Significance
Toll-like receptors (TLRs) are highly conserved innate receptors that form homo- or heterodimers to detect microbial danger signals and activate the immune system. TLR5 detects flagellin of bacteria and functions as a homodimeric receptor complex. A crystallized fragment of TLR5b of the zebrafish (Danio rerio) serves as a model structure for the homodimeric TLR5–flagellin interaction. Here we report that zebrafish TLR5 unexpectedly functions as a heterodimeric flagellin receptor composed of the duplicated gene products TLR5b and TLR5a. The unique heterodimeric nature of zebrafish TLR5 indicates important receptor differences between species, contributes to a deeper understanding of the activation mechanism of TLRs, and provides an illustrative example of the functional coevolution of duplicated genes.
Keywords: TLR5, heterodimer, flagellin, subfunctionalization, zebrafish
Abstract
Toll-like receptor 5 (TLR5) of mammals, birds, and reptiles detects bacterial flagellin and signals as a homodimeric complex. Structural studies using truncated TLR5b of zebrafish confirm the homodimeric TLR5–flagellin interaction. Here we provide evidence that zebrafish (Danio rerio) TLR5 unexpectedly signals as a heterodimer composed of the duplicated gene products drTLR5b and drTLR5a. Flagellin-induced signaling by the zebrafish TLR5 heterodimer increased in the presence of the TLR trafficking chaperone UNC93B1. Targeted exchange of drTLR5b and drTLR5a regions revealed that TLR5 activation needs a heterodimeric configuration of the receptor ectodomain and cytoplasmic domain, consistent with ligand-induced changes in receptor conformation. Structure-guided substitution of the presumed principal flagellin-binding site in human TLR5 with corresponding zebrafish TLR5 residues abrogated human TLR5 activation, indicating a species-specific TLR5–flagellin interaction. Our findings indicate that the duplicated TLR5 of zebrafish underwent subfunctionalization through concerted coevolution to form a unique heterodimeric flagellin receptor that operates fundamentally differently from TLR5 of other species.
Toll-like receptors (TLRs) are evolutionarily highly conserved membrane-bound receptors that initiate activation of the immune system upon detection of microbial ligands (1, 2). TLRs are composed of a cytoplasmic Toll–interleukin-1 (TIR) signaling domain, a single transmembrane (TM) segment, and a leucine-rich repeat (LRR)–containing ectodomain (3). Microbe-driven diversification of the ectodomain has resulted in a family of distinct TLRs across species that recognize a wide variety of conserved microbial structures (4). For example, TLR4 detects bacterial LPS (5), TLR15 is activated by microbial proteases (6), while microbial nucleic acids are sensed by TLRs 3, 7–9, and 21 (7–11). Bacterial flagellin, the major constituent of the flagellum of motile bacteria including pathogens such as Salmonella spp. and Pseudomonas aeruginosa, is recognized by TLR5 (12, 13). Irrespective of the type of ligand, TLRs need to form receptor dimers to exert their function (14). Whereas TLR1 and TLR6 function in a heterodimeric complex with TLR2, other TLRs, including TLR3, TLR4, TLR7–9, and TLR5, act as homodimers. In all cases, binding of ligand to the TLR complex induces a conformational change, which positions the intracellular TIR domains of the dimer into close proximity, thereby forming a docking station for intracellular adapter molecules. Binding of adapter molecules initiates further signaling that activates transcription factors such as NF-κB, which translocate to the nucleus to start proinflammatory gene expression (14).
Structural analyses of crystallized TLR-ligand complexes have fundamentally contributed to a detailed understanding of TLR–ligand interaction and dimerization. To date, the ectodomains of many mammalian TLRs have been crystallized (15–18), with the exception of TLR5. For this receptor, only the protein structure of the N-terminal part of the ectodomain of TLR5b of zebrafish (Danio rerio) in complex with Salmonella flagellin has been resolved. The structure indicates that bacterial flagellin binds to homodimers of zebrafish TLR5b (19), consistent with the homodimeric nature of mammalian TLR5. However, direct evidence that binding of flagellin to zebrafish TLR5b initiates signaling is lacking.
Zebrafish is a widely used model animal and belongs to the teleostei or bony fish. Teleost fish underwent an additional round of whole-genome duplication. The duplication and reorganization of TLR genes have led to a higher diversity of TLR gene repertoires among teleosts compared with nonteleost vertebrates (20–24). The common carp (Cyprinus carpio) has two paralogs of TLR2 (20, 25), and the Atlantic cod (Gadus morhua) has lost TLR5 but has massively expanded TLR8 genes (22). Zebrafish carries two paralogs of TLR4 (26) and retained both CpG DNA receptors, TLR9 (present in mammals) and TLR21 (present in birds) (27). In addition, the zebrafish has duplicated TLR5, resulting in the two paralogs: TLR5a and TLR5b. The functional consequences of these TLR duplications in teleosts are poorly understood.
In the present study we provide evidence that zebrafish TLR5b does not signal as a conventional TLR5 homodimer but instead cooperates with its paralog TLR5a to form a unique heterodimeric TLR5 that responds to flagellin. Structure-guided amino acid substitution and functional analyses of the N-terminal part of TLR5 indicate species-specific functional differences between zebrafish and human TLR5. Targeted exchange of multiple defined regions of zebrafish TLR5a and TLR5b indicates that a transregional heterodimeric configuration is required for receptor activation and signal initiation, which is consistent with ligand-induced changes in receptor conformation.
Results
drTLR5a and drTLR5b Are Both Required for Functional Flagellin Recognition.
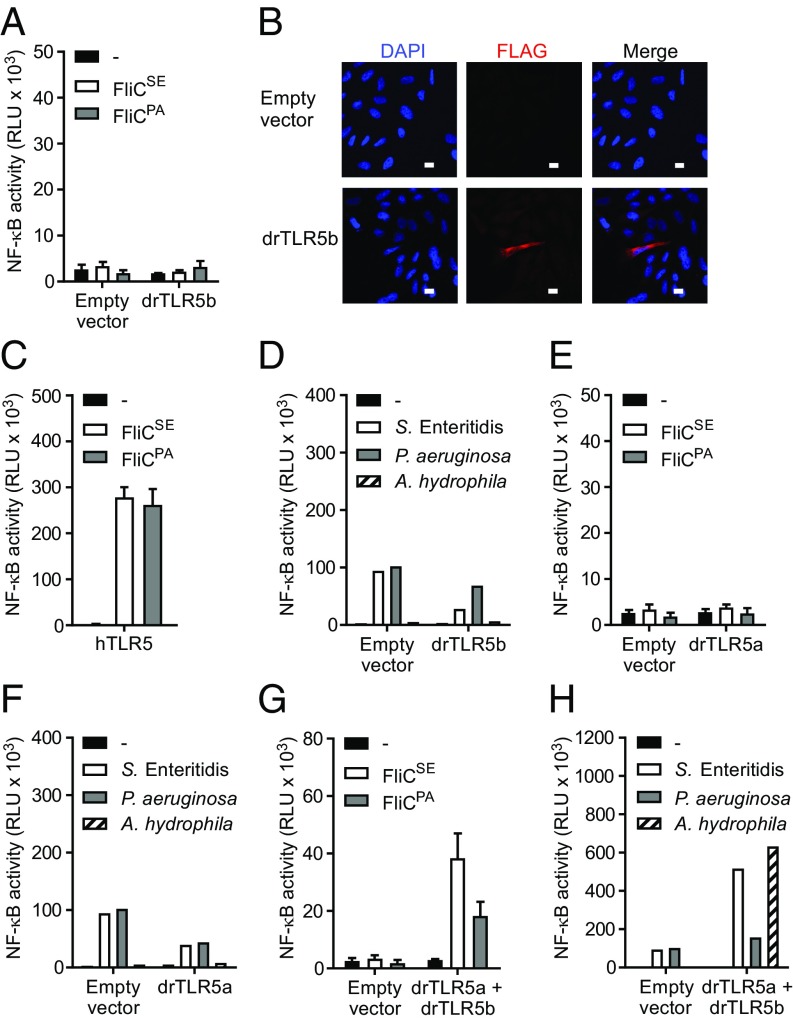
Since the published crystal structure of the zebrafish TLR5b (drTLR5b) ectodomain indicates binding of flagellin to a receptor homodimer (19), we investigated whether stimulation of full-length drTLR5b with bacterial flagellin activates the canonical TLR5 inducible NF-κB signaling pathway. The corresponding tlr5b gene was amplified from zebrafish and cloned in front of the C-terminal FLAG tag of an expression vector. The plasmid was transfected into human HeLa-57A cells that do not express endogenous TLRs but stably express an NF-κB–inducible luciferase reporter gene. Stimulation of drTLR5b transfected cells with recombinant flagellin of Salmonella enterica serovar Enteritidis (FliCSE) or P. aeruginosa (FliCPA) (28) failed to activate NF-κB (Fig. 1A), although confocal microscopy confirmed expression of FLAG-tagged drTLR5b in transfected cells (Fig. 1B). Both FliCSE and FliCPA did activate NF-κB in HeLa-57A cells transfected with human TLR5 (hTLR5), indicating that TLR expression and signaling are functional in these cells and that the flagellins displayed TLR5 activating capacity (Fig. 1C). To test whether drTLR5b might recognize native instead of recombinantly produced flagellin, we stimulated transfected cells with whole-cell lysates of highly motile S. Enteritidis, P. aeruginosa, and of the fish pathogen Aeromonas hydrophila. None of the lysates activated cells in a drTLR5b-dependent fashion (Fig. 1D).
