Significance
Analyses of 10 cell lines from which individual genes associated with innate immunity had been knocked out revealed the following: knockout of some genes is associated with increases in the expression of overlapping networks of genes and significant loss of ability to support the replication of HSV-1; knockout of other genes is associated with decreases in the expression of overlapping networks of genes and, overall, no effect on viral replication; the phenotype of cells from which a gene was deleted reflects the sum total of the effects of genes up- or down-regulated as a consequence of the deletion; and key functions associated with innate immunity are normally repressed and must be activated in response to infection.
Keywords: knockout, gene networks, innate immunity
Abstract
Analyses of the levels of mRNAs encoding IFIT1, IFI16, RIG-1, MDA5, CXCL10, LGP2, PUM1, LSD1, STING, and IFNβ in cell lines from which the gene encoding LGP2, LSD1, PML, HDAC4, IFI16, PUM1, STING, MDA5, IRF3, or HDAC 1 had been knocked out, as well as the ability of these cell lines to support the replication of HSV-1, revealed the following: (i) Cell lines lacking the gene encoding LGP2, PML, or HDAC4 (cluster 1) exhibited increased levels of expression of partially overlapping gene networks. Concurrently, these cell lines produced from 5 fold to 12 fold lower yields of HSV-1 than the parental cells. (ii) Cell lines lacking the genes encoding STING, LSD1, MDA5, IRF3, or HDAC 1 (cluster 2) exhibited decreased levels of mRNAs of partially overlapping gene networks. Concurrently, these cell lines produced virus yields that did not differ from those produced by the parental cell line. The genes up-regulated in cell lines forming cluster 1, overlapped in part with genes down-regulated in cluster 2. The key conclusions are that gene knockouts and subsequent selection for growth causes changes in expression of multiple genes, and hence the phenotype of the cell lines cannot be ascribed to a single gene; the patterns of gene expression may be shared by multiple knockouts; and the enhanced immunity to viral replication by cluster 1 knockout cell lines but not by cluster 2 cell lines suggests that in parental cells, the expression of innate resistance to infection is specifically repressed.
In a preceding publication, we reported that depletion of PUM1 by siRNA resulted in a sequential activation of subsets of genes associated with innate immune responses (1). Thus, within 12 and 48 h after transfection of the siRNA targeting PUM1, there was an upsurge (phase 1) in the accumulation of LGP2, CXCL10, IL6, and PKR mRNAs. This was followed 24 h later (phase 2) by a significant accumulation of mRNAs encoding RIG-I, SP100, MAD5, IFIT1, PML, STING, and IFN β (IFNβ). Phase 1 and phase 2 transcriptional activation was traced to activation of LGP2 secondary to depletion of PUM1 (1). The data did not exclude the possibility that LGP2 acted by activation of as-yet-unidentified intermediaries. The results suggested that the activated network persists for a limited time, inasmuch as one end product of the activated network, IFNβ, was reduced to background levels by day 5 after transfection of the siRNA. The results of these studies raised several questions. The foremost questions that arose were whether networks of activated genes persist in cell lines in which individual genes associated with various aspects of innate immune responses are knocked out, whether the networks differ depending on knocked out gene, and whether knockouts define permissivity of HSV replication in knocked-out cell lines.
In the studies reported here, we examined the accumulation of mRNAs of 10 genes encoded for proteins (IFIT1, IFI16, RIG-I, MDA5, CXCL10, LGP2, PUM1, LSD1, STING, and IFNβ) involved in various aspects of gene regulation and in innate immune responses in 10 cell lines in which LGP2, LSD1, PML, HDAC4, PUM1, IFI16, STING, MDA5, IRF3, or HDAC1 had been knocked out. We also tested the ability of knocked-out cell lines to support the replication of HSV-1. The findings reported here suggest that the knocked-out cell lines form three clusters. Cluster 1 consists of cell lines in which partially overlapping networks of genes were up-regulated. Cluster 2 comprises knockout cell lines in which partially overlapping networks of genes were down-regulated. The third cluster comprises two cell lines in which none of the tested genes was significantly affected. The replication of HSV was significantly diminished in several knockout cell lines. In none of the knockout cell lines have the virus yields exceeded to a significant level those obtained in the parental cell line.
The results reported here challenge the hypothesis that the phenotype of a cell can be specifically linked to the absence of a gene product that had been depleted by siRNA or whose gene had been knocked out.
Results
Analyses of Gene Expression and HSV-1 Replication in Knocked-Out Cell Lines.
The objectives of the first series of experiments were to examine the constitutive expression of 10 genes in parent and in 10 knockout cell lines. Both the probed gene and the knocked-out genes have been reported to play a role in innate immune responses. To further relate the knocked-out genes to innate immunity, we examined the replication of HSV-1 in the various cell lines. The knocked-out cell lines (i.e., ΔLGP2, ΔLSD1, ΔPML, ΔHDAC4, ΔPUM1, ΔIFI16, ΔSTING, ΔMDA5, ΔIRF3, and ΔHDAC1) were derived from HEp-2 cells and probed for the accumulation of mRNAs encoding IFNβ, STING, LSD1 PUM1, LGP2, CXCL10, MDA5, RIG-I, IFI16, and IFIT1. The procedures and reagents designed to knock out the targeted genes, and the DNA probes used to quantify the various mRNAs, are described in Materials and Methods.
