Significance
Recent findings from in vivo fluorescence and immunolabeling measurements hinted at the possible role of outer membrane (OM) extensions as Shewanella oneidensis MR-1 nanowires. However, a detailed understanding of the architecture and electron transport mechanism along OM extensions was lacking. In this work, we report a unique setup for correlative light and electron microscopy of Shewanella OM extensions and demonstrate that they are chains of interconnected outer membrane vesicles with densities, consistent with periplasmic and OM cytochromes, distributed along their length. We propose, based on the packing density of cytochromes measured from electron cryotomograms, that the electron transport mechanism involves a combination of direct electron hopping and diffusion of electron carriers.
Keywords: extracellular electron transport, electron cryotomography, membrane cytochromes, bacterial nanowires, Shewanella
Abstract
Bacterial nanowires have garnered recent interest as a proposed extracellular electron transfer (EET) pathway that links the bacterial electron transport chain to solid-phase electron acceptors away from the cell. Recent studies showed that Shewanella oneidensis MR-1 produces outer membrane (OM) and periplasmic extensions that contain EET components and hinted at their possible role as bacterial nanowires. However, their fine structure and distribution of cytochrome electron carriers under native conditions remained unclear, making it difficult to evaluate the potential electron transport (ET) mechanism along OM extensions. Here, we report high-resolution images of S. oneidensis OM extensions, using electron cryotomography (ECT). We developed a robust method for fluorescence light microscopy imaging of OM extension growth on electron microscopy grids and used correlative light and electron microscopy to identify and image the same structures by ECT. Our results reveal that S. oneidensis OM extensions are dynamic chains of interconnected outer membrane vesicles (OMVs) with variable dimensions, curvature, and extent of tubulation. Junction densities that potentially stabilize OMV chains are seen between neighboring vesicles in cryotomograms. By comparing wild type and a cytochrome gene deletion mutant, our ECT results provide the likely positions and packing of periplasmic and outer membrane proteins consistent with cytochromes. Based on the observed cytochrome packing density, we propose a plausible ET path along the OM extensions involving a combination of direct hopping and cytochrome diffusion. A mean-field calculation, informed by the observed ECT cytochrome density, supports this proposal by revealing ET rates on par with a fully packed cytochrome network.
Redox reactions are essential to all biological energy conversion strategies (1). In respiratory organisms, free energy is harvested from the environment as electrons extracted from an electron donor are transferred through the cellular electron transport (ET) chain to a terminal electron acceptor (EA). While most eukaryotes, including humans, are dependent on molecular oxygen (O2) as their terminal EA, anaerobic prokaryotes can acquire energy by employing a wide variety of alternative EAs. Like O2, many of these EAs can diffuse inside the cell, where they participate in redox reactions with intracellular ET chain components. However, dissimilatory metal-reducing bacteria (DMRB) can also utilize insoluble EAs such as metal oxide minerals that are inaccessible to the electron transport chain components at the inner membrane, by transporting electrons across the cell envelope (2–6). This extracellular electron transport (EET) process has important implications in renewable energy technologies, wastewater treatment, bioremediation, and global biogeochemical cycles (3, 7–9).
The gram-negative bacteria Geobacter and Shewanella are two of the best-studied DMRB model systems (2, 5, 6, 10, 11) and are known to produce extracellular appendages proposed to act as bacterial nanowires, transporting electrons over micrometer-long distances to terminal extracellular EAs. Geobacter nanowires are type IV pili (12) and their electron conductivity has been attributed to either an incoherent electron hopping mechanism along a path of aromatic residues (13–15) or a coherent “metallic-like” mechanism facilitated by proposed π stacking of aromatic residues (16–18). These pili may also interact with separate extracellular redox proteins, possibly working in concert to allow EET, with the pili playing a larger role at cellular layers more distant from electrode surfaces (19). Electrochemical gating signatures of transverse conduction through Geobacter biofilms that span interdigitated electrodes appear consistent with a network of redox cofactors such as the hemes of cytochromes abundant in DMRB (20), but these measurements do not necessarily preclude a role for pili in vertical charge transport, especially at biofilm layers farther away from the underlying electrodes as described by Steidl et al. (19). Transport through the extracellular appendages of Shewanella requires the presence of multiheme cytochromes as the electron carriers (21), but a detailed analysis of the underlying mechanism and extent to which it may allow EET under physiological conditions requires a better understanding of the cytochrome distribution and structure of the appendages under native conditions.
Previous electrochemical, biochemical, genetic, and structural studies of Shewanella have identified an intricate network of redox proteins that traffic electrons from the inner membrane quinone pool through the periplasm and across the outer membrane (OM) (6, 7, 11). A critical electron transfer module is the Mtr pathway, in which electrons are transferred from the periplasmic decaheme cytochrome MtrA to the outer membrane decaheme cytochrome MtrC through the transmembrane porin MtrB (22, 23). Under conditions of direct cell surface contact with minerals or electrodes, MtrC (and a partnering decaheme cytochrome OmcA) can transfer electrons directly to these solid EAs (24). The EET rate from the surface-exposed cytochromes to such external surfaces can also be enhanced by interactions with secreted flavins that function either as cytochrome-bound cofactors (25–27) or soluble shuttles capable of interacting with even more distant EA surfaces (28, 29).
Recent findings from live fluorescence light microscopy (fLM) have hinted at the possible role of Shewanella OM extensions as bacterial nanowires that transport respiratory electrons to EAs micrometers away from the cell (30). First, the production of OM extensions has been shown to correlate with an increase in the cellular reductase activity (30). Second, the thickness of dried OM extensions (two collapsed, 5-nm–thick lipid bilayers) matches the thickness of dried and fixed conductive appendages from Shewanella oneidensis (∼10 nm) (21). Third, immunofluorescence measurements have shown that the S. oneidensis multiheme cytochromes MtrC and OmcA localize along these OM extensions (30). Importantly, the same multiheme cytochromes have been shown to be essential for the solid-state conductance of dried and fixed S. oneidensis appendages (21). Although these multiple lines of evidence point to the ability of S. oneidensis OM extensions to play a role in ET, direct conductance measurements were demonstrated only on dry samples where the distribution and conformation of the ET components may not be the same as in vivo (21). Additionally, outer membrane vesicles (OMVs), structures similar to OM extensions, have been found to be involved in various other functions including pathogenesis, microbial interactions, and survival during stress conditions (31). Therefore, to understand the extent to which S. oneidensis OM extensions can carry electrons will require direct in vivo ET measurements, challenging experiments due to the difficulty in controlling growth and positioning of OM extensions to interface electrodes. However, ultrastructural studies of the native configuration of ET components, such as presented in this paper, can provide useful information on the potential ET properties of OM extensions.
So far, the diffraction-limited resolution of fLM has precluded visualization of the macromolecular details of the OM extension and its cytochrome distribution (30). Many other details remain unclear, including formation and stabilization mechanisms, as well as the processes underlying the large morphological variation and dynamic nature of these filaments. Furthermore, it has been challenging to distinguish OM extensions from other filaments (flagella, pili, and dehydrated extracellular polymeric substances) (32, 33). Here, we use electron cryotomography (ECT) to capture near-native images of OM extensions from S. oneidensis MR-1. ECT can deliver high-resolution 3D structural details of cellular structures. By capturing the specimen in a thin layer of vitreous ice, structures of interest are preserved in a fully hydrated and essentially native state (34).
We have developed a unique experimental setup allowing bacteria to form OM extensions on an electron microscopy (EM) grid inside a perfusion flow imaging platform. Using fluorescent membrane staining, we monitored OM extension growth in real time by fLM and subsequently located and imaged the same structures by ECT. We discuss the challenges involved in retaining the fragile OM extensions for EM imaging and the methodology we developed to address these sample preparation issues. Our fLM and ECT results reveal the vesicular nature of S. oneidensis OM extensions and shed light on a potential mechanism for their stabilization as OMV chains. The high resolution of ECT reveals the positions of periplasmic and OM multiheme cytochromes under near-native conditions. We discuss how these structural measurements inform and help refine proposed models (30, 35, 36) for long-distance ET.
Results
Conditions for Reliable OM Extension Production for ECT.
While OMVs and OM extensions have previously been described in both planktonic and surface-attached Shewanella cultures using various methods such as EM, atomic force microscopy (AFM), and fLM (21, 30, 37, 38), there has not been an extensive exploration of the optimal culturing and sample preparation workflows most suitable for detection of these structures. Here we utilized negative stain transmission electron microscopy (TEM) and ECT to assess both culturing and sample preparation steps that lead to robust formation, preservation, and detection of OM extensions. These steps are summarized in Fig. S1.
We first tested liquid cultures of S. oneidensis MR-1, either from continuous-flow bioreactors (chemostats) operated under O2-limited conditions (21, 30, 37) or from batch cultures (SI Materials and Methods) by visually assaying for OM extension formation by EM. Despite the presence of membrane blebs and OMVs, longer OM extensions were rarely detected by either negative stain TEM or ECT under our cultivation conditions, even when fixed with glutaraldehyde to potentially stabilize the structures (Figs. S2 and S3). Separate imaging with scanning electron microscopy (SEM) revealed an abundance of filaments, but SEM’s lower level of structural detail makes it difficult to distinguish the target OM extensions from other filaments such as pili, flagella, and filamentous polymeric substances.