Fig. 1.
Induction of NF-κB by zebrafish TLR5b (drTLR5b) and drTLR5a upon stimulation with purified flagellins and bacterial lysates. (A and C–H) HeLa-57A cells were transfected with drTLR5b, drTLR5a, hTLR5 or drTLR5b and drTLR5a combined as indicated. Control cells were transfected with empty vector. Cells were stimulated (5 h) with vehicle (–) or 1 µg mL−1 of purified recombinant FliCSE or FliCPA flagellin (A, C, E, and G) or 2 µg mL−1 of total protein from lysates of S. Enteritidis, P. aeruginosa,or A. hydrophila (D, F, and H). Note that the NF-κB response of the drTLR5 heterodimer is higher when stimulated with bacterial lysate compared with purified recombinant flagellin. (B) Fluorescence microscopy of empty vector or FLAG-tagged drTLR5b transfected HeLa-57A cells stained with M2 α-FLAG and DAPI for nuclear visualization. (White scale bars: 10 µm.) NF-κB activity is represented by luciferase activity in RLUs. Values are the mean ± SEM of three independent experiments (A, C, E, and G) or a representative of three independent experiments (D, F, and H) all performed in duplicate.
Since, in zebrafish, the tlr5 gene is duplicated, we questioned whether its paralog, annotated as TLR5a (drTLR5a), could be the functional receptor to flagellin. drtlr5a and drtlr5b are positioned in tandem on chromosome 20. Both genes translate into proteins of 881 amino acids that have characteristic TLR5 features, including a signal peptide, N-terminal LRR (NTLRR), 22 consecutive LRRs, C-terminal LRR (CTLRR), TM domain (TM), and an intracellular TIR signaling domain (Fig. S1). Overall, the protein sequences of drTLR5b and drTLR5a are 83% similar, with the highest similarity in the TM and TIR domains and the lowest similarity in the LRR ligand-binding ectodomain, suggesting a possible different interaction of flagellin with drTLR5a than with drTLR5b. We cloned drTLR5a and expressed the receptor as described for drTLR5b. Stimulation of drTLR5a-transfected cells with FliCSE and FliCPA failed to activate NF-κB (Fig. 1E). drTLR5a also did not respond to bacterial lysates (Fig. 1F). Since both receptors separately were not functional, we explored the possibility that duplicated drTLR5b and drTLR5a may have coevolved to form a functional receptor. Stimulation of cells cotransfected with drTLR5a and drTLR5b indeed yielded robust activation of NF-κB when exposed to FliCSE and FliCPA (Fig. 1G). Likewise, drTLR5a and drTLR5b together activated NF-κB in response to lysate of motile bacteria, especially A. hydrophila (Fig. 1H). These data strongly suggest that drTLR5a and drTLR5b do not function as conventional homodimers but instead form functional TLR5 heterodimers.
Zebrafish UNC93B1 Facilitates Formation of Functional drTLR5a and drTLR5b Heterodimers.
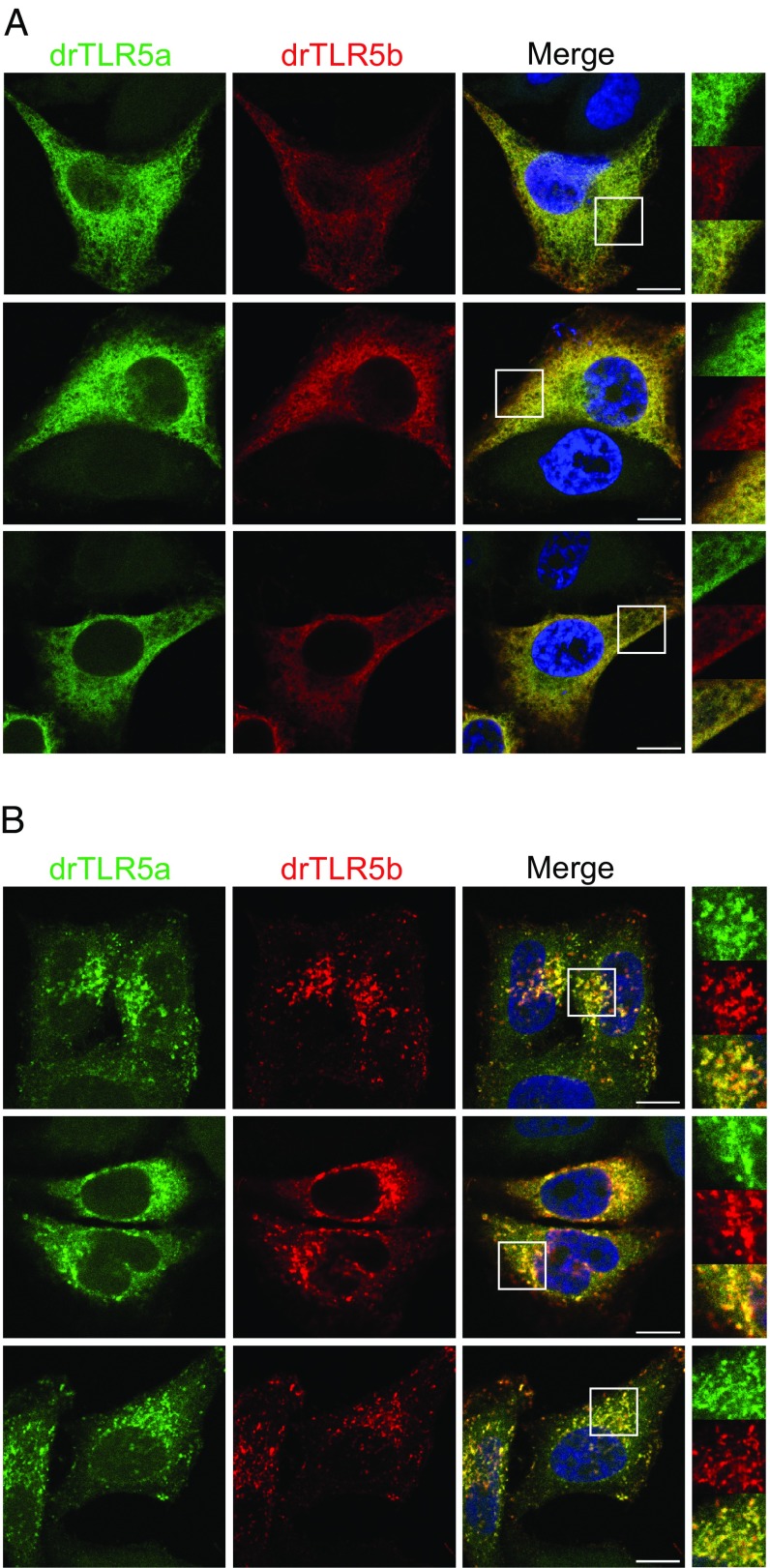
To gain additional evidence for drTLR5a and drTLR5b acting as heterodimeric receptors, we constructed drTLR5a with a C-terminal HA tag and visualized receptor localization in drTLR5a-HA– and drTLR5b-FLAG–transfected HeLa-57A cells. Confocal microscopy revealed overlapping expression of drTLR5a and drTLR5b throughout cells rather than at a specific cellular location (Fig. 2A). Previously, human TLR5 was found to localize at the cell surface only in the presence of the TLR trafficking chaperone UNC-93 homolog B1 (UNC93B1) (29). UNC93B1 interacts with TLRs via acidic amino acids in the TLR extracellular juxtamembrane region (EJM) (29, 30) and is required for TLRs 3, 7, 8, and 9 to traffic to endolysosomal compartments (31–33). Inspection of the amino acid sequences of drTLR5a and drTLR5b revealed identical EJMs that contained multiple acidic residues (Fig. S1), suggesting that drTLR5a and drTLR5b may interact with UNC93B1. Cotransfection of the zebrafish homolog of UNC93B1 (drUNC93B1) resulted in relocation of drTLR5a-HA and drTLR5b-FLAG to the same vesicle-like compartments (Fig. 2B). Separate cotransfection of FLAG-tagged drUNC93B1 with drTLR5a-HA or drTLR5b-HA showed an independent interaction of drUNC93B1 with both receptors (Fig. S2A). Staining of the drTLR5a- and drTLR5b-containing vesicles using antibodies against early endosome antigen 1 (EEA-1) or lysosomal associated membrane protein 1 (LAMP-1) demonstrated stronger association of drTLR5a and drTLR5b with LAMP-1– than with EEA-1–positive vesicles (Fig. S2B).
