Significance
RNAs contain over 100 types of chemical modifications, and N6-methyladenosine (m6A) is the most common internal modification in eukaryotic mRNA. m6A is involved in a variety of important biological processes, including sex determination in Drosophila, by modifying Sxl pre-mRNA and regulating its alternative splicing. m6A is installed by a large methyltransferase complex called the m6A “writer.” We have identified xio as a component of the Drosophila sex determination pathway based on its female-to-male transformation phenotypes. Xio interacts with other m6A writer subunits, and its loss of function shows typical phenotypes associated with other m6A factors, such as Sxl splicing misregulation, adult defects, and reduced m6A levels. Therefore, we conclude that Xio is a member of the m6A writer complex.
Keywords: CG7358, Xio, sex determination, m6A writer complex, alternative splicing
Abstract
N6-methyladenosine (m6A), the most abundant chemical modification in eukaryotic mRNA, has been implicated in Drosophila sex determination by modifying Sex-lethal (Sxl) pre-mRNA and facilitating its alternative splicing. Here, we identify a sex determination gene, CG7358, and rename it xio according to its loss-of-function female-to-male transformation phenotype. xio encodes a conserved ubiquitous nuclear protein of unknown function. We show that Xio colocalizes and interacts with all previously known m6A writer complex subunits (METTL3, METTL14, Fl(2)d/WTAP, Vir/KIAA1429, and Nito/Rbm15) and that loss of xio is associated with phenotypes that resemble other m6A factors, such as sexual transformations, Sxl splicing defect, held-out wings, flightless flies, and reduction of m6A levels. Thus, Xio encodes a member of the m6A methyltransferase complex involved in mRNA modification. Since its ortholog ZC3H13 (or KIAA0853) also associates with several m6A writer factors, the function of Xio in the m6A pathway is likely evolutionarily conserved.
Sex determination is one of the most fundamental problems in biology and affects all aspects of life, such as morphology, metabolism, aging, and behavior (1). For more than 90 y, Drosophila has remained a major model organism to study sex determination genes and mechanisms (2). Similar to humans, Drosophila males have XY chromosomes, and females have XX chromosomes. Sex-lethal (Sxl), the master regulatory gene in the Drosophila sex determination pathway, is activated in females by the X-chromosome counting system while it is not expressed in males. Once activated, Sxl maintains its own expression by controlling the alternative splicing of its own pre-mRNA. Sxl also regulates the alternative splicing of the downstream gene transformer (tra), which, together with transformer2 (tra2), controls the alternative splicing of doublesex (dsx) and fruitless (fru), generating male- and female-specific transcription factors (3). In addition, Sxl prevents the activation of the male-specific dosage compensation system by repressing male-specific lethal 2 (msl-2) at the level of splicing and translational control (4).
In addition to these genes, three factors encoded by female-lethal-2-d (fl(2)d), virilizer (vir), and spenito (nito) have been shown to be involved in the sex determination pathway and to be required for Sxl alternative splicing regulation (5–9). Recently, Fl(2)d, Vir, and Nito were shown to encode components of the N6-methyladenosine (m6A) methyltransferase complex, revealing that the m6A pathway modulates sex determination in Drosophila (10–12). m6A is the most abundant chemical modification in mRNA, and its level is dynamically regulated (13). m6A pathway factors include the methyltransferase complex (or writers), demethylases (or erasers), and readers. Both in Drosophila and mammals, known writer complex subunits include METTL3 (or Ime4) (14), METTL14, Fl(2)d (WTAP), Vir (KIAA1429), and Nito (Rbm15/15B) (15). These writer components, as well as the reader YT521-B, are required for Drosophila sex determination and Sxl splicing regulation. Further, m6A modification sites have been mapped to Sxl introns, thus facilitating Sxl pre-mRNA alternative splicing. Importantly, m6A methylation is required in human dosage compensation by modifying the long noncoding RNA XIST, suggesting that m6A-mediated gene regulation is an ancient mechanism for sex determination (16).
Recent emerging studies suggest that m6A is involved in numerous key biological processes, such as development, disease, stem cell differentiation, immunity, and behavior, by controlling various aspects of RNA metabolism, such as splicing, stability, folding, export, and translation (17). Although many m6A methylated mRNAs have been identified, Sxl pre-mRNA is arguably one of the best understood examples for m6A modification and is useful for mechanistic studies. Importantly, Drosophila sex determination provides a unique system to screen for new components as all previously identified writers and readers show unambiguous sex transformation phenotypes (18).
