Significance
Bacteria live in surface-attached communities known as biofilms, where spatial structure is tightly linked to community function. We have developed a genetically encoded biofilm patterning tool (“Biofilm Lithography”) by engineering bacteria such that the expression of membrane adhesion proteins responsible for surface attachment is optically regulated. Accordingly, these bacteria only form biofilm on illuminated surface regions. With this tool, we are able to use blue light to pattern Escherichia coli biofilms with 25 μm spatial resolution. We present an accompanying biophysical model to understand the mechanism behind light-regulated biofilm formation and to provide insight on related natural biofilm processes. Overall, this biofilm patterning tool can be applied to study natural microbial communities as well as to engineer living biomaterials.
Keywords: biofilm, optogenetics, bacterial patterning, lithography, pDawn-Ag43
Abstract
Bacterial biofilms represent a promising opportunity for engineering of microbial communities. However, our ability to control spatial structure in biofilms remains limited. Here we engineer Escherichia coli with a light-activated transcriptional promoter (pDawn) to optically regulate expression of an adhesin gene (Ag43). When illuminated with patterned blue light, long-term viable biofilms with spatial resolution down to 25 μm can be formed on a variety of substrates and inside enclosed culture chambers without the need for surface pretreatment. A biophysical model suggests that the patterning mechanism involves stimulation of transiently surface-adsorbed cells, lending evidence to a previously proposed role of adhesin expression during natural biofilm maturation. Overall, this tool—termed “Biofilm Lithography”—has distinct advantages over existing cell-depositing/patterning methods and provides the ability to grow structured biofilms, with applications toward an improved understanding of natural biofilm communities, as well as the engineering of living biomaterials and bottom–up approaches to microbial consortia design.
Biofilms are surface-attached communities of microbes and represent the predominant mode of life for bacteria on earth (1). While well known for their role in biofouling and infections (2, 3), biofilms can also be harnessed as biotechnological tools, such as those used in wastewater treatment (4). Recent research has highlighted their potential as living biomaterials in applications including the prevention of biofouling (5), nanoparticle templating, protein immobilization, and bioelectricity (6, 7), as well as a promising platform upon which to engineer synthetic microbial communities (8, 9).
A key feature of natural biofilms is distinct spatial patterning coupled to ecological relationships within the microbial community, such as metabolic division of labor between colocalized strains (10). In some cases, this type of structure allows simultaneous biochemical reactions to occur that would be incompatible within single cells (11). Clearly, the full biotechnological capabilities of engineered beneficial biofilms cannot be realized without reliable tools to control biofilm structure.
Such patterning tools should ideally be able to structure stable/viable biofilms with high spatial resolution, in a variety of environments without necessarily requiring substrate pretreatment or directly exposed surfaces. Many techniques exist to pattern cells to surfaces, including (but not limited to) inkjet printing (12, 13), microcontact printing (14, 15), polydimethylsiloxane (PDMS) stenciling (16), patterned substrate modification (17–19), microfluidics (20), photoactivated antibiotic (21), light-switchable adhesion proteins (22), and optogenetic cyclic-di-GMP regulation (23, 24). Optogenetic approaches have also been used to control gene expression on preexisting bacterial lawns (25). However, to our knowledge, no existing method comprehensively fulfills the listed requirements for biofilm patterning.
Here we presenta technique termed “Biofilm Lithography” to structure bacterial biofilms by projecting optical patterns that induce a planktonic-to-biofilm phenotypic switch in engineered bacteria. Our method rests on two key elements: (i) Light-regulated transcriptional elements have been developed for bacteria, such as pDawn, which uses a light–oxygen–voltage domain to regulate gene expression according to blue light illumination (26). (ii) The switch from planktonic to biofilm phenotype in bacteria can be controlled by the expression of membrane proteins that promote cell-substrate attachment, such as antigen 43 (Ag43), a homodimerizing autotransporter adhesin that has been demonstrated to induce both biofilm formation as well as cell–cell adhesion (27, 28). Here we bring Ag43 under the control of pDawn, allowing us to pattern biofilms with light.
Results
pDawn-Ag43 Expression Drives Light-Regulated Biofilm Formation in E. coli.
We postulated that E. coli cells expressing Ag43 from the blue light-responsive promoter pDawn should transition from a planktonic to biofilm phenotype when illuminated by blue light. To test our hypothesis, we designed a construct (termed pDawn-Ag43) where a ribosomal binding site and the Ag43 coding sequence have been inserted downstream of the pDawn transcriptional control element (Fig. 1A). We then transformed pDawn-Ag43 into the MG1655 strain of E. coli, a weak native biofilm former (29) that is known to form biofilm with Ag43 overexpression (28).