Fig. 2.
Effect of zebrafish UNC93B1 (drUNC93B1) on localization of drTLR5a and drTLR5b. Confocal microscopy on HeLa-57A cells transfected with (A) drTLR5-HA and drTLR5b-FLAG or (B) drTLR5a-HA and drTLR5b-FLAG and untagged drUNC93B1. Merge images show nuclei stained with DAPI (blue). White boxes in merge images indicate the magnified area shown for each channel on the right of merge images. Images were selected from three independent experiments, and three representative images are shown for each transfected group. (Scale bars in merge images: 10 µm.)
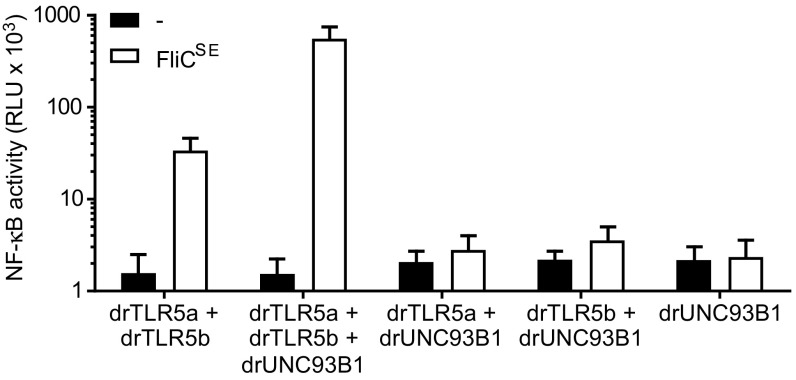
Flagellin stimulation of cells expressing drUNC93B1 and both drTLR5 receptors induced a potent (>10-fold) increase in NF-κB activity compared with cells lacking drUNC93B1 (Fig. 3). drUNC93B1 alone or in combination with either drTLR5a or drTLR5b did not induce NF-κB activity toward FliCSE (Fig. 3). Exposure of cells to FliCSE for 3 h did not alter the localization of either drTLR5a or drTLR5b at LAMP-1–positive vesicles (Fig. S3). Together, these findings indicate that zebrafish UNC93B1 operates as a trafficking chaperone to both drTLR5a and drTLR5b and that despite accumulative localization in vesicles, separate receptors cannot function as a typical TLR5 homodimer, whereas drUNC93B1-mediated relocation of both receptors facilitates the formation of functional drTLR5a and drTLR5b heterodimers.
Fig. 3.
Effect of drUNC93B1 on drTLR5a- and drTLR5b-mediated NF-κB activation. HeLa-57A cells expressing drUNC93B1, drTLR5a, and/or drTLR5b in the indicated combinations were stimulated (5 h) with vehicle (–) or 1 µg mL−1 FliCSE. Data shows NF-κB activity represented by luciferase activity in RLUs. Values are the mean ± SEM of three independent experiments performed in duplicate.
The N-Terminal Fragment of drTLR5b but Not drTLR5a Is Functional as a Homodimer.
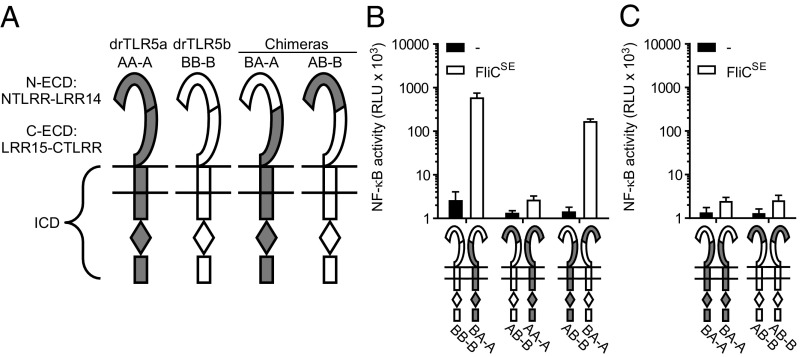
Our finding that full-length drTLR5b cannot signal as a homodimer was unexpected as the reported crystallized protein structure indicated that the N-terminal fragment (NTLRR-LRR14) of drTLR5b binds flagellin as a homodimeric complex (19). To investigate this discrepancy between binding and signaling in response to flagellin, we first determined whether the N-terminal region of the drTLR5b ectodomain can function in homodimeric configuration when all other regions of the receptor complex are heterodimeric. For this, we replaced the N-terminal part of the ectodomain (NTLRR-LRR14) of drTLR5a for the corresponding drTLR5b sequence. The exchanged protein fragment matches exactly with the published crystallized structure of drTLR5b (Fig. S1). The resulting chimeric receptor was named “BA-A.” In this terminology, “BA” indicates the combination of the N-terminal part of the ectodomain (N-ECD) of drTLR5b and the C-terminal part of the ectodomain (C-ECD) of drTLR5a; “–” indicates the TM region; and “A” indicates the drTLR5a intracellular domain (ICD) (Fig. 4A). Exposure of cells cotransfected with chimera BA-A and WT drTLR5b (BB-B) to FliCSE (Fig. 4B) or FliCPA (Fig. S4) strongly activated NF-κB. This suggests that flagellin can bind to the homodimeric drTLR5b N-ECD, as the crystal structure dictates, and is capable of inducing functional dimerization when other regions of the receptor are in a heterodimeric configuration. To determine whether the N-ECD of drTLR5a can act in a similar way, we constructed the inverse chimera “AB-B” and cotransfected this chimera with WT drTLR5a (AA-A). Interestingly, stimulation of this receptor combination with FliCSE (Fig. 4B) or FliCPA (Fig. S4) failed to activate NF-κB. To verify that the AB-B chimeric construct was expressed and functional, cells were cotransfected with chimeras AB-B and BA-A, which would reinstate a fully complementary heterodimeric complex. FliCSE stimulation of these cells yielded NF-κB activation (Fig. 4B). Cells transfected with only the BA-A or AB-B chimeras did not respond to FliCSE (Fig. 4C). These results indicate that the interaction with flagellin differs between drTLR5a and drTLR5b and that heterodimerization is required for TLR5 signal transduction.
Fig. 4.
Functionality of the N-terminal part of the ectodomain of drTLR5b and drTLR5a. (A) Schematic representation of WT and chimeric TLR5 constructs. N-ECD indicates the N-terminal part of the ectodomain ranging from the NTLRR to LRR14; C-ECD indicates the C-terminal part of the ectodomain ranging from LRR15 to the CTLRR. (B and C) HeLa-57A cells transfected with drUNC93B1 and the indicated receptor combinations were stimulated (5 h) with vehicle (–) or 1 µg mL−1 FliCSE. Data show NF-κB activity represented by luciferase activity in RLUs. Values are the mean ± SEM of three independent experiments performed in duplicate.
Transfer of N-ECD Amino Acids from drTLR5b to drTLR5a and Human TLR5.