Here, we identified a component in the Drosophila sex determination pathway as well as m6A modification pathway. As this gene, CG7358, has not been studied before, we named it Xiong (Xio, Chinese character for maleness) since its loss of function shows female-to-male transformations. We demonstrate that Xio interacts with other methyltransferase factors and that loss of Xio activity phenocopies other methyltransferase mutants in terms of Sxl splicing regulation and adult defects. Altogether, our study identifies and characterizes a conserved component of the m6A writer complex.
Results
Xio Interacts and Colocalizes with Known m6A Writer Complex Subunits.
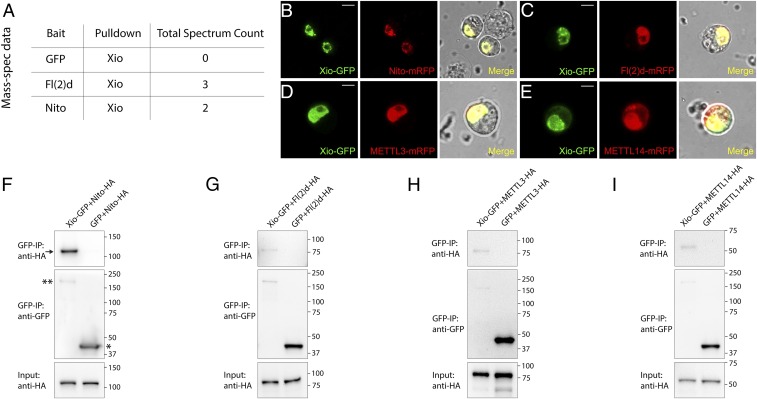
From the Drosophila protein–protein interaction database (DPiM), we identified one interesting protein, CG7358/Xio, which, as a bait, can pull down Nito, Vir, and Fl(2)d in affinity purification and mass spectrometry (mass-spec) experiments (Fig. S1) (19). Similarly, in our own mass-spec studies, Nito or Fl(2)d as a bait can reciprocally pull down Xio (Fig. 1A and Dataset S1). As these proteins are core components of the RNA m6A methyltransferase complex, we hypothesized that Xio is a new factor of the m6A writer complex. To confirm these interactions, we performed both colocalization and coimmunoprecipitation (co-IP) experiments. Xio-GFP localized to nuclei in live S2 cells and colocalized with Nito-mRFP, Fl(2)d-mRFP, METTL3-mRFP, and METTL14-mRFP (Fig. 1 B–E). Next, we transfected Xio-GFP and different HA-tagged constructs in S2 cells and used GFP alone as a control. Although GFP was expressed at a much higher amount, only Xio-GFP was able to pull down Nito-HA, Fl(2)d-HA, METTL3-HA, and METTL14-HA (Fig. 1 F–I). Interestingly, the pull-down between Xio and Nito is particularly strong compared with other factors, suggesting that Xio may directly interact with Nito, which is consistent with the published mass-spec data where most Nito peptides were pulled down using Xio as a bait (Fig. S1B). Altogether, these data suggest that Xio is a new component of the m6A writer complex.
Fig. 1.
Xio colocalizes and interacts with other m6A writer components. (A) From mass-spec experiments, Fl(2)d and Nito, but not GFP, can pull down Xio. (B–E) Xio-GFP and Nito-mRFP, Fl(2)d-mRFP, METTL3-mRFP, or METTL14-mRFP were cotransfected into S2 cells, and their subcellular localization was examined in live cells. (Scale bars: 5 μm.) (F–I) Xio-GFP or GFP and Nito-HA, Fl(2)d-HA, METTL3-HA, METTL14-HA were cotransfected into S2 cells. Cell lysates were immunoprecipitated using a GFP nanobody and analyzed by Western blot. Although GFP (asterisk) is expressed at a much higher amount than Xio-GFP (double asterisk), only Xio-GFP can pull down Nito-HA, Fl(2)d-HA, METTL3-HA, and METTL14-HA. Note that much more Nito-HA (arrow) was co-IPed than other factors.