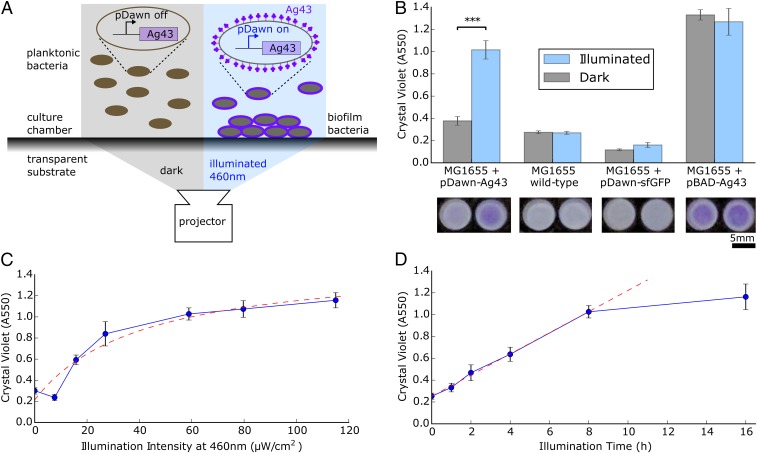
Fig. 1.
pDawn control of the adhesin Ag43 enables light-controlled deposition of biofilm onto surfaces. (A) Ag43 was inserted downstream of pDawn control. Escherichia coli with pDawn-Ag43 can be optically stimulated with a projector to express Ag43 and to form biofilm at surfaces. (B) Cells engineered with the pDawn-Ag43 construct form biofilm contingent upon illumination (***P 0.001). Wild-type and pDawn controls fail to form significant biofilm, and pBAD-Ag43 expression forms biofilm regardless of illumination. Representative 96-well crystal violet stains are shown below. (C) Increasing illumination intensity increases biofilm formation, saturating past ∼41 W/cm2. Dashed curve, fit with Monod model. (D) Increasing illumination time increases biofilm formation up to approximately 8 h. Dashed curve, linear fit. (B–D, error bars represent SD, n = 4 wells.)
We seeded cultures of MG1655/pDawn-Ag43 inside a polystyrene well plate with M63 growth media and used a portable LED projector (Ivation Pro4) to provide blue light illumination during an overnight growth incubation (Fig. 1A). Using crystal violet staining for quantification (30), we found that MG1655 transformed with pDawn-Ag43 formed robust biofilm when grown under illumination, compared with weak biofilm when grown in the dark (Fig. 1B). We further verified using qPCR that light stimulation of MG1655/pDawn-Ag43 increased Ag43 mRNA transcript levels approximately 15-fold (Ag43 mRNA Transcript Levels Measured by qRT-PCR) and that this elevated gene expression did not cause a growth rate defect (MG1655+pDawn-Ag43 Growth Curves: Dark vs. Illuminated).
To verify that the pDawn-regulated Ag43 expression is in fact responsible for this light-dependent phenotype, we ran a series of controls: (i) untransformed MG1655; (ii) MG1655 transformed with pDawn-sfGFP—a pDawn plasmid expressing superfolder green fluorescent protein (sfGFP) instead of Ag43; and (iii) MG1655 transformed with pBAD-Ag43—a plasmid expressing Ag43 under the control of the promoter, induced with 10 mM arabinose (Fig. 1B). As expected, native MG1655 and MG1655 transformed with pDawn-sfGFP both failed to form strong biofilm regardless of dark or illuminated growth conditions. This indicates that at the intensity used (40 W/cm2), blue light illumination on its own is unable to stimulate biofilm growth in MG1655, even with stimulation of the YF1/fixJ two-component sensor involved in pDawn. We verified that pDawn works as expected in MG1655/pDawn-sfGFP by confirming sfGFP expression (pDawn-sfGFP Control). On the other hand, MG1655 transformed with pBAD-Ag43 induced with arabinose formed biofilm regardless of illumination intensity, indicating that Ag43 expression is sufficient to induce biofilm formation. We conclude that pDawn-regulated expression of Ag43 is responsible for the light-dependent biofilm phenotype in MG1655/pDawn-Ag43.
Biofilm Formation Can Be Tuned by Illumination Intensity and Time.
We characterized the influence of illumination intensity and time on the extent of biofilm formation. By stimulating cells overnight for 16 h across a range of intensities from 0 to 115 W/cm2, we found that biofilm formation increases with brighter illumination in a saturating manner with a characteristic illumination intensity constant of 41 W/cm2 when fit with the Monod equation (Fig. 1C; see fit/model choice discussion in Fitting Biofilm Formation vs. Illumination Intensity/Time). When illuminating with 40 W/cm2, we found a linear increasing relationship between biofilm formation and illumination time up to ∼8 h, beyond which biofilm formation appears to slow down (Fig. 1D). For the remainder of this paper, we use standard illumination conditions of 40 W/cm2 for 16 h unless stated otherwise.
Biofilm Can Be Patterned Using Light in Various Environments.
Next, we developed a protocol to form and visualize patterned biofilms based on pDawn-Ag43. We cotransformed MG1655/pDawn-Ag43 with a red fluorescent protein (mRFP) expression plasmid. Applying the same culture conditions as before, we set up the projector using Microsoft Powerpoint to illuminate various patterns (e.g., stripes, polka dots, pictures) on bacterial cultures in polystyrene wells and subsequently verified that the patterns were recapitulated as bacterial biofilms using fluorescence microscopy (Fig. 2A).
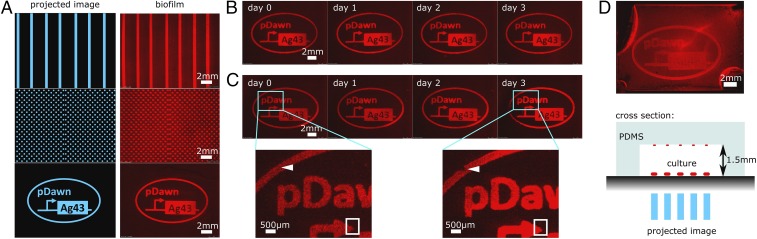
Fig. 2.