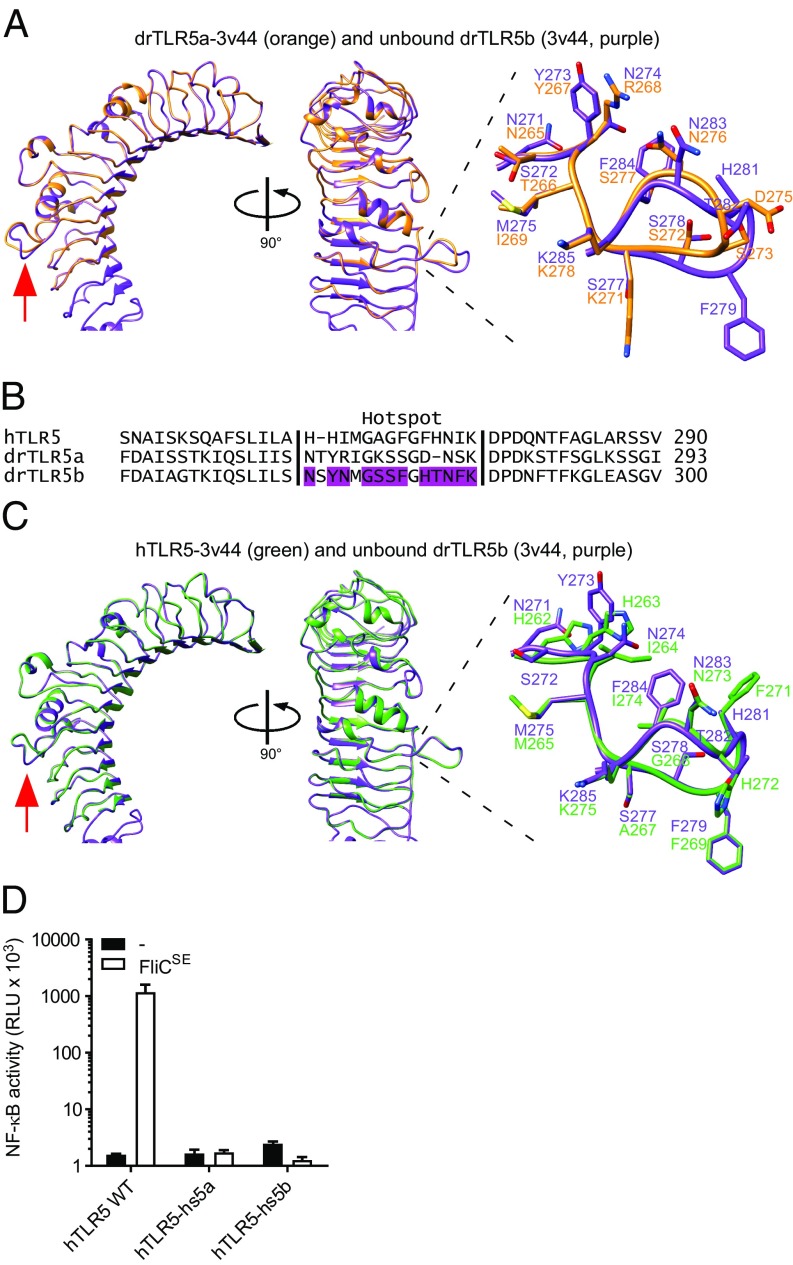
The N-ECD protein sequences of drTLR5a and drTLR5b vary in 108 of 369 amino acids (excluding the signal peptide). To analyze the possible structural implications of this difference, we modeled the N-ECD sequence of drTLR5a onto the crystallized structure of the ligand-free [Protein Data Bank (PDB) ID: 3v44] and the flagellin-bound drTLR5b N-ECD (PDB ID: 3v47). Superposition of the structures predicted that the drTLR5 N-ECDs are structurally highly similar both in the absence (Fig. 5A) or presence (Fig. S5A) of flagellin, except for a laterally protruding loop that connects LRR9 and LRR10. This loop was found to change conformation upon binding of flagellin (19). Yoon et al. (19) marked this loop as the flagellin-binding “hotspot” since its deletion severely impaired flagellin binding. The hotspot forms part of 45 residues in the drTLR5b N-ECD crystal structure that are involved in dimerization and flagellin binding. Twenty-one of the 45 residues in drTLR5b, including six residues in the hotspot, are different at the same position in drTLR5a (Fig. S5B). In an attempt to reconstitute a functional homodimeric drTLR5a N-ECD, we adapted chimera AB-B by replacing the deviant drTLR5a N-ECD residues for their corresponding drTLR5b residues. We first tested the effect of replacing 14 of the 21 residues that were least similar by constructing chimera A14B-B. Although the chimera was expressed (Fig. S5C), cotransfection of cells with chimera A14B-B and AA-A (drTLR5a) did not restore responsiveness to FliCSE. When all 21 deviant residues were replaced with their drTLR5b counterparts, functionality was still not restored, as shown by the lack of FliCSE-induced activation of chimera A21B-B coexpressed with AA-A. Cotransfection of chimeras A14B-B or A21B-B with chimera BA-A also did not result in an FliCSE-responsive receptor complex (Fig. S5D). These results indicate that the transfer of amino acids involved in flagellin binding and dimerization in drTLR5b to drTLR5a is not sufficient to reconstitute a functional N-terminal domain.
Fig. 5.
Structural modeling and functionality of the flagellin-binding hotspot in hTLR5, drTLR5a, and drTLR5b. (A) Superposition of unbound drTLR5b (PDB ID: 3v44; purple) and a model of drTLR5a based on 3v44 (drTLR5a-3v44; orange). The red arrow indicates the flagellin-binding hotspot that forms a loop between LRR9 and LRR10. (B) Alignment of the putative flagellin-binding hotspot of hTLR5 and drTLR5a with drTLR5b. Purple-colored residues in drTLR5b are involved in flagellin binding (see ref. 19). (C) Superposition of unbound drTLR5b (PDB ID: 3v44; purple) and a model of hTLR5 based on 3v44 (hTLR5-3v44; green). (D) HeLa-57A cells transfected with WT hTLR5 (hTLR5 WT), hTLR5 containing the hotspot of drTLR5a (hTLR5-hs5a), or the hotspot of drTLR5b (hTLR5-hs5b) were stimulated (5 h) with vehicle (–) or 1 µg mL−1 FliCSE. Data show NF-κB activity represented by luciferase activity in RLUs. Values are the mean ± SEM of three independent experiments performed in duplicate.
Of the 45 interacting residues in drTLR5b, the hotspot is the only cluster of residues that is somewhat evolutionarily conserved in TLR5 of avian, reptilian, and mammalian species, including human TLR5 (Fig. S6). Although the loop of hTLR5 contains different residues (Fig. 5B), modeling of the hTLR5 N-ECD onto drTLR5b predicted no structural difference of the LRR9 loop, neither in the absence (Fig. 5C) or the presence (Fig. S7A) of flagellin. To determine whether the predicted different loop conformation of drTLR5a versus drTLR5b and hTLR5 would affect receptor function, we replaced the loop in homodimeric hTLR5 for the structurally dissimilar drTLR5a hotspot loop (hTLR5-hs5a). hTLR5 carrying the structurally similar drTLR5b hotspot loop (hTLR5-hs5b) was constructed as a control. While receptors were expressed (Fig. S7B) and WT hTLR5 activated NF-κB in response to FliCSE, hTLR5-hs5a as well as hTLR5-hs5b failed to respond to FliCSE (Fig. 5D). These findings suggest that the LRR9 loop is crucial for hTLR5 activation and that, despite predicted structural similarity, this flagellin-binding hotspot cannot be functionally exchanged between drTLR5b and hTLR5.
Functional drTLR5 Requires Heterodimerization of Either the N- or C-Terminal Ectodomain.
