Significance
The plasma membrane of eukaryotic cells is compartmentalized into domains enriched in specific lipids and proteins. However, our understanding of the mechanisms and functions of this lateral segregation remains incomplete. Here, we show that the clustering of the yeast Can1 arginine transporter into domains is dictated by its conformation and requires sustained biogenesis of complex sphingolipids. Furthermore, this clustering confers to Can1 and other transporters protection from ubiquitin-dependent endocytosis. Under nutrient-starvation conditions, this protective role is reinforced, thereby allowing cells to preserve a fraction of their nutrient transporters from bulk endocytosis and to more efficiently resume growth when replenishing compounds are available. Our study reveals nutrient-regulated protection from endocytosis as an important role for protein partitioning into membrane domains.
Keywords: ubiquitin, transporter, membrane domain, yeast, endocytosis
Abstract
The eukaryotic plasma membrane is compartmentalized into domains enriched in specific lipids and proteins. However, our understanding of the molecular bases and biological roles of this partitioning remains incomplete. The best-studied domain in yeast is the membrane compartment containing the arginine permease Can1 (MCC) and later found to cluster additional transporters. MCCs correspond to static, furrow-like invaginations of the plasma membrane and associate with subcortical structures named “eisosomes” that include upstream regulators of the target of rapamycin complex 2 (TORC2) in the sensing of sphingolipids and membrane stress. However, how and why Can1 and other nutrient transporters preferentially segregate in MCCs remains unknown. In this study we report that the clustering of Can1 in MCCs is dictated by its conformation, requires proper sphingolipid biosynthesis, and controls its ubiquitin-dependent endocytosis. In the substrate-free outward-open conformation, Can1 accumulates in MCCs in a manner dependent on sustained biogenesis of complex sphingolipids. An arginine transport-elicited shift to an inward-facing conformation promotes its cell-surface dissipation and makes it accessible to the ubiquitylation machinery triggering its endocytosis. We further show that under starvation conditions MCCs increase in number and size, this being dependent on the BAR domain-containing Lsp1 eisosome component. This expansion of MCCs provides protection for nutrient transporters from bulk endocytosis occurring in parallel with autophagy upon TORC1 inhibition. Our study reveals nutrient-regulated protection from endocytosis as an important role for protein partitioning into membrane domains.
The plasma membrane (PM), as the boundary between the cell and its environment, is a platform for selective nutrient exchange, signaling events, and cell–cell interactions. Understanding the mechanisms that coordinate the numerous functions of the PM is a major challenge. It has been well documented that the PM is compartmentalized into domains of various sizes and with distinct protein and/or lipid compositions (1, 2). Much remains to be learned, however, about the mechanisms and biological roles of this organization. Yeasts and other fungi are established systems for studying PM compartmentalization (3–6). They display multiple coexisting membrane domains (7–9), ranging from static patches to dynamic, network-like compartments. The best-studied domain is the membrane compartment containing the Arg transporter Can1 (MCC, originally named after the founding member) (10, 11), corresponding to static, furrow-like invaginations of the PM associated with subcortical structures called “eisosomes” (Fig. S1) (12–14). This domain, hereafter called the “eisosome membrane compartment” (EMC), is distinct from other membrane compartments, such as the transient sites of endocytosis (13, 15, 16) and the highly dynamic domains postulated to house the target of rapamycin complex 2 (TORC2) membrane compartment (MCT) (17, 18).
The core components of eisosomes are two self-assembling BAR-domain proteins, Pil1 and Lsp1 (19, 20). Pil1 is the main organizer of eisosomes, since pil1Δ cells lack the furrow-like invaginations. The role of Lsp1 remains elusive (12, 14). EMC-resident proteins include the Sur7 tetraspan protein found exclusively in the EMCs and several proteins that localize there only dynamically, such as the cytoplasmic Slm1/2 proteins, the Pkh1/2 kinases, several nutrient transporters, and the tetraspan Nce102 (7, 13, 17, 21). In addition to proteins, EMCs are proposed to be enriched in ergosterol (13, 22) and sphingolipids (SLs) (17, 21). Eisosome assembly is controlled by the SL level via a negative feedback loop, through phosphorylation of Pil1 (Fig. S1) (17, 18). In addition, Nce102 has been identified as the only nonessential transmembrane protein required for EMC organization, acting upstream of the Pkh1/2 kinases in SL sensing in both yeast and Aspergillus nidulans (13, 21, 23).
In addition to Can1, several other transporters (Lyp1, Fur4, Mup1, and Tat2) cluster in the EMCs. So does the heterologously expressed HUP1 transporter, while other permeases, such as Gap1, are more uniformly distributed at the PM (7, 13, 22). How and why specific transporters concentrate in EMCs remains undetermined. It has been proposed that the role of this partitioning might be protection from degradation, as Can1 displays accelerated turnover in the absence of eisosomes, especially in the presence of its substrate (13). This view has been challenged, however, in a report asserting that eisosomes remain enriched in Can1 during Arg-induced endocytosis (15).
We recently investigated the mechanism of substrate-transport–elicited Can1 endocytosis (24, 25). This endocytosis is initiated by ubiquitylation of the transporter by the Rsp5/Nedd4 ubiquitin (Ub) ligase, an E3 HECT enzyme transferring Ub to Lys residues of target proteins (26, 27). Rsp5-dependent ubiquitylation of transporters typically requires members of the α-arrestin family acting as adaptors for the ligase (28, 29). The main α-arrestin involved in Arg-induced ubiquitylation of Can1 is Art1. The Bul1/2 α-arrestins contribute to the process as well (25). Arg-induced Can1 ubiquitylation requires a shift of the transporter to the inward-facing (IF) conformation of the transport cycle. This unveils a short N-terminal tail sequence recognized by Art1. This sequence is normally masked when the transporter adopts a substrate-free outward-facing (OF) conformation. This masking requires an intact 87-ELK-89 tripeptide present in the N-terminal tail of Can1, just upstream from the first transmembrane (TM) segment. Furthermore, to gain access to the unmasked sequence of Can1, Art1 needs to be stimulated via TORC1, and this occurs upon Arg uptake into the cells (Fig. 1A). Importantly, the inactive Can1(E184Q) mutant is stabilized in the IF state and constitutively exposes the Art1-binding site (25).
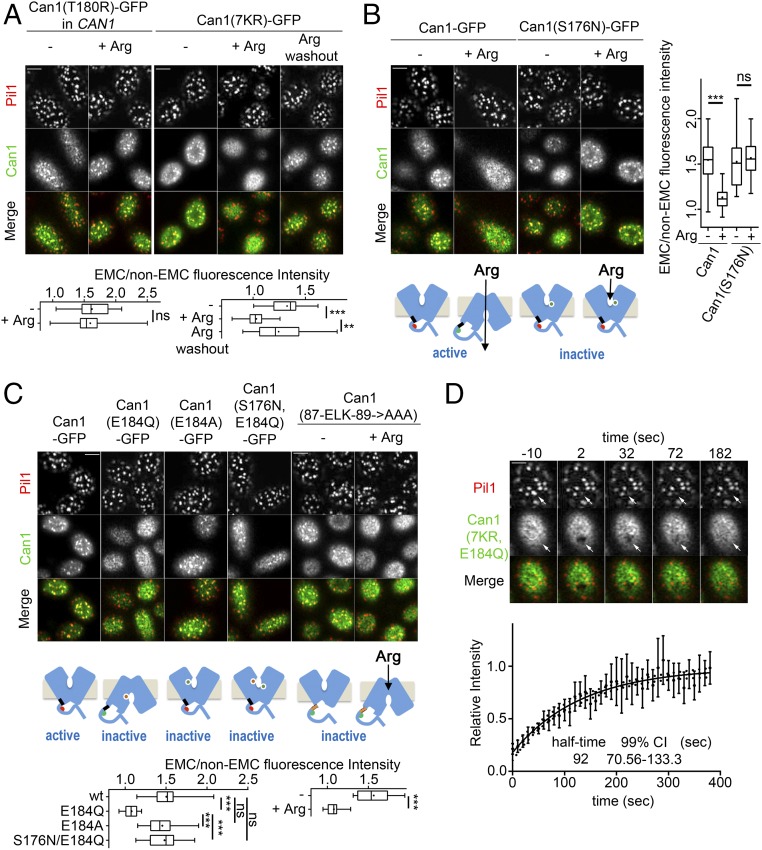
Fig. 1.