Biofilm Lithography enables the structured patterning of viable biofilms over multiple days and inside closed-culture chambers. (A) Various illumination images (stripes, polka dots, pictures) are recapitulated as biofilm patterns; patterns (shown on Left for reference) were illuminated from below using a projector (Fig. 1A). (B) Patterned biofilms remain stable for at least 3 d under ambient conditions in PBS buffer. (C) In culture media, growth and eventual detachment of viable biofilm can be observed over 3 d. Growth regions with increased mRFP signal and instances of biofilm detachment are marked by white rectangles and triangles, respectively. (D) Biofilms can be patterned with light inside sealed, transparent culture chambers, without requiring direct physical access to the substrate. Image contrast appears reduced compared with B and C due to a secondary biofilm image that forms at the PDMS ceiling, and planktonic bacteria were harder to wash out from the chamber without perturbing the patterned biofilm on the polystyrene.
We next investigated the long-term stability of the patterned biofilms. After overnight incubation, samples were removed from optical illumination and rinsed with PBS, and subsequent daily imaging demonstrated that the biofilm pattern remained stable over a period of 3 d in PBS (Fig. 2B). When biofilm was maintained in M63 growth medium over the same time span, we observed growth and shedding of viable biofilm (Fig. 2C), analogous to expansion and dispersal processes in natural biofilms (29). Using qPCR, we confirmed that Ag43 mRNA levels remain stable over the course of these long-term experiments (Ag43 mRNA Transcript Levels Measured by qRT-PCR).
Next, we tested this tool’s ability to pattern other materials besides polystyrene. Using glass coverslips and PDMS coupons placed into well-plates, we confirmed that our engineered cells are able to form patterned biofilms on both glass and PDMS (Patterned Biofilm Formation on Other Surfaces). For all tested substrates, no surface pretreatment or patterning was required.
Furthermore, we tested whether our method works on samples inside completely enclosed environments. This is in contrast to the requirement of direct surface access for other patterning methods such as inkjet-based printers or microcontact printing (12–15). We used molded PDMS cavities (31) bonded to polystyrene to create enclosed culture chambers with dimensions 19 mm × 13 mm × 1.5 mm (Fig. 2D). Bacteria were cultured in this chamber using the same illumination conditions as before. We found that patterned biofilms form as expected on the polystyrene substrate, with a faint secondary biofilm image present on the PDMS ceiling (Fig. 2D). Hence, our method can pattern biofilm inside enclosed environments.
Biofilms Can Be Optically Patterned with 25 m Resolution.
To more quantitatively characterize pDawn-Ag43–mediated biofilm patterning, we collected volumetric biofilm data via confocal laser scanning microscopy (Fig. 3A) and measured the biofilm’s average thickness to be 14.4 m (Quantifying Biofilm Thickness and Density from Confocal Biofilm Images). Assuming an average E. coli cell volume of 1 m3 (32) and that cells constitute 10 of total biofilm volume (33), this approximately corresponds to an average thickness of seven cells and a surface density of 1.4 × 106 cells per mm2. From this, we can estimate an average biofilm deposition rate on the order of 25 cells per mm2s over the course of a 16 h incubation (Quantifying Biofilm Thickness and Density from Confocal Biofilm Images).
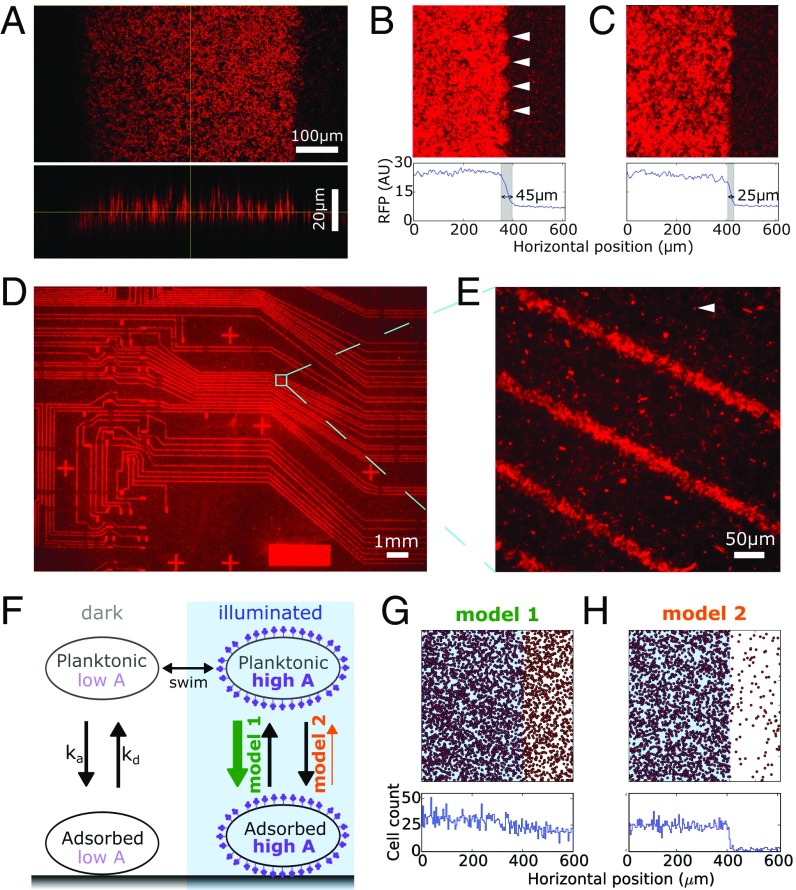
Fig. 3.