Because OM extensions in liquid cultures were only rarely observed by both ECT and negative stain TEM, we next tested surface-attached cultures. Building on our previous work utilizing coverslip-attached cultures to reveal the composition of S. oneidensis OM extensions (30), we developed a method for monitoring their growth directly on EM grids inside a perfusion flow imaging platform by fLM (Fig. 1). While extensions were seen abundantly by fLM, very few structures remained intact until the final step of either negative stain TEM or ECT workflow, whether unfixed or fixed with formaldehyde (Figs. S4 and S5). This suggests that OM extensions are fragile structures that need to be stabilized for TEM imaging. Fortunately, we found that fixation with glutaraldehyde stabilized the extensions, enabling us to reliably visualize the structures by correlative light and electron microscopy (CLEM) (Fig. S6 and Movies S1 and S2). We conclude that (i) OM extensions are more frequent and consistently present in surface-attached cultures compared with liquid cultures under our experimental conditions, and (ii), although abundantly produced in surface-attached samples, OM extensions are fragile structures that are easily disrupted unless preserved by glutaraldehyde fixation for TEM imaging.
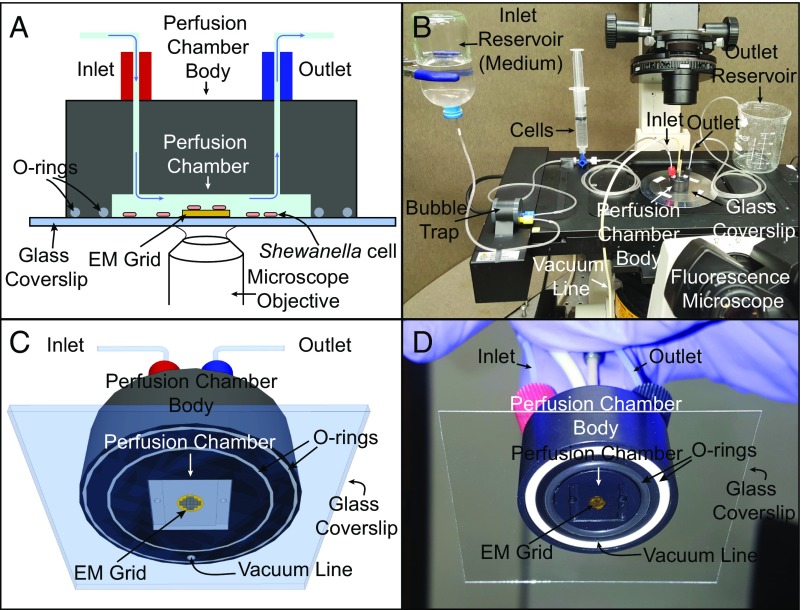
Fig. 1.
Schematic and actual images of the perfusion flow imaging platform (objects not drawn to scale). (A and B) Cross-sectional (A) and 3D (B) views of the perfusion flow imaging platform. An electron microscopy (EM) grid is glued to a glass coverslip that seals the perfusion chamber. S. oneidensis cells injected into the sealed chamber attach to the grid surface and are sustained by a continuous flow of the medium. Cells are labeled with the fluorescent membrane dye FM 4-64FX and monitored in real time for OM extension growth using an inverted fluorescent microscope placed under the perfusion chamber. (C and D) A 3D schematic (C) and image (D) of the perfusion chamber interior with an attached EM grid.
Live Fluorescence Microscopy of OM Extension Growth on EM Grids.
Building on our previous work, we developed an optimized perfusion flow imaging platform setup consisting of a microliter-volume laminar perfusion flow chamber placed on an inverted fluorescence microscope, with an EM grid-attached glass coverslip sealing the chamber (Fig. 1). S. oneidensis cells are then introduced into the chamber, where they attach to the surface of the EM grid, and sterile media are flowed into the chamber throughout the experiment. Using this setup, we observed the formation of OM extensions live on the EM grid surface with the fluorescent membrane dye FM 4-64FX. Cells were located relative to grid holes by fLM (Fig. 2 and Movie S3) to allow registration with subsequent EM imaging and thus enabling CLEM.
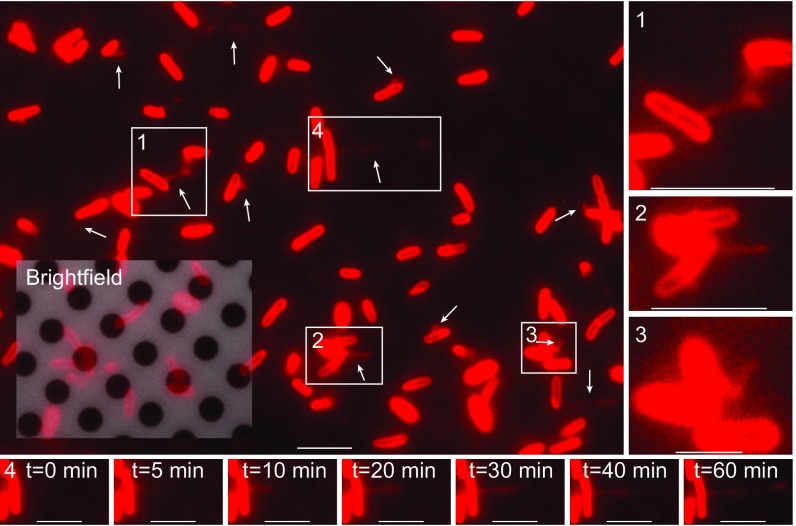
Fig. 2.
Live in vivo observation of the formation of S. oneidensis OM extensions (white arrows) on an EM grid. (Scale bar: 5 µm.) Inset is an overlap of red fluorescence and reflective brightfield channels, revealing both the holey carbon film coating the EM grid and the fluorescently labeled cells attached to it. Movie S3 is a time-lapse movie of this. (1, 2, and 3) Enlarged views of boxed regions from the main panel. (Scale bars in 1, 2, and 3: 5 µm, 5 µm, and 2 µm, respectively.) (4) Time-lapse images of the growth of a single OM extension from boxed region 4 in the main panel. t = 0 min is an arbitrary starting time point. (Scale bar: 5 µm.)
ECT Reveals OM Extensions Are Dynamic Chains of Interconnected OMVs.
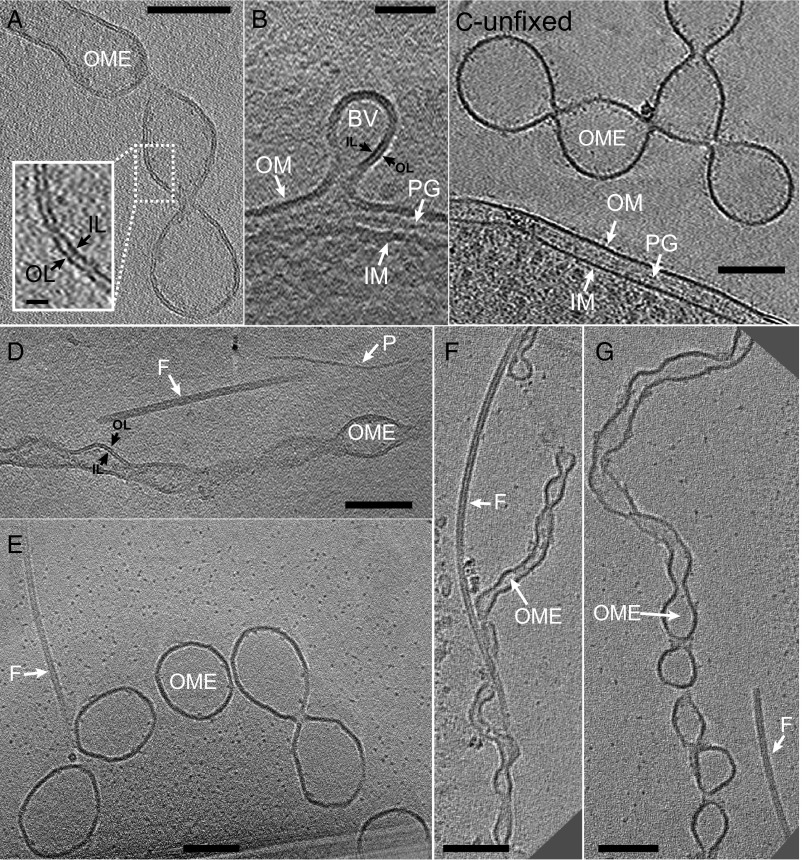
For ECT, grids from the perfusion flow imaging platform were removed, plunge frozen, and transferred to the electron microscope, where the fLM-identified OM extensions were located and imaged (Fig. 3). ECT images confirmed that appendages observed in fLM are in fact OM extensions, with the two leaflets of the lipid bilayer clearly resolved along their length (Fig. 4 A and B). Cryotomograms revealed OM extensions to be chains of interconnected OMVs in both unfixed (Fig. 4C) and fixed samples (Fig. 4 D–G). Previous fLM and AFM work showed that OM extensions cover a range of morphologies from apparently smooth tubes to clearly distinguishable OMV chains (30). Here, with the higher resolution of ECT, we observed that, with the exception of one smooth structure (Fig. S7), all OM extensions including those that appeared smooth in fLM were distinguishable as OMV chains (Figs. 3 and 4). The images also captured vesicle budding (Fig. 4B), a process that underlies the initial stage of OMV production (39). Importantly, ECT allowed us to clearly distinguish between pili, flagella, and OM extensions—the three known extracellular appendages in S. oneidensis (Fig. 4 D–G and Movies S4 and S5).