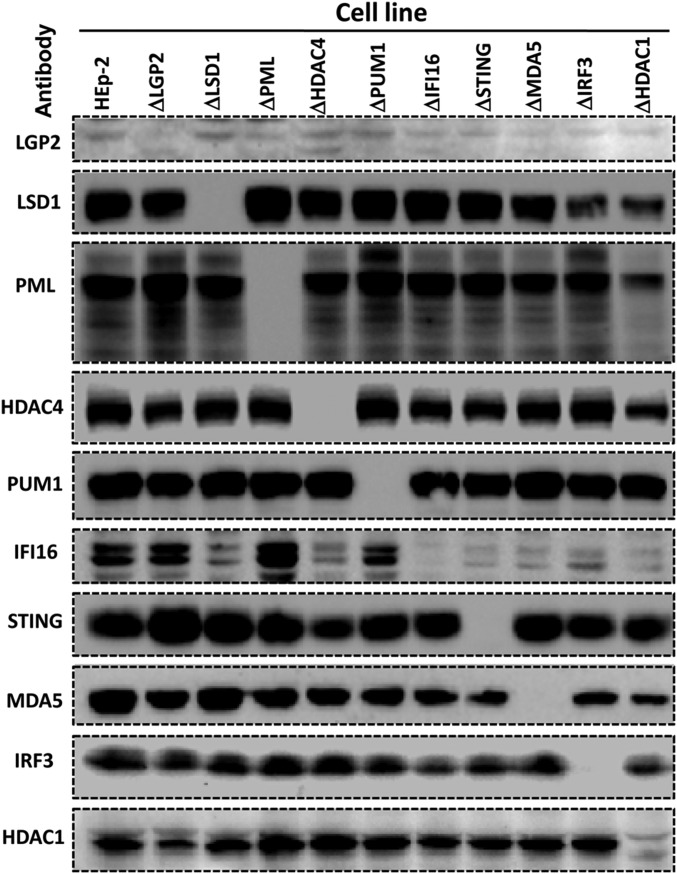
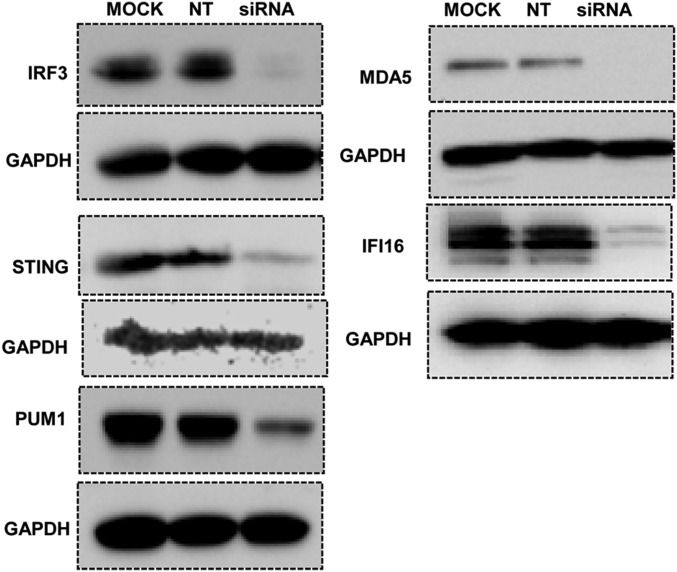
The results of the first series of experiments may be summarized as follows: Fig. 1 shows an immunoblot of the 10 cell lines probed with antibodies to the proteins specified by the knocked-out genes. The sources of the antibodies used to probe for the specific proteins are listed in Materials and Methods. The immunoblots presented in this figure show that the protein targeted in each of the targeted cell lines is no longer detectable. Note that the ΔPML cell line was described elsewhere and included here for consistency.
Fig. 1.
Analyses of lysates of knocked-out cell lines for accumulation of protein products of the knocked-out genes. Cultures of HEp-2 cells or of the knockout cell lines each containing 3 × 105 of cells were exposed to IFNβ (200 ng/mL, for LGP2 or MDA5 antibody immunoblotting) or mock treated (for other antibodies, immunoblotting except LGP2 and MDA5). After 24 h, the cells were solubilized subjected to electrophoresis in denaturing gels and immunoblotted with respective antibodies. The sources of antibodies used in these studies are listed in Materials and Methods.
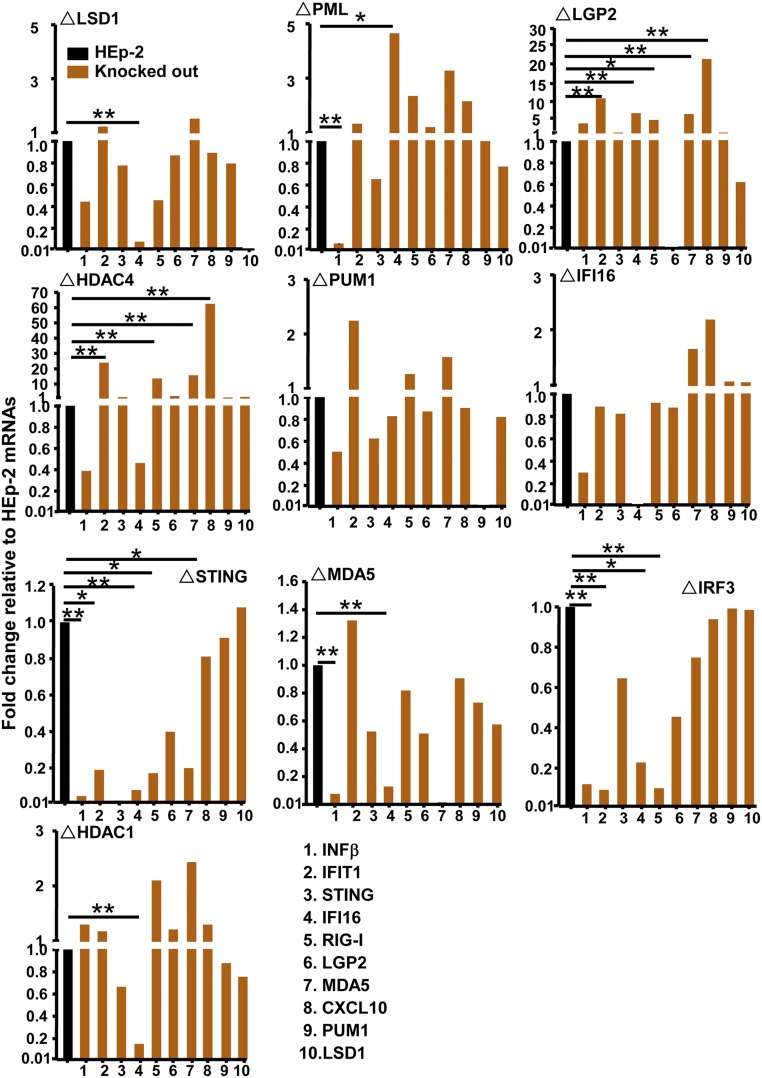
Fig. 2 shows the amounts of mRNAs detected by the 10 probes in the 10 knockout cell lines relative to those detected in HEp-2 cells. The analyses were performed in triplicate. The results show that the ratios of mRNAs detected in knockout cell lines to those in HEp-2 cells ranged from >10-fold lower than the levels detected in parental HEp-2 cells to >60-fold higher. On the basis of statistical analyses, we deemed as significant mRNA levels that were at least fourfold higher or fourfold lower than those obtained in the parental cell line.
