, rapid increases in both the affordability and adoption rate of next-generation sequencing will catalyze the eventual systematic characterization of genomic mutations that enable cells to evade the selective pressures imposed upon them by both natural and artificial systems. Our method will enable experimentalists to capitalize upon this expansive characterization, and in turn, manipulate it to prevent undesired outcomes.
This study focused on preventing gain-of-function mutations within microbial systems, in which cutting leads to loss of episomal elements or lethality when the target is present within the genome (41–44). When applying similar strategies to higher eukaryotes the effect of nonhomologous end joining-based mutagenesis (NHEJ) must be considered, as noninactivating mutations have been shown to occur that can themselves lead to gain-of-function phenotypes (45, 46). As the outcome of NHEJ-based repair has been shown to be nonrandom, a potential strategy to mitigate Cas9 derived gain-of-function alleles would be to prescreen the selected target locus and tgRNA to determine the spectrum of observed repair profiles, post Cas9-mediated cleavage.
The design of sufficiently discriminatory tgRNAs currently necessitates library screens in the spirit of those described within this text. That is, it is necessary to empirically test a small library of candidate tgRNAs to find the guide sequence that is both effective at cutting the target allele while exhibiting minimal cutting against the desired allele. Fortunately, these screens are easy to perform with standard laboratory equipment and do not require costly high-throughput oligo synthesis. In some cases, more extensive screens (or further system optimization) may be required due to lingering levels of system toxicity, as is evidenced by the mild decreases in cfu/mL that we saw with the NM and ST1 tgRNA libraries (SI Appendix, Fig. S3). We do not, at this time, have sufficient screening data to propose an in silico model for forward design of tgRNAs; however, we do anticipate that, given sufficient adoption of this technique, such an approach may be possible in the future.
Even in its nascent form, the described mutation prevention system provides the scientific community with a powerful means to directly manipulate evolutionary outcomes and in doing so gain fundamental insights into evolutionary plasticity and adaptive mechanisms. Furthermore, as our ability to engineer targeted nucleases such as Cas9 continues to rapidly improve, one can imagine the eventual generation of increasingly complex mutation prevention strategies. We further expect that the generalizable principle of guide RNA tuning will exhibit synergistic performance improvements with newly engineered, high-specificity variants of Cas9 (26, 27, 29), as well as orthogonal DNA binding proteins like Cpf1 (47, 48). As a result, applications requiring true single-nucleotide specificity, such as the disabling of diseased alleles delineated by SNPs, will finally be realized.
Materials and Methods
Experiments were performed in an E. coli MG1655-mutS::kan background (received as a gift from Dr. Harris Wang, Columbia University, New York); all gRNA/tgRNA screening experiments were performed in a wild-type E. coli MG1655 background. Overnight liquid cultures were grown in lysogeny broth (LB) supplemented with the appropriate antibiotic(s) and incubated at 37 °C for 12 h, with shaking. Overnight solid cultures (including spot assays and mutation assays) were grown on LB agar plates supplemented with the appropriate antibiotic(s) and incubated at 37 °C for 12–16 h. The mutation rates for experiments were determined using either resistant colony-forming units (TEM-1 based assay) or through next generation sequencing (endogenous mutation prevention experiments). All mouse experiments were performed in accordance with and as approved by the Harvard Medical School Institutional Animal Care and Use Committee (protocol no. 04957) and Harvard Medical School Committee on Microbiological Safety (approval no. 12-085). Detailed protocols are contained within SI Appendix.
Supplementary Material
Acknowledgments
We thank L. Certain, X. Guo, A. Tsou, A. Tung, S. Vora, N. C. Yeo, and J. Zhu for helpful discussions and technical assistance. We acknowledge research support from the US National Institutes of Health National Human Genome Research Institute Grant P50 HG005550, and the Wyss Institute for Biologically Inspired Engineering. A.C. was funded by the National Cancer Institute Grant 5T32CA009216-34 and a Career Award for Medical Scientists from the Burroughs Wellcome Fund. J.J.C. was supported by the Defense Threat Reduction Agency Grant HDTRA1-15-1-0051 and the Paul G. Allen Frontiers Group.
Footnotes
Conflict of interest statement: G.M.C. is a founder and advisor for Editas Medicine. G.M.C. has equity in Editas, Enevolv, and Caribou Biosciences. A patent has been filed by Harvard University related to the work within this article.