During Arg transport, Can1 EMC clustering is abolished in a ubiquitylation-independent way. (A) Model of substrate transport-induced ubiquitylation and endocytosis of the Can1 permease. The binding site for Art1 (Art1BS) in the N-terminal tail is hidden (red hemicycle) when Can1 is in the substrate-free OF conformation. In the presence of Arg, the Art1BS is exposed (green hemicycle). A tripeptide sequence (87-ELK-89) in the N-terminal tail close to TMS1 (black rectangle) is required to mask the Art1BS when Can1 is in the OF conformation. (See text for details.) (B, Left) Shown are surface and middle section (also see Fig. S2B) confocal microscopy images of WT and art1Δ bul1/2Δ strains (gap1Δ can1Δ) expressing Pil1-mCherry and Can1-GFP or Can1(7KR)-GFP. Growth conditions are as described in Fig. S2A. (Right) Quantifications: Can1-GFP EMC/non-EMC fluorescence intensity ratios are plotted (n = 32–50 cells). The horizontal midline and the cross represent the median and average values, respectively. Each box is bounded by the upper and lower quartiles; the whiskers denote the maximal and minimal ratios. (C) Shown are surface section confocal images from representative FRAP experiments, of Can1(7KR)-GFP Pil1-mCherry–expressing cells, inside or outside the EMC in the absence of Arg (Upper Left and Upper Right, respectively) or inside the EMC in the presence of Arg (Lower Left). Specific regions (white arrows) were photobleached with or without the prior addition of 5 mM Arg for 30 min. The times stated refer to the time of the bleaching at t = 0 s. (Lower Right) Quantifications: Points and error bars respectively represent the mean and SD of the relative intensity for each time point (n = 8 per condition). FRAP curves are fitted models obtained by regression analysis. Dotted curves show 99% CI inside (black curve) and outside (blue curve) EMCs or inside EMCs in the presence of Arg (red curve). The probability that the calculated K values do not differ (F-pvalue of K): black and blue curves < 0.0001; black and red curves, 0.0023; blue and red curves, 0.7173. (D) Shown are surface section confocal microscopy images of an art1Δ bul1/2Δ gap1Δ can1Δ PIL1-mCherry strain expressing Can1-GFP, Can1(T180R)-GFP, or Can1(S176N,T456S)-GFP. Conditions and quantifications (n = 32–42) are as in B. ***P < 0.001; ns, nonsignificant, P > 0.05. (Scale bar: 2 μm.)
Here we exploited recently accumulated tools and data about conformation-dependent endocytosis of Can1 and other transporters (24, 25, 30) to study the mechanism and physiological significance of the partitioning of these proteins into EMCs. Our results show that in the substrate-free OF conformation Can1 clusters in EMCs because of slower diffusion due to complex SLs and is protected there from ubiquitylation, endocytosis, and degradation. When Can1 molecules shift to an IF conformation, as during transport of their substrate, they exit the EMCs and become accessible to the ubiquitylation machinery. Additionally, we show that Can1 EMC clustering and the EMCs themselves increase during nutrient starvation and that this requires Lsp1. We thus reveal a role for EMCs as nutrient-starvation–protective PM reservoir domains.
Results
Preferential Segregation of the Can1 Permease into the EMCs Is Relieved upon Substrate Transport.
In the course of previous work (25) we noticed that when Can1 ubiquitylation is impaired (Fig. S2A) the transporter stabilized at the PM changes from patchy to more homogeneous distribution after the addition of Arg. To study this, we set up a quantitative assay to monitor the clustering of proteins in EMCs (Materials and Methods). The method was based on confocal microscopy coupled to Airyscan detection, allowing higher resolution and an increased signal-to-noise ratio (31). Using this approach, we confirmed that Can1 preferentially but not exclusively clusters in the EMCs (Fig. S2B). In keeping with previous studies, Gap1 appeared not to segregate preferentially into the EMCs (Fig. S2B). We then exploited this experimental set-up to examine the effect of Arg on the EMC clustering of Can1 under conditions impairing Can1 ubiquitylation and endocytosis (Fig. 1 B and C). More specifically, we examined the localization of the permease in the hypomorphic npi1-1 mutant (affected in Npi1/Rsp5 Ub ligase) (26) and in an art1Δ bul1/2Δ triple mutant lacking all the α-arrestins involved in substrate-induced ubiquitylation of Can1. We also analyzed the distribution of Can1(7KR), a fully functional mutant permease where all seven Ub-acceptor Lys residues of the N-terminal tail have been replaced with Arg (25). In all three situations, remarkably, the addition of Arg abolished Can1 clustering in the EMCs (Fig. 1B) without affecting eisosome integrity (Fig. S2C). We further analyzed this redistribution using fluorescence recovery after photobleaching (FRAP) (Fig. 1C) and found Arg to cause a dramatic reduction in the half-time of recovery of EMC-located Can1. Thus, in the presence of Arg, Can1 diffuses through the EMCs more dynamically, behaving essentially as it does outside the EMCs (Fig. 1C and Movies S1–S3). These results suggest that in the absence of its substrate, Can1 preferentially segregates into the EMCs because of slower diffusion within them, and Arg addition relieves this effect.
Because in the presence of Arg Can1 no longer clusters in the EMCs, we next determined whether this depends on Can1 transport activity. We analyzed Can1(T180R), a loss-of-function mutant with a substitution at the substrate-binding site, previously shown to be unable to bind Arg (32). After Arg addition, Can1(T180R) remained in the EMCs (Fig. 1D). We also tested Can1(S176N,T456S), a mutant converted into a Lys-specific transporter (32). We found the EMC clustering of this mutant to be abolished upon Lys rather than Arg addition (Fig. 1D).
Taken together, these observations demonstrated that substrate transport abolishes Can1 EMC clustering. Furthermore, as this phenomenon was observed under conditions where Can1 is stabilized at the PM because of impaired substrate transport-induced ubiquitylation, we deduce that the exit of Can1 molecules from the EMCs precedes its ubiquitylation.
Transport-Elicited Relief of Can1 EMC Clustering Involves a Shift of the Transporter to an IF Conformation.
Transport-elicited relief of Can1 EMC clustering could, in principle, be caused by intracellular accumulation of the transported amino acid. We observed, however, that the inactive Can1(T180R) remained clustered in the EMCs even when coexpressed with a nontagged Can1 transporting Arg into the cell (Fig. 2A). This indicates that Can1 diffuses more quickly out of the EMCs when it is catalyzing transport, probably because of a conformational change coupled to transport catalysis. In support of this view, the phenomenon proved to be reversible: Upon removal of Arg, EMC clustering of preexisting nonubiquitylatable Can1(7KR) was restored (Fig. 2A).
Fig. 2.
Substrate-triggered abolition of Can1 EMC clustering is induced by transition to the IF state. (A, Upper) Shown are surface section confocal microscopy images of an art1Δ bul1/2Δ gap1Δ PIL1-mCherry strain expressing Can1(T180R)-GFP or Can1(7KR)-GFP. For Arg washout, cells were washed and resuspended in Arg-free medium for 30 min. (Lower) Quantifications (n = 32–49) are as in Fig. 1B. (B) Shown are surface section confocal microscopy images (Left) and quantification (Right) of an art1Δ bul1/2Δ gap1Δ can1Δ PIL1-mCherry strain expressing Can1-GFP or Can1(S176N)-GFP. Conditions and quantifications (n = 34–42) are as in Fig. 1B. Representations are as in Fig. 1A. (C) Shown are surface section confocal microscopy images of an art1Δ bul1/2Δ gap1Δ can1Δ PIL1-mCherry strain expressing Can1-GFP or the indicated mutant. Conditions and quantifications (n = 22–38) as in Fig. 1B. The cartoons summarize previous knowledge (25, 32). The E184Q substitution stabilizes the transporter in an IF state, permanently exposing the Art1BS even in the absence of substrate. The S176N substitution is epistatic to E184Q and blocks Can1 in an OF state. Art1BS is no longer exposed. Can1(ELK89-AAA) is inactive and permanently exposes Art1BS. Representations are as in Fig. 1A. (D) Shown are surface section confocal images of one representative FRAP experiment in EMCs of a gap1Δ can1Δ PIL1-mCherry strain expressing Can1(7KR,E184Q)-GFP. Conditions (n = 9), analysis, and representations are as in Fig. 1C. ***P < 0.001; **0.001 < P < 0.01; ns, nonsignificant, P > 0.05. (Scale bar: 2 μm.)
To gain more insight into the link between Can1 conformation and its EMC clustering, we first focused on the inactive mutant Can1(S176N), predicted to bind Arg but to be prevented by steric hindrance from shifting to the IF state (25, 32). Consistently, we observed no Arg-induced relief of Can1(S176N) EMC clustering (Fig. 2B). We next examined Can1(E184Q), another inactive mutant previously shown to be stabilized in an IF state by additional H-bonds between Gln184 and residues of other TMs (25). Importantly, Can1(E184Q) was found not to segregate preferentially into the EMCs. This was visible in the art1Δ bul1/2Δ strain and in WT cells expressing a mutant Can1 combining the E184Q and 7KR substitutions in the N-terminal tail (Fig. 2C and Fig. S3). This phenotype was not observed with Can1(E184A) (Fig. 2C), which is also inactive. It is thus specific to the form with Gln at position 184, which can form the potential extra H-bonds stabilizing the IF conformation. To investigate this further, we took advantage of the fact that S176N is epistatic to E184Q, as it inhibits the transition to the IF state. The Can1(S176N/E184Q) double mutant, thus inactive and stabilized in the OF conformation (25), was found to cluster normally in the EMCs (Fig. 2C). The above findings were confirmed by FRAP analysis of Can1(7KR,E184Q): Even in the absence of Arg, the diffusion of this mutant inside the EMCs (Fig. 2D and Movie S4) was as dynamic as that of the WT protein in the presence of substrate (Fig. 1C). Can1(E184Q) is thus, to our knowledge, a unique case where a single amino acid substitution specifically affects the dynamics of lateral PM segregation of a membrane protein.