Fig. 2.
Accumulation of mRNAs of selected cellular genes in parental and knockout cell lines. The parental and knocked-out cells were seeded in amounts of 3 × 105 cells per culture. After 24 h of incubation, the cells were harvested and total RNA was extracted, and 0.5 μg of each RNA was reverse-transcribed to cDNA, quantified and normalized with respect to 18S RNA as described in Materials and Methods. In this figure, the amounts of mRNAs encoding IFNβ (1), IFIT1 (2), STING (3), IFI16 (4), RIG-I (5), LGP2 (6), MDA5 (7), CXCL10 (8), PUM1 (9), and LSD1 (10) are shown normalized with respect to mRNA levels in HEp-2 cells (black bar) and presented as means ± SD. Student’s t test was performed to calculate the P values (n = 3). *P ≤ 0.05; **P ≤ 0.02.
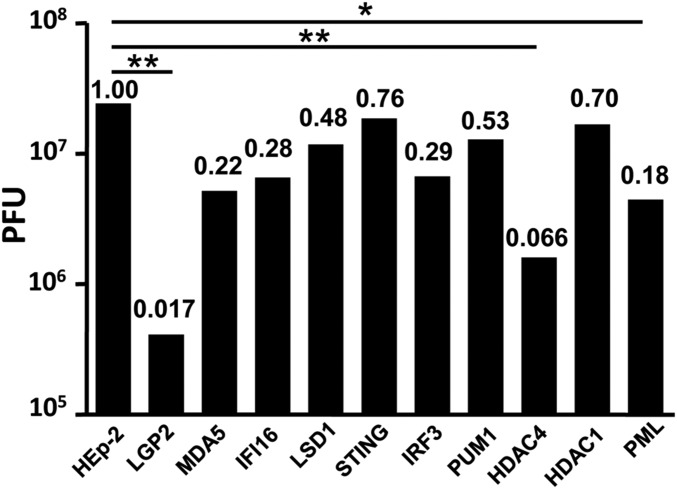
Fig. 3 shows the yields of HSV-1(F) in HEp-2 cells and in the knockout cell lines done in parallel at the same time. In these experiments, the cell lines were exposed to 0.01 PFU per cell and harvested 48 h after infection. The figure shows the ratios of virus yields in knocked-out cells relative to those obtained in HEp-2 cells. The results were that none of the cell lines produced significantly more virus than the parental HEp-2 cell. Yields fivefold or more lower than those obtained in HEp-2 cells were significant at 95% or greater confidence levels. These included ΔPML, ΔHDAC4, and ΔLGP2. The decrease in the yields of HSV-1 in ΔPML and ΔHDAC4 cell lines obtained in this study were consistent with the earlier reports (2, 3).
Fig. 3.
Replication of HSV-1(F) in HEp-2 and knockout cell lines. Replicate cultures of HEp-2 or indicated knockout cells were exposed to 0.01 PFU of virus per cell for 2 h. The inoculum was then replaced with fresh medium. Virus progeny were harvested at 48 h after infection and titered on Vero cells. The numbers above the bars show the ratios of virus yield obtained in the knocked-out cell lines relative to those obtained in HEp-2 cells. Student’s t test was used to calculate the P values (n = 3). *P ≤ 0.05; **P ≤ 0.02.
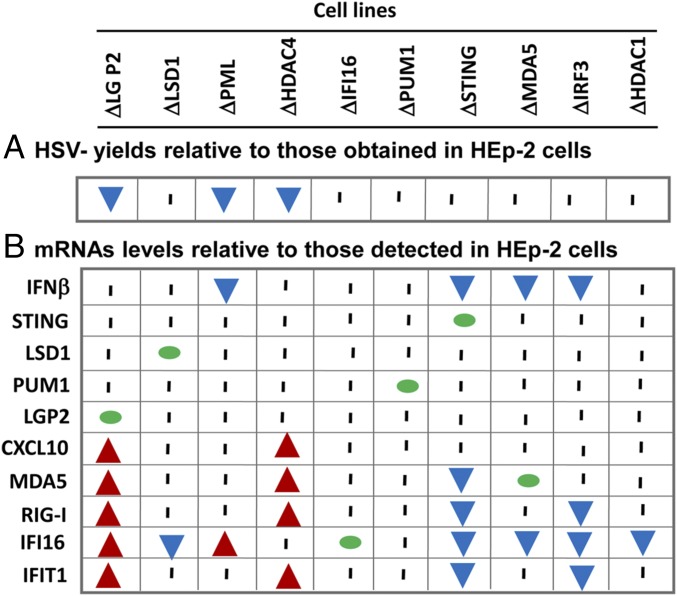
To facilitate analyses of the data and to highlight the key features of the results, the data presented in Figs. 2 and 3 are summarized in Fig. 4. Specifically, the results suggest that the 10 cell lines form three clusters. The first cluster comprises the cell lines ΔLGP2, ΔPML, and ΔHDAC4. The key features of these cell lines are significant decreases in virus yields and increased accumulation of at least one mRNA.
Fig. 4.
Summary of the accumulation of mRNAs (B) and HSV-1(F) (A) yields in knockout cell lines. This diagram summarizes in simplified form the significant data shown in Figs. 2 and 3. The probed mRNAs are listed on the y axis. The knockout cell lines are listed on the x axis. The numbers at the top of each cell line are the ratios of virus yields obtained in knockout cell lines to yields obtained in HEp-2 cells. Green dots indicate that the probed gene had been deleted. Red arrows indicate that the probed mRNA increased significantly relative to the levels measured in HEp-2 cells; blue arrows indicate that the amounts of mRNA detected in the knockout cell lines were significantly lower than the amount detected in HEp-2 cells. A vertical bar indicates that the values obtained in knockout cell lines were not significantly different from those measured in HEp-2 cells.
The second cluster comprises cell lines ΔLSD1, ΔSTING, ΔMDA5, ΔIRF3, and ΔHDAC1. A characteristic of these cell lines is the significant decreases in the accumulation of mRNAs encoded by at least one gene and no effect on virus yields. The striking feature of the data is that for the most part, the genes whose expression is down-regulated in cell lines comprising the second cluster are up-regulated in the first cluster. Moreover, although the number of parameters detailed in this study is relatively small, no two cell lines exhibited identical responses to the knockouts.