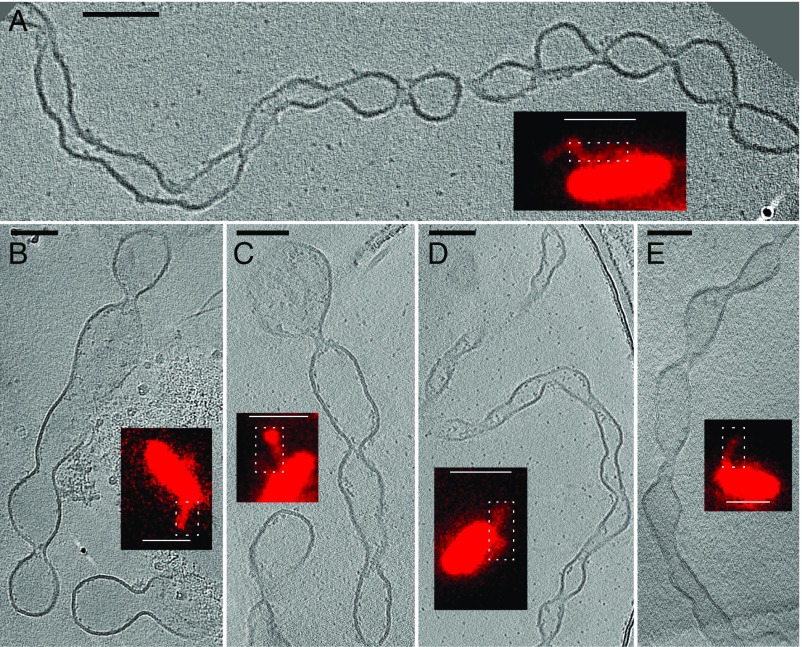
Fig. 3.
Targeting dynamic OM extensions of S. oneidensis for ECT using correlative light and electron microscopy. Target locations on fixed and plunge-frozen electron microscopy grids, from the perfusion flow imaging platform, were imaged by ECT, revealing the OMV chain morphology of the OM extensions. (A–E) Representative images from ECT, with corresponding fLM image (Insets). (ECT scale bars, 100 nm; fLM scale bars, 2 µm.) White dotted boxes in the fLM images indicate the corresponding approximate regions imaged in ECT. The ECT images shown are tomographic slices from 3D reconstructions (Fig. S6 and Movies S1 and S2).
Fig. 4.
ECT images of S. oneidensis OM extensions. (A) OM extension membrane bilayer is clearly resolved. Inset is enlarged view of boxed region with the inner and outer leaflets indicated with arrows. (Scale bar, 100 nm; Inset scale bar, 10 nm.) (B) A budding vesicle emerging as an extension of the cellular outer membrane. A similar process perhaps underlies the initial stages of OM extension formation. (Scale bar: 50 nm.) (C) OM extension from an unfixed chemostat sample exhibits identically branched OMV chain morphology as observed in both unfixed and fixed samples from the perfusion flow imaging platform. (Scale bar: 100 nm.) (Figs. S5 and S9 and Movie S13.) (D) An OM extension, a flagellum, and a pilus next to each other, allowing direct comparison of their sizes and morphologies, indicating that ECT facilitates the identification and distinguishability of different extracellular appendages in S. oneidensis. (Scale bar: 100 nm.) (Movie S4.) (E–G) ECT reveals OM extensions are of varying thicknesses and degrees of tubulation. Next to each OM extension is a flagellum that can act as a molecular marker for comparison of varying OM extension dimensions. (Scale bar: 100 nm.) (Movie S5 corresponds to F.) BV, budding vesicle; F, flagellum; IL, inner leaflet; IM, inner membrane; OL, outer leaflet; OME, OM extension; P, pilus; PG, peptidoglycan.
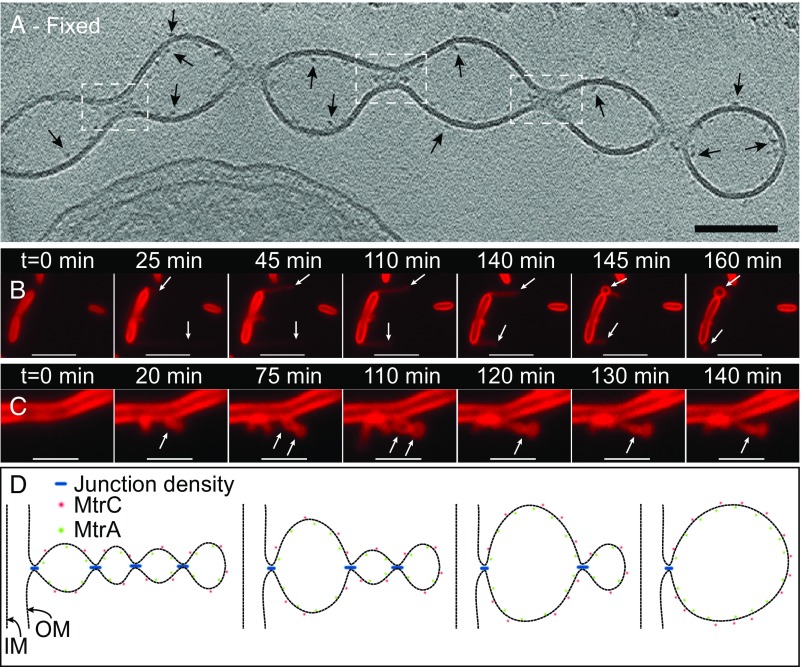
Electron-dense regions were observed at the junctions connecting neighboring vesicles throughout the length of the OM extensions in both fixed and unfixed samples (Fig. 5A, Fig. S8, and Movie S6). This finding points to yet unknown molecules that potentially facilitate the constriction of the membrane to allow OMV connections and is consistent with the fLM observations of OM extensions as dynamic structures capable of growth, shrinking, and reversible transition between OMV chain and individual vesicle morphologies (Fig. 5 B and C and Movies S7–S10). Fig. 5D provides a model to visualize how the junction densities seen in ECT, when added or removed, may account for the dynamic transitions in vesicle chains observed in fLM.
Fig. 5.
Proposed model for the formation and stabilization of OMV chains. (A) ECT image of a chemically fixed OM extension reveals the presence of densities at junctions that connect one vesicle to the next along the OMV chain (white dashed boxes). While all of the junction densities are not visible in the tomographic slice in A, Movie S6 is a 3D reconstruction of the same OM extension revealing the densities present at every junction. In addition, densities possibly related to decaheme cytochromes can be observed on the interior and exterior of the OM along the extension (arrows). (Scale bar: 100 nm.) (Fig. S8.) (B and C) Time-lapse fluorescence images recorded in real time in the perfusion flow imaging platform monitoring the growth and transformation of an OM extension from an apparently long filament (OMV chain morphology) to a single large vesicle (B, indicated by arrows) in S. oneidensis Δflg (a mutant strain lacking flagellin genes). (Movie S7.) Movie S8 shows OM extensions from wild-type cells also exhibiting a similar behavior to Δflg and a large vesicular morphology to an apparently smoother filament (OMV chain morphology) (C, indicated by arrows) in wild-type S. oneidensis MR-1 cells. (Movies S9 and S10). The cells and the OM extensions in B and C are stained by the membrane stain FM 4-64FX. (Scale bars in B and C: 5 µm and 2 µm, respectively.) (D) Schematic depicting a hypothesis for the formation and stabilization mechanism of OMV chains: Junction densities on the interior of the OM extension facilitate the constriction of the membrane, enabling the formation of an OMV chain. These constriction densities can be removed or added to facilitate transformation of an OMV chain to a large vesicle or vice versa as observed in B and C, respectively.
Distribution of Multiheme Cytochromes Along OM Extensions.
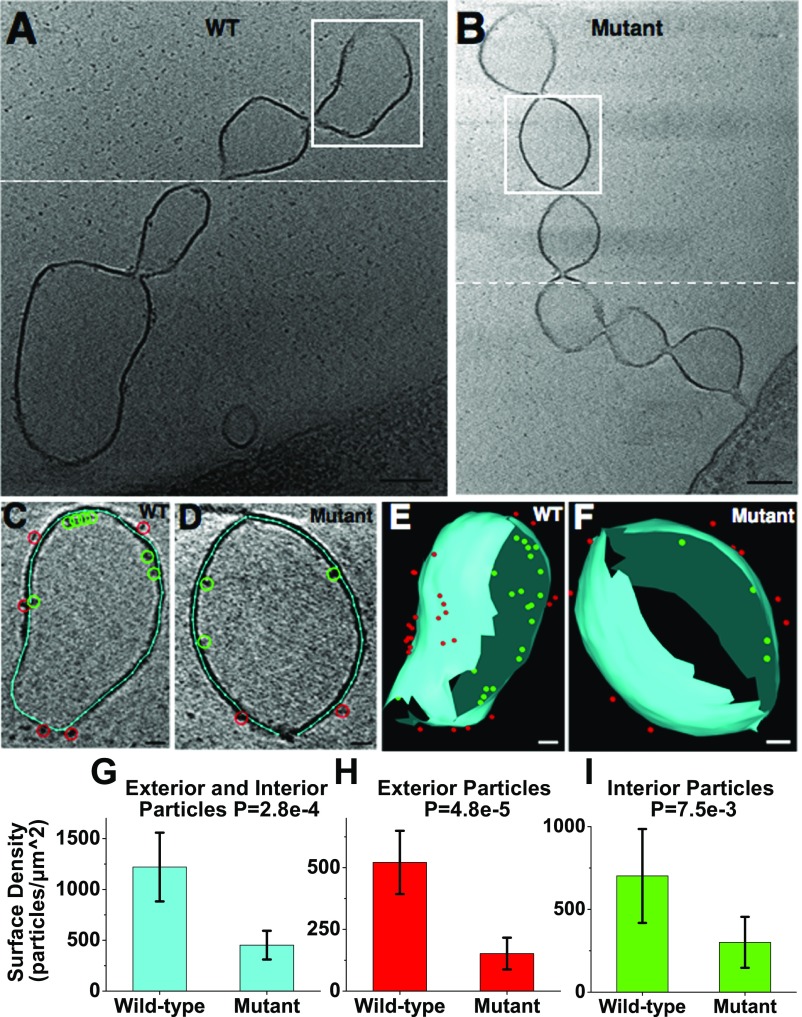
In S. oneidensis, previous immunofluorescence measurements have shown that the OM cytochromes MtrC and OmcA localize along the length of OM extensions (30). Additionally, the same cytochromes were shown to be essential for solid-state conductance of fixed and dried appendages consistent with OM extensions (21, 30). The packing density of these cytochromes is crucial in determining the potential mechanism of ET along OM extensions, but has remained unknown. Here, using ECT, we observed electron-dense particles on the interior and exterior of the OM extensions. We confirmed that the observed particles correspond to periplasmic and OM cytochromes by imaging OM extensions from a mutant, ΔMtr/ΔmtrB/ΔmtrE (40), lacking genes encoding eight identified functional S. oneidensis periplasmic and OM cytochromes. Our results showed a significantly higher interior and exterior particle density in the wild type compared with the mutant (Fig. 6 and Movie S11), confirming that a majority of the densities in wild-type OM extensions are indeed cytochromes. In addition, utilizing the OM extension with the highest number of densities (Fig. 7A), we overlaid available structures of the decaheme cytochromes MtrA (41) and MtrC (26) on representative interior and exterior densities, respectively, and found a similarity in overall shape and size of these structures to the observed EM densities (Fig. 7B).