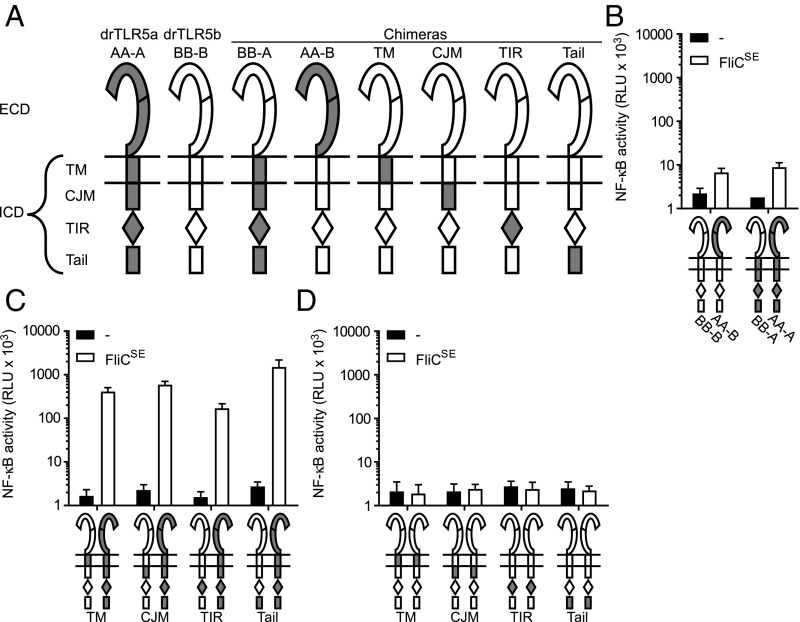
To identify other (non–N-ECD) region(s) in drTLR5a and drTLR5b that must operate in a heterodimeric configuration to establish a functional TLR5 dimer, we systematically exchanged domains or regions between drTLR5a and drTLR5b. Expression of different combinations of these chimeric and WT receptors enabled us to determine the contribution of the different domains to the formation of functional heterodimeric TLR5. Reduced or loss-of-function upon homodimerization would indicate that the substituted domain must be in heterodimeric configuration to maintain functionality. Having established that the drTLR5b N-ECD can function in homodimeric configuration (see above), we next tested the effect of homodimerization of the full drTLR5b ectodomain while keeping the ICD heterodimeric. For this, we constructed and expressed the combination of the chimeric receptor “BB-A” (consisting of the entire drTLR5b ectodomain linked to the drTLR5a ICD) (Fig. 6A) and WT drTLR5b (BB-B). Exposure of the transfected cells to FliCSE did not result in NF-κB activity. Likewise, the reverse chimera AA-B in combination with WT drTLR5a (AA-A) failed to respond to FliCSE. The combined expression of BB-A and AA-B, which restored complete heterodimerization, did activate NF-κB upon FliCSE exposure, ensuring that chimeras were expressed and functional (Fig. 6B). These findings indicate that the full ectodomains of drTLR5a and drTLR5b cannot function in homodimeric configuration.
Fig. 6.
Heterodimerization of the ectodomain of drTLR5a and drTLR5b. (A) Schematic representation of WT and chimeric TLR5 constructs. N-ECD indicates the N-terminal part of the ectodomain ranging from NTLRR to LRR14. C-ECD indicates the C-terminal part of the ectodomain ranging from LRR15 to the CTLRR. (B–D) HeLa-57A cells transfected with drUNC93B1 and the indicated receptor combinations were stimulated (5 h) with vehicle (–) or 1 µg mL−1 FliCSE. Data show NF-κB activity represented by luciferase activity in RLUs. Values are the mean ± SEM of three independent experiments performed in duplicate.
As the N-terminal part of the drTLR5b ectodomain but not the full ectodomain did signal in homodimeric configuration (Fig. 4B), we anticipated that the remaining C-terminal part (LRR15-CTLRR) must be heterodimeric to allow dimer activation. To verify this, chimera AB-A was constructed (Fig. 6A) and cotransfected with WT BB-B. Unexpectedly, stimulation of these cells with FliCSE still activated NF-κB, despite the homodimeric configuration of the drTLR5b C-ECD. Similarly, homodimerization of the drTLR5a C-ECD tested by expressing chimera BA-B in combination with WT AA-A conferred NF-κB activation upon FliCSE stimulation (Fig. 6C). All ectodomain chimeras transfected individually did not respond to FliCSE stimulation (Fig. 6D). Taken together, these results show that the full-length drTLR5a and drTLR5b ectodomains cannot function in homodimeric configuration, but that heterodimerization of either the N- or C-terminal LRR region is required to allow receptor activation.
Zebrafish TLR5 Signaling Requires Heterodimerization Across Multiple Regions in the ICD.
After delineating the extracellular needs for functional TLR5 heterodimerization, we determined the configurational requirements for the formation of functional intracellular receptor domains (ICDs). At the amino acid level, the ICDs of drTLR5a and drTLR5b are very similar (Fig. S1), suggesting that the ICDs of drTLR5a and drTLR5b may initiate signaling in homodimeric configuration. However, cells cotransfected with WT BB-B and chimera AA-B, forming a heterodimeric ECD but a homodimeric ICD of drTLR5b, did not respond to FliCSE. Likewise, coexpression of WT AA-A and chimera BB-A failed to activate NF-κB in response to FliCSE (Fig. 7B). To explore which distinct part of the ICD must be heterodimeric to allow signal initiation, we subdivided the ICD into four regions: TM, cytoplasmic juxtamembrane (CJM), TIR, and the Tail regions and constructed chimeras of drTLR5b in which these regions were substituted with their drTLR5a counterparts (Fig. 7A and Fig. S1). Coexpression of WT drTLR5a and chimeric drTLR5b carrying the drTLR5a TM region still activated NF-κB, indicating that the TM region does not have to be heterodimeric to form a functional dimer. Interestingly, receptor combinations forming homodimeric CJM, TIR, or Tail regions also still activated NF-κB when stimulated with FliCSE (Fig. 7C). As expected from their fully homodimeric configuration, the ICD chimeras transfected individually were unresponsive to FliCSE stimulation (Fig. 7D). These results indicate that while separate regions of the ICD can have identical sequences and still form a functional dimer, an entirely homodimeric ICD blocks the ability to initiate signaling. This suggests that heterodimeric interactions of multiple regions between the TLR5 ICDs are required to constitute a functional zebrafish TLR5 receptor.
Fig. 7.
Heterodimerization of the ICD of drTLR5a and drTLR5b. (A) Schematic representation of WT and chimeric TLR5 constructs. (B–D) HeLa-57A cells transfected with drUNC93B1 and the indicated receptor combinations were stimulated (5 h) with vehicle (–) or 1 µg mL−1 FliCSE. Data show NF-κB activity represented by luciferase activity in RLUs. Values are the mean ± SEM of three independent experiments performed in duplicate.
Discussion
The detection of flagellin by TLR5 is important to initiate immune responses against motile pathogenic bacteria (34–36). Vertebrate animals, including mammals, birds, and reptiles, have one TLR5 ortholog that detects flagellin as a homodimeric complex (12, 13, 37). The crystal structure of drTLR5b in complex with flagellin provided structural support for the homodimeric mode of action of other TLR5 species. However, direct evidence that drTLR5b is functional as a homodimeric receptor has been lacking. Here we show that zebrafish TLR5 only responds to flagellin as a heterodimeric complex composed of the products of the duplicated tlr5 genes: drTLR5a and drTLR5b. When expressed separately, drTLR5b and drTLR5a were unable to activate NF-κB in response to purified flagellins or lysates of motile bacteria. Even after expression of drUNC93B1, which led to redistribution and more robust NF-κB activation by the heterodimeric receptor, individual drTLR5a and drTLR5b did not operate as homodimers. Additional evidence for the requirement to form a heterodimeric complex came from the use of chimeric receptors that were only functional in combination with other chimeras composed of swapped complementary regions (that allowed full heterodimerization). Given that TLRs universally function via dimerization (3, 38), our data firmly indicate that drTLR5a and drTLR5b detect flagellin as a heterodimeric complex. Our results provide the molecular basis for the observation that knockdown of either drtlr5a or drtlr5b in zebrafish embryos hampers up-regulation of proinflammatory genes after flagellin challenge (39, 40).
Zebrafish UNC93B1 had a profound effect on the trafficking of drTLR5a and drTLR5b and signaling by the TLR5 heterodimer. drUNC93B1 enhanced relocalization of both receptors toward LAMP-1–specific compartments and this coincided with a strongly increased capacity of cells to respond to flagellin. UNC93B1 is a membrane protein that complexes in the endoplasmic reticulum (ER) to TLRs 3, 7, 8, and 9 and TLR5 via acidic residues in the TLR EJM (29, 41, 42). The UNC93B1-TLR complex then exits the ER to transit to the Golgi apparatus. Further trafficking of TLRs to endosomes, lysosomes, or the cell surface involves other trafficking partners (41, 42). In the EJM of both drTLR5a and drTLR5b, multiple acidic residues have been conserved and the ability of drUNC93B1 to facilitate relocalization of drTLR5a and drTLR5b independently identifies drUNC93B1 as a trafficking chaperone for both zebrafish TLR5 receptors.