The Can1(E184Q) mutant stabilized in the IF conformation constantly exposes the Art1-binding site to the cytosol (25). It shares this property with the inactive Can1(ELK89-AAA) mutant, where three consecutive residues close to the Art1-binding site are each replaced by alanine (25). Remarkably, Can1(ELK89-AAA) clustered normally in EMCs in the absence of Arg (Fig. 2C). This means that exposure of the Art1-binding site, as occurs in WT Can1 during Arg transport, is not sufficient to accelerate exit of the permease from EMCs. Upon Arg addition, importantly, the EMC clustering of Can1(ELK89-AAA) was abolished (Fig. 2C), despite its lack of transport activity (25). This result, although expected [as Can1(ELK89-AAA) has an intact substrate binding site and thus should be able to recognize Arg], confirms that Arg uptake is not needed to promote loss of Can1 EMC clustering. It also suggests that Can1(ELK89-AAA) binds Arg properly and then undergoes a change of conformation, for instance a shift to the IF state, affecting its lateral PM segregation.
In conclusion, substrate transport-induced relief of Can1 EMC clustering correlates with adoption by the permease of an IF conformation occurring naturally during transport catalysis. This conformation is stabilized as a result of the E184Q substitution unless the transition is impaired by an additional S176N.
Sustained SL Biogenesis Is Crucial for Can1 Preferential Segregation into the EMCs.
EMCs have been proposed to accumulate ergosterol and SLs (13, 17, 21, 22). Furthermore, SLs are known to affect eisosome assembly (33), and specific EMC-resident proteins are involved in sensing SL levels and regulating SL biosynthesis (Fig. S1) (17, 21). We thus speculated that SLs could influence the conformation-dependent partitioning of Can1 in this compartment.
To evaluate our hypothesis, we first examined whether Can1 EMC clustering depends on the SL level (Fig. 3A). For this, we monitored the localization of Can1 in cells treated with the specific SL biosynthesis inhibitors myriocin (Myr) or aureobasidin A (AbA) (Fig. 3B). As such treatments typically cause eisosome disassembly (Fig. S4A), we performed the experiment in a strain expressing as sole Pil1 the mutant Pil1(4A)-GFP, which renders eisosomes insensitive to SL depletion-driven disassembly (21, 33). As previously reported, inhibition of SL biosynthesis did not affect the localization of Sur7-mRFP in Pil1(4A)-GFP–containing EMCs (Fig. 3A and Fig. S4B). In remarkable contrast, Can1(7KR)-mCherry clustering was abolished in these EMCs upon Myr treatment and was rescued when phytosphingosine (PHS) was supplied simultaneously with Myr to restore SL biogenesis (Fig. 3A and Fig. S4B). Can1 EMC clustering was also abolished when the conversion of phytoceramide into complex SLs was inhibited with AbA (Fig. 3A and Fig. S4B). This shows that Can1 EMC clustering requires the pool of complex SLs. Additionally, FRAP analysis showed that the half-time of Can1 recovery in EMCs was dramatically reduced when the biogenesis of complex SLs was inhibited (Fig. 3C and Movies S5 and S6) in the same manner as when Arg was added (Fig. 1C). These results show that sustained complex SL biogenesis is necessary for the slower diffusion, and thus the preferential segregation, of Can1 in the EMCs. We next sought to determine whether Myr-induced loss of Can1 clustering in EMC correlates with a different propensity of the protein to be extracted by the detergent Triton X-100. This property of membrane proteins is related to their association with membranes enriched in sterols and SLs (34, 35). We first analyzed Can1(7KR)-GFP in membranes of cells where Myr-induced eisosome disassembly was prevented by the expression of Pil1(4A)-MARS as sole Pil1 [as shown in Fig. S4C, this mutant behaves like Pil1(4A)-GFP, used in Fig. 3A]. Although the extraction yield seemed to increase after Myr treatment (Fig. S4D), variations between experiments made it difficult to ascertain it. We thus turned to a simpler experimental scheme taking advantage of the inability of Can1(E184Q) to cluster in EMCs. This experiment was performed after repressing GAL1-driven expression of the mutant CAN1 genes to restrict the analysis to the Can1 molecules located at the PM and not in the secretory or endocytic pathway. Can1(7KR,E184Q) was found in independent experiments to be much more readily extractable with Triton X-100 than Can1(7KR), i.e., to require a lower concentration of detergent for its solubilization (Fig. 3D and Fig. S4E). Can1(7KR,E184Q) behaved similarly to Gap1 or Pma1, membrane proteins respectively not enriched in or even excluded from EMCs (Fig. S2A) (22). Our results thus suggest a correlation between the EMC clustering of Can1 and its decreased solubilization in detergent. They show that both require sustained SL biogenesis. This supports the view either that Can1, at least in its substrate-free OF conformation, avidly binds SLs or that SLs are indirectly required for the EMC partitioning of Can1.
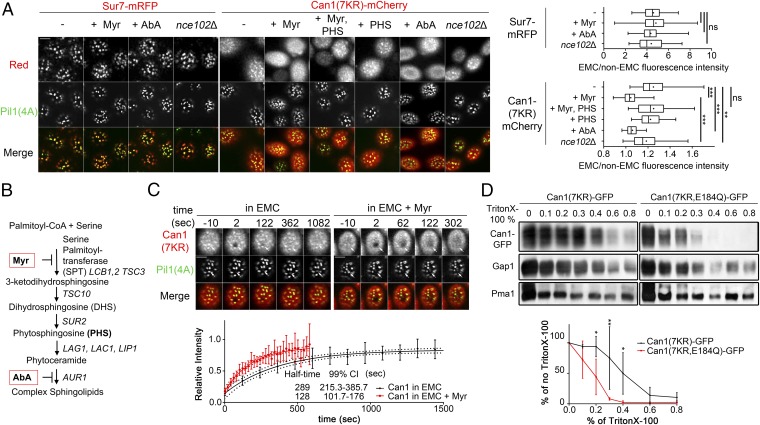
Fig. 3.
Can1 EMC clustering requires SLs. (A, Left) Shown are surface and middle section (see also Fig. S4B) confocal microscopy images of gap1Δ can1Δ and gap1Δ can1Δ nce102Δ strains expressing Sur7-mRFP, Pil1(4A)-GFP as the sole Pil1, and Can1(7KR)-mCherry, grown in YNB Gal Am medium. Glu was added for 90 min and then 10 μM Μyr, 10 μM Myr + 10 μM PHS, 10 μM PHS, or 1 μg/mL ΑbA was added for 90 min. (Right) Quantifications (n = 41–67 for Sur7, 34–100 for Can1) are as in Fig. 1B. (B) The SL biosynthesis pathway of Saccharomyces cerevisiae. The metabolic intermediates and the genes encoding the enzymes involved are named. The steps inhibited by Myr and AbA are also highlighted. (C) FRAP experiments on EMCs carried out, analyzed, and represented as in Fig. 1C on a gap1Δ can1Δ strain expressing Can1(7KR)-mCherry and Pil1(4A)-GFP treated (red, n = 14) or not (black, n = 11) with 10-μΜ Myr for 90 min. F-pvalue of K < 0.0001. (D, Upper) Detergent resistance of Can1(7KR) and Can1(7KR,E184Q). can1Δ strains expressing either Can1(7KR)-GFP or Can1(7KR,E184Q)-GFP under the GAL1 promoter were grown in Gal Pro medium, and Glu was added for 1.5 h. Thirty-microgram aliquots of membrane-enriched protein extracts were treated with increasing concentrations of Triton X-100. Following centrifugation and washing, the detergent-resistant insoluble pellet was resuspended in sample buffer and immunoblotted. (Lower) Points and error bars respectively represent the mean and SD of the percentage relative intensity for each concentration (n = 3 biological replicates) of Can1-GFP in comparison with the extracts treated in the absence of Triton X-100. Statistically significant points are shown. See Fig. S4E for quantification of Gap1 and Pma1 signals. ***P < 0.001; **0.001 < P < 0.01; *P < 0.05; ns, nonsignificant, P > 0.05. (Scale bar: 2 μm.)