Biofilm Lithography enables patterning with a lateral resolution down to 25 m, which is explained by transient, light-independent cell–surface attachment followed by light-activated adhesin expression. (A) Confocal microscopy reveals average biofilm thickness of 15 m, surface roughness coefficient of 0.33, and characteristic roughness length scale of 5.4 m. (B) Step-response analysis across light–dark boundary with high-resolution microscopy of striped illumination sample indicates m spatial resolution; white triangles point to artifacts due to projector pixels. (C) Using electrical tape as field stop (as opposed to a projector) enables a spatial resolution of m. (D) Biofilm patterned over large area and at high resolution are possible with photomask (shown example was originally designed for patterning microfluidic channels). (E) Higher magnification imaging of D confirms feature sizes of m; white triangle points to an example of a spurious individual cell. (F) Schematic of Monte-Carlo simulation with cell swimming, adsorption/desorption, and light-regulated levels of adhesin . In model 1, adhesin level is directly proportional to the adsorption rate of planktonic cells, whereas in model 2, adhesin level is inversely proportional to the desorption rate of adsorbed cells. (G) No clear biofilm boundary can be observed at the light–dark border in model 1 where optically regulated adhesin production increases adsorption rate . (H) Clear biofilm boundary can be observed in model 2 where optically regulated adhesin production decreases desorption rate . (For G and H, adsorbed cells are labeled red, illuminated region is marked in blue, and histogram of cell positions plotted below.)
The confocal images also reveal that the biofilm surface is not smooth, and we determine a surface roughness coefficient (34) of ∼0.33 (Quantifying Biofilm Roughness from Confocal Biofilm Images). This is in general agreement with the value of 0.31 estimated by assuming biofilm deposition to be a purely Poisson process (Quantifying Biofilm Roughness from Confocal Biofilm Images). Using autocorrelation analysis, we derive an approximate length scale for the surface roughness on the order of 5.7 m (Quantifying Biofilm Roughness from Confocal Biofilm Images). We speculate that clustering on this length scale may be a result of cell division and intercellular Ag43 homodimerization leading to a different effective affinity for cell–cell vs. cell–surface binding.
Next, we determined the spatial resolution (smallest feature size) that could be patterned with this method by measuring the step response of the red fluorescence signal across a light–dark illumination boundary. Using the striped illumination sample (Fig. 2A, Top), we estimate resolution of ∼45 m (Fig. 3B and Determining Width of Transition Region Between Dark and Illuminated Regions). This is on the order of the optical resolution limit of the projector setup, which has a pixel–pixel distance of ∼80 m; the effect is visible in corresponding periodic structural artifacts at this length scale (Fig. 3B, white triangles). To improve this resolution, we applied electrical tape to the bottom of the well plate to act as a field stop and repeated the step-response analysis across the tape boundary. Based on this analysis, we estimate the spatial resolution to be 25 m, measured as the width of the transition region across the light–dark boundary (Fig. 3C and Determining Width of Transition Region Between Dark and Illuminated Regions).
To validate this point further, we taped a film photomask [originally designed for microfluidic circuit fabrication (35)] to the bottom of the culture chamber. This structure was faithfully recapitulated over the area of multiple mm2 (Fig. 3D) with feature sizes at the scale of 25 m (Fig. 3E and Profile Plot of High-Resolution Biofilm Patterning). By comparing the peak RFP intensity in this image to basal RFP expression in nonilluminated regions, we estimate a contrast of 3.3 (Profile Plot of High-Resolution Biofilm Patterning). We noted that the feature sizes of the final patterned biofilm are reduced compared with the underlying photomask—that is, 25 m-wide stripes were observed in the biofilm corresponding to 75 m transparent stripes in the photomask (Images of Photomask). This reduction is roughly consistent with the 25 m light–dark transition region measured earlier and suggests that the transition region lies on the illuminated side of the light–dark boundary. We also noted spurious cells attached to the surface (Fig. 3E, white triangle), which along with the transition region and surface roughness length scales constitute a spatial resolution limit for Biofilm Lithography as currently implemented.
Biophysical Modeling of Biofilm Formation Explains High Spatial Resolution.
Finally, we sought to understand mechanistically how MG1655/pDawn-Ag43 biofilm patterning can achieve high spatial resolution across illumination boundaries, given the inherent motility of the MG1655 strain.
Our initial working hypothesis (model 1, Fig. 3F) was that pDawn-Ag43 works by optically regulating Ag43 expression in freely swimming planktonic cells. The expression of surface appendages on cell membranes can help overcome cell-substrate electrostatic repulsion (36–38), which could lead to increased adsorption rate and higher levels of biofilm formation in illuminated regions. However, a back-of-the-envelope calculation suggests that this model is inconsistent with the observed 25 m spatial resolution. Motile E. coli have an effective diffusivity due to motility of /s (39), while a very conservative lower bound on pDawn-Ag43 stimulation delay based only on gene expression time can be estimated at (40–42). Bacterial swimming during the time delay between stimulation and attachment would then blur features below a length scale of at least . A realistic estimate for spatial resolution would be even larger due to additional delays related to pDawn-signaling and protein accumulation/export, suggesting that model 1 is inconsistent with the observed spatial resolution given cell motility (Estimating Spatial Resolution Limits Due to Diffusion).