The use of an extensive set of chimeric receptors revealed that the N-ECD of drTLR5b but not drTLR5a could act in homodimeric configuration to activate the receptor complex. In the drTLR5b N-ECD, 45 residues are involved in dimerization and flagellin binding (19). Of these 45 residues, 21 are different in the drTLR5a N-ECD; and of these 21 residues, 14 differ significantly. Our attempts to restore homodimeric functionality of the drTLR5a N-ECD by replacing these differing residues in the AB-B chimera for their drTLR5b counterparts and combining this chimera with WT drTLR5a (AA-A) failed. In addition, replacement of the differing residues in an otherwise heterodimeric complex (by combining chimeras A14B-B or A21B-B with BA-A) abrogated function. These findings indicate that the 45 residues involved in flagellin binding and dimerization of the drTLR5b N-ECD are either not sufficient for the formation of a functional homodimeric drTLR5a N-ECD or that epistatic interactions within the chimeric N-ECD prevent the formation of a functional receptor. Information as to whether this difference between the drTLR5a and drTLR5b N-ECD is caused by a lack of interaction of drTLR5a with flagellin or drTLR5a signaling defects awaits elucidation of the flagellin-drTLR5a N-ECD crystal structure.
Our finding that the N-ECD of drTLR5b has maintained the ability to function as a homodimer is fully in line with the homodimeric binding of flagellin in the drTLR5b N-ECD crystal structure and indicates that drTLR5b more closely resembles homodimeric TLR5 of other species. Nonetheless, the drTLR5b ectodomain was only functional if at least one of the ECD regions (N- or C-ECD) was in heterodimeric configuration with drTLR5a. The requirement for heterodimerization in the ECD was not specifically linked to the N- or C-ECD regions. This suggests that flagellin-induced receptor complex activation depends on the ectodomains of drTLR5a and drTLR5b to have different conformations rather than on specific amino acid residues. Structural information on the C-ECDs of TLR5 is needed to elucidate such different conformations in the ectodomains.
Modeling of hTLR5 onto drTLR5b predicted very high structural similarity of the N-ECD, including at the loop between LRR9 and LRR10, which forms the flagellin-binding hotspot. However, substitution of amino acids that form this loop in hTLR5 for those of drTLR5b completely abrogated hTLR5 activation. According to Yoon et al. (19), >60% of interactions in the drTLR5b loop would be conserved in hTLR5 and the loop would thus also form a flagellin-binding hotspot in hTLR5. Our findings confirm that the hotspot is critical for hTLR5 activation but also indicate that the hTLR5 and drTLR5b hotspots are functionally distinct. The flagellin-binding hotspot forms a pocket that accommodates a highly conserved arginine residue in flagellin (R90). Flagellin binding and thus insertion of R90 in the hotspot pocket alters the conformation of the LRR9 loop (19). Mutation of R90 preserves binding of flagellin to hTLR5 and mouse TLR5 (mTLR5) but reduces receptor activation (43, 44), suggesting that insertion of R90 into the hotspot pocket initiates a conformational change that is necessary for receptor activation (44). Although the hotspot pockets of mTLR5 and hTLR5 differ only by three residues (Fig. S6), R90 mutations differentially affect mTLR5 and hTLR5 activation (43, 44). In addition, substitution of the proline residue (P268) in the mTLR5 hotspot for the alanine residue (A267) in hTLR5 changed mTLR5 flagellin recognition specificity toward the pattern of hTLR5 (43). Homology models constructed of hTLR5 and mTLR5 based on drTLR5b predicted differential binding affinities between hTLR5 and mTLR5 to flagellin residue E93. Flagellin residue E93 interacts extensively with drTLR5b, but mutation of E93 had no functional effect on both human and mouse TLR5 (44). In the present study, substitution of the hTLR5 hotspot with corresponding drTLR5b residues effectively changed eight residues and inserted one residue. This alteration was sufficient to completely preclude hTLR5 activation, suggesting that the conformational change in the drTLR5b hotspot upon flagellin binding differs from what is necessary to induce functional homodimerization of hTLR5. Our findings corroborate the published work and provide additional evidence that TLR5–flagellin recognition is species-specific. This is important to consider when using TLR5 of nontarget species as a model to define the exact mechanism of flagellin-binding and flagellin-induced changes required for TLR5 activation.
Our data demonstrate that the cytoplasmic domains of drTLR5a and drTLR5b initiated signaling in a hetero- but not a homodimeric configuration. This was unexpected since the drTLR5a and drTLR5b cytoplasmic domains are highly similar. When tested separately, all of the defined cytoplasmic regions, including the TIR domain, could function in homodimeric configuration only if other regions were heterodimeric. Since a fully homodimeric cytoplasmic domain could not signal, these findings suggest functional interdependence of the distinct regions (i.e., a requirement for transregional heterodimeric configuration). This may be best explained by different, conformational changes occurring in hetero- versus homodimeric cytoplasmic domains. TLR activation involves ligand-induced conformational changes in the ectodomain that transit through the TM region to result in dimerization of the cytoplasmic TIR domains (14, 45). The TLR TIR domain consists of five β-sheets surrounded by five α-helixes (46). These secondary structures are connected by loops, and the BB loops of two TIR domains have to align correctly to form a dimerization interface, which is necessary for the TIR domains to recruit signaling adapters (14, 47). Based on our findings, we postulate that activation of the drTLR5 heterodimer starts with a flagellin-binding–induced conformational change in the ectodomain, probably in the LRR9 loop, which, in turn, induces a conformational change (e.g., rotational) across multiple regions in the heterodimeric cytoplasmic domain to correctly align the TIR BB loops to form an appropriate dimerization interface. Such a ligand-induced rotational conformational change in drTLR5 could be similar to those occurring in other type I membrane receptors such as the epidermal growth factor and thrombopoietin receptors (14, 48, 49) and may also be required for activation of other heterodimeric TLRs such as the TLR1/2 and TLR2/6 combinations. Further research using structural and biophysical experiments is necessary to validate this scenario.
The drTLR5a and drTLR5b receptors respond to flagellin as a complementary heterodimer. Whereas in zebrafish (a teleost fish) the tlr5a and tlr5b paralogs have likely arisen through a tandem gene duplication, non-teleost vertebrates, including mammals, birds, reptiles, and also the spotted gar (Lepisosteus occulatus), a holostean fish (50), all have one tlr5 ortholog. This implies that the common ancestor of teleosts and non-teleost vertebrates likely had one TLR5 ortholog that may have operated as a homodimer. Partitioning of the ancestral function (i.e., flagellin detection) over the drTLR5a and drTLR5b paralogs points to subfunctionalization of drTLR5a and drTLR5b that may be explained by the duplication-degeneration-complementation (DDC) model (51, 52). According to this model, duplicated genes randomly acquire different degenerative mutations to the extent that protein paralogs must complement each other to maintain the ancestral function. drTLR5a and drTLR5b form an interesting example of protein subfunctionalization since the receptors maintain flagellin detection by physical complementation. Although drTLR5a and drTLR5b clearly detect flagellin as a heterodimer, we cannot exclude that homodimeric drTLR5a or drTLR5b may detect ligands other than flagellin. However, such ligands will likely not include common and abundant molecules from gram-negative bacteria since neither drTLR5a nor drTLR5b responded to lysates of such bacteria. To our knowledge, zebrafish TLR5 is the first example of TLR subfunctionalization in teleost fish. It is likely that in other species at least some of the duplicated TLR genes also evolved through DDC to maintain ancestral functions. This may explain the conservation in mammals of the TLR1, TLR2, and TLR6 heterodimeric partners.
In conclusion, the duplicated TLR5 genes of the zebrafish model organism encode two functional receptors that have subfunctionalized via concerted coevolution to detect bacterial flagellins as a unique TLR5 heterodimer. The zebrafish TLR5 heterodimer expands our understanding of the mechanism of ligand-induced TLR activation and provides an illustrative example of the functional coevolution of duplicated genes.
Materials and Methods
DNA Isolation and cDNA Synthesis.
Zebrafish tissue was kindly provided by J. den Hertog (Hubrecht Institute, Utrecht, The Netherlands). Genomic DNA was isolated from zebrafish tissue using the high pure template kit (Roche) according to the manufacturer’s instructions. RNA was extracted from tissue lysed with RLT buffer (Qiagen) (1% β-mercaptoethanol) in 1.4-mm Fastprep lysing matrix tubes (MPbio) in a Magna Lyser centrifuge [6,500 × g, 40 s, room temperature (RT)] (Roche). Total RNA was isolated using the RNeasy mini kit (Qiagen), treated with DNase I (1 U mg−1 RNA; Thermo Fisher Scientific), and reverse transcribed into cDNA using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Recombinant DNA Techniques.
Phusion high-fidelity DNA polymerase, dNTPs, fast digest restriction endonucleases, T4 DNA ligase, and primers were purchased from Thermo Fisher Scientific. PCR products were extracted from agarose gel using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific). Plasmids were isolated with Nucleobond Xtra Midi prep kit (Macherey-Nagel).