The behavior of Can1 under these conditions is reminiscent of that of Nce102, the tetraspan putative SL sensor required for eisosome assembly (Fig. S1) (21, 23). We thus hypothesized that Nce102 might play an active role in Can1 recruitment to the EMCs and in its dissipation from them upon SL depletion. To test this hypothesis, we again used cells expressing Pil1(4A)-GFP, in which the eisosomes do not disassemble upon deletion of NCE102 (Fig. S4B) (21). As previously reported, we found Sur7-mRFP to localize normally to the EMCs of a Pil1(4A)-GFP nce102Δ strain (Fig. 3A). Can1 EMC clustering was slightly reduced in this strain but clearly was not abolished (Fig. 3A). This shows that Can1 remains able to segregate preferentially into the EMCs in the absence of Nce102. We next evaluated whether Nce102 might play a role in SL depletion-induced dissipation of Can1 from the EMCs, as it behaves like Can1 under these conditions. For this we fused the GFP-binder protein (GB) (36) to Sur7 (Sur7-GB) to trap Nce102-GFP in the EMCs and thus maintain its inhibitory effect on Pkh1/2 even upon SL depletion (Fig. S5A). In the presence of Sur7-GB, Nce102-GFP was indeed trapped in the EMCs and remained clustered there upon Myr treatment, while the (Pil1-mtagBFP2–labeled) eisosomes remained intact (Fig. S5 B and C). Most interestingly, the same cells displayed significantly reduced clustering of Can1 (Fig. S5 B and C). This clearly shows that Nce102 is not sufficient to retain Can1 in the EMCs upon SL depletion. Together, the above results indicate that Nce102 is not essential for the EMC localization of Can1, although it might somehow affect its clustering there (discussed below).
EMCs Protect Transporters from Ubiquitylation and Endocytosis.
We next investigated the physiological significance of Can1 clustering in the EMCs. It has been debated whether this partitioning might affect the Arg-induced endocytosis of the transporter (13, 15). According to our recent findings (Fig. 1A), an Arg-induced shift to the IF state is required for Can1 ubiquitylation. However, as such a shift is also required to facilitate Can1 exit from the EMCs (Fig. 2), we first investigated whether this exit might, in fact, be sufficient for Can1 down-regulation. For this we created a strain lacking EMCs by deleting the PIL1 gene. Can1 endocytosis did not occur in these cells unless Arg was added, whereas the inactive Can1(T180R) remained at the cell surface even in the presence of functional Can1 (Fig. 4A). These results show that to promote Can1 ubiquitylation and endocytosis, it is not sufficient to prevent it artificially from clustering in the EMCs, even when Art1 is activated via TORC1 (as cells were grown in the presence of NH4+). They confirm, furthermore, that a substrate-induced transition of the transporter to the IF state is essential for triggering Can1 ubiquitylation and endocytosis (25).
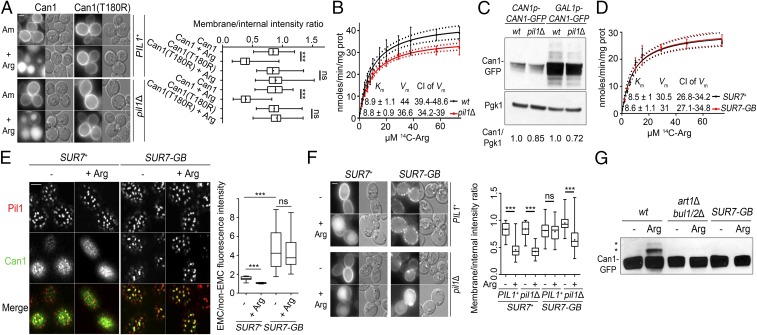
Fig. 4.
Can1 in EMCs is protected from ubiquitylation and endocytosis. (A, Left) Epifluorescence microscopy of Arg-induced endocytosis of Can1-GFP and Can1(T180R)-GFP in PIL1 CAN1 and pil1Δ CAN1 cells. (Right) Quantifications (n = 91–104) are as in Fig. S2A. (B) Concentration-dependent kinetics of [14C]Arg uptake into gap1Δ and gap1Δ pil1Δ cells. Error bars indicate the SD; n = 3. Curve fitting and calculation of the Vm and Km values (in micromoles), 99% CI (dotted curves), and F-pvalues were carried out by Michaelis–Menten analysis. F-pvalue of Vm = 0.0003, F-pvalue of Km = 0.9245. (C) gap1Δ can1Δ and gap1Δ can1Δ pil1Δ cells expressing Can1-GFP from the native or GAL1 promoter were grown in Glu Pro or Gal Pro medium, respectively. To the latter, Glu was added for 90 min before cells were collected. Total protein extracts were immunoblotted with anti-GFP and anti-Pgk1. Quantifications shown below the blots are the ratios of Can1-GFP/Pgk1 intensities. Ratios in the PIL1+ (WT) strain are set at 1. (D) Concentration-dependent kinetics of [14C]Arg uptake in gap1Δ can1Δ Can1-GFP cells expressing Sur7 or Sur7-GB and grown in Raf Pro medium. Gal was added for 1 h, and then Glu was added for 90 min. Error bars indicate SD; n = 2. Analysis and representations are as in B. F-pvalue of Vm = 0.7976; F-pvalue of Km = 0.9492. (E, Left) Shown are surface section confocal images of SUR7-GB and SUR7 strains (gap1Δ can1Δ) expressing Pil1-mCherry and Can1(7KR)-GFP grown as in D. The cells were then treated or not with 5 mM Arg for 30 min. (Right) Quantifications (n = 22–37) are as in Fig. 1B. (F, Left) Epifluorescence microscopy of Arg-induced endocytosis of Can1-GFP in PIL1 and pil1Δ cells (gap1Δ can1Δ) expressing Sur7 or Sur7-GB, grown as in D. (Right) Quantifications (n = 74–106) are as in Fig. S2A. (G) Western blotting of total protein extracts of Can1-GFP–expressing strains (gap1Δ can1Δ) grown as in D and probed with anti-GFP. Asterisks indicate Ub-Can1-GFP conjugates. ***P < 0.001; ns, nonsignificant, P > 0.05. (Scale bar: 2 μm.)
It has been suggested, on the basis of growth tests for canavanine sensitivity, that Can1 is more active inside than outside the EMCs (7). We sought to confirm this by directly measuring the uptake of radiolabeled Arg in WT and pil1Δ cells. We did find lower Can1 activity in pil1Δ cells, this being due to a significantly reduced maximal velocity (Vm) without any change in the apparent Km (Fig. 4B). This 20% decrease in Vm, however, correlated very well with a decrease in the Can1 protein level observed in pil1Δ cells independently of the promoter used to express CAN1 (Fig. 4C). This suggests that localization of Can1 to the EMCs does not, in fact, alter the kinetic characteristics of the transporter. To investigate this further and to take into account that only a fraction of the Can1 is present in the EMCs at a given time, we measured the kinetic characteristics of the transporter (Fig. 4D) after tethering Can1-GFP to the EMCs by coexpressing it with Sur7-GB (as in Fig. S5A). To ensure that there would be enough Sur7-GB molecules to trap all the Can1-GFP molecules in the EMCs, we examined cells after a 1-h CAN1 induction pulse. Under these conditions, Can1 was found almost exclusively in the EMCs, as illustrated by an increase of the EMC/non-EMC intensity ratio from 1.4 to 4.4 (Fig. 4E). Importantly, EMC-trapped Can1-GFP was active, displayed unaltered kinetic characteristics (Fig. 4D), and did not dissipate from EMCs upon Arg addition (Fig. 4E). Consistently, Sur7-GB–bound Can1-GFP was protected from Arg-induced endocytosis and ubiquitylation in cells possessing eisosomes (Fig. 4 F and G). Furthermore, its endocytosis was largely restored in a pil1 mutant lacking eisosomes (Fig. 4F). This rules out the possibility of an artifact caused by the binding of GB to GFP. In conclusion, these results indicate that EMC-trapped Can1 cannot be ubiquitylated. As the transporter remains fully active, this is not due to the failure to adopt an IF conformation. The results, rather, are fully compatible with the view that TORC1-activated Art1 and Rsp5 cannot gain access to EMC-located Can1.
EMCs as Starvation-Protective Transporter Reservoirs.