For another hypothesis (model 2, Fig. 3F), we note that pDawn-Ag43–mediated patterning occurs within the context of other biofilm-related processes—in particular, bacteria at a liquid–solid interface naturally exhibit reversible adsorption and desorption (43), switching between planktonic and transiently adsorbed subpopulations. Instead of only stimulating the planktonic subpopulation (by increasing their adsorption, as discussed above), pDawn-Ag43 could also have a significant effect on the desorption rate of transiently adsorbed cells. Left unstimulated in the dark, these cells readily desorb from the surface (43), but in illuminated regions, membrane-expressed Ag43 reduces the desorption rate of adsorbed cells, effectively anchoring them to the surface (38). Model 2 is consistent with high spatial resolution patterning that is not limited by bacterial motility since the adsorbed cells are immotile.
To quantitatively confirm these models, we developed a Monte-Carlo simulation of cell swimming and adsorption/desorption (Fig. 3F; see also Monte-Carlo Modeling). Individual cells are simulated to swim in a virtual culture chamber, one side of which is “illuminated,” causing increased production of adhesin . Bacterial cells adsorb and desorb from the surface with respective rates and , which are functions of their adhesin expression level . We simulated model 1 by setting constant and an increasing function of and observed no clear transition at the light–dark boundary (Fig. 3G). We simulated model 2 by setting constant and a decreasing function of and observed a clear increase in adsorbed cells in the illuminated region, with a sharp transition across the light–dark boundary (Fig. 3H). Notably, this model did not need to incorporate additional features such as increased adsorption, 3D cell–cell interactions, or a permanently attached cell state. Therefore, we establish that a biophysical model with just two key features, (i) optically controlled adhesin expression and (ii) adhesin decreasing desorption rate of adsorbed cells, is sufficient to explain light-regulated biofilm formation with high spatial resolution despite bacterial motility. Additionally, the light–dark transition region in the simulations is less than 1 m (Higher Resolution Analysis of Light–Dark Boundary in Monte-Carlo Simulations)—smaller than the experimentally observed 25 m transition region. This discrepancy points to unaccounted-for experimental sources of noise such as colony growth/clustering, optical scattering, and 3D effects (e.g., cell stacking), which may inform future experimental strategies to improve spatial resolution even further.
Discussion
In conclusion, we have developed a method (Biofilm Lithography) that uses light-regulated adhesin expression (pDawn-Ag43) to quantitatively control biofilm formation and patterning with high spatial resolution. Compared with existing cell deposition and patterning approaches such as inkjet printing (12, 13), microcontact printing (14, 15), microfluidics (20), PDMS stenciling (16), and patterned substrate modification (17–19), this method can pattern on a variety of surfaces, without the need for surface prepatterning or pretreatment, within enclosed chambers, over large areas, and at high spatial resolution. Rapid prototyping of different biofilm geometries is possible with low-cost digital projectors at resolutions of 45 m; resolutions down to 25 m can be reached with photomasks and likely also with more advanced optical projector setups (44, 45). This resolution represents an important step toward the engineering of biofilm communities, as natural biofilm microcolonies exist around this length scale (10). Our biophysical model suggests that the pDawn-Ag43 patterning mechanism works alongside natural surface adsorption/desorption in bacteria and involves the stimulation of transiently adsorbed cells on the biofilm substrate toward a more permanently attached state. This insight gives additional support to the proposed role of Ag43 as an adhesin involved in biofilm maturation as opposed to initial surface adsorption (46). Ultimately, optogenetic patterning tools such as pDawn-Ag43 can be applied toward an improved understanding of naturally existing biofilms (47, 48), the design of synthetic microbial consortia (8), distributed metabolic engineering (49), and new types of integrated diagnostic and microfluidic devices (50), with impact and trajectory that may potentially parallel that of silicon photolithography in the semiconductor industry (51, 52).
Materials and Methods
Plasmids and Bacterial Strains.
MG1655 was obtained from the Coli Genetic Stock Center (Yale University, New Haven, CT; CGSC strain ). pDawn-Ag43 was constructed using standard cloning techniques using pDawn (a gift from Andreas Moeglich, Universität Bayreuth, Bayreuth, Germany; Addgene Plasmid ) plasmid as a starting point. First, Gibson Assembly was used to swap out the kanamycin marker for a less commonly used spectinomycin resistance marker. BamHI/XhoI restriction digest was then used to linearize the resulting plasmid at the multiple cloning site downstream of the promoter to create the backbone for pDawn-Ag43.
The coding sequence for Ag43 was obtained from the BioBricks iGEM distribution, part number (iGEM Foundation, Cambridge, MA). We noted during construction that several PstI restriction enzyme sites remained in the coding sequence, making the part incompatible with the BioBrick Standard. Using a combination of site-directed mutagenesis and DNA synthesis (IDT gBlock synthesis), these PstI sites were removed and replaced with silent mutations so as to not alter the final amino acid sequence. Using this now BioBrick-compatible part, we used standard BioBrick prefix/suffix assembly to add a medium strength ribosomal binding site ( rbs) upstream of the Ag43 coding sequence. We then used BamHI/XhoI digestion/ligation to insert rbs-Ag43 into the backbone prepared earlier to create pDawn-Ag43.