DNA Constructs.
Zebrafish tlr5b and tlr5a were amplified from genomic DNA with forward primers CV039 and CV095 (Table S1), respectively, to add a BamHI restriction site and Kozak sequence at the start of the gene and reverse primers CV054 and CV096, respectively, to add an NotI restriction site at the end of the gene. Both drtlr5b and drtlr5a PCR products were digested with BamHI and NotI and ligated into pTracer-CMV2ΔGFP/3×FLAG (11), yielding drTLR5b and drTLR5a carrying a C-terminal 3×FLAG tag. To create HA (hemagglutinin-epitope)-tagged drTLR5a and drTLR5b, pTracer-CMV2ΔGFP/3×FLAG was modified by inserting a 3×HA sequence (YPYDVPDYA) ending with a stop codon in the 3×FLAG sequence using NotI and ClaI, yielding pTracer-CMV2ΔGFP/3×HA. drtlr5a and drtlr5b were cut from pTracer-CMV2ΔGFP/3×FLAG using BamHI and NotI and inserted in pTracer-CMV2ΔGFP/3×HA, yielding drTLR5b and drTLR5a carrying a C-terminal 3×HA tag. Zebrafish unc93b1 was amplified from cDNA with forward primer CV159 and reverse primer CV160 or reverse primer CV180 to maintain the stop codon. The PCR products were cloned into pTracer-CMV2ΔGFP/3×FLAG using KpnI and NotI restriction enzymes, yielding drUNC93B1 with a C-terminal 3×FLAG tag or untagged drUNC93B1. Human TLR5 was cloned from pUNO-hTLR5-GFP (Invivogen) with primers CV257 and CV259, and ligated into pTracer-CMV2ΔGFP/3×FLAG using BamHI and NotI to yield hTLR5 with a C-terminal 3×FLAG tag. Chimeric DNA constructs were created by standard overlap extension PCR technique. The DNA fragments encoding the N-terminal ECD in chimeras A14B-B, A21B-B, hTLR5-hs5a, and hTLR5-hs5b were purchased from GeneArt (Thermo Fisher Scientific) and fused to the receptor genes by overlap extension PCR. Chimeric PCR products were ligated in pTracer-CMV2ΔGFP/3×FLAG using BamHI and NotI. All DNA constructs were verified by sequencing (Macrogen). The sequences were deposited in GenBank with the following accession numbers: drTLR5b: MF983797; drTLR5a: MF983798; and drUNC93B1: MF983799.
Protein Sequence Analysis and TLR5 Modeling.
Protein sequences were aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) (53) with default settings. LRRs were identified by manual sequence inspection according to Matsushima et al. (54) and with use of the Leucine Rich Repeat Finder (www.lrrfinder.com/lrrfinder.php) (55). The TM domain was predicted with TMHMM Server version 2.0 (www.cbs.dtu.dk/services/TMHMM/) (56). The TIR domain was predicted by identifying five alternating β-sheets and α-helices (46) using the Proteus Protein Structure Prediction server (www.proteus2.ca/proteus2/) (57). Structural models of the ECD N-terminal part of drTLR5a and hTLR5 were created using SWISS-MODEL (default settings; https://swissmodel.expasy.org/) (58) with the drTLR5b crystal structure (PDB: 3v44) or the drTLR5b-flagellin crystal structure (PDB: 3v47) as templates. UCSF (University of California, San Francisco) Chimera (www.rbvi.ucsf.edu/chimera) (59) was used to view protein models.
Isolation of Recombinant Flagellins and Bacterial Lysate.
Construction and purification of recombinant HIS-tagged FliC of FliCSE (strain 90-13-706 CVI; Lelystad, The Netherlands) and FliCPA (reptile clinical isolate; Utrecht University) have been described previously (13, 37). Purified flagellins were stored in buffer: 4 M urea, 10 mM NaH2PO4, and 100 mM Tris (all Sigma) at −20 °C. For lysate isolation, S. Enteritidis, P. aeruginosa, and A. hydrophila (fish clinical isolate; Utrecht University) were grown (24 h) on sheep blood agar plates (Biotrading) at 28 °C. Single colonies were grown (28 °C, 16 h) in 5 mL heart infusion (HI) broth (Biotrading) at 160 rpm and placed on ice. Microscopic examination confirmed that all bacteria were highly motile. Cultures were normalized to an OD550 of 3, pelleted by centrifugation (30 min, 5,000 × g, 4 °C), washed with 1 mL Dulbecco’s PBS (DPBS; Sigma), vortexed, and collected by centrifugation (15 min, 5,000 × g, 4 °C). Pellets were redissolved in 2 mL DPBS and placed at 70 °C for 1 h. Heat-killed bacteria were sonicated (6 × 15 s; Vibra-cell, Sonics) and centrifuged (14,000 × g, 40 min, 4 °C). Supernatants were concentrated with Pierce protein concentrators, 10 kDa (Thermo Fisher Scientific), for 15 min at 5,000 × g, RT. Total protein concentration of lysates was determined by BCA assay (Thermo Fisher Scientific). Lysate supernatants were stored at −20 °C until use.
Cell Culture and Transient Transfection.
Human HeLa-57A cells [that stably carry a NF-κB luciferase reporter construct (60)] were cultured in DMEM (Thermo Fisher Scientific) with 5% FCS (Bodinco) at 37 °C and 10% CO2. For transfection, cells were grown to 80% confluency in a 12-well plate and transfected with 670 ng of each plasmid using Fugene HD (Promega) at a DNA to Fugene ratio of 1:3 following the manufacturer’s instructions. Empty pTracer 3×FLAG plasmid was used to equalize the total amount (2 µg) of transfected plasmid added to each well.
Luciferase Assay.
Twenty-four hours after transfection, cells were redistributed in a 96-well plate. After 24 h, cells were washed twice with medium without FCS and stimulated with purified flagellin or bacterial lysate in 100 μL medium without FCS. After 5 h at 37 °C, cells were washed twice with DPBS (Sigma) and lysed with reporter lysis buffer (50 μL; Promega) at −80 °C for ≥1 h. After thawing, 15 μL lysate was mixed with 37 μL luciferase reagent (Promega), and luciferase activity was measured with a TriStar2 luminometer (Berthold). Results were expressed in relative light units (RLUs). Receptors were assigned to be unresponsive when luciferase activity after flagellin stimulation did not exceed 10 RLUs (×103).
Confocal Laser Microscopy.
Twenty-four hours after transfection, cells were seeded onto glass coverslips. After 24 h, cells were prepared for confocal microscopy. Cells were washed with TBS and fixed (30 min) with TBS/1.5% paraformaldehyde (Affimetrix). Cells were permeabilized and blocked (30 min) with TBS containing 0.1% saponin and 0.2% BSA (both Sigma). Next, cells were incubated (1 h) with mouse M2-α-FLAG (F3165; Sigma), mouse α-EEA-1 (610456; BD Bioscience), or rabbit α-LAMP-1 (24170; Abcam); washed with TBS; incubated (1 h) with Alexa Fluor-568 goat-α-mouse IgG (A11031; Thermo Fisher Scientific) or Alexa Fluor-568 goat-α-rabbit IgG (A11036; Thermo Fisher Scientific); washed with TBS; and incubated (1 h) with Alexa Fluor-488 mouse-α-HA (A21287; Thermo Fisher Scientific). After antibody staining, cells were washed with TBS and MilliQ and embedded in Prolong diamond mounting solution (Thermo Fisher Scientific). Cells were viewed on a Leica SPE-II or Nikon A1R/STORM laser confocal microscope, and images were processed using Leica LAS AF or Nikon NIS software.
Supplementary Material
Acknowledgments
We thank Dr. R. Wubbolts (Center for Cell Imaging, Department of Biochemistry and Cell Biology, Utrecht University, Utrecht, The Netherlands) for acquisition of confocal microscope data and Dr. R. T. Hay (Institute of Biomolecular Sciences, School of Biomedical Sciences, University of St. Andrews, Scotland, UK) for providing the HeLa-57A cell line.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.E.V. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. drTLR5b: MF983797; drTLR5a: MF983798; drUNC93B1: MF983799).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719245115/-/DCSupplemental.