Xio Is a Ubiquitously Expressed Nuclear Protein, and Its Expression Patterns Mimic Other m6A Pathway Members.
xio is located on the X chromosome, and its transcript is alternatively spliced, producing proteins of 1,150, 1,139, and 842 amino acids, respectively (Fig. S2A). Xio protein has no obvious domains, and its human ortholog is ZC3H13. The biological function of Xio and its ortholog have not been studied in any organism. xio expression patterns are very similar to those of other m6A writers and readers, with highest expression in the CNS and high expression in the ovary, imaginal discs, and fat tissues (Fig. S3A) (modENCODE developmental and tissue expression database) (20). Developmentally, xio expression was enriched in early embryos, decreased during larval stages, and rose again at pupal stages (Fig. S3A), which coincides with the reported m6A levels (11). xio expression showed enrichment in the neuroectoderm at later stages of embryogenesis (Fig. S3B) (FlyExpress) (21), which is highly similar to known m6A writers and readers (11).
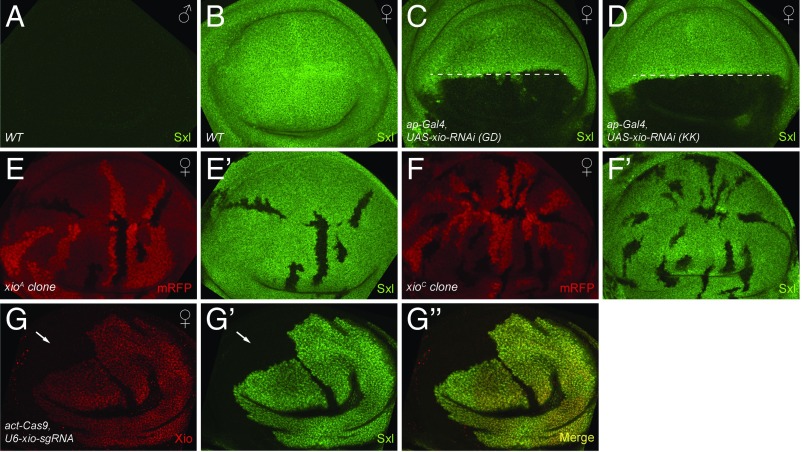
To study Xio function, we generated and evaluated tools. We raised an antibody against Xio, and, as expected, Xio was a ubiquitously expressed nuclear protein that colocalizes with Fl(2)d (Fig. S2 A–B″). Two nonoverlapping RNAi lines effectively knocked down Xio when induced by ap-Gal4 (Fig. S2 C and D). In addition, we constructed a transgenic xio-sgRNA under U6:3 promoter (22), and, when crossed with actin-Cas9, this line can generate random clones that showed no detectable Xio protein (Fig. 3G). Finally, we obtained two ethyl methanesulfonate (EMS) mutants of xio previously generated in a mosaic screen of lethal mutations on the X chromosome (23). xioC/Y flies died at the pupal stage while xioA/Y flies died as pharate, with some flies half way out of the pupal case (Fig. S4C). We induced mosaic clones of these mutants and found that Xio proteins are significantly reduced (Fig. S2 E–F′).
Fig. 3.
Xio is required for Sxl levels in somatic tissues. Sxl stainings in WT male (A) and female (B) wing discs. (C and D) Expressing xio RNAi in the dorsal half of the disk (below the dashed line) using ap-Gal4 results in strong reduction of Sxl stainings. (C) GD35212; (D) KK110253. (E–F′) Sxl staining is abolished in xioA (E′) or xioC (F′) mitotic mutant clones that are marked by the absence of mRFP (E and F). (G and G′) Both Xio and Sxl staining are completely abolished in xio mutant clones (arrows) generated by actin-Cas9/U6-xio-sgRNA.
Xio Is Required for Sex Determination in Drosophila.