Last, the ΔPUM1 and ΔIFI16 cell lines form the third cluster. These cell lines did not differ significantly from the parent HEp-2 cell with respect to the parameters analyzed in this study.
Identification of Effector Genes Responsible for Changes in Expression of Select Genes in Knockout Cell Lines.
The key features of the results reported here is that in each of two groups, knockout of individual genes results in either up-regulation or down-regulation of a group of seemingly unrelated genes. Thus, cells lacking intact LGP2, PML, or HDAC4 each accumulated significantly higher levels of mRNAs encoded by one to five different genes. Conversely, cells devoid of LSD1, STING, MDA5, IRF3, or HDAC1 accumulated significantly lower levels of mRNAs encoded by at least four genes. There are at least two hypotheses that could explain these results. The first hypothesis is that LGP2, PML, and HDAC4 each act independently as transcriptional repressors or significantly affect the stability of the mRNAs. Consistent with this hypothesis, LSD1, STING, MDA5, IRF3, or HDAC1 act independently to induce the synthesis or stabilize the mRNAs encoding MDA5, RIG-I, IFI16, or IFIT1. An alternative hypothesis is that the up-regulation or down-regulation is the consequence of a cascade of events initiated by one or a small number of effector gene products.
To test these hypotheses, we selected three group 1 knocked-out cell lines. They were ΔLGP2, ΔPML, and ΔLSD1. Each cell line was mock transfected or transfected with IRF3, STING, IFI16, MDA5, or PUM1 siRNA. The control and siRNA-transfected cells were harvested 60 h after transfection and analyzed with respect to accumulations of mRNAs encoding RIG-I, IFIT1, IFI16, STING, LGP2, or MDA5 mRNAs. The results may be summarized as follows: The specificity of the IRF3, STING, IFI16, and MDA5 siRNAs is shown in Fig. 5. In each instance, at 60 h after transfection, there was a significant reduction of targeted mRNA, whereas the NT siRNA had no effect. The specificity of the PUM1 siRNAs used in this study was reported in the preceding publication (1).
Fig. 5.
Efficiency of siRNAs in depleting targeted proteins after transfection of HEp-2 cells. HEp-2 cells seeded on six-well plates were either mock treated or transfected with 100 nM IRF3, MDA5, STING, IFI16, or PUM1 siRNA or nontarget siRNA (siNT). The cells were harvested 60 h after transfection. Cell lysates were separated on denaturing 10% polyacrylamide gels, electrically transferred to nitrocellulose sheets, and immunoblotted for indicated proteins. GAPDH was used as a loading control. For confirmation of siMDA5 depletion, mock-treated, nontarget, or siMDA5-transfected cells were exposed to IFNβ (100 ng/mL) at 36 h after transfection; the cells were harvested after 24 h of additional incubation and processed as described earlier.
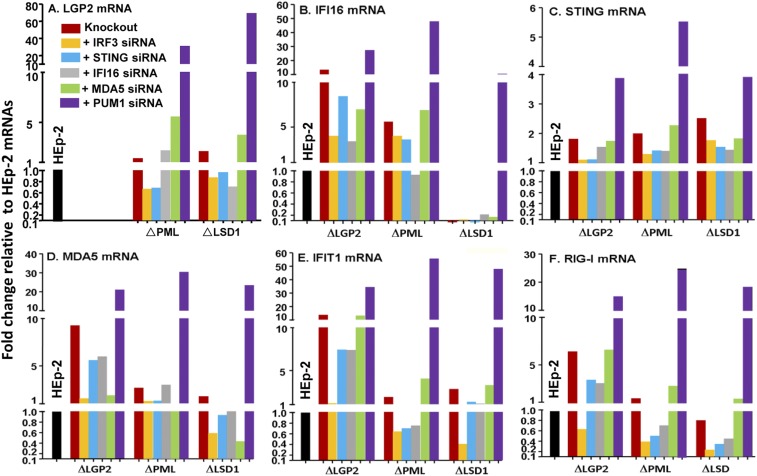
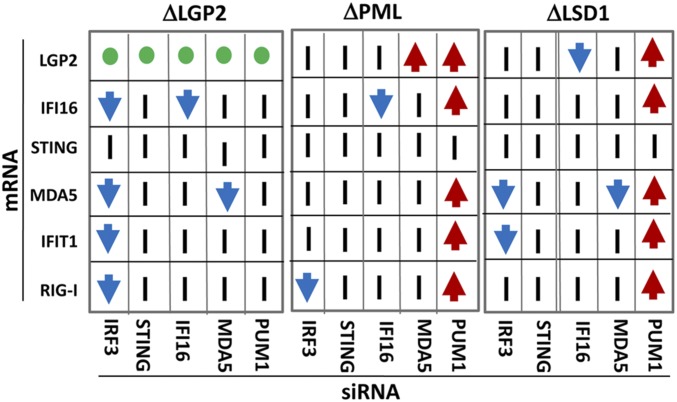
The results of the transfection of the three cell lines with each of the five siRNAs are shown in Fig. 6. In this study, we monitored the values of six mRNAs in four cell lines after transfection of five siRNAs. To fit the massive amounts of data in a simplified format, all the mRNA values are shown relative to the corresponding values obtained in parental HEp-2 cells (black bars with values set to 1). A further simplification of the data is shown in Fig. 7. In this figure, the data are grouped for each cell line normalized with respect to the mRNA level in mock-treated knocked-out cells. The red and blue arrows indicate that in consequence of transfection of siRNAs, the mRNA levels increased or decreased at least fourfold relative to the levels observed in mock-treated knocked-out cells.
Fig. 6.
Accumulation of mRNAs of selected cellular genes in ΔLGP2, ΔPML, and ΔLSD1 cells after mock transfection or transfection of IRF3, STING, IFI16, MDA5, or PUM1 siRNAs. HEp-2 or knockout cells grown in six-well plates were mock-treated or transfected with 100 nM IRF3, STING, IFI16, MDA5, or PUM1 siRNA. The cells were harvested at 60 h after transfection, and 0.5 μg total RNA was reverse-transcribed to cDNA, as described in Materials and Methods. mRNAs encoding LGP2 (A), IFI16 (B), STING (C), MDA-5 (D), IFIT1 (E), and RIG-I (F) were normalized with respect to 18s RNA and are shown as fold change compared with the mRNA levels measured in HEp-2 cells.