Fig. 6.
Difference in surface density along OM extensions between S. oneidensis wild type and a mutant lacking all identified functional OM and periplasmic cytochromes (∆Mtr/∆mtrB/∆mtrE) (40). (A and B) Representative ECT images of OM extensions from wild-type and mutant strains, respectively, revealing electron-dense particles on the interior and exterior of the membrane. White dashed line indicates two different slices have been combined to provide the best possible view of OM extension. (Scale bar: 100 nm.) (C and D) Enlarged views of the vesicle from the boxed regions in A and B, respectively, with membrane (cyan line), interior particles (green circles), and exterior particles (red circles) labeled as model points. (Scale bar: 20 nm.) (E and F) The 3D reconstructions of the vesicles in C and D, respectively. Meshed view of the membrane is generated and all of the observed interior and exterior densities are shown as model points in 3D. (Scale bar: 20 nm.) (Movie S11.) (G–I) Surface density (in particles/µm2) of total, exterior, and interior particles in the wild-type (n = 8) and cytochrome mutant (n = 5) OM extensions (one vesicle analyzed per OM extension). Statistical significance is determined by P values from unpaired one-tailed Student’s t tests. Error bars represent one SD around the mean.
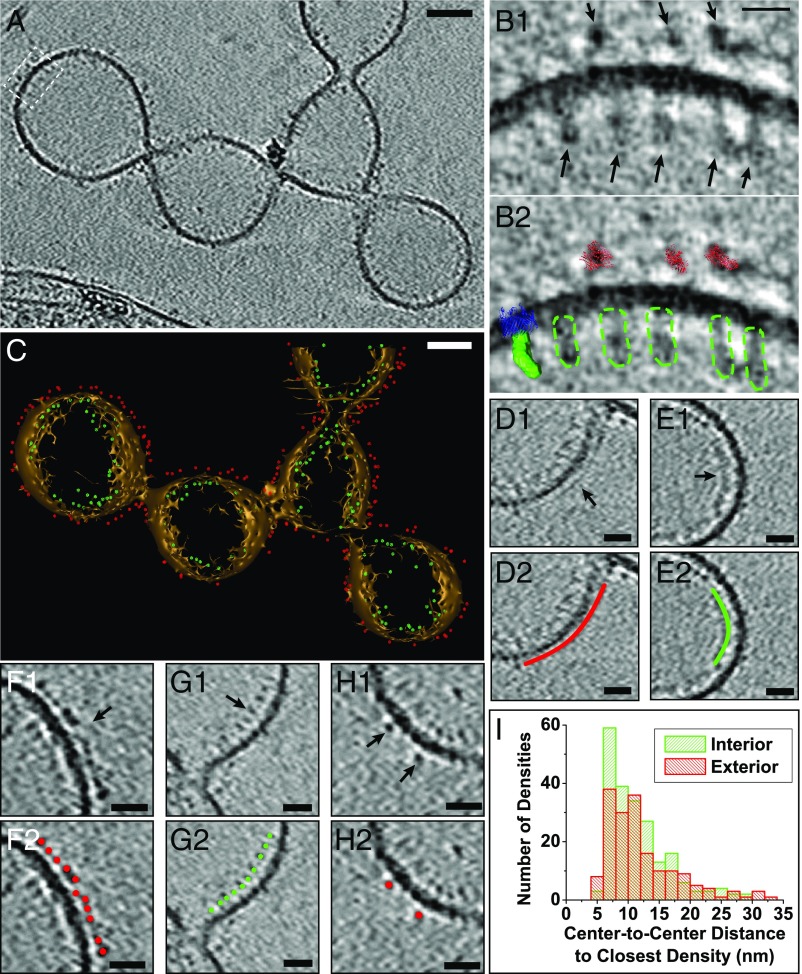
Fig. 7.
Positions and packing of decaheme cytochromes along the OM extension length in S. oneidensis. (A) ECT image of an unfixed OM extension showing densities on both the interior and exterior of the OM corresponding to putative MtrA and MtrC cytochromes, respectively. (Scale bar: 50 nm.) (B1) Enlarged view of boxed area from A. (B2) Comparison of EM densities in B1 with the crystal structure of MtrC (26), low-resolution SAXS model of MtrA (41), and the MtrB homolog LptD (72, 73) (only the LptD structure was used for this model from the LptD-LptE two-protein crystal structure), highlighting the similarity in overall shape and size of these structures to the observed EM densities. Red, MtrC crystal structure; green, surface view of MtrA SAXS model; blue, LptD crystal structure; dotted green, outline of putative MtrA densities on the EM map. (Scale bar: 10 nm.) (C) A 3D isosurface view of the OM extension in A with all of the interior and exterior densities (putative MtrA and MtrC, respectively) represented as model points in green and red, respectively. (Scale bar: 50 nm.) (Movie S12.) (D–H) Representative regions from the OM extension in A demonstrating differences in packing density of MtrA and MtrC. (D1 and E1) Continuous exterior (D1) and interior (E1) densities that may be related to tightly packed MtrC and MtrA, respectively. (F1) Relatively closely packed exterior densities of putative MtrC with an average center-to-center interdensity distance of 7.3 nm (SD = 2.1 nm). (G1) Relatively closely packed interior densities of putative MtrA with an average center-to-center interdensity distance of 8.9 nm (SD = 2.0 nm). (H1) Isolated exterior densities of putative MtrC. D2, E2, F2, G2, and H2 are duplicates of D1, E1, F1, G1, and H1, respectively, with model points or lines highlighting the interior (green) and exterior (red) densities. (Scale bars: 20 nm.) (I) Histogram showing distribution of center-to-center distances to closest densities for all observed putative MtrAs (in green) and MtrCs (in red).
We marked all of the observed interior and exterior densities along the OM extension as model points and reconstructed 3D models of both the OM extension and the cytochromes (Fig. 7C and Movie S12). The model allowed us to calculate the distance of each cytochrome from its nearest neighbor and thus investigate the possible ET mechanism along OM extensions. The observed density distribution fell in one of three categories: patches where the densities were almost continuous and indistinguishable from one another (Fig. 7 D and E), sections where the exterior and interior densities clustered closely but were distinguishable from one another (Fig. 7 F and G), and regions where the densities were farther apart (Fig. 7H). In summary, we did not observe a continuous crystalline-like packing of densities along the entire OM extension length. Instead, the OM and periplasmic densities were distributed over a range of center-to-center spacings, from 4.9 nm to 32.5 nm and from 5.0 nm to 29.0 nm, respectively (Fig. 7I). This distribution of densities suggested an ET model that supplements direct electron hopping between close cytochromes in tightly packed sections with physical diffusion of cytochromes to bridge larger gaps.
Calculations Suggest Maximum Overall ET Rate Is Achieved with a Combination of Cytochrome Physical Diffusion and Direct Electron Hopping.