When located inside the EMCs, Can1 thus appears to be protected from Ub-dependent down-regulation. However, it efficiently dissipates out of EMCs and undergoes ubiquitylation and endocytosis in the presence of Arg. What, then, could be the biological role of Can1 EMC clustering? We speculated that preferential segregation into the EMCs could protect Can1 from endocytosis under particular conditions and when its substrate Arg is not present. We have recently reported that inhibition of TORC1 by rapamycin (Rap) promotes efficient Ub-dependent down-regulation of multiple permeases, including Can1 (37). Interestingly, TORC1 inhibition also occurs under nitrogen starvation. In nitrogen-starved cells, bulk transporter degradation caused by TORC1 inhibition might, like autophagy (also stimulated under nitrogen starvation), enable cells to retrieve free amino acids (38). We speculated that under such conditions it might be advantageous for a cell to preserve a fraction of its transporter molecules for use when their substrates became available again, and the role of their clustering in EMCs might be to preserve those molecules. To assess this model, we compared the extent of Rap-induced Can1 down-regulation in WT and pil1Δ cells and found it to be more pronounced in the latter (Fig. 5A). This was in agreement with uptake measurements showing that Rap addition caused a greater reduction of Can1 activity in pil1Δ cells (Fig. 5B). Other EMC-resident transporters (Lyp1, Fur4, and Mup1) showed a similar behavior, unlike Gap1, which does not cluster in the EMCs (Fig. 5 A and B). Actually, Gap1 appeared to be slightly but significantly protected in pil1Δ cells. This calls to mind a previous report on Ste3 endocytosis (12) and the fact that in pil1Δ cells the cortical endoplasmic reticulum is more in contact with the PM and reduces the formation of endocytic sites (39). This general endocytic defect might thus limit the extent of transporter internalization we observed in pil1Δ cells after Rap addition (Fig. 5 A and B). As both Rap treatment and extreme starvation cause complete inactivation of TORC1, we next subjected WT and pil1Δ cells to nitrogen starvation for 4 h, added Arg, and monitored growth recovery of WT and pil1Δ cells to see if the former, where a fraction of the Can1 was protected against down-regulation, might recover faster. No difference in recovery was observed (Fig. 5C), most likely because Arg is efficiently incorporated via the general amino acid permease Gap1, which is induced under nitrogen-starvation conditions (40). This experiment, however, revealed two interesting phenotypes in pil1Δ cells. First, their growth was slightly but significantly slower, an effect largely relieved if NH4+ was used instead of Arg to restore growth. This suggests that their growth rate on Arg is limited by the level of Can1 at the PM, which is 20% lower in pil1Δ than in WT cells (Fig. 4 B and C). Second and more importantly, pil1Δ cells reached the stationary phase earlier than WT cells, an effect again relieved if NH4+ was used instead of Arg to restore growth (Fig. 5C). This observation suggests that Can1 partitioning into the EMCs could be important to protect a fraction of the Can1 molecules from the down-regulation reported to occur in the late logarithmic and stationary phases of growth (38). In support of this view, we confirmed by microscopy and Western blotting that, in the absence of Arg, the pil1Δ cells showed more pronounced Can1-specific degradation than the WT cells as the cultures gradually reached the stationary phase (Fig. 5D and Fig. S6 A and B). Most importantly, intact Can1-GFP was barely detectable at the PM of pil1Δ cells that had remained in the stationary phase for 12 h, whereas WT cells still displayed a clear signal corresponding to intact, PM-located Can1-GFP (Fig. 5D and Fig. S6 A and B). In agreement with these results, pil1Δ gap1Δ cells showed significantly slower recovery than PIL1+ gap1Δ cells when transferred from a stationary-phase culture to solid Arg medium (Fig. 5E), a phenotype not observed when the cells were collected during exponential growth (Fig. S6C). These results show that during the late logarithmic and stationary phase EMCs protect a fraction of the transporter from bulk starvation-induced endocytosis and degradation, allowing more efficient recovery when nutrients become available again.
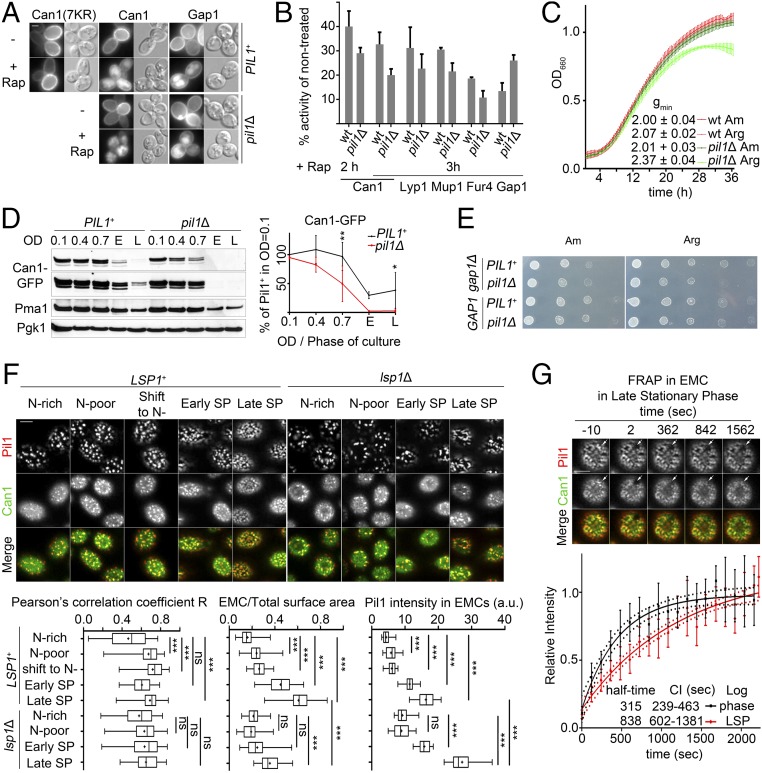
Fig. 5.
The EMC as a reservoir of transporters for nutrient starvation. (A) Epifluorescence microscopy of PIL1 and pil1Δ cells (gap1Δ can1Δ) expressing Can1-GFP, Gap1-GFP, or Can1(7KR)-GFP, treated or not with 200 ng/mL Rap for 3 h. (B) [14C]-amino acid uptake measurements (Arg for Can1, Lys for Lyp1, Met for Mup1, uracil for Fur4, citrulline for Gap1) on PIL1 and pil1Δ cells (gap1Δ when not measuring citrulline) treated or not with 200 ng/mL Rap for 2 or 3 h. Error bars indicate SD; n = 3. (C) Growth curves of WT and pil1Δ cells starved for nitrogen for 4 h in minimal medium containing Arg or Am as the sole nitrogen source. Equal amounts of cells were seeded into 24-well plates. Error bars indicate the SD of two technical replicates. The experiment is representative of three biological replicates. (D, Left) Western blotting of total protein extracts (probed with anti-GFP, stripped and probed with anti-Pma1 and anti-Pgk1) of PIL1 and pil1Δ (gap1Δ can1Δ) cells expressing Can1-GFP and grown in glutamine plus Am at different ODs or having reached (E) or remained for 12 h in (L) the stationary phase. (Right) The Can1-GFP/Pgk1 intensity ratio, normalized to 1 for the PIL1+ in OD = 0.1 from two to three independent biological replicates shown in Fig. S6B. Quantifications are as in Fig. 3D. (E) Spot dilution tests of PIL1 and pil1Δ cells (± gap1Δ) that had remained in the stationary phase for 12 h. (F, Upper) Shown are surface section confocal images of gap1Δ can1Δ and gap1Δ can1Δ lsp1Δ strains expressing Pil1-mCherry and Can1(7KR)-GFP taken from early log-phase cultures on Gal Pro (N-poor) or Gal Am + Gln (N-rich) medium. Cells grown on N-rich medium and then shifted to medium lacking any nitrogen source (Shift to N−) for 2 h, were also observed at the beginning of the stationary phase (Early SP) and after 12 h in the stationary phase (Late SP). Pil1-mCherry images were differentially treated for brightness and contrast to improve the visibility of details and do not reflect differences in the intensity measured. (Lower) Quantifications (n = 33–79) are as in Fig. 1B (see also Materials and Methods and Fig. S6D). (G, Upper) FRAP experiments on EMCs of a gap1Δ can1Δ strain expressing Can1(7KR)-GFP and Pil1-mCherry from cells in the late stationary phase carried out, analyzed, and represented as in Fig. 1C. (Lower) Quantifications: the fitted curve (n = 11) is compared with the one of Can1 in EMCs (log phase, n = 8) from Fig. 1C. F-pvalue of K < 0.0001. ***P < 0.001; **0.001 < P < 0.01; *P < 0.05; ns, nonsignificant, P > 0.05. (Scale bar: 2 μm.)