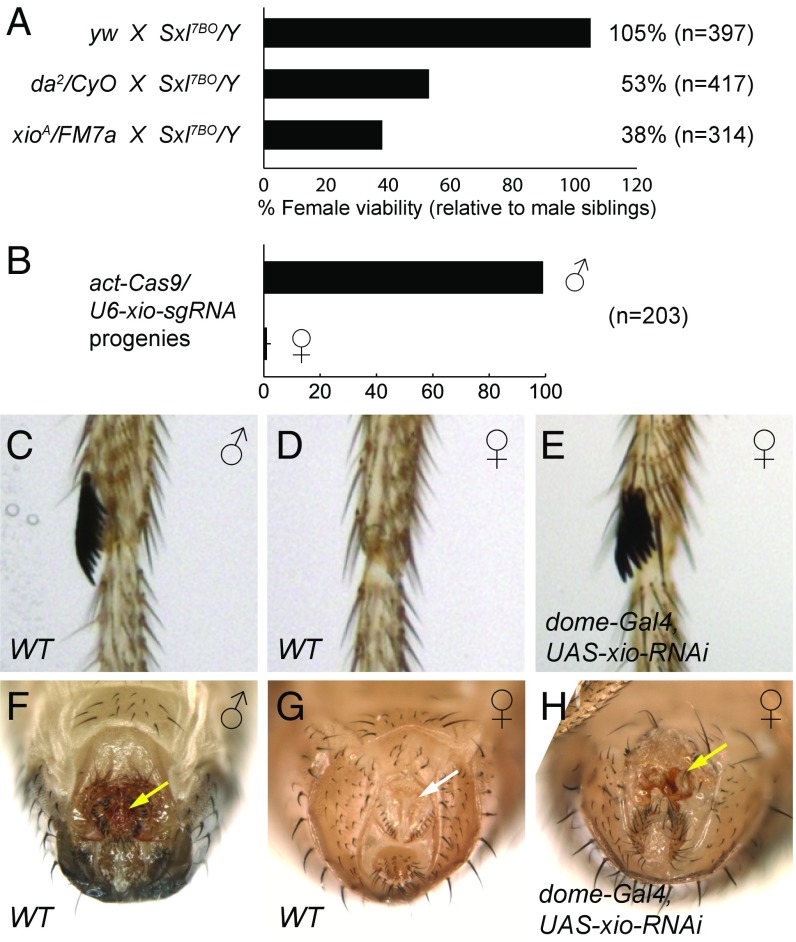
One of the major targets regulated by m6A modification is the Drosophila sex determination gene Sxl (10–12). Activation of Sxl in female embryos requires the coordination of two promoters, the establishment promoter SxlPe and the maintenance promoter SxlPm. SxlPe is transiently activated by maternal factors in females, and Sxl produced from SxlPe subsequently drives the autoregulatory splicing loop for SxlPm-derived transcripts (2). Any perturbation of these processes can lead to female-specific lethality, as manifested by progeny sex imbalance. This system is particularly sensitive when one copy of Sxl is removed (24): as shown, for example, for daughterless (da), which encodes a maternal factor required for Sxl activation. When da2 heterozygous mothers were crossed to Sxl7BO/Y (a null allele of Sxl) fathers, only 53% of the expected Sxl7BO/+ females survived to adulthood (Fig. 2A). Importantly, when xioA heterozygous mothers were crossed to Sxl7BO/Y fathers, only 38% of the expected Sxl7BO/+ female adults were observed (Fig. 2A), clearly implicating Xio in Sxl regulation.
Fig. 2.
Xio is required for sex determination in Drosophila. (A) Females of the indicated genotypes were crossed to Sxl7BO/Y males, and the resulting progeny were scored. For yw and da2/CyO crosses, the viability of total female adults relative to sibling male adults was quantified. For the xioA/FM7a cross, the viability of Sxl7BO/FM7a females relative to FM7a/Y males was quantified. n, total number of adult progeny counted. (B) actin-Cas9 flies were crossed to U6-xio-sgRNA flies, and the number of male and female progeny was counted. n, total number of adult progeny counted. (C) Foreleg of a WT male showing the sex combs. (D) Foreleg of a WT female. (E) Foreleg of a female fly expressing xio RNAi driven by dome-Gal4. Genitalia of WT male (F) and female (G) flies show distinct morphology. (H) xio RNAi driven by dome-Gal4 transforms female genital morphology into male-like. The white arrow indicates vaginal bristles, and the yellow arrows indicate penis apparatus.