References
- 1.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 2.Leulier F, Lemaitre B. Toll-like receptors–Taking an evolutionary approach. Nat Rev Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 3.Gay NJ, Gangloff M. Structure and function of Toll receptors and their ligands. Annu Rev Biochem. 2007;76:141–165. doi: 10.1146/annurev.biochem.76.060305.151318. [DOI] [PubMed] [Google Scholar]
- 4.Roach JC, et al. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci USA. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 6.de Zoete MR, Bouwman LI, Keestra AM, van Putten JPM. Cleavage and activation of a Toll-like receptor by microbial proteases. Proc Natl Acad Sci USA. 2011;108:4968–4973. doi: 10.1073/pnas.1018135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 8.Hemmi H, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 9.Jurk M, et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol. 2002;3:499. doi: 10.1038/ni0602-499. [DOI] [PubMed] [Google Scholar]
- 10.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 11.Keestra AM, de Zoete MR, Bouwman LI, van Putten JPM. Chicken TLR21 is an innate CpG DNA receptor distinct from mammalian TLR9. J Immunol. 2010;185:460–467. doi: 10.4049/jimmunol.0901921. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi F, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 13.Voogdt CGP, Bouwman LI, Kik MJL, Wagenaar JA, van Putten JPM. Reptile Toll-like receptor 5 unveils adaptive evolution of bacterial flagellin recognition. Sci Rep. 2016;6:19046. doi: 10.1038/srep19046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gay NJ, Symmons MF, Gangloff M, Bryant CE. Assembly and localization of Toll-like receptor signalling complexes. Nat Rev Immunol. 2014;14:546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 15.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanji H, Ohto U, Shibata T, Miyake K, Shimizu T. Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands. Science. 2013;339:1426–1429. doi: 10.1126/science.1229159. [DOI] [PubMed] [Google Scholar]
- 17.Ohto U, et al. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature. 2015;520:702–705. doi: 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, et al. Structural analysis reveals that Toll-like receptor 7 is a dual receptor for guanosine and single-stranded RNA. Immunity. 2016;45:737–748. doi: 10.1016/j.immuni.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Yoon SI, et al. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palti Y. Toll-like receptors in bony fish: From genomics to function. Dev Comp Immunol. 2011;35:1263–1272. doi: 10.1016/j.dci.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Christoffels A, et al. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol Biol Evol. 2004;21:1146–1151. doi: 10.1093/molbev/msh114. [DOI] [PubMed] [Google Scholar]
- 22.Solbakken MH, et al. Evolutionary redesign of the Atlantic cod (Gadus morhua L.) Toll-like receptor repertoire by gene losses and expansions. Sci Rep. 2016;6:25211. doi: 10.1038/srep25211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 24.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 25.Fink IR, et al. Molecular and functional characterization of Toll-like receptor (Tlr)1 and Tlr2 in common carp (Cyprinus carpio) Fish Shellfish Immunol. 2016;56:70–83. doi: 10.1016/j.fsi.2016.06.049. [DOI] [PubMed] [Google Scholar]
- 26.Sullivan C, et al. The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J Immunol. 2009;183:5896–5908. doi: 10.4049/jimmunol.0803285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeh D-W, et al. Toll-like receptor 9 and 21 have different ligand recognition profiles and cooperatively mediate activity of CpG-oligodeoxynucleotides in zebrafish. Proc Natl Acad Sci USA. 2013;110:20711–20716. doi: 10.1073/pnas.1305273110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song WS, Yoon SI. Crystal structure of FliC flagellin from Pseudomonas aeruginosa and its implication in TLR5 binding and formation of the flagellar filament. Biochem Biophys Res Commun. 2014;444:109–115. doi: 10.1016/j.bbrc.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Huh J-W, et al. UNC93B1 is essential for the plasma membrane localization and signaling of Toll-like receptor 5. Proc Natl Acad Sci USA. 2014;111:7072–7077. doi: 10.1073/pnas.1322838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J, et al. Acidic amino acid residues in the juxtamembrane region of the nucleotide-sensing TLRs are important for UNC93B1 binding and signaling. J Immunol. 2013;190:5287–5295. doi: 10.4049/jimmunol.1202767. [DOI] [PubMed] [Google Scholar]
- 31.Kim Y-M, Brinkmann MM, Paquet M-E, Ploegh HL. UNC93B1 delivers nucleotide-sensing Toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 32.Itoh H, et al. UNC93B1 physically associates with human TLR8 and regulates TLR8-mediated signaling. PLoS One. 2011;6:e28500. doi: 10.1371/journal.pone.0028500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi R, Singh D, Kao CC. Proteolytic processing regulates Toll-like receptor 3 stability and endosomal localization. J Biol Chem. 2012;287:32617–32629. doi: 10.1074/jbc.M112.387803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carvalho FA, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijay-Kumar M, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–3921. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen-Nissen E, et al. Cutting edge: Tlr5-/- mice are more susceptible to Escherichia coli urinary tract infection. J Immunol. 2007;178:4717–4720. doi: 10.4049/jimmunol.178.8.4717. [DOI] [PubMed] [Google Scholar]
- 37.Keestra AM, de Zoete MR, van Aubel RAMH, van Putten JPM. Functional characterization of chicken TLR5 reveals species-specific recognition of flagellin. Mol Immunol. 2008;45:1298–1307. doi: 10.1016/j.molimm.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Bryant CE, et al. Advances in Toll-like receptor biology: Modes of activation by diverse stimuli. Crit Rev Biochem Mol Biol. 2015;50:359–379. doi: 10.3109/10409238.2015.1033511. [DOI] [PubMed] [Google Scholar]
- 39.Stockhammer OW, Zakrzewska A, Hegedûs Z, Spaink HP, Meijer AH. Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J Immunol. 2009;182:5641–5653. doi: 10.4049/jimmunol.0900082. [DOI] [PubMed] [Google Scholar]
- 40.Yang S, Marín-Juez R, Meijer AH, Spaink HP. Common and specific downstream signaling targets controlled by Tlr2 and Tlr5 innate immune signaling in zebrafish. BMC Genomics. 2015;16:547. doi: 10.1186/s12864-015-1740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee BL, et al. UNC93B1 mediates differential trafficking of endosomal TLRs. eLife. 2013;2:e00291. doi: 10.7554/eLife.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelka K, Shibata T, Miyake K, Latz E. Nucleic acid-sensing TLRs and autoimmunity: Novel insights from structural and cell biology. Immunol Rev. 2016;269:60–75. doi: 10.1111/imr.12375. [DOI] [PubMed] [Google Scholar]
- 43.Andersen-Nissen E, Smith KD, Bonneau R, Strong RK, Aderem A. A conserved surface on toll-like receptor 5 recognizes bacterial flagellin. J Exp Med. 2007;204:393–403. doi: 10.1084/jem.20061400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forstnerič V, Ivičak-Kocjan K, Ljubetič A, Jerala R, Benčina M. Distinctive recognition of flagellin by human and mouse Toll-like receptor 5. PLoS One. 2016;11:e0158894. doi: 10.1371/journal.pone.0158894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Latz E, et al. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat Immunol. 2007;8:772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- 46.Xu Y, et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 47.Nyman T, et al. The crystal structure of the human Toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J Biol Chem. 2008;283:11861–11865. doi: 10.1074/jbc.C800001200. [DOI] [PubMed] [Google Scholar]
- 48.Endres NF, et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matthews EE, et al. Thrombopoietin receptor activation: Transmembrane helix dimerization, rotation, and allosteric modulation. FASEB J. 2011;25:2234–2244. doi: 10.1096/fj.10-178673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braasch I, et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 2016;48:427–437. doi: 10.1038/ng.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Innan H, Kondrashov F. The evolution of gene duplications: Classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 53.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsushima N, et al. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate Toll-like receptors. BMC Genomics. 2007;8:124. doi: 10.1186/1471-2164-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Offord V, Coffey TJ, Werling D. LRRfinder: A Web application for the identification of leucine-rich repeats and an integrative Toll-like receptor database. Dev Comp Immunol. 2010;34:1035–1041. doi: 10.1016/j.dci.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 57.Montgomerie S, Sundararaj S, Gallin WJ, Wishart DS. Improving the accuracy of protein secondary structure prediction using structural alignment. BMC Bioinformatics. 2006;7:301. doi: 10.1186/1471-2105-7-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biasini M, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez MS, Thompson J, Hay RT, Dargemont C. Nuclear retention of IkappaBalpha protects it from signal-induced degradation and inhibits nuclear factor kappaB transcriptional activation. J Biol Chem. 1999;274:9108–9115. doi: 10.1074/jbc.274.13.9108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.