A similar protocol was used to create pDawn-sfGFP, with a superfolderGFP coding sequence used in place of Ag43. pBAD-Ag43 was created by using standard BioBrick prefix/suffix assembly to insert the Ag43 coding sequence downstream of an araC–pBAD expression vector (BioBricks part ). Sequences for pDawn-sfGFP, pDawn-Ag43, and pBAD-Ag43 are available from Addgene.
The plasmid for red fluorescent protein expression was obtained from the BioBricks iGEM distribution and expresses mRFP from the Lac promoter ().
Biofilm Formation and Patterning.
E. coli strains were cultured to late log phase in LB broth under dark conditions (OD600 1.4, ∼6 h with shaking at 37 °C after 1:1,000 dilution of overnight culture). Media was supplemented with antibiotics as appropriate (50 g/mL for kanamycin and spectinomycin, 10 g/mL for tetracycline, and 100 g/mL for ampicillin). These cultures were then seeded onto non-tissue culture-treated polystyrene well plates at 1:100 dilution into M63 media supplemented with glucose and casamino acids.
For assays characterizing biofilm formation, 96-well black-walled plates (Corning Costar 3631) were used. Patterning assays were performed in 6- or 12-well plates (Corning Falcon 351146/351143).
Well plates containing the biofilm cultures were taped to the ceiling of a 37 °C incubator, ensuring the bottom surface of the well plates remained uncovered. An Ivation Pro4 Wireless Pocket Projector (IVPJPRO4) was secured below the ceiling of the incubator, pointing upward toward the well plate on the incubator ceiling. The projector was connected via HDMI cable to a laptop through the incubator’s side access port, and Microsoft PowerPoint software was used to project blue light patterns.
Global illumination intensity was tuned by placing an adjustable neutral density filter ( concept AMSKU0124) at the aperture of the projector. Local illumination intensity was further tuned by taping thin neutral density filters from the Lee’s Filters Designer’s Edition Swatchbook (Lee’s Filters part SWB) to the bottom surface of the well plates over specific wells to subject bacterial samples to a wide range of illumination intensities. Intensity of the projected pattern was measured using a Newport optical power meter with UV-vis photodetector (Newport 840C/818-UV). Illumination time was adjusted on the software end through Microsoft PowerPoint.
For biofilm characterization experiments, biofilm cultures were placed in the incubator overnight (16 h). Media was subsequently aspirated, and wells were gently washed twice with PBS. Wells were then stained with 0.1 crystal violet (Acros Organics 212120250) for 10 min, before another 2× wash with PBS. Wells were then allowed to dry before imaging, followed by A550 nm quantification using 30 acetic acid solubilization as previously detailed (30).
To prepare samples for fluorescence microscopy, biofilm cultures were prepared using bacteria cotransformed with the plasmid for mRFP expression. To pattern optical illumination for biofilm patterning experiments, various patterns were generated on the software end as PowerPoint illustrations, or alternatively, a film photomask was taped directly under the well plate. Cultures were incubated with patterned illumination as described above and rinsed twice with PBS before imaging under a wide-field fluorescence microscope. For the long-term culture experiments in PBS, cultures were prepared and patterned overnight, before media was aspirated, followed by PBS rinse. The sample was then imaged under a wide-field fluorescence microscope and left in PBS under ambient dark conditions for 3 d with daily imaging. The same protocol was used for long culture experiments in media, except cells were maintained in M63 media with daily PBS rinse.
qRT-PCR.
See Ag43 mRNA Transcript Levels Measured by qRT-PCR for details.
Confocal Microscopy.
To prepare cultures for confocal microscopy, a drop of self-hardening Shandon immunomount (Thermo Scientific 9990402) was dropped onto the cultured sample, before being covered with a glass coverslip (FisherScientific 12-545-81). The sample was then allowed to harden overnight at room temperature in the dark. The following day, the samples were imaged through the glass coverslip using a Leica Upright confocal microscope (Leica DMRXE), using a 20×/0.50 water immersion objective with an excitation line at 543 nm for mRFP.
Monte-Carlo Modeling.
The Monte-Carlo bacterial adhesion simulation was implemented in MATLAB using a forward Euler numerical approach, time discretized in dt = 100 ms timesteps (repeats run with dt = 50 ms timesteps produced qualitatively identical results). Simulations were run for 16 h over an area of (600 μm)2. Within this area, a 400 m-wide stripe on the left is illuminated with blue light. Bacterial cells were initialized with a random position and velocity direction, with velocity magnitude in the range m/s (39). Cells were also initialized with a basal adhesin level of . Adhesin level is regulated by an ordinary differential equation.
| [1] |
where I is a boolean variable representing whether a cell is being illuminated or not (calculated based on cell position). and represent the basal and light-activated production rates of adhesin protein, respectively. These proteins are in turn degraded at a rate . The parameter values s−1, s−1, and s−1 were used so that in the dark cells revert to their original basal adhesin level with characteristic protein turnover time on the order of tens of minutes (53), and in the light, they increase their expression by two orders of magnitude (from to )—approximately the reported dynamic range of pDawn (26). This ordinary differential equation was numerically solved within the same forward Euler loop as the overall simulation.
Simultaneously during each simulation time step, cell positions were updated based on velocity, and cells tumbled with a probability of based on a tumbling frequency of s−1 (39). During a tumbling event, a cell’s velocity vector was reoriented randomly. Also at each time step, planktonic cells were adsorbed to the surface with a probability , while adsorbed cells were desorbed with a probability . The rates of adsorption and desorption were dictated by adhesin level .