To investigate the impact of cytochrome density on ET along OM extensions, we used the Blauch–Saveant model (42) that accounts for mobility of redox carriers in addition to direct electron hopping between redox carriers in the membrane. The relative contribution from redox carrier physical diffusion and direct hopping to the overall ET rate is determined by the ratio te/tp (42), where te and tp are the time constants for electron hopping and physical motion of redox carriers, respectively. With decaheme OM cytochromes as the redox carriers in OM extensions, and using 3 μm2/s as a representative value for the physical diffusion coefficient of integral membrane proteins of similar size (Dphys) (43), tp is estimated to be ∼3 × 10−6 s (SI Materials and Methods). In addition, using the electron residence time in the heme chains of the individual cytochromes, estimated from calculated and measured electron flux through MtrF (104 s−1) (44, 45) and MtrCAB (24), te can be estimated to be ∼10−4 s and hence te/tp to be ∼30. This relatively high value of te/tp (i.e., te/tp >> 1) justifies a mean-field approach developed by Blauch and Saveant (42), leading to a simple expression for the apparent diffusion coefficient (Dap) (42),
| [1] |
where Dphys is the redox carrier physical diffusion coefficient, De is the electron hopping diffusion coefficient which can be calculated using te (SI Materials and Methods), fc is the correlation factor, and X is the fractional loading of redox carriers in the membrane which can be calculated using particle densities extracted from the cryotomograms (SI Materials and Methods). Therefore, for OM extensions, Dap is estimated to be ∼3 × 10−8 cm2/s. In addition, the electron flux through an OM extension (J) can be calculated by (42)
| [2] |
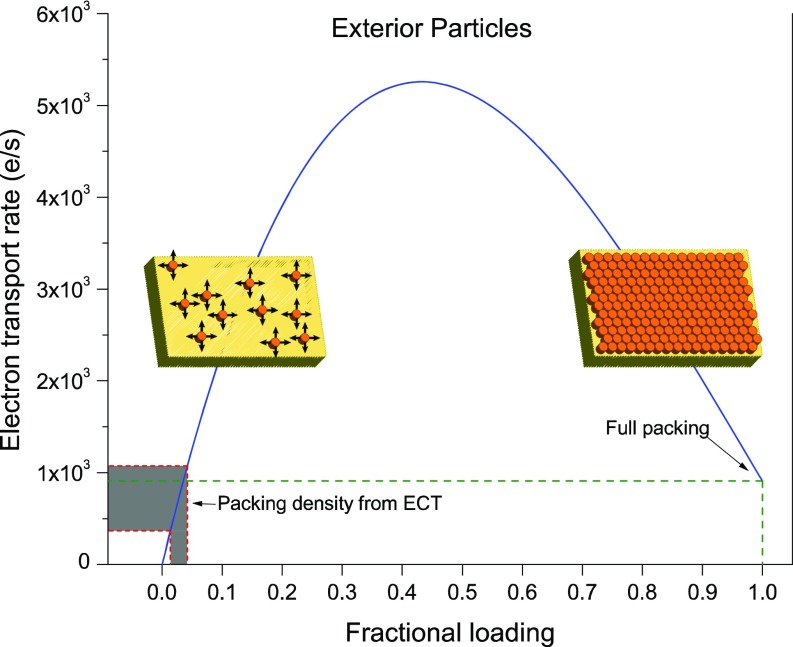
where C is the concentration of the reduced redox carriers and x is the position along the length of the OM extension. The resulting overall ET rate for an idealized 1-µm–long, 100-nm–diameter OM extension is shown in Fig. 8, where MtrC molecules are assumed to be the electron carriers (SI Materials and Methods).
Fig. 8.
Calculated ET rates along S. oneidensis OM extensions based on the Blauch–Saveant model (42). With MtrC as the main electron carrier, and accounting for both electron hopping and cytochrome physical mobility, ET rates are plotted (blue curve) as a function of cytochrome fractional loading (ratio of measured to the maximum possible cytochrome density). The gray-shaded area shows the range of calculated ET rates for OM extensions using cytochrome densities found in ECTs (one vesicle per tomogram from nine tomograms analyzed). The model predicts transport rates at full packing cytochrome density (fractional loading = 1) comparable with rates calculated from ECT data.
Discussion
Here we show high-resolution images of OM extensions in S. oneidensis, using ECT. We found the OM extensions to be OMV chains possibly stabilized by constriction densities at the junctions. Bacterial membrane extensions have been reported in multiple organisms: “nanopods” in Comamonadaceae including Delftia (46), “outer membrane tubes” in Francisella novicida (47), “periplasmic tubules” in Chlorochromatium aggregatum (48), “membrane tubules” in Salmonella typhimurium (49), “nanotubes” in Bacillus subtilis (50) and connecting Escherichia coli cells to each other and to Acinetobacter baylyi cells (51), and “connecting structures” that allow exchange of material between Clostridium acetobutylicum and Desulfovibrio vulgaris cells (52). However, membrane extensions in the form of OMV chains, similar to those reported here, have only recently been discovered and much remains unknown about their formation mechanism and specific function (53). In the gram-negative Shewanella vesiculosa (54) and Myxococcus xanthus (55, 56) and the gram-positive B. subtilis (57), membrane extensions in the form of OMV chains have been observed using cryo-EM with implications for cell–cell connections in the latter two examples. While the S. oneidensis OM extensions are proposed to function as electron conduits (30), their structural similarity to these previous reports highlights the significance of imaging these structures as a model system to study the formation of OMV chains.
To find a condition that consistently and frequently produced intact OM extensions for ECT imaging, we systematically tested different methods of growth and sample preparation conditions, as summarized in Fig. S1. We found that our optimized perfusion setup (Fig. 1) was best suited for the formation (Fig. 2), subsequent CLEM (Fig. 3), and high-resolution cryotomography of OM extensions (Fig. 4). The OMV chain morphology exhibited by these OM extensions is unlikely to be an artifact of fixation since we also observed a similar OMV chain architecture in OM extensions from unfixed samples (Fig. 7A and Fig. S5). While flagella and pili were identified as smooth filaments measuring ∼10 nm and ∼3 nm in thickness, respectively, OM extensions varied in thickness typically from ∼20 nm to 200 nm (Fig. 4 D–G), depending on the size and extent of tubulation of the constituent OMVs. Typically, there was an inverse relationship between OM extension length and its constituent OMV size. The measured thickness of hydrated OM extensions in ECT is different from the previously reported AFM measurements of ∼10 nm for air-dried conductive appendages (21, 58, 59). This is consistent with the finding that the appendages are OM extensions (30) because, in AFM, dehydration causes OM extensions to collapse to an ∼10-nm thickness, roughly corresponding to two lipid bilayers, while ECT preserves samples in a frozen-hydrated state, leading to more accurate estimates of native thickness. In addition to changing the OM extension thickness, dehydration will alter the cytochrome conformation and packing along OM extensions, which could significantly impact their electron-carrying capabilities. An interesting feature we observed is the ability of the vesicle chains to branch (Fig. 7A, Fig. S9, and Movie S13), which may offer the advantage of increasing the likelihood of contacting terminal solid-phase EAs in the environment. To our knowledge this is a unique report of branching reported in bacterial membrane extensions. OM extensions were also found to be flexible (Fig. S10), potentially improving their ability to contact solid-phase EAs.
Our ECT images of S. oneidensis OM extensions reveal that individual vesicles open into each other, share a continuous lumen, and thus form a chain of vesicles that are internally connected. This OMV architecture is reminiscent of the “pearls on a string” morphology caused by the pearling instability that transforms membrane tubes into a string of interconnected vesicles (60, 61). It has been shown that this transformation may be caused by an increase in membrane tension that can be stimulated in multiple ways, including osmotic gradient (62), mechanical perturbation (60, 61), elongational flow (63), electric field (64), bilayer asymmetry (65), nanoparticle adsorption onto the inner leaflet (66), or polymer anchorage onto a membrane (67, 68). Our observation of densities in ECT, at the junctions of neighboring vesicles in both fixed (Fig. 5A and Movie S6) and unfixed (Fig. S8) OM extensions, is consistent with the latter mechanism of polymer anchorage onto a membrane in which “constriction densities” or “junction densities” interact with the OM extension membrane, resulting in the formation of the OMV morphology (Fig. 5A, boxed regions). As schematized in Fig. 5D, addition and removal of such constriction densities may also explain the dynamic behavior observed in fLM, where OM extensions transition to and from individual vesicles (Fig. 5 B and C and Movies S7–S10). Although the identity of these junction densities is yet to be established, we hypothesize their potential role in the formation and stabilization of OMV chains based on our ECT and fLM observations.
The OM decaheme cytochrome MtrC, the periplasmic decaheme cytochrome MtrA, and the porin MtrB form the MtrCAB complex (23, 69) that is proposed to form a contiguous EET conduit from the periplasm to the cellular exterior (23). The presence of MtrC and its homolog OmcA has been linked to the solid-state conductance of S. oneidensis appendages consistent with OM extensions (21). These cytochromes are localized along the length of S. oneidensis OM extensions and are thought to mediate ET by a multistep redox hopping mechanism (30, 35). While the intraprotein hemes’ arrangement within MtrC and OmcA allows sequential tunneling (multistep hopping) through the heme chain (44, 45), the packing density and orientation of these cytochromes are critical parameters that determine the mechanism of putative interprotein electron transfer along the entire OM extension. However, before this work, little was known about the packing density of MtrC and OmcA molecules along OM extensions.
The OM extensions in ECT showed densities on both the inside and the outside of the membrane (Figs. 6A and 7A), features consistent with periplasmic and OM proteins, respectively. To examine whether these densities correspond to cytochromes, we imaged OM extensions from a S. oneidensis cytochrome mutant lacking eight known functional periplasmic and OM cytochromes (40). The mutant showed a significantly lower number of densities compared with the wild type. While the remaining densities seen in the ECT map must correspond to other OM and periplasmic proteins, our mutant studies confirm that the majority of the observed densities are cytochromes. It is worth noting that the exterior particle surface density on OM extensions (∼500 proteins/µm2, Fig. 6H) is roughly equal to or less than the expected surface density of MtrC and OmcA on the S. oneidensis cell surface based on previous estimates of these proteins per cell (1,000–30,000 proteins/µm2) (70, 71).