To gain more insight into the mechanisms by which EMCs act and are regulated during nutrient starvation, we monitored EMC assembly under different nutrient supply conditions. For this we used confocal microscopy to quantify four EMC-related parameters: Can1 clustering, the intensity of Pil1-mCherry fluorescence, the number of EMCs per square micrometer, and the proportion of the total membrane area occupied by EMCs. These parameters were measured during the early log phase of growth on either nitrogen-rich [ammonium (Am) + glutamine] or nitrogen-poor medium (proline, Pro) after a shift from nitrogen-rich medium to nitrogen-starvation conditions and in cells that had reached the early stationary phase or that had remained in the stationary phase for 12 h (Fig. 5F and Fig. S6D). Can1 EMC clustering was found to be low in nitrogen-rich conditions and significantly increased in nitrogen-poor medium or after a shift to nitrogen-starvation conditions. This effect correlated with an increased fluorescence intensity of Pil1 in the EMCs. Most importantly, this increase was even more pronounced in the stationary phase and was accompanied by a twofold increase in the number of EMCs and a nearly fourfold increase in the area of the PM covered by EMCs. Similarly, the enrichment of EMCs in Can1 also increased during the stationary phase, while under these conditions we could observe by FRAP analysis a nearly threefold increase in the halftime of Can1 recovery in EMCs (Fig. 5G and Movie S7). Our results in total show that during the stationary phase, although Can1 is still able to diffuse out of EMCs, the increase of EMCs provides protection from starvation-induced endocytosis for a significant subpopulation of transporter molecules, which is enough to sustain efficient recovery once the nutrients become available again. Importantly, we also found the global increase of EMCs during the stationary phase to be largely dependent on Lsp1 (Fig. 5F and Fig. S6D), the paralog of Pil1. The function of Lsp1 in the organization of eisosomes has so far remained elusive, probably because lsp1Δ cells have been examined mostly during the early log phase of growth. During this phase, we observed no EMC assembly defect (Fig. 5F and Fig. S6D), in keeping with previous reports (12). The present results thus show that Can1 EMC clustering and the EMCs themselves are regulated by nutrient availability and suggest a role of EMCs as starvation-protective membrane domains.
Discussion
Partitioning of proteins into specific PM compartments is reported to have various biological functions, such as preferential targeting to apical/basolateral membranes (41), optimal coordination of signaling processes in raft-like entities, and allosteric regulation of the intrinsic function of membrane proteins (1, 2, 42). The present study, focused on yeast EMCs, reveals another role for such partitioning: protecting proteins from endocytosis. Furthermore, membrane-protein clustering in the EMCs is globally reinforced under conditions of nutrient starvation to preserve the EMC-located proteins from bulk endocytosis. Our data thus highlight a general, previously unforeseen mechanism for controlling protein abundance at the cell surface, acting in addition to the more specific mechanisms which regulate the endocytosis and turnover of individual proteins.
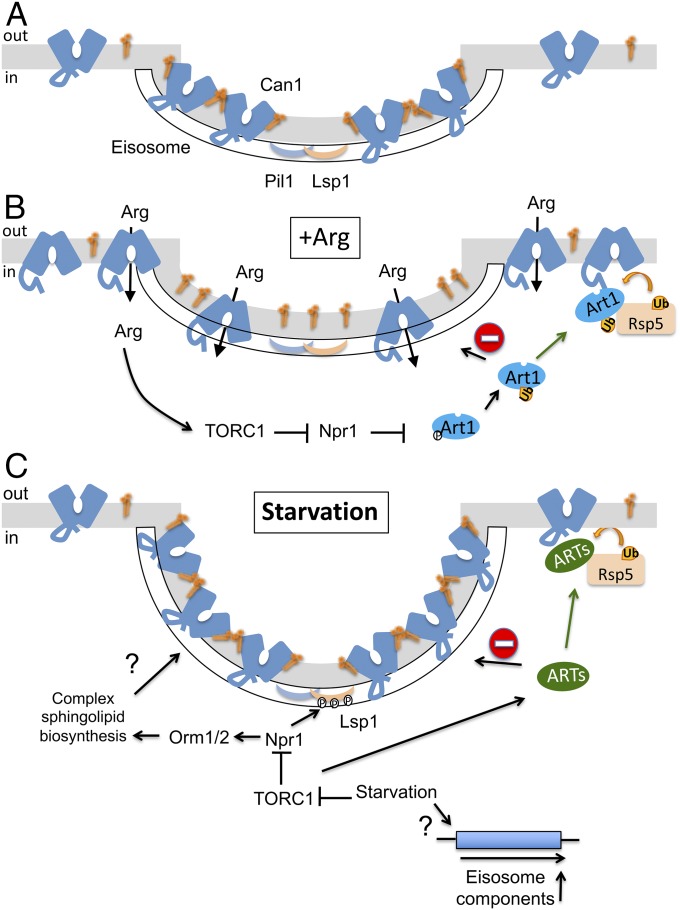
Our main conclusions regarding the role of yeast EMCs are in keeping with previous studies showing that these compartments do not colocalize with sites of clathrin-dependent endocytosis (13, 15, 16) and that the turnover rates of the Can1 and Fur4 transporters are slightly increased in mutants lacking key EMC components (13). It has been proposed that in the presence of excess substrate Can1 is actively released from the EMCs before undergoing endocytosis (13). This view has been challenged in another study suggesting that Ub might act as a signal for Can1 exit from the EMCs (15). Our recent work on the 3D structure of Can1 (32) and on the mechanisms eliciting its ubiquitylation and endocytosis in response to substrate transport (24, 25) has provided a framework for further investigating how and why this protein and other transporters preferentially segregate into the EMCs and exit from them before their down-regulation. We propose a model (Fig. 6) in which the permease, in the absence of its substrate, diffuses more slowly through the EMCs, and this depends on complex SL biogenesis. This segregation behavior would be a feature of Can1 in the OF conformation. When present in the EMCs, Can1 and other EMC-resident transporters would be protected from ubiquitylation mediated by the Rsp5 Ub ligase via adaptors of the α-arrestin family. Our results show that this protection requires intact eisosomes, suggesting that the subcortical protein complex beneath the invagination could potentially exclude the ART/Rsp5 complex from the PM of the EMCs. Alternatively, EMC-localized transporters could be prevented from interacting with the ART/Rsp5 complex when found in the OF conformation due to conformational restrictions or interactions with other EMC components. Nevertheless, the molecular details of this putative exclusion mechanism remain to be investigated. Upon substrate transport, the protein transiently shifts to an IF conformation. In this state, Can1 would diffuse out of the EMCs and become accessible to the ubiquitylation machinery (Fig. 6 A and B). We propose that this model applies also to other EMC-located transporters known to undergo Ub-dependent endocytosis in the presence of excess substrate (30). This model is consistent with the previously reported loss of Can1 EMC partitioning upon partial depolarization of the membrane potential (ΔΨ) (22). Can1, like many other transporters, displays an asymmetric distribution of charged residues on both sides of the membrane (32). Consequently, disruption of the ΔΨ could affect the conformation adopted by Can1, even in the absence of substrate, and thus its EMC partitioning. Our findings are in keeping with previous reports suggesting that EMCs could potentially be enriched in complex SLs (17, 18, 21, 23, 33). We have envisaged the possibility that the Nce102 protein involved in SL sensing might contribute to Can1 clustering, but we show that in nce102Δ mutants the permease still segregates preferentially into the EMCs, although less efficiently. Although SLs play, via Nce102, an important role in eisosome assembly, the molecular function of Nce102 and the mechanism of SL level sensing remain unclear. The apparent partial contribution of Nce102 to Can1 clustering and our finding that this clustering requires sustained SL biogenesis prompt us to propose that Nce102, rather than sensing SLs, might be involved in active recruitment of SLs to the EMCs.
Fig. 6.
Model of the mechanisms of Can1 lateral PM segregation and regulated protection from ubiquitylation by starvation-protective EMCs. (A) In the absence of substrate, Can1 in an OF conformation concentrates in EMCs (which associate subcortically with Pil1- and Lsp1-containing eisosomes), probably because it interacts with particular lipids (for instance SLs). (B) In the presence of Arg, a transient shift of the transporter to an IF conformation results in the loss of clustering in EMCs because its potential interaction with lipids (illustrated) or its partitioning in highly ordered membrane domains is reduced. Meanwhile, Arg activates Art1 via the TORC1-Npr1 pathway. Art1 recruits the Rsp5 Ub-ligase only to Can1 molecules that have adopted the IF conformation and are outside the EMCs. (C) Upon nutrient starvation, the eisosomes are induced at transcriptional and posttranslational levels; inhibition of TORC1 leads to Npr1 stimulation, which phosphorylates Lsp1 and also Orm1/2 at the Golgi, inducing the biogenesis of complex SLs. These events correlate with deeper eisosomal invaginations capable of hosting more proteins, protecting them from ubiquitylation and endocytosis promoted by certain uncharacterized α-arrestins (ARTs) that somehow are activated by nutrient starvation and/or TORC1 inhibition.