In the first model, where cell adsorption is increased upon illumination, the relationships were set as s and s−1. In the second model, where cell desorption was decreased upon illumination, the relationships were set as s−1 and s−1 /A. The basal values for adsorption and desorption rate, s−1 and s−1, were derived from quantitative bacterial adsorption/desorption time measurements (43) and approximately correspond to planktonic cells adsorbing every few hours to the surface, after which they natively remain adsorbed for a few minutes if not stimulated, or a few hours if stimulated. Simulations were run for 576,000 timesteps (16 h real time). The final state of the simulation was plotted with red dots marking the position of the attached cells.
Supplementary Material
Acknowledgments
The authors thank D. Glass, H. Kim, A. Spormann, D. Endy, M. Covert, N. Cira, A. Choksi, S. Rajan, and A. Dvorak for helpful suggestions and the Spormann laboratory for access to its confocal microscope and furthermore acknowledge the support from Stanford Bio-X Bowes and Natural Sciences and Engineering Research Council of Canada-Postgraduate Scholarship fellowships and the American Cancer Society (RSG-14-177-01).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in Addgene [ID codes 107741 (pBAD-Ag43), 107742 (pDawn-Ag43), and 107743 (pDawn-sfGFP)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720676115/-/DCSupplemental.
References
- 1.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 2.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 3.Flemming HC. Biofouling in water systems–Cases, causes and countermeasures. Appl Microbiol Biotechnol. 2002;59:629–640. doi: 10.1007/s00253-002-1066-9. [DOI] [PubMed] [Google Scholar]
- 4.Grady CPL, Daigger GT, Love NG, Filipe CDM. Biological Wastewater Treatment. 3rd Ed CRC Press; Boca Raton, FL: 2011. [Google Scholar]
- 5.Wood TL, et al. Living biofouling-resistant membranes as a model for the beneficial use of engineered biofilms. Proc Natl Acad Sci USA. 2016;113:E2802–E2811. doi: 10.1073/pnas.1521731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen AY, et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat Mater. 2014;13:515–523. doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen PQ, Botyanszki Z, Tay PKR, Joshi NS. Programmable biofilm-based materials from engineered curli nanofibres. Nat Commun. 2014;5:4945. doi: 10.1038/ncomms5945. [DOI] [PubMed] [Google Scholar]
- 8.Brenner K, You L, Arnold FH. Engineering microbial consortia: A new frontier in synthetic biology. Trends Biotechnol. 2008;26:483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Mee MT, Wang HH. Engineering ecosystems and synthetic ecologies. Mol BioSyst. 2012;8:2470–2483. doi: 10.1039/c2mb25133g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen BB, Haagensen JA, Heydorn A, Molin S. Metabolic commensalism and competition in a two-species microbial consortium. Appl Environ Microbiol. 2002;68:2495–2502. doi: 10.1128/AEM.68.5.2495-2502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terada A, Hibiya K, Nagai J, Tsuneda S, Hirata A. Nitrogen removal characteristics and biofilm analysis of a membrane-aerated biofilm reactor applicable to high-strength nitrogenous wastewater treatment. J Biosci Bioeng. 2003;95:170–178. doi: 10.1016/s1389-1723(03)80124-x. [DOI] [PubMed] [Google Scholar]
- 12.Xu T, et al. Construction of high-density bacterial colony arrays and patterns by the ink-jet method. Biotechnol Bioeng. 2004;85:29–33. doi: 10.1002/bit.10768. [DOI] [PubMed] [Google Scholar]
- 13.Merrin J, Leibler S, Chuang JS. Printing multistrain bacterial patterns with a piezoelectric inkjet printer. PLoS One. 2007;2:e663. doi: 10.1371/journal.pone.0000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerf A, Cau JC, Vieu C. Controlled assembly of bacteria on chemical patterns using soft lithography. Colloids Surf B Biointerfaces. 2008;65:285–291. doi: 10.1016/j.colsurfb.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 15.Xu L, et al. Microcontact printing of living bacteria arrays with cellular resolution. Nano Lett. 2007;7:2068–2072. doi: 10.1021/nl070983z. [DOI] [PubMed] [Google Scholar]
- 16.Eun YJ, Weibel DB. Fabrication of microbial biofilm arrays by geometric control of cell adhesion. Langmuir. 2009;25:4643–4654. doi: 10.1021/la803985a. [DOI] [PubMed] [Google Scholar]
- 17.Palacios-Cuesta M, Cortajarena AL, Garcia O, Rodriguez-Hernandez J. Patterning of individual Staphylococcus aureus bacteria onto photogenerated polymeric surface structures. Polym Chem. 2015;6:2677–2684. [Google Scholar]
- 18.Suo Z, Avci R, Yang X, Pascual DW. Efficient immobilization and patterning of live bacterial cells. Langmuir. 2008;24:4161–4167. doi: 10.1021/la7038653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu H, Hou S, Yongyat C, Detore S, Ren D. Patterned biofilm formation reveals a mechanism for structural heterogeneity in bacterial biofilms. Langmuir. 2013;29:11145–11153. doi: 10.1021/la402608z. [DOI] [PubMed] [Google Scholar]
- 20.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA. 2008;105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velema WA, van der Berg JP, Szymanski W, Driessen AJM, Feringa BL. Bacterial patterning controlled by light exposure. Org Biomol Chem. 2014;13:1639–1642. doi: 10.1039/c4ob02483d. [DOI] [PubMed] [Google Scholar]