Data deposition: The sequences reported in this paper have been deposited with the National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA329016 (accession nos. SRR3909289, SRR3909312–SRR3909322, SRR3909346, SRR3909347, SRR3909359, SRR3909371–SRR3909373, SRR3909397–SRR3909399, SRR3909417, SRR3909422, SRR3909435, SRR3909456, SRR3909457, SRR3909458, SRR3909460, SRR3909483, SRR3909484, SRR3909507, SRR3909508, SRR3909531, SRR3909532, SRR3909553, SRR3909554, SRR3909578, SRR3909579, SRR3909580, SRR3909597, SRR3909603, SRR3909621, SRR3909634, SRR3909638, SRR3909654, SRR3909655, SRR3909678, SRR3909694, SRR3909695, SRR3909711, SRR3909719, SRR3909731, SRR3909747, SRR3909770, SRR3909786, SRR3909795, SRR3909813, SRR3909820, SRR3909821, SRR3909844, SRR3909848, SRR3909866, SRR3909887, SRR3909892, SRR3909907, SRR3909923, SRR3909924, SRR3909949, SRR3909950, SRR3909973, SRR3909974, SRR3909997, SRR3909998, SRR3910021, SRR3910022, SRR3910045, SRR3910062, SRR3910072, SRR3910084, SRR3910085, SRR3910086, SRR3910087, SRR3910088, SRR3910089, SRR3910090, SRR3910091, SRR3910092, SRR3910093, SRR3910094, SRR3910095, SRR3910096, SRR3910117, SRR3910118, SRR3910139, and SRR3910153).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718148115/-/DCSupplemental.
References
- 1.Andersson DI, Hughes D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 2.Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nat Rev Genet. 2013;14:703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köser CU, Ellington MJ, Peacock SJ. Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 2014;30:401–407. doi: 10.1016/j.tig.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vockley JG, Niederhuber JE. Diagnosis and treatment of cancer using genomics. BMJ. 2015;350:h1832. doi: 10.1136/bmj.h1832. [DOI] [PubMed] [Google Scholar]
- 5.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semenova E, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33:175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 20.Kim D, et al. Digenome-seq: Genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243, 1, 243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 21.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyvekens N, Topkar VV, Khayter C, Joung JK, Tsai SQ. Dimeric CRISPR RNA-guided FokI-dCas9 nucleases directed by truncated gRNAs for highly specific genome editing. Hum Gene Ther. 2015;26:425–431. doi: 10.1089/hum.2015.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen JS, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegel EC, Bryson V. Mutator gene of Escherichia coli B. J Bacteriol. 1967;94:38–47. doi: 10.1128/jb.94.1.38-47.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox EC. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- 32.Drawz SM, Bonomo RA. Three decades of β-lactamase inhibitors. Clin Microbiol Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou Z, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi R, Itagaki N, Sugawara I. Overview of anti-tuberculosis (TB) drugs and their resistance mechanisms. Mini Rev Med Chem. 2007;7:1177–1185. doi: 10.2174/138955707782331740. [DOI] [PubMed] [Google Scholar]
- 36.Schatz A, Bugle E, Waksman S. Streptomycin, a substance exhibiting antibiotic activity against Gram-positive and Gram-negative bacteria. Exp Biol Med. 1944;55:66–69. doi: 10.1097/01.blo.0000175887.98112.fe. [DOI] [PubMed] [Google Scholar]
- 37.Timms AR, Steingrimsdottir H, Lehmann AR, Bridges BA. Mutant sequences in the rpsL gene of Escherichia coli B/r: Mechanistic implications for spontaneous and ultraviolet light mutagenesis. Mol Gen Genet. 1992;232:89–96. doi: 10.1007/BF00299141. [DOI] [PubMed] [Google Scholar]
- 38.Sensi P, Greco AM, Ballotta R. Rifomycin. I. Isolation and properties of rifomycin B and rifomycin complex. Antibiot Annu. 1959–1960;7:262–270. [PubMed] [Google Scholar]
- 39.Jin DJ, Gross CA. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 40.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiCarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol. 2014;32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bikard D, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Overbeek M, et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol Cell. 2016;63:633–646. doi: 10.1016/j.molcel.2016.06.037. [DOI] [PubMed] [Google Scholar]
- 46.Ipsaro JJ, et al. Rapid generation of drug-resistance alleles at endogenous loci using CRISPR-Cas9 indel mutagenesis. PLoS One. 2017;12:e0172177. doi: 10.1371/journal.pone.0172177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shmakov S, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.