Despite the wealth of evidence that SLs play an important role in membrane protein function, only a few clear-cut cases of protein–SL interactions have been reported (43). One such example is the highly specific interaction of a particular SL species with a conserved SL-binding motif in the unique TM segment of p24, demonstrated to be important for p24 function (44). Similarly, a potential interaction of EMC-located transporters with SLs could also be mediated by TMs rather than cytoplasmic tails. In support of this view, the lateral segregation patterns of membrane proteins are reported to be determined by their TMs (7). Furthermore, in an Ala-scanning mutagenesis of the Can1 cytosolic N-terminal tail (25), we found no mutant displaying an EMC-partitioning defect, nor does Can1(ELK89-AAA), a variant in which the normally hidden Art1-binding site is constitutively exposed to the cytosol (Fig. 2). Alternatively, intrinsic physicochemical properties of the TMs could promote the EMC partitioning of certain transporters, as has been recently proposed for the recruitment of membrane proteins in membrane rafts (45). Given that EMCs share many similarities with membrane rafts, differences in the length or the surface area of TMs could contribute to EMC partitioning. Based on this hypothesis, a speculative mechanism for the conformation-dependent partitioning of Can1 in EMCs could be envisaged, based on the differential exposure of TMs with varying length and/or surface area in the OF and IF conformations to some putative highly ordered (sterol- and SL-enriched) lipid bilayer of EMCs.
The role of EMCs in SL homeostasis has been well documented (17, 18, 21, 23). Our data reveal a previously unidentified additional role for these compartments under nutrient-starvation conditions: protecting protein molecules inside the EMCs from down-regulation. This finding is in keeping with the reported increase in complex SL biogenesis observed upon TORC1 inhibition (46). As both eisosome assembly and Can1 EMC clustering require sustained SL biogenesis, it is tempting to speculate that the increase in SL level occurring during nutrient starvation reinforces the role of EMCs as starvation-protective membrane areas (Fig. 6C). Our data also indicate that Lsp1, previously viewed as nonessential to eisosome assembly (12, 19, 20), participates in EMC expansion during starvation. Thus Lsp1 could potentially play an essential role in forming new eisosomes or in duplicating preexisting ones during the stationary phase. Interestingly, Lsp1 has been identified as a potential substrate of the TORC1-regulated Npr1 kinase known to be stimulated under starvation conditions (29). These findings suggest a tight regulation of EMCs by TORC1 (Fig. 6C), in addition to their well-established regulation by TORC2 (17, 18, 33). Several reports are also consistent with our finding that the number of EMCs and the area they occupy increase during nutrient limitation. For instance, the core eisosomal proteins (Pil1, Lsp1, Nce102, and Sur7) display a threefold increase in PM extracts from cells shifted to a poor nitrogen source (47). It has also long been shown by electron microscopy, in both budding and fission yeast, that in the stationary phase the PM invaginations increase in both number and depth (48, 49), as is consistent with our observed increase in Pil1-mCherry intensity in the EMCs. Eisosomes also seem important in nitrogen stress-induced yeast filamentation, as Pun1, a protein essential to this process, is a Sur7-like component of EMCs (50). Last, the main mRNA decay enzyme, exoribonuclease Xrn1, is reported to accumulate in EMCs specifically after glucose (Glu) exhaustion, and this accumulation negatively regulates the activity of the enzyme during the stationary phase (51, 52). Two other EMC-resident proteins, Fmp45 and Pst2, are essential for recovery from the stationary phase (53). Thus, EMC expansion seems important in various regulatory mechanisms promoting cell survival during and recovery from nutrient starvation. It could also explain why EMCs are necessary for the pathogenicity of fungi in animals and plants (54, 55). Since eisosomes are fungus-specific structures (56), they appear to be attractive targets for the development of novel, highly specific antifungals.
Materials and Methods
Strains, Plasmids, Growth Media, and Epifluorescence Microscopy.
The yeast strains and plasmids are listed in Tables S1 and S2. Cells were grown at 29 °C on a minimal buffered medium, pH 6.1 (57), with galactose (Gal) (3%), raffinose (Raf) (3%), or Glu (3%) as the carbon source, and with (NH4)2SO4 (10 mM), proline (Pro) (10 mM), glutamine (10 mM), or Arg (5 mM) as the sole nitrogen source. Difco Yeast Nitrogen Base (YNB) was used in the experiments shown in Fig. 3A. The CAN1-GFP and GAP1-GFP genes were expressed under the control of the GAL promoter or their native promoter. pGAL1-driven expression was repressed by the addition of 3% Glu 0.5 or 1.5 h before treatment, cell filtration, or microscopic observation. The tagged versions of SUR7, PIL1, and NCE102 were expressed under their respective native promoters. Comparative analysis of growth in liquid cultures was performed by growing cells in a 24-well Greiner microplate incubator coupled to a SYNERGY multi-mode reader (BioTek Instruments). For epifluorescence and confocal microscopy, cells were grown as previously described (25). Details and a description of fluorescence signal quantification are available in Supporting Information.
Confocal Microscopy, FRAP, and Quantifications.
Images were acquired on a Zeiss LSM710 microscope equipped with Airyscan and a 100× differential interference contrast, numerical aperture (NA) 1.46 Alpha-Plan-Apochromat objective, with ZEN 2.1 SP2 software. For quantifications images were analyzed with a custom-made FIJI macro, calculating the EMC/non-EMC fluorescence intensity, the density of EMCs, the intensity of Pil1 in EMCs, and the EMC/total surface area ratio. Details are given in Supporting Information. In each figure only a few cells representative of the whole cell population, observed in at least two independent biological replicate experiments, are shown.
Statistical Analysis of Quantifications.
Prism software, one-way ANOVA with the nonparametric Kruskal–Wallis test, and Dunn’s multiple-comparison post hoc analyses were used to assess the significance of the value differences of all measurements.
Protein Extracts and Western Blotting.
For immunoblot analyses, crude cell extracts were prepared and analyzed by SDS/PAGE as previously described (26). Membrane-enriched extracts and determination of Can1 detergent resistance were performed per Grossmann et al. (13, 22). Detailed descriptions are given in Supporting Information.
Permease Activity Assays and Determination of Can1 Kinetic Characteristics.
Permease transport activities were determined by measuring the initial uptake rate (20–60 s) of 14C-labeled amino acids (Perkin-Elmer) corrected for protein concentration, as previously described (32). Error bars indicate SEs determined for at least two independent biological replicates. Details are given in Supporting Information.
Supplementary Material
Acknowledgments
We thank Catherine Jauniaux and Simon Lissoir for excellent technical assistance, Christine Decaestecker for useful discussions about the statistical analyses, Tobias Walther for providing the Pil1(4A) plasmids, Pierre Courtoy for critically reading the manuscript, and Roland Wedlich-Söldner for providing plasmids with the Sur7-GB and mKate2 sequences and for fruitful discussions, together with his laboratory members Jon V. Busto and Daniel Haase. C.G. is a postdoctoral researcher of Fonds de la Recherche Scientifique de Belgique. S.G. was supported by an Erasmus+ Framework studentship. M.C. is a fellow of the fonds pour La Formation a la Recherche dans l’Industrie et dans L’Agriculture. The Center for Microscopy and Molecular Imaging is supported by the European Regional Development Fund and the Walloon Region. This work was supported by Fonds National de la Recherche Scientifique PDR Grant 23655065.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719462115/-/DCSupplemental.