Next, we used the CRISPR/Cas9 system to perturb xio function. Surprisingly, crossing actin-Cas9 with U6-xio-sgRNA produced progeny that are almost exclusively males (Fig. 2B). This result, together with the genetic interaction experiments, implies that Xio is involved in sex determination. To directly look at the sexual phenotype, we expressed xio RNAi using dome-Gal4 and observed striking transformation of female tissues into males, as evidenced by the appearance of sex combs in the forelegs of xio RNAi females (Fig. 2 C–E). In addition, dome-Gal4/xio-RNAi females showed strong abnormalities in the genitalia. Typical female structures, such as vaginal bristles (Fig. 2 G, white arrow), disappear, and structures resembling the male penis apparatus can be found (Fig. 2 F and H, yellow arrow). Together, these data suggest that Xio is a new component of the Drosophila sex determination pathway.
Xio Regulates Sxl Protein Levels in the Soma and Germline.
The phenotype associated with loss of Xio function suggests that Xio may regulate Sxl activity. In somatic tissues such as wing discs, Sxl protein was expressed ubiquitously in females but absent in males (Fig. 3 A and B). We used three different approaches to inactivate Xio function for examination of Sxl expression. First, expression of xio RNAi in the dorsal half of the wing disk using ap-Gal4 led to a significant reduction of Sxl levels (Fig. 3 C and D). Second, Sxl levels were almost absent from xioA or xioC mitotic mutant clones (Fig. 3 E–F′). Third, crossing of actin-Cas9 with U6-xio-sgRNA generated random xio loss-of-function clones. These clones were marked by the loss of Xio staining, and Sxl was completely depleted in these clones as well (Fig. 3 G–G″, arrows).
Finally, we asked whether Xio regulates Sxl levels in ovaries by inducing mitotic clones in both germline cells and follicle cells. Consistent with the disk results, Sxl was strongly reduced in either xio mutant germline or follicle cell clones (in Fig. S5, arrows indicate follicle cell clones, and asterisks indicate germline clones). Altogether, we conclude that Xio regulates Sxl levels in both germline and somatic tissues.
Xio Controls Sxl Alternative Splicing both in Vivo and in Cell Lines.
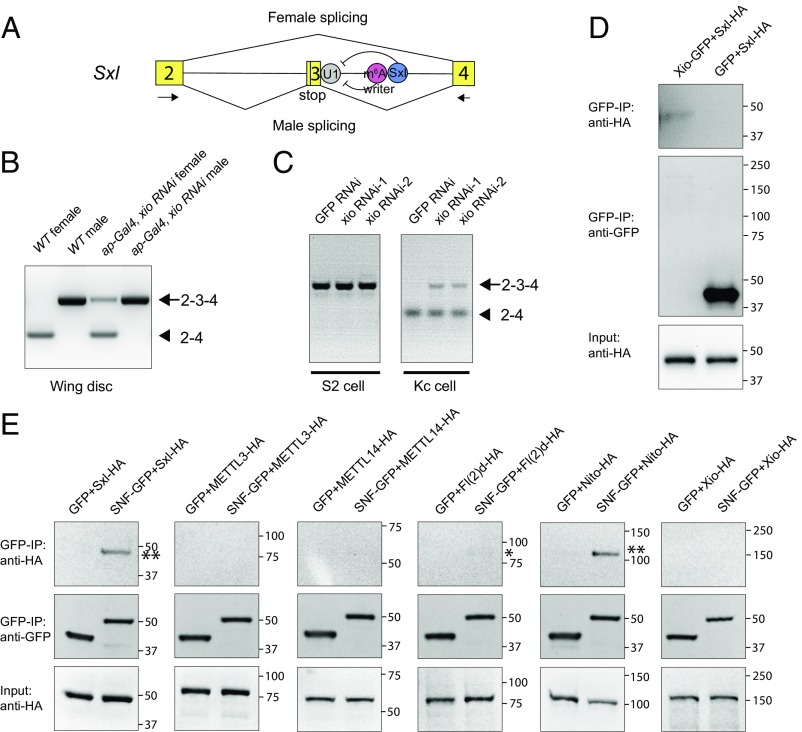
Sxl transcripts are alternatively spliced. While the male form includes exon 3 that contains a stop codon and leads to early termination of Sxl protein, the female form skips exon 3 and thus produces a functional Sxl protein (Fig. 4A) (25). To monitor Sxl splicing pattern, we used a pair of primers flanking exon 3 that detects the small female and large male spliced Sxl products by RT-PCR (Fig. 4A) (24). In ap-Gal4/xio RNAi female wing discs, a large band corresponding to the male-specific spliced form was clearly detected (Fig. 4B). We further analyzed Sxl splicing regulation in Drosophila cell lines. The S2 cell is a male cell line, and Sxl is spliced in the male form, while the Kc cell is a female cell line, and Sxl is spliced in the female form (Fig. 4C). While xio dsRNA had no effect on the male-specific splicing of Sxl in S2 cells, treating Kc cells with xio dsRNAs led to Sxl splicing shifted from the female form to the male form (Fig. 4C).