Fig. 7.
Summary of the accumulation of mRNAs in knockout cells transfected with siRNAs targeted to IRF3, STING, IFI16, MDA5, or PUM1. The three panels summarize the results of analyses ΔLGP2, ΔPML, or ΔLSD1 cell lines. For each panel, the x axis shows the transfected siRNA. The y axis identifies the mRNA. Red arrows indicate that the siRNA caused at least a fourfold increase in the mRNA level relative to that of mock-transfected cells. Blue arrows indicate that the siRNA caused at least a fourfold decrease in the mRNA relative levels in mock-transfected cells. A vertical bar indicates no significant changes. The green circle indicates that as expected, no LGP2 mRNA could be detected in ΔLGP2 cells. Last, the black circles identify mRNAs specifically targeted by corresponding mRNAs, and therefore expected to be down-regulated.
The key features of the results presented in Figs. 6 and 7 are as follow: (i) Depletion of PUM1 resulted in multifold up-regulation of mRNAs encoded by LGP2, IFI16, MDA5, IFIT1, and RIG-I in ΔPML and ΔLSD1 cell lines, but not in ΔLGP2 cell line. The results are consistent with the hypothesis that PUM1 acts as a master repressor of multiple genes, but that it requires LGP2 for its action. (ii) Depletion of IRF3 resulted in significant decrease in the accumulation of RIG-I, IFIT1, MDA5, and IFI16 mRNAs in the ΔLGP2 cell line. The down-regulation was limited to RIG-I in ΔPML cell line and to MDA5 and IFIT1 in the ΔLSD1 cell line. The results suggest that the function of IRF3 as an up-regulator of gene expression is defined by the expression of multiple genes. (iii) Of the siRNAs tested, STING siRNA had no effect on the accumulation of any of the mRNAs tested. Other siRNAs had an effect in one cell line, but not in the other two. For example, MDA5 siRNA enhanced the accumulation of LGP2 mRNA in the ΔPML cell line, but not in the other two cell lines.
Discussion
As noted in the introduction, in a preceding publication (1), we reported that transfection of HEp-2 cells with PUM1 siRNA resulted in a biphasic upsurge in the expression of genes associated with innate immune responses. Thus, during phase 1 between 12 and 48 h after transfection, there was a significant increase in the accumulation of LGP2, CXCL10, IL6, and PKR mRNAs. This was followed 24 h later (phase 2) by a significant increase in the accumulation of mRNAs encoding RIG-I, SP100, MDA5, IFIT1, PML, STING, and IFNβ. The results suggested that PUM1, but not PUM2, acted directly or via one or more intermediaries to activate LGP2, and this in turn activated directly or through intermediaries the expression of at least some phase 1 and phase 2 genes. The results revealed that suppression of PUM1 results in the activation of a network in a sequence that can be monitored in real time. Moreover, the data suggested that the effect of transfection was likely transient. The surprising finding was that depletion of PUM1 resulted in a sequence of events that led to production of IFN β and reduction in HSV-1 gene expression. The role of PUM1 could not be related to its function as a posttranscriptional regulator of mRNA (1, 4–6).
The key questions addressed in this report were whether gene knockouts also result in changes in the expression of multiple genes, whether the networks of modified gene responses in different knockouts overlap, and whether enhancement of antiviral activity is a common consequence of gene knockouts.
It should be noted that there is a vast difference between depletion of targeted mRNAs by siRNA and gene knockouts. Depletion results in a stress response that dissipates over time, as illustrated by the disappearance of INFβ on the fifth day after transfection of PUM1 siRNA. In principle, depletion of mRNA does not result in the selection of survivors altered in perpetuity. In contrast, the stable cell lines generated by gene knockouts reflect the results of genetic and/or epigenetic changes that compensate the cells for gene loss and enable them to survive and divide.
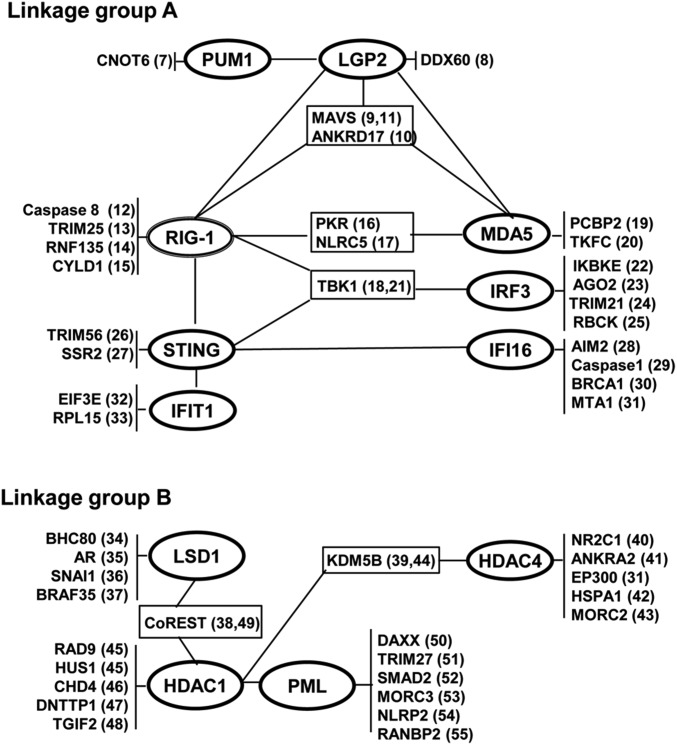
In the studies reported here, we measured HSV replication and the accumulation of mRNAs encoded by 10 genes associated with various aspects of innate immunity in 10 knockout cell lines. A characteristic of the genes selected for these studies is that their products perform a wide spectrum of functions ranging from chromatin modifiers, transcriptional regulators, sensors of nucleic acids, and so on, and have been reported to interact with multiple proteins (7–55). As illustrated in Fig. 8, they form two distinct linkage groups.
Fig. 8.
Linkage maps showing reported interactions of the products of genes investigated in this study among themselves and products of other genes. The numbers in parentheses refer to references.