Next, to determine whether the outside densities match the size of MtrC, we overlaid the crystal structure of MtrC (26) onto three of these densities, illustrated in Fig. 7B. Since these densities did not appear symmetric on the EM map, and since the orientation of MtrC at the cellular OM is unknown, for each EM density, we overlaid the MtrC crystal structure in the orientation that best matched that specific density. Using this approach, the size of the OM features was found to be consistent with MtrC [noting, however, that this approach cannot distinguish between MtrC and other Shewanella OM proteins of similar size, including the structurally homologous decaheme cytochromes MtrF and OmcA (26)]. We applied a similar approach to compare the OM extension interior densities with the periplasmic decaheme cytochrome MtrA. The interior densities were more oblong than their outside counterparts, an observation consistent with the rod-like shape of MtrA previously revealed by small-angle X-ray scattering (SAXS) (41). By overlaying this low-resolution SAXS model on the EM map, the internal densities were found to be consistent in size and shape with MtrA (Fig. 7B). While the structure of MtrB is not yet known, we overlaid the crystal structure of a similarly sized protein [LptD from Salmonella enterica (72, 73)] in Fig. 7B and found that the size of the porin matches the width of the bilayer as expected. Taken collectively, our analyses highlight the similarity in overall shape and size between multiheme cytochromes and the observed EM densities.
The isosurface representation of the OM extensions, including the placement of the detected periplasmic and OM proteins (Fig. 7C and Movie S12), allows a holistic evaluation of different interprotein electron transfer mechanisms. Remarkably, we observed OM (Fig. 7F) and periplasmic (Fig. 7G) proteins clustering closely only over segments of the OM extension and not along its entire length. These observed tightly packed sections of up to ∼70 nm and ∼75 nm had center-to-center distances of 7.3 nm (SD = 2.1 nm) and 8.9 nm (SD = 2.0 nm) between neighboring proteins for the OM and periplasmic proteins, respectively. Taking the overall dimensions of MtrC (26) (∼9 × 6 × 4 nm) and the locations of the hemes (including terminal hemes at the protein edges) into account (26), the center-to-center distances point to the possibility of direct electron tunneling [requiring <2 nm separation (1)] between terminal hemes of neighboring OM cytochromes within these segments. However, such a crystalline-like packing of cytochromes was not observed over the micrometer lengths of whole OM extensions (Fig. 7). Instead, we observed a wide distribution of center-to-center spacings, presented for both the OM and the periplasmic densities as shown in Fig. 7I. Since center-to-center spacings beyond 11 nm and 7 nm for MtrC and MtrA, respectively, do not allow direct electron transfer between neighboring cytochromes (SI Materials and Methods), intermediate diffusive events are required to link the hemes of neighboring proteins beyond such distances. This may be accomplished by lateral physical diffusion of the multiheme cytochromes, resulting in collisions and electron exchange between neighboring cytochromes. Thus, the cytochrome distribution in ECT suggests a model of ET that involves both electron hopping and physical diffusion. Even though physical diffusion is known to enhance ET rates in assemblies of redox carriers (42), it has been typically ignored in studying EET, with the exception of a few recent studies. Paquete et al. (74) suggested that OmcA, which interacts with MtrC and is attached only by a lipidated cysteine at the N terminus, is mobile on the surface of Shewanella. Similarly, Zhang et al. (75) recently noted the need to consider the molecular motion of ET components in live biofilms, rather than a hypothetical static model of immobilized redox cofactors.
We therefore performed calculations to investigate the role of cytochrome physical diffusion in ET properties of OM extensions. Using surface densities of OM and periplasmic cytochromes found from the cryotomograms, and following the Blauch–Saveant (42) approach for calculating ET rates in assemblies of mobile redox carriers (Eq. 1), we built a model for OM extension ET that accounts for both electron hopping and cytochrome physical diffusion. We calculated an apparent diffusion coefficient (Dap) of up to ∼3 × 10−8 cm2/s, which is on par with the lower range of Dap measured in electroactive biofilms (75, 76) and even higher than some redox polymers (75). We also calculated the overall ET rate along an average-sized OM extension as a function of cytochrome fractional loading (Fig. 8). An interesting feature of the Blauch–Saveant model is that the physical diffusion of cytochromes in the membrane could significantly enhance the ET rate along OM extensions and that, counterintuitively, a less-than-full packing density of cytochromes will lead to the maximum overall ET rate (Fig. 8 and SI Materials and Methods) (42). As shown in Fig. 8, cytochrome densities extracted from cryotomograms predict a comparable ET rate to that of a fully packed array of cytochromes and that an increase in cytochrome density from the observed values could even enhance the ET rate above that of a fully packed configuration. It is important to note that our calculation leading to Fig. 8 takes only the physical diffusion of OM cytochromes into account and that the electron transfer rate may be further enhanced by diffusion of small redox-active molecules between cytochromes. In this context, it is important to note that the Shewanella decaheme cytochromes have flavin-binding sites (26), and flavins are known to enhance EET (27). Overall, our calculations show that a combination of physical diffusion and direct hopping may enhance ET beyond direct hopping alone. The extent of this enhancement, however, will depend on whether diffusion of additional molecules beyond MtrC/OmcA can contribute to ET (e.g., periplasmic cytochromes or small molecules such as flavins) and the precise values of the physical diffusion coefficients (e.g., for MtrC/OmcA proteins in the membrane or the likely faster MtrA diffusion within the periplasm). The preceding analysis is therefore intended for heuristic reasons and to motivate future studies targeting the diffusive dynamics of electron carriers in redox-active OM extensions. It is important to note that the calculations here may not be relevant to the results of previous high-conductivity measurements on dried and fixed appendages (21), because dehydration and fixation will alter the conformation, packing, and order of cytochromes along OM extensions. A recent study reported measurements of S. oneidensis nanofilaments under various relative humidity conditions and concluded that these filaments are capable of a hybrid electron and ion conductivity (77). While it is unclear if the latter nanofilaments are the same as the cytochrome-containing membrane extensions described here, we note that EET must be generally accompanied by cation transport to maintain charge neutrality. It is important to note that the model proposed here does not preclude counter-ion flow. Indeed, Okamoto et al. (78) recently reported evidence for proton transport associated with EET in the S. oneidensis MtrC and OmcA multiheme cytochromes.
In summary, our ECT imaging revealed particles consistent in size and morphology with decaheme cytochromes and their distribution along OM extensions. We do not expect all of the densities observed on the inside and the outside of the membrane to correspond to MtrA and MtrC, respectively, since, for example, we cannot distinguish between MtrC and other structural homologs, and there are other membrane proteins as well. However, it is already clear that cytochromes are not tightly packed along the entire length of OM extensions, even when all of the densities are treated as cytochromes. This irregular packing of cytochromes means that EET along whole OM extensions likely requires a combination of direct electron hopping and physical molecular diffusion by EET proteins or shuttles. Our calculations, based on the ECT data, show that such a model involving cytochrome diffusion can enhance ET rates to values comparable to a fully packed cytochrome configuration.
Materials and Methods
Perfusion Flow Imaging Platform.
The perfusion flow imaging platform was used as described previously (30), with some modifications. S. oneidensis MR-1, Δflg (79), or Δcrp (80) cells (Table S1) were grown overnight in Luria–Bertani (LB) broth at 30 °C up to an OD600 of 2.4–2.8, washed twice in a defined medium (30), and used in the perfusion flow imaging experiments. For experiments where ECT densities along OM extensions in wild-type and cytochrome mutant [ΔMtr/ΔmtrB/ΔmtrE (40), Table S1] strains were quantified, after the initial LB growth, 5 mL of the washed culture was transferred to an anaerobic sealed serum bottle with 100 mL of a defined medium (30) supplemented with 30 mM sodium fumarate. This anaerobic culture was placed in an incubator at 30 °C, shaking at 150 rpm, and was grown to an OD600 of 0.25 (∼24 h). The culture was then washed in a defined medium (30) and used for the perfusion flow imaging experiments. A glow-discharged, X-thick carbon-coated, R2/2, Au NH2 London finder Quantifoil EM grid (Quantifoil Micro Tools) was glued to a 43 mm × 50 mm no. 1 glass coverslip using waterproof silicone glue (General Electric Company); applied to two opposite edges of the grid; and let dry for ∼30 min. Using a vacuum line, the perfusion chamber (model VC-LFR-25; C&L Instruments) was sealed against the grid-attached glass coverslip and placed on an inverted microscope (Nikon Eclipse Ti-E) that continually imaged the grid surface. A total of ∼10 mL of the washed culture was injected into the chamber slowly to allow cells to settle on the grid surface, followed by a flow of sterile defined medium from an inverted serum bottle through a bubble trap (model 006BT-HF; Omnifit) into the perfusion chamber inlet. The serum bottle was pressurized by N2 in the headspace to sustain a flow rate of 5 ± 1 µL/s. After ∼2 h of perfusion flow, cells on the grid surface began to produce OM extensions. Cells and OM extensions were visualized by the fluorescent membrane stain FM 4-64FX that was present in the flow medium throughout the experiment (25 µg in 100 mL of medium). Subsequently, the flow of medium was stopped and the perfusion chamber was opened under sterile medium. When fixing, the sample (cells on EM grid-attached coverslip) was treated with either 2.5% glutaraldehyde for 15 min or 4% formaldehyde for 60 min. The grid was then detached from the coverslip by scraping off the silicone glue at the grid edges using a 22-gauge needle and rinsed by transferring three times in deionized water, before using for TEM imaging.
ECT.