References
- 1.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 2.Sezgin E, Levental I, Mayor S, Eggeling C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malinsky J, Opekarová M, Tanner W. The lateral compartmentation of the yeast plasma membrane. Yeast. 2010;27:473–478. doi: 10.1002/yea.1772. [DOI] [PubMed] [Google Scholar]
- 4.Olivera-Couto A, Aguilar PS. Eisosomes and plasma membrane organization. Mol Genet Genomics. 2012;287:607–620. doi: 10.1007/s00438-012-0706-8. [DOI] [PubMed] [Google Scholar]
- 5.Douglas LM, Konopka JB. Fungal membrane organization: The eisosome concept. Annu Rev Microbiol. 2014;68:377–393. doi: 10.1146/annurev-micro-091313-103507. [DOI] [PubMed] [Google Scholar]
- 6.Malinsky J, Opekarová M. New insight into the roles of membrane microdomains in physiological activities of fungal cells. Int Rev Cell Mol Biol. 2016;325:119–180. doi: 10.1016/bs.ircmb.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Spira F, et al. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat Cell Biol. 2012;14:640–648. doi: 10.1038/ncb2487. [DOI] [PubMed] [Google Scholar]
- 8.Kock C, Arlt H, Ungermann C, Heinisch JJ. Yeast cell wall integrity sensors form specific plasma membrane microdomains important for signalling. Cell Microbiol. 2016;18:1251–1267. doi: 10.1111/cmi.12635. [DOI] [PubMed] [Google Scholar]
- 9.Murley A, et al. Sterol transporters at membrane contact sites regulate TORC1 and TORC2 signaling. J Cell Biol. 2017;216:2679–2689. doi: 10.1083/jcb.201610032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malínská K, Malínský J, Opekarová M, Tanner W. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol Biol Cell. 2003;14:4427–4436. doi: 10.1091/mbc.E03-04-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malinska K, Malinsky J, Opekarova M, Tanner W. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J Cell Sci. 2004;117:6031–6041. doi: 10.1242/jcs.01493. [DOI] [PubMed] [Google Scholar]
- 12.Walther TC, et al. Eisosomes mark static sites of endocytosis. Nature. 2006;439:998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
- 13.Grossmann G, et al. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J Cell Biol. 2008;183:1075–1088. doi: 10.1083/jcb.200806035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strádalová V, et al. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J Cell Sci. 2009;122:2887–2894. doi: 10.1242/jcs.051227. [DOI] [PubMed] [Google Scholar]
- 15.Brach T, Specht T, Kaksonen M. Reassessment of the role of plasma membrane domains in the regulation of vesicular traffic in yeast. J Cell Sci. 2011;124:328–337. doi: 10.1242/jcs.078519. [DOI] [PubMed] [Google Scholar]
- 16.Athanasopoulos A, Boleti H, Scazzocchio C, Sophianopoulou V. Eisosome distribution and localization in the meiotic progeny of Aspergillus nidulans. Fungal Genet Biol. 2013;53:84–96. doi: 10.1016/j.fgb.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Berchtold D, et al. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat Cell Biol. 2012;14:542–547. doi: 10.1038/ncb2480. [DOI] [PubMed] [Google Scholar]
- 18.Niles BJ, Mogri H, Hill A, Vlahakis A, Powers T. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc Natl Acad Sci USA. 2012;109:1536–1541. doi: 10.1073/pnas.1117563109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olivera-Couto A, Graña M, Harispe L, Aguilar PS. The eisosome core is composed of BAR domain proteins. Mol Biol Cell. 2011;22:2360–2372. doi: 10.1091/mbc.E10-12-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziółkowska NE, Karotki L, Rehman M, Huiskonen JT, Walther TC. Eisosome-driven plasma membrane organization is mediated by BAR domains. Nat Struct Mol Biol. 2011;18:854–856. doi: 10.1038/nsmb.2080. [DOI] [PubMed] [Google Scholar]
- 21.Fröhlich F, et al. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J Cell Biol. 2009;185:1227–1242. doi: 10.1083/jcb.200811081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossmann G, Opekarová M, Malinsky J, Weig-Meckl I, Tanner W. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 2007;26:1–8. doi: 10.1038/sj.emboj.7601466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Athanasopoulos A, Gournas C, Amillis S, Sophianopoulou V. Characterization of AnNce102 and its role in eisosome stability and sphingolipid biosynthesis. Sci Rep. 2015;5:15200. doi: 10.1038/srep15200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghaddar K, et al. Substrate-induced ubiquitylation and endocytosis of yeast amino acid permeases. Mol Cell Biol. 2014;34:4447–4463. doi: 10.1128/MCB.00699-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gournas C, et al. Transition of yeast Can1 transporter to the inward-facing state unveils an α-arrestin target sequence promoting its ubiquitylation and endocytosis. Mol Biol Cell. 2017;28:2819–2832. doi: 10.1091/mbc.E17-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hein C, Springael J-Y, Volland C, Haguenauer-Tsapis R, André B. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 27.Galan JM, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 28.Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 29.MacGurn JA, Hsu PC, Smolka MB, Emr SD. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 2011;147:1104–1117. doi: 10.1016/j.cell.2011.09.054. [DOI] [PubMed] [Google Scholar]
- 30.Gournas C, Prévost M, Krammer EM, André B. Function and regulation of fungal amino acid transporters: Insights from predicted structure. Adv Exp Med Biol. 2016;892:69–106. doi: 10.1007/978-3-319-25304-6_4. [DOI] [PubMed] [Google Scholar]
- 31.Huff J. The Airyscan detector from ZEISS: Confocal imaging with improved signal-to-noise ratio and super-resolution. Nat Methods. 2015;12:i–ii. [Google Scholar]
- 32.Ghaddar K, et al. Converting the yeast arginine can1 permease to a lysine permease. J Biol Chem. 2014;289:7232–7246. doi: 10.1074/jbc.M113.525915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walther TC, et al. Pkh-kinases control eisosome assembly and organization. EMBO J. 2007;26:4946–4955. doi: 10.1038/sj.emboj.7601933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder R, London E, Brown D. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc Natl Acad Sci USA. 1994;91:12130–12134. doi: 10.1073/pnas.91.25.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanada K, Nishijima M, Akamatsu Y, Pagano RE. Both sphingolipids and cholesterol participate in the detergent insolubility of alkaline phosphatase, a glycosylphosphatidylinositol-anchored protein, in mammalian membranes. J Biol Chem. 1995;270:6254–6260. doi: 10.1074/jbc.270.11.6254. [DOI] [PubMed] [Google Scholar]
- 36.Rothbauer U, et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Crapeau M, Merhi A, André B. Stress conditions promote yeast Gap1 permease ubiquitylation and down-regulation via the arrestin-like Bul and Aly proteins. J Biol Chem. 2014;289:22103–22116. doi: 10.1074/jbc.M114.582320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones CB, et al. Regulation of membrane protein degradation by starvation-response pathways. Traffic. 2012;13:468–482. doi: 10.1111/j.1600-0854.2011.01314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stradalova V, et al. Distribution of cortical endoplasmic reticulum determines positioning of endocytic events in yeast plasma membrane. PLoS One. 2012;7:e35132. doi: 10.1371/journal.pone.0035132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jauniaux J-C, Grenson M. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur J Biochem. 1990;190:39–44. doi: 10.1111/j.1432-1033.1990.tb15542.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S, et al. Dopaminergic and glutamatergic microdomains in a subset of rodent mesoaccumbens axons. Nat Neurosci. 2015;18:386–392. doi: 10.1038/nn.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laganowsky A, et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ernst AM, Brügger B. Sphingolipids as modulators of membrane proteins. Biochim Biophys Acta. 2014;1841:665–670. doi: 10.1016/j.bbalip.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 44.Contreras F-X, et al. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature. 2012;481:525–529. doi: 10.1038/nature10742. [DOI] [PubMed] [Google Scholar]
- 45.Lorent JH, et al. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat Commun. 2017;8:1219. doi: 10.1038/s41467-017-01328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimobayashi M, Oppliger W, Moes S, Jenö P, Hall MN. TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis. Mol Biol Cell. 2013;24:870–881. doi: 10.1091/mbc.E12-10-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villers J, et al. Study of the plasma membrane proteome dynamics reveals novel targets of the nitrogen regulation in yeast. Mol Cell Proteomics. 2017;16:1652–1668. doi: 10.1074/mcp.M116.064923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeo K, Shigeta M, Takagi Y. Plasma membrane ultrastructural differences between the exponential and stationary phases of Saccharomyces cerevisiae as revealed by freeze-etching. J Gen Microbiol. 1976;97:323–329. doi: 10.1099/00221287-97-2-323. [DOI] [PubMed] [Google Scholar]
- 49.Walther P, Müller M, Schweingruber ME. The ultrastructure of the cell surface and plasma membrane of exponential and stationary phase cells of Schizosaccharomyces pombe, grown in different media. Arch Microbiol. 1984;137:128–134. [Google Scholar]
- 50.Xu T, et al. A profile of differentially abundant proteins at the yeast cell periphery during pseudohyphal growth. J Biol Chem. 2010;285:15476–15488. doi: 10.1074/jbc.M110.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grousl T, Opekarová M, Stradalova V, Hasek J, Malinsky J. Evolutionarily conserved 5′-3′ exoribonuclease Xrn1 accumulates at plasma membrane-associated eisosomes in post-diauxic yeast. PLoS One. 2015;10:e0122770. doi: 10.1371/journal.pone.0122770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaškovičová K, et al. mRNA decay is regulated via sequestration of the conserved 5′-3′ exoribonuclease Xrn1 at eisosome in yeast. Eur J Cell Biol. 2017;96:591–599. doi: 10.1016/j.ejcb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 53.Martinez MJ, et al. Genomic analysis of stationary-phase and exit in Saccharomyces cerevisiae: Gene expression and identification of novel essential genes. Mol Biol Cell. 2004;15:5295–5305. doi: 10.1091/mbc.E03-11-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Douglas LM, Wang HX, Konopka JB. The MARVEL domain protein Nce102 regulates actin organization and invasive growth of Candida albicans. MBio. 2013;4:e00723-13. doi: 10.1128/mBio.00723-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang LB, Tang L, Ying SH, Feng MG. Two eisosome proteins play opposite roles in autophagic control and sustain cell integrity, function and pathogenicity in Beauveria bassiana. Environ Microbiol. 2017;19:2037–2052. doi: 10.1111/1462-2920.13727. [DOI] [PubMed] [Google Scholar]
- 56.Scazzocchio C, Vangelatos I, Sophianopoulou V. Eisosomes and membrane compartments in the ascomycetes: A view from Aspergillus nidulans. Commun Integr Biol. 2011;4:64–68. doi: 10.4161/cib.4.1.13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacobs P, Jauniaux J-C, Grenson M. A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. J Mol Biol. 1980;139:691–704. doi: 10.1016/0022-2836(80)90055-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.