The results of the studies reported here may be summarized as follows: With two exceptions (ΔIFI16 and ΔPUM1), the knocked-out cell lines exhibited changes in the expression of at least 1 (ΔHDAC1) and, in most cases, changes in the expression of multiple genes. These were either predominantly up-regulated (cluster 1) or predominantly down-regulated (cluster 2).
Second, in both clusters, the same genes were up-regulated or down-regulated in more than one knockout cell line. Nevertheless the cell lines differed with respect to the overall pattern of response. A key question that remains unresolved is whether all the changes in gene expression are essential for the function of the knocked-out cell lines and whether some changes are covariant but do not contribute to the phenotype of the cells. Irrespective of the role of each individual changes, the phenotype of each cell line reflects the sum total of alterations in gene expression, and not merely the function of the deleted gene.
Third, none of the 10 knockout cell lines produced more virus than the parental HEp-2 cells. A curious finding was that the clusters of cell lines in which one or more genes were up-regulated produced 5–100-fold less virus than HEp-2 cells. These cell lines were ΔLGP2, ΔPML, and ΔHDAC4. PML and HDAC4 were reported elsewhere to be required for HSV-1 replication (2, 3). Conversely, the cell lines exhibiting decreases in gene expression did not differ significantly from the parental cell line with respect to virus yields. The significance of this finding is twofold. Foremost, implicit in this finding is that the genetic and or epigenetic changes that confer survival and growth of some knocked-out cell lines may also confer a heightened immunity against infection. Equally significant, the innate antiviral state of some but not all knockout cell lines suggests that in unstressed parental cells, the antiviral state is repressed and must be activated by specific sensors.
Fourth, the role of STING in defining antiviral immunity depends on the context in which it is probed. An earlier publication from one of the reporting laboratories showed that depletion of STING by siRNA significantly increased HSV-1 yields (56). This report shows no perceptible decrease in virus yields in ΔSTING cells. The discrepancy in the results obtained by the two methods is consistent with the hypothesis noted earlier in the text, in that transient depletion of a gene product in contrast to the deletion of the gene may affect different cohorts of cellular genes and yield divergent results.
Fifth, perhaps the major conflict to emerge from the comparison of the two methods of silencing the expression of a gene emerged from analyses of the role of LGP2. Specifically, in cells depleted of PUM1, the expression of LGP2 was enhanced and was required for subsequent enhancement of expression of several genes, including INFβ, and down-regulation of viral gene expression (1). In the studies reported here, we noted a dramatic decrease in virus yields and up-regulation of genes associated with innate immunity. One hypothesis that could explain the data is that the function of LGP2 is defined by one or more cellular gene products that remain to be identified and that are regulated differently in depleted and knocked-out cells.
Last, the question arose of whether we could identify master regulators among the cluster of up- or down-regulated genes. To test one approach, we selected three knockout cell lines (i.e., ΔLGP2, ΔPML, and ΔLSD1) that exhibited multiple up-regulated genes. These were transfected with IRF3, IFI16, STING, MDA5, or PUM1 siRNAs. The key finds were that IRF3 down-regulated mRNAs encoding IFI16, MDA5, IFI1, and RIG-I in the ΔLGP2 cell line, but not in the ΔPML or ΔLSD1 cell line. Conversely, transfection of the three cell lines with PUM1 siRNA resulted in the up-regulation of LGP2, IFI16, MDA5, IFIT1, and RIG-1 in the ΔLSD1 and ΔPML cell line, but not, as expected, in the ΔLGP2 cell line. On the basis of this approach, IRF3 up-regulates a network of genes, but its function is down-regulated by LGP2. However, as previously reported, PUM1 siRNA up-regulates LGP2 and indirectly up-regulates a network of genes. Consistent with earlier results, we detected up-regulation of multiple genes after transfection of PUM1 siRNA in ΔPML and ΔLSD1 cell lines, but not in cells in which LGP2 was knocked out. The data suggest that the regulatory functions of PUM1 are transient. In these studies, IRF3 and PUM1 appear to control the expression of a network in which the primary effector is LGP2.
The significance of this report stems from three conclusions. Foremost, multiple single-gene knocked-out cell lines exhibit covariant up- or down-regulation of partially overlapping clusters of genes. Implicit in this finding is that the phenotype of knocked-out cell lines cannot be ascribed to a single gene. Moreover, in the studies reported here, up-regulation of innate immunity appears to be associated with up-regulation of expression of partially overlapping gene clusters and is not a general property of knocked out cells. Last, the term “stress” inadequately portrays the range of stimuli to which cells respond in many and diverse ways.
Materials and Methods
Cell Lines and Virus Strains.
HEp-2 cells obtained from the American Type Culture Collection and the knockout cell lines were routinely cultured in DMEM (Life Technologies) containing 5% FBS (Life Technologies). HSV-1(F) is the prototype HSV-1 strain used in this laboratory (57).
RNA Interference.
Gene knockdown was achieved using siRNAs directed against human IRF3, STING, IFI16, MDA5, or PUM1 genes (all from GenePharma); the sequences of the siRNA were as follows: siPUM1(5′-GCUGCUUACUAUGACCAAATT-3′), siIRF3(5′-GGAGGAUUUCGGAAUCUUCTT-3′), siSTING(5′-GCAACAGCAUCUAUGAGCUUCUGGAGAAC-3′), siIFI16(5′-CACGUUGAAACCAAGACUGAATT-3′), and siMDA5(5′-GGAUUGUGCAGAAAGAAAATT-3′).
The NT siRNA(5′-UUCUCCGAACGUGUCACGUTT-3′) was used as negative control. For siRNA transfection, cells (3 × 105 per well) seeded in six-well plates were transfected with siRNA at a final concentration of 100 nM. At 60 h posttransfection, cells were harvested for further analysis. All transfections were carried out using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions.
Generation of Knockout Cell Lines Using CRISPR.
Parental HEp-2 cells were cotransfected with the sgRNA vector plasmid, targeting indicated sequences (Table 1), and a donor vector containing a selection cassette expressing GFP was inserted into the targeted site. Selected cells (GFP+) were serially diluted to form single cell-derived colonies. The knockouts were further verified by genomic DNA sequencing and immunoblotting (Fig. 1).
Table 1.