ECT samples were prepared as described previously (81) with minor modifications. Cells from batch cultures and chemostats were mixed with BSA-treated 10-nm colloidal gold solution and 4 μL of this mixture was applied to a glow-discharged, X-thick carbon-coated, R2/2, 200 mesh copper Quantifoil grid (Quantifoil Micro Tools) in a Vitrobot chamber (FEI). Excess liquid was blotted off with a blot force of 6, blot time of 3 s, and drain time of 1 s and the grid was plunge frozen for ECT imaging. All perfusion samples were on glow-discharged, X-thick carbon-coated, R2/2, Au NH2 London finder Quantifoil EM grids (Quantifoil Micro Tools) and were blotted either manually or automatically using the Vitrobot after addition of 1.5 μL of 10-nm gold fiducial markers. Imaging of all ECT samples was performed on an FEI Polara 300-keV field emission gun electron microscope equipped with a Gatan image filter and K2 Summit counting electron-detector camera (Gatan). Data were collected using the UCSFtomo software (82), with each tilt series ranging from −60° to 60° in 1° increments, an underfocus of ∼5–10 µm, and a cumulative electron dose of ∼130–160 e/A2 for each individual tilt series. The IMOD software package was used to calculate 3D reconstructions (83).
Supplementary Material
Acknowledgments
We thank Dr. Yi-Wei Chang and Dr. Matthew Swulius for help with preparing Fig. 7 B and C, respectively. We are grateful to Dr. Sean J. Elliott for providing the SAXS model file for MtrA (41) used in Fig. 7B and to Dr. Jeffrey A. Gralnick for providing the cytochrome mutant strain. We thank Dr. Catherine Oikonomou for helping edit the manuscript. P.S. acknowledges support by the Caltech Center for Environmental Microbial Interactions. Work in the laboratory of G.J.J. is supported by the Howard Hughes Medical Institute. The in vivo OM extension imaging platform and mapping of EET proteins are funded by the Air Force Office of Scientific Research Presidential Early Career Award for Scientists and Engineers (FA955014-1-0294, to M.Y.E.-N.). Modeling of ET kinetics and partial support for S.P. are funded by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through Grant DE-FG02-13ER16415 (to M.Y.E.-N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718810115/-/DCSupplemental.
References
- 1.Gray HB, Winkler JR. Electron tunneling through proteins. Q Rev Biophys. 2003;36:341–372. doi: 10.1017/s0033583503003913. [DOI] [PubMed] [Google Scholar]
- 2.Myers CR, Nealson KH. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science. 1988;240:1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- 3.Nealson KH, Belz A, McKee B. Breathing metals as a way of life: Geobiology in action. Antonie Van Leeuwenhoek. 2002;81:215–222. doi: 10.1023/a:1020518818647. [DOI] [PubMed] [Google Scholar]
- 4.Gralnick JA, Newman DK. Extracellular respiration. Mol Microbiol. 2007;65:1–11. doi: 10.1111/j.1365-2958.2007.05778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fredrickson JK, et al. Towards environmental systems biology of Shewanella. Nat Rev Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 6.Shi L, et al. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat Rev Microbiol. 2016;14:651–662. doi: 10.1038/nrmicro.2016.93. [DOI] [PubMed] [Google Scholar]
- 7.Bretschger O, et al. Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl Environ Microbiol. 2007;73:7003–7012. doi: 10.1128/AEM.01087-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Logan BE. Exoelectrogenic bacteria that power microbial fuel cells. Nat Rev Microbiol. 2009;7:375–381. doi: 10.1038/nrmicro2113. [DOI] [PubMed] [Google Scholar]
- 9.Rabaey K, Rozendal RA. Microbial electrosynthesis - Revisiting the electrical route for microbial production. Nat Rev Microbiol. 2010;8:706–716. doi: 10.1038/nrmicro2422. [DOI] [PubMed] [Google Scholar]
- 10.Hau HH, Gralnick JA. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol. 2007;61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 11.Breuer M, Rosso KM, Blumberger J, Butt JN. Multi-haem cytochromes in Shewanella oneidensis MR-1: Structures, functions and opportunities. J R Soc Interface. 2015;12:20141117. doi: 10.1098/rsif.2014.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reguera G, et al. Extracellular electron transfer via microbial nanowires. Nature. 2005;435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 13.Feliciano GT, Steidl RJ, Reguera G. Structural and functional insights into the conductive pili of Geobacter sulfurreducens revealed in molecular dynamics simulations. Phys Chem Chem Phys. 2015;17:22217–22226. doi: 10.1039/c5cp03432a. [DOI] [PubMed] [Google Scholar]
- 14.Lampa-Pastirk S, et al. Thermally activated charge transport in microbial protein nanowires. Sci Rep. 2016;6:23517. doi: 10.1038/srep23517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosert KM, Steidl RJ, Castro-Forero A, Worden RM, Reguera G. Electronic characterization of Geobacter sulfurreducens pilins in self-assembled monolayers unmasks tunnelling and hopping conduction pathways. Phys Chem Chem Phys. 2017;19:11163–11172. doi: 10.1039/c7cp00885f. [DOI] [PubMed] [Google Scholar]
- 16.Malvankar NS, et al. Tunable metallic-like conductivity in microbial nanowire networks. Nat Nanotechnol. 2011;6:573–579. doi: 10.1038/nnano.2011.119. [DOI] [PubMed] [Google Scholar]
- 17.Malvankar NS, Yalcin SE, Tuominen MT, Lovley DR. Visualization of charge propagation along individual pili proteins using ambient electrostatic force microscopy. Nat Nanotechnol. 2014;9:1012–1017. doi: 10.1038/nnano.2014.236. [DOI] [PubMed] [Google Scholar]
- 18.Malvankar NS, et al. Structural basis for metallic-like conductivity in microbial nanowires. MBio. 2015;6:e00084. doi: 10.1128/mBio.00084-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steidl RJ, Lampa-Pastirk S, Reguera G. Mechanistic stratification in electroactive biofilms of Geobacter sulfurreducens mediated by pilus nanowires. Nat Commun. 2016;7:12217. doi: 10.1038/ncomms12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yates MD, et al. Measuring conductivity of living Geobacter sulfurreducens biofilms. Nat Nanotechnol. 2016;11:910–913. doi: 10.1038/nnano.2016.186. [DOI] [PubMed] [Google Scholar]
- 21.El-Naggar MY, et al. Electrical transport along bacterial nanowires from Shewanella oneidensis MR-1. Proc Natl Acad Sci USA. 2010;107:18127–18131. doi: 10.1073/pnas.1004880107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartshorne RS, et al. Characterization of an electron conduit between bacteria and the extracellular environment. Proc Natl Acad Sci USA. 2009;106:22169–22174. doi: 10.1073/pnas.0900086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richardson DJ, et al. The ‘porin-cytochrome’ model for microbe-to-mineral electron transfer. Mol Microbiol. 2012;85:201–212. doi: 10.1111/j.1365-2958.2012.08088.x. [DOI] [PubMed] [Google Scholar]
- 24.White GF, et al. Rapid electron exchange between surface-exposed bacterial cytochromes and Fe(III) minerals. Proc Natl Acad Sci USA. 2013;110:6346–6351. doi: 10.1073/pnas.1220074110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto A, Hashimoto K, Nealson KH, Nakamura R. Rate enhancement of bacterial extracellular electron transport involves bound flavin semiquinones. Proc Natl Acad Sci USA. 2013;110:7856–7861. doi: 10.1073/pnas.1220823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards MJ, et al. Redox linked flavin sites in extracellular decaheme proteins involved in microbe-mineral electron transfer. Sci Rep. 2015;5:11677. doi: 10.1038/srep11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu S, Jangir Y, El-Naggar MY. Disentangling the roles of free and cytochrome-bound flavins in extracellular electron transport from Shewanella oneidensis MR-1. Electrochim Acta. 2016;198:49–55. [Google Scholar]
- 28.Marsili E, et al. Shewanella secretes flavins that mediate extracellular electron transfer. Proc Natl Acad Sci USA. 2008;105:3968–3973. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coursolle D, Baron DB, Bond DR, Gralnick JA. The Mtr respiratory pathway is essential for reducing flavins and electrodes in Shewanella oneidensis. J Bacteriol. 2010;192:467–474. doi: 10.1128/JB.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pirbadian S, et al. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc Natl Acad Sci USA. 2014;111:12883–12888. doi: 10.1073/pnas.1410551111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat Rev Microbiol. 2015;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray R, Lizewski S, Fitzgerald LA, Little B, Ringeisen BR. Methods for imaging Shewanella oneidensis MR-1 nanofilaments. J Microbiol Methods. 2010;82:187–191. doi: 10.1016/j.mimet.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Dohnalkova AC, et al. Imaging hydrated microbial extracellular polymers: Comparative analysis by electron microscopy. Appl Environ Microbiol. 2011;77:1254–1262. doi: 10.1128/AEM.02001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gan L, Jensen GJ. Electron tomography of cells. Q Rev Biophys. 2012;45:27–56. doi: 10.1017/S0033583511000102. [DOI] [PubMed] [Google Scholar]
- 35.Pirbadian S, El-Naggar MY. Multistep hopping and extracellular charge transfer in microbial redox chains. Phys Chem Chem Phys. 2012;14:13802–13808. doi: 10.1039/c2cp41185g. [DOI] [PubMed] [Google Scholar]