Fig. 4.
Xio regulates Sxl alternative splicing and interacts with Sxl. (A) Model showing female- or male-specific Sxl alternative splicing mediated by the m6A writer and Sxl protein. Only the regulatory events in the intron downstream of exon 3 are shown. The arrows indicate the primers used for RT-PCR. (B) Sxl splicing was analyzed by RT-PCR using RNA extracted from wing discs of indicated genotypes. Male-specific bands: 2-3-4. Female-specific bands: 2-4. (C) S2 or Kc cells were treated with xio RNAi or GFP RNAi, and Sxl splicing was analyzed by RT-PCR. (D) Sxl-HA, Xio-GFP, or GFP were transfected into S2 cells. Cell lysates were IPed and analyzed by Western blot. (E) SNF-GFP or GFP and Sxl-HA, METTL3-HA, METTL14-HA, Fl(2)d-HA, Nito-HA, and Xio-HA were cotransfected into S2 cells. Cell lysates were IPed and analyzed by Western blot. While large amounts of Sxl-HA and Nito-HA were co-IPed by SNF-GFP (double asterisk), a very low amount of Fl(2)d-HA was observed (asterisk), and there are no detectable METTL3-HA, METTL14-HA, and Xio-HA from the pull down.
We then analyzed how the m6A writer complex contributes to Sxl splicing regulation. In females, Sxl itself is the key protein that binds its pre-mRNA and inhibits splicing of the male-specific exon 3 by interacting with components of the spliceosome, such as SNF, a protein component of the U1 and U2 small nuclear ribonucleoproteins (snRNPs) (26, 27). Since the m6A writer components Fl(2)d and Nito interact with Sxl (6, 8), we examined whether Xio interacts with Sxl using a co-IP assay in S2 cells. As shown in Fig. 4D, GFP-Xio, but not GFP alone, can pull down Sxl-HA. These data are consistent with recent findings that m6A sites have been mapped in introns on both sides of exon 3 and in the vicinity of Sxl binding sites (10, 12). We next asked whether the m6A writer complex physically interacts with the spliceosome. In S2 cells where Sxl is absent, we performed co-IP experiments between SNF and five m6A writer subunits (Fig. 4E). As a positive control, SNF-GFP pulled down a large amount of Sxl-HA, in agreement with a previous report (27). Interestingly, we found that even more Nito-HA was co-IPed by SNF-GFP, indicating that these two proteins interact strongly with each other. However, a very low amount of Fl(2)d-HA was pulled down, and there were no detectable METTL3-HA, METTL14-HA, and Xio-HA from the co-IP. These results indicate that the m6A writer complex can interact with the spliceosome independent of Sxl and Nito serves as a bridge between the spliceosome and other m6A subunits. Together, we propose a model that both Sxl and the m6A writer complex interact with the spliceosome and they also interact with each other to repress the inclusion of the male-specific exon (Fig. 4A).
xio Mutant Phenocopies Mettl3 Adult Defects, and xio Is Required for m6A Levels.