Sequences of sgRNAs used to knockout target genes
| Gene | sgRNA targeting sequences | |
| Probe 1 | Probe 2 | |
| LGP2 | sgRNA#1: 5′-GCGCATGCTGGATGGACGC-3′ | #2: 5′-GGGACATGGGACCACGTGC-3′ |
| LSD1 | sgRNA#1: 5′-CGCAAGAAAGAGCCTCCGCG-3′ | #2: 5′-GGCGGAACCGCCGGGGTCCGC-3′ |
| PML | sgRNA#1: 5′-CTCGGAGATCGGGCGGGTGC-3′ | #2: 5′-GGCCTTCAGAGGGGGTCTCG-3′ |
| HDAC4 | sgRNA#1: 5′-GTGGTCCAGGCGCAGGTCCA-3′ | #2: 5′-GCCTCTGGAACTCAGCGATG-3′ |
| HDAC1 | sgRNA#1: 5′-TGAGTCATGCGGATTCGGTG-3′ | #2: 5′-CTATGGTCTCTACCGAAAAA-3′ |
| PUM1 | sgRNA#1: 5′-TCTTAGACAGGAATCGCCCG-3′ | #2: 5′-GTCCCCCACTCTCCGATCGT-3′ |
| IFI16 | sgRNA#1: 5′-GACCAGCCCTATCAAGAAAG-3′ | #2: 5′-GTCGGACACCTTACTCCCTTT-3 |
| STING | sgRNA#1: 5′-AGAGCACACTCTCCGGTACC-3′ | #2: 5′-CTGGGACTGCTGTTAAACG-3′ |
| MDA-5 | sgRNA#1: 5′-TGGTTGGACTCGGGAATTCG-3′ | #2: 5′-TCATGAGCGTTCTCAAACGA-3′ |
| IRF3 | sgRNA#1: 5′-GCTGGTGCATATGTTCCCGGG-3′ | #2: 5′-GCAACCCTTCTTTGCGGTTG-3′ |
RNA Extraction and Real-Time Quantitative PCR.
Total RNA was isolated using TRI Reagent solution (Thermo Scientific) and treated with DNase I (Takara). cDNA was synthesized from 0.5 μg RNA with the aid of the Rever Tra Ace q-pcr RT Kit (Toyobo) in accordance with instructions provided by the suppliers. Gene expression was measured by real-time quantitative PCR analysis using SYBR Green Realtime PCR master mix (Toyobo) in Step one plus Real-Time PCR system (AB Applied Biosystems), using the following primers: IFI16-F AAAGTTCCGAGGTGATGC, IFI16-R TGACAGTGCTGCTTGTGG, PUM1-F CTTGCATTTGGACAAGGTCTG, PUM1-R CATTCACTACAAGGGCACCAG, LSD1-F CCGCTCCACGAGTCAAAC, and LSD1-R ATCCCAGAACACCCGATC, sequences of other primers were as described previously (1). Transcript expression was normalized to the 18SRNA, and relative expression changes was determined using the 2−ΔΔCT method.
Immunoblot Assays.
Cells were collected 60 h after transfection. The procedures for harvesting, solubilization, protein quantification, SDS/PAGE, and transfer to nitrocellulose membranes were as described previously (58).
Antibodies.
The antibodies used in this study included rabbit polyclonal anti-LGP2 (sc-134667; Santa Cruz), rabbit monoclonal anti-PUM1 (ab92545; Abcom), rabbit monoclonal anti-MDA5 (#5321; CST), rabbit polyclonal anti-STING (19851–1-AP; Proteintech), rabbit monoclonal anti-IFI16 (#14970; CST), rabbit monoclonal anti-PML (ab179466; Abcom), rabbit polyclonal anti-LSD1 (#2139; CST), rabbit monoclonal anti-HDAC1 (#34589; CST), rabbit monoclonal anti-HDAC4 (#7628; CST), rabbit monoclonal anti-IRF3 (ab76409; Abcom), and rabbit monoclonal anti-GAPDH (#2118; CST).
Virus Titration.
A total of 1 × 106 of HEp-2 or knockout cells were seeded onto six-well plates, After 24 h of incubation, the cells were exposed to 0.01 PFU HSV-1(F) per cell. The cells were harvested at 48 h after infection. Viral progeny were titrated on Vero cells.
Acknowledgments
These studies were supported by Shenzhen Science and Innovation Commission Project Grant JCYJ20170411094933138 to Shenzhen International Institute for Biomedical Research, National Nature Science Foundation of China Grant NSFC 81472826, Guangdong Nature Science Foundation Grant 2016A030308007, and Guangzhou Science Technology and Innovation Commission Project Grant 201504010016 to Guangzhou Medical University.
Footnotes
The authors declare no conflict of interest.