- 36.Polizzi NF, Skourtis SS, Beratan DN. Physical constraints on charge transport through bacterial nanowires. Faraday Discuss. 2012;155:43–62, discussion 103–114. doi: 10.1039/c1fd00098e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorby YA, et al. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc Natl Acad Sci USA. 2006;103:11358–11363. doi: 10.1073/pnas.0604517103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorby Y, et al. Redox-reactive membrane vesicles produced by Shewanella. Geobiology. 2008;6:232–241. doi: 10.1111/j.1472-4669.2008.00158.x. [DOI] [PubMed] [Google Scholar]
- 39.Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- 40.Coursolle D, Gralnick JA. Reconstruction of extracellular respiratory pathways for iron(III) reduction in Shewanella oneidensis strain MR-1. Front Microbiol. 2012;3:56. doi: 10.3389/fmicb.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Firer-Sherwood MA, Ando N, Drennan CL, Elliott SJ. Solution-based structural analysis of the decaheme cytochrome, MtrA, by small-angle X-ray scattering and analytical ultracentrifugation. J Phys Chem B. 2011;115:11208–11214. doi: 10.1021/jp203603r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blauch D, Saveant J. Dynamics of electron hopping in assemblies of redox centers. Percolation and diffusion. J Am Chem Soc. 1992;114:3323–3332. [Google Scholar]
- 43.Ramadurai S, et al. Lateral diffusion of membrane proteins. J Am Chem Soc. 2009;131:12650–12656. doi: 10.1021/ja902853g. [DOI] [PubMed] [Google Scholar]
- 44.Breuer M, Rosso KM, Blumberger J. Electron flow in multiheme bacterial cytochromes is a balancing act between heme electronic interaction and redox potentials. Proc Natl Acad Sci USA. 2014;111:611–616. doi: 10.1073/pnas.1316156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byun HS, Pirbadian S, Nakano A, Shi L, El-Naggar MY. Kinetic Monte Carlo simulations and molecular conductance measurements of the bacterial decaheme cytochrome MtrF. ChemElectroChem. 2014;1:1932–1939. [Google Scholar]
- 46.Shetty A, Chen S, Tocheva EI, Jensen GJ, Hickey WJ. Nanopods: A new bacterial structure and mechanism for deployment of outer membrane vesicles. PLoS One. 2011;6:e20725. doi: 10.1371/journal.pone.0020725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCaig WD, Koller A, Thanassi DG. Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J Bacteriol. 2013;195:1120–1132. doi: 10.1128/JB.02007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wanner G, Vogl K, Overmann J. Ultrastructural characterization of the prokaryotic symbiosis in “Chlorochromatium aggregatum”. J Bacteriol. 2008;190:3721–3730. doi: 10.1128/JB.00027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galkina SI, et al. Membrane tubules attach Salmonella Typhimurium to eukaryotic cells and bacteria. FEMS Immunol Med Microbiol. 2011;61:114–124. doi: 10.1111/j.1574-695X.2010.00754.x. [DOI] [PubMed] [Google Scholar]
- 50.Dubey GP, Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell. 2011;144:590–600. doi: 10.1016/j.cell.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 51.Pande S, et al. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun. 2015;6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 52.Benomar S, et al. Nutritional stress induces exchange of cell material and energetic coupling between bacterial species. Nat Commun. 2015;6:6283. doi: 10.1038/ncomms7283. [DOI] [PubMed] [Google Scholar]
- 53.Bohuszewicz O, Liu J, Low HH. Membrane remodelling in bacteria. J Struct Biol. 2016;196:3–14. doi: 10.1016/j.jsb.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pérez-Cruz C, et al. New type of outer membrane vesicle produced by the Gram-negative bacterium Shewanella vesiculosa M7T: Implications for DNA content. Appl Environ Microbiol. 2013;79:1874–1881. doi: 10.1128/AEM.03657-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Remis JP, et al. Bacterial social networks: Structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ Microbiol. 2014;16:598–610. doi: 10.1111/1462-2920.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei X, Vassallo CN, Pathak DT, Wall D. Myxobacteria produce outer membrane-enclosed tubes in unstructured environments. J Bacteriol. 2014;196:1807–1814. doi: 10.1128/JB.00850-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dubey GP, et al. Architecture and characteristics of bacterial nanotubes. Dev Cell. 2016;36:453–461. doi: 10.1016/j.devcel.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 58.El-Naggar MY, Gorby YA, Xia W, Nealson KH. The molecular density of states in bacterial nanowires. Biophys J. 2008;95:L10–L12. doi: 10.1529/biophysj.108.134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leung KM, et al. Shewanella oneidensis MR-1 bacterial nanowires exhibit p-type, tunable electronic behavior. Nano Lett. 2013;13:2407–2411. doi: 10.1021/nl400237p. [DOI] [PubMed] [Google Scholar]
- 60.Bar-Ziv R, Moses E. Instability and “pearling” states produced in tubular membranes by competition of curvature and tension. Phys Rev Lett. 1994;73:1392–1395. doi: 10.1103/PhysRevLett.73.1392. [DOI] [PubMed] [Google Scholar]
- 61.Bar-Ziv R, Tlusty T, Moses E. Critical dynamics in the pearling instability of membranes. Phys Rev Lett. 1997;79:1158–1161. [Google Scholar]
- 62.Sanborn J, Oglecka K, Kraut RS, Parikh AN. Transient pearling and vesiculation of membrane tubes under osmotic gradients. Faraday Discuss. 2013;161:167–176, discussion 273–303. doi: 10.1039/c2fd20116j. [DOI] [PubMed] [Google Scholar]
- 63.Kantsler V, Segre E, Steinberg V. Critical dynamics of vesicle stretching transition in elongational flow. Phys Rev Lett. 2008;101:048101. doi: 10.1103/PhysRevLett.101.048101. [DOI] [PubMed] [Google Scholar]
- 64.Sinha K, Gadkari S, Thaokar R. Electric field induced pearling instability in cylindrical vesicles. Soft Matter. 2013;9:7274–7293. [Google Scholar]
- 65.Chaieb S, Rica S. Spontaneous curvature-induced pearling instability. Phys Rev E. 1998;58:7733–7737. [Google Scholar]
- 66.Yu Y, Granick S. Pearling of lipid vesicles induced by nanoparticles. J Am Chem Soc. 2009;131:14158–14159. doi: 10.1021/ja905900h. [DOI] [PubMed] [Google Scholar]
- 67.Tsafrir I, et al. Pearling instabilities of membrane tubes with anchored polymers. Phys Rev Lett. 2001;86:1138–1141. doi: 10.1103/PhysRevLett.86.1138. [DOI] [PubMed] [Google Scholar]
- 68.Campelo F, Hernández-Machado A. Model for curvature-driven pearling instability in membranes. Phys Rev Lett. 2007;99:088101. doi: 10.1103/PhysRevLett.99.088101. [DOI] [PubMed] [Google Scholar]
- 69.Ross DE, et al. Characterization of protein-protein interactions involved in iron reduction by Shewanella oneidensis MR-1. Appl Environ Microbiol. 2007;73:5797–5808. doi: 10.1128/AEM.00146-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borloo J, et al. A kinetic approach to the dependence of dissimilatory metal reduction by Shewanella oneidensis MR-1 on the outer membrane cytochromes c OmcA and OmcB. FEBS J. 2007;274:3728–3738. doi: 10.1111/j.1742-4658.2007.05907.x. [DOI] [PubMed] [Google Scholar]
- 71.Ross DE, Brantley SL, Tien M. Kinetic characterization of OmcA and MtrC, terminal reductases involved in respiratory electron transfer for dissimilatory iron reduction in Shewanella oneidensis MR-1. Appl Environ Microbiol. 2009;75:5218–5226. doi: 10.1128/AEM.00544-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong H, et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature. 2014;511:52–56. doi: 10.1038/nature13464. [DOI] [PubMed] [Google Scholar]
- 73.Poole RK. Advances in Bacterial Electron Transport Systems and Their Regulation. 1st Ed Academic; Cambridge, MA: 2016. [Google Scholar]
- 74.Paquete CM, et al. Exploring the molecular mechanisms of electron shuttling across the microbe/metal space. Front Microbiol. 2014;5:318. doi: 10.3389/fmicb.2014.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang X, et al. Rapid and quantitative assessment of redox conduction across electroactive biofilms by using double potential step chronoamperometry. ChemElectroChem. 2017;4:1026–1036. [Google Scholar]
- 76.Liu Y, Bond DR. Long-distance electron transfer by G. sulfurreducens biofilms results in accumulation of reduced c-type cytochromes. ChemSusChem. 2012;5:1047–1053. doi: 10.1002/cssc.201100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grebenko A, et al. Impedance spectroscopy of single bacterial nanofilament reveals water-mediated charge transfer. PLoS One. 2018;13:e0191289. doi: 10.1371/journal.pone.0191289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okamoto A, Tokunou Y, Kalathil S, Hashimoto K. Proton transport in the outer-membrane flavocytochrome complex limits the rate of extracellular electron transport. Angew Chem Int Ed Engl. 2017;56:9082–9086. doi: 10.1002/anie.201704241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bouhenni R, et al. The role of Shewanella oneidensis MR-1 outer surface structures in extracellular electron transfer. Electroanalysis. 2010;22:856–864. [Google Scholar]
- 80.Charania MA, et al. Involvement of a membrane-bound class III adenylate cyclase in regulation of anaerobic respiration in Shewanella oneidensis MR-1. J Bacteriol. 2009;191:4298–4306. doi: 10.1128/JB.01829-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cornejo E, Subramanian P, Li Z, Jensen GJ, Komeili A. Dynamic remodeling of the magnetosome membrane is triggered by the initiation of biomineralization. MBio. 2016;7:e01898-15. doi: 10.1128/mBio.01898-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng SQ, et al. UCSF tomography: An integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol. 2007;157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 83.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.