- 22.Chen F, Wegner SV. Blue light switchable bacterial adhesion as a key step towards the design of biofilms. ACS Synth Biol. 2017;6:2170–2174. doi: 10.1021/acssynbio.7b00197. [DOI] [PubMed] [Google Scholar]
- 23.Ryu MH, Fomicheva A, Moskvin OV, Gomelsky M. Optogenetic module for dichromatic control of c-di-GMP signaling. J Bacteriol. 2017;199:e00014-17. doi: 10.1128/JB.00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pu L, Yang S, Xia A, Jin F. Optogenetics manipulation enables prevention of biofilm formation of engineered Pseudomonas aeruginosa on surfaces. ACS Synth Biol. 2017;7:200–208. doi: 10.1021/acssynbio.7b00273. [DOI] [PubMed] [Google Scholar]
- 25.Levskaya A, et al. Synthetic biology: Engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 26.Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Möglich A. From dusk till dawn: One-plasmid systems for light-regulated gene expression. J Mol Biol. 2012;416:534–542. doi: 10.1016/j.jmb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Klemm P, Hjerrild L, Gjermansen M, Schembri MA. Structure-function analysis of the self-recognizing Antigen 43 autotransporter protein from Escherichia coli. Mol Microbiol. 2004;51:283–296. doi: 10.1046/j.1365-2958.2003.03833.x. [DOI] [PubMed] [Google Scholar]
- 28.Da Re S, Le Quéré B, Ghigo JM, Beloin C. Tight modulation of Escherichia coli bacterial biofilm formation through controlled expression of adhesion factors. Appl Environ Microbiol. 2007;73:3391–3403. doi: 10.1128/AEM.02625-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reisner A, Haagensen JAJ, Schembri MA, Zechner EL, Molin S. Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol. 2003;48:933–946. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- 30.O’Toole GA. Microtiter dish biofilm formation assay. J Visualized Exp. 2011:e2437. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia Y, Whitesides GM. Soft lithography. Annu Rev Mater Sci. 1998;28:153–184. [Google Scholar]
- 32.Phillips R, Kondev J, Theriot J, Orme N. Physical Biology of the Cell. Garland Science; New York: 2013. [Google Scholar]
- 33.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–33. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 34.Murga R, Stewart PS, Daly D. Quantitative analysis of biofilm thickness variability. Biotechnol Bioeng. 1995;45:503–510. doi: 10.1002/bit.260450607. [DOI] [PubMed] [Google Scholar]
- 35.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 36.Donlan RM. Biofilms: Microbial life on surfaces. Emerging Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunne WM., Jr Bacterial adhesion: Seen any good biofilms lately? Clin Microbiol Rev. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berg HC, Brown DA. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 40.Henderson IR, Owen P. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving dam and OxyR. J Bacteriol. 1999;181:2132–2141. doi: 10.1128/jb.181.7.2132-2141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel U, Jensen KF. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J Bacteriol. 1994;176:2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young R, Bremer H. Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem J. 1976;160:185–194. doi: 10.1042/bj1600185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meinders JM, Van der Mei HC, Busscher HJ. Physicochemical aspects of deposition of Streptococcus thermophilus B to hydrophobic and hydrophilic substrata in a parallel plate flow chamber. J Colloid Interf Sci. 1994;164:355–363. [Google Scholar]
- 44.Stirman JN, Crane MM, Husson SJ, Gottschalk A, Lu H. A multispectral optical illumination system with precise spatiotemporal control for the manipulation of optogenetic reagents. Nat Protoc. 2012;7:207–220. doi: 10.1038/nprot.2011.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SA, et al. Proceedings of the 2015 CHI Conference on Human Factors in Computing Systems. ACM; New York: 2015. Trap it! A playful human-biology interaction for a museum installation; pp. 2593–2602. [Google Scholar]
- 46.Beloin C, Roux A, Ghigo JM. Escherichia coli biofilms. Curr Top Microbiol Immunol. 2008;322:249–289. doi: 10.1007/978-3-540-75418-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kreft JU. Biofilms promote altruism. Microbiology. 2004;150:2751–2760. doi: 10.1099/mic.0.26829-0. [DOI] [PubMed] [Google Scholar]
- 48.Rudge TJ, Steiner PJ, Phillips A, Haseloff J. Computational modeling of synthetic microbial biofilms. ACS Synth Biol. 2012;1:345–352. doi: 10.1021/sb300031n. [DOI] [PubMed] [Google Scholar]
- 49.Chen AH, Silver PA. Designing biological compartmentalization. Trends Cell Biol. 2012;22:662–670. doi: 10.1016/j.tcb.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 51.Burbank DP. The near impossibility of making a microchip. Am Heritage Invent Technol. 1999;15:44–53. [Google Scholar]
- 52.Garner CM. Lithography for enabling advances in integrated circuits and devices. Phil Trans A Math Phys Eng Soc. 2012;370:4015–4041. doi: 10.1098/rsta.2011.0052. [DOI] [PubMed] [Google Scholar]
- 53.Nath K, Koch AL. Protein degradation in Escherichia coli. I. Measurement of rapidly and slowly decaying components. J Biol Chem. 1970;245:2889–2900. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.