References
- 1.Liu Y, Qu L, Liu Y, Roizman B, Zhou GG. PUM1 is a biphasic negative regulator of innate immunity genes by suppressing LGP2. Proc Natl Acad Sci USA. 2017;114:E6902–E6911. doi: 10.1073/pnas.1708713114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu P, Mallon S, Roizman B. PML plays both inimical and beneficial roles in HSV-1 replication. Proc Natl Acad Sci USA. 2016;113:E3022–E3028. doi: 10.1073/pnas.1605513113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lomonte P, et al. Functional interaction between class II histone deacetylases and ICP0 of herpes simplex virus type 1. J Virol. 2004;78:6744–6757. doi: 10.1128/JVI.78.13.6744-6757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gennarino VA, et al. Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type Ataxin1 levels. Cell. 2015;160:1087–1098. doi: 10.1016/j.cell.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 6.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Etten J, et al. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem. 2012;287:36370–36383. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komuro A, Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol. 2006;80:12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Ankrd17 positively regulates RIG-I-like receptor (RLR)-mediated immune signaling. Eur J Immunol. 2012;42:1304–1315. doi: 10.1002/eji.201142125. [DOI] [PubMed] [Google Scholar]
- 11.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 12.Rajput A, et al. RIG-I RNA helicase activation of IRF3 transcription factor is negatively regulated by caspase-8-mediated cleavage of the RIP1 protein. Immunity. 2011;34:340–351. doi: 10.1016/j.immuni.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 14.Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-β induction during the early phase of viral infection. J Biol Chem. 2009;284:807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 15.Friedman CS, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pham AM, et al. PKR transduces MDA5-dependent signals for type I IFN induction. PLoS Pathog. 2016;12:e1005489. doi: 10.1371/journal.ppat.1005489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui J, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng W, et al. Negative regulation of virus-triggered IFN-β signaling pathway by alternative splicing of TBK1. J Biol Chem. 2008;283:35590–35597. doi: 10.1074/jbc.M805775200. [DOI] [PubMed] [Google Scholar]
- 19.You F, et al. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 20.Diao F, et al. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc Natl Acad Sci USA. 2007;104:11706–11711. doi: 10.1073/pnas.0700544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Gu L, Fullam A, Brennan R, Schröder M. Human DEAD box helicase 3 couples IκB kinase ε to interferon regulatory factor 3 activation. Mol Cell Biol. 2013;33:2004–2015. doi: 10.1128/MCB.01603-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, et al. AGO2 negatively regulates type I interferon signaling pathway by competition binding IRF3 with CBP/p300. Front Cell Infect Microbiol. 2017;7:195. doi: 10.3389/fcimb.2017.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura T, et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol. 2015;210:973–989. doi: 10.1083/jcb.201503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang M, et al. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 2008;18:1096–1104. doi: 10.1038/cr.2008.277. [DOI] [PubMed] [Google Scholar]
- 26.Tsuchida T, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veeranki S, Duan X, Panchanathan R, Liu H, Choubey D. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS One. 2011;6:e27040. doi: 10.1371/journal.pone.0027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aglipay JA, et al. A member of the pyrin family, IFI16, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene. 2003;22:8931–8938. doi: 10.1038/sj.onc.1207057. [DOI] [PubMed] [Google Scholar]
- 31.Kang HJ, et al. Differential regulation of estrogen receptor α expression in breast cancer cells by metastasis-associated protein 1. Cancer Res. 2014;74:1484–1494. doi: 10.1158/0008-5472.CAN-13-2020. [DOI] [PubMed] [Google Scholar]
- 32.Fensterl V, Sen GC. Interferon-induced Ifit proteins: Their role in viral pathogenesis. J Virol. 2015;89:2462–2468. doi: 10.1128/JVI.02744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu Y-A, et al. A novel interaction between interferon-inducible protein p56 and ribosomal protein L15 in gastric cancer cells. DNA Cell Biol. 2011;30:671–679. doi: 10.1089/dna.2010.1149. [DOI] [PubMed] [Google Scholar]
- 34.Iwase S, et al. Characterization of BHC80 in BRAF-HDAC complex, involved in neuron-specific gene repression. Biochem Biophys Res Commun. 2004;322:601–608. doi: 10.1016/j.bbrc.2004.07.163. [DOI] [PubMed] [Google Scholar]
- 35.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 36.Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010;29:4896–4904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hakimi MA, et al. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y-J, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 39.Barrett A, et al. Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int J Cancer. 2007;121:265–275. doi: 10.1002/ijc.22673. [DOI] [PubMed] [Google Scholar]
- 40.Franco PJ, Farooqui M, Seto E, Wei LN. The orphan nuclear receptor TR2 interacts directly with both class I and class II histone deacetylases. Mol Endocrinol. 2001;15:1318–1328. doi: 10.1210/mend.15.8.0682. [DOI] [PubMed] [Google Scholar]
- 41.Nie J, et al. Ankyrin repeats of ANKRA2 recognize a PxLPxL motif on the 3M syndrome protein CCDC8. Structure. 2015;23:700–712. doi: 10.1016/j.str.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Seo JH, et al. ARD1-mediated Hsp70 acetylation balances stress-induced protein refolding and degradation. Nat Commun. 2016;7:12882. doi: 10.1038/ncomms12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao Y, et al. Involvement of histone deacetylation in MORC2-mediated down-regulation of carbonic anhydrase IX. Nucleic Acids Res. 2010;38:2813–2824. doi: 10.1093/nar/gkq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein BJ, et al. The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 2014;6:325–335. doi: 10.1016/j.celrep.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai RL, Yan-Neale Y, Cueto MA, Xu H, Cohen D. HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J Biol Chem. 2000;275:27909–27916. doi: 10.1074/jbc.M000168200. [DOI] [PubMed] [Google Scholar]
- 46.Weiss K, et al. DDD Study De novo mutations in CHD4, an ATP-dependent chromatin remodeler gene, cause an intellectual disability syndrome with distinctive dysmorphisms. Am J Hum Genet. 2016;99:934–941. doi: 10.1016/j.ajhg.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itoh T, et al. Structural and functional characterization of a cell cycle associated HDAC1/2 complex reveals the structural basis for complex assembly and nucleosome targeting. Nucleic Acids Res. 2015;43:2033–2044. doi: 10.1093/nar/gkv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melhuish TA, Gallo CM, Wotton D. TGIF2 interacts with histone deacetylase 1 and represses transcription. J Biol Chem. 2001;276:32109–32114. doi: 10.1074/jbc.M103377200. [DOI] [PubMed] [Google Scholar]
- 49.Humphrey GW, et al. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 50.Li H, et al. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol Cell Biol. 2000;20:1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao T, Duprez E, Borden KLB, Freemont PS, Etkin LD. Ret finger protein is a normal component of PML nuclear bodies and interacts directly with PML. J Cell Sci. 1998;111:1319–1329. doi: 10.1242/jcs.111.10.1319. [DOI] [PubMed] [Google Scholar]
- 52.Lin H-K, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431:205–211. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- 53.Mimura Y, Takahashi K, Kawata K, Akazawa T, Inoue N. Two-step colocalization of MORC3 with PML nuclear bodies. J Cell Sci. 2010;123:2014–2024. doi: 10.1242/jcs.063586. [DOI] [PubMed] [Google Scholar]
- 54.Lo Y-H, et al. Selective inhibition of the NLRP3 inflammasome by targeting to promyelocytic leukemia protein in mouse and human. Blood. 2013;121:3185–3194. doi: 10.1182/blood-2012-05-432104. [DOI] [PubMed] [Google Scholar]
- 55.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalamvoki M, Roizman B. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci USA. 2014;111:E611–E617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ejercito PM, Kieff ED, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 58.Zhou G, Te D, Roizman B. The CoREST/REST repressor is both necessary and inimical for expression of herpes simplex virus genes. MBio. 2010;2:e00313-10. doi: 10.1128/mBio.00313-10. [DOI] [PMC free article] [PubMed] [Google Scholar]