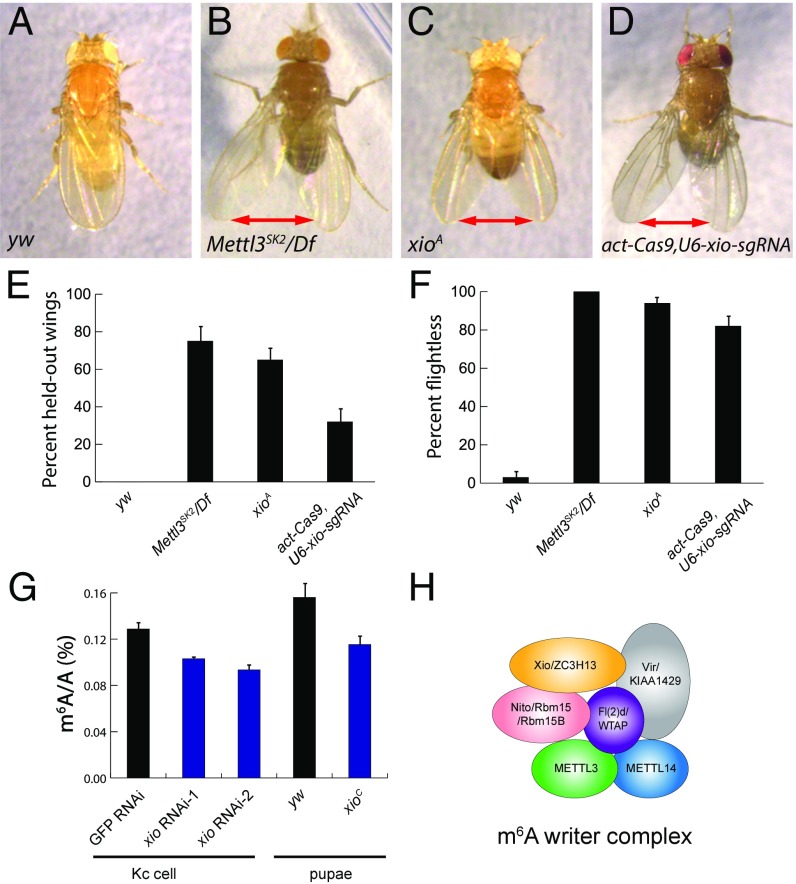
Other than sex determination phenotype, m6A writer and reader mutants exhibit characteristic adult defects. The most prominent ones are held-out wings and flightless phenotypes in both adult males and females (10, 12), likely due to functions of m6A modifications in the nervous system (11). If Xio is a component of the m6A writer complex, one would expect to see similar defects associated with xio mutant. By crossing xioA heterozygous females to males of different backgrounds, we were able to recover xioA hemizygous males and analyze their adult phenotypes (Fig. S4C). WT flies normally keep their wings in a folded position (Fig. 5 A and E); however, the majority of Mettl3 mutant flies cannot fold their wings correctly and exhibited held-out wings (Fig. 5 B and E). Interestingly, xioA mutant flies showed strong held-out wing phenotypes (Fig. 5 C and E). In addition, progeny of actin-Cas9 crossed with U6-xio-sgRNA exhibited similar held-out wings (Fig. 5 D and E), further confirming that this defect is specifically due to loss of xio.
Fig. 5.
xio mutants show adult defects, and Xio is required for proper m6A levels. (A) yw flies have their wings properly folded. (B) Mettl3SK2/Df, (C) xioA, (D) actin-Cas9/U6-xio-sgRNA flies cannot fold their wings and exhibit a held-out wing phenotype (marked by the double arrows). The frequency of flies that show held-out wings was quantified in E; error bars represent SEM. (F) Flies of the indicated genotypes were tested for their flight abilities, and the number of flightless flies was quantified; error bars represent SEM. All flies used from A–F are males. (G) Quantifications of m6A relative to A in Kc cells and pupae. Compared with controls, xio RNAi cells and xioC mutant pupae showed reduced m6A levels in their RNA after one round of polyA purification. Error bars represent SD. (H) A model of the m6A writer complex comprised of six core components.
We further tested these flies for their flight ability. While 100% of Mettl3 mutant flies could not fly, the majority of xioA mutant flies, as well as actin-Cas9/U6-xio-sgRNA flies, were flightless (Fig. 5F). Together, these results indicate that xio mutants resemble m6A pathway mutants in terms of sex determination, held-out wings, and flight abilities, strongly arguing that Xio is a key component of the m6A methylation pathway.
Next, we directly measured N6-methyladenosine levels in xio mutants by quantitative liquid chromatography–mass spectrometry (LC-MS). We used an external calibration curve prepared with A and m6A standards to determine the absolute quantities of each ribonucleoside (Fig. S6). After one round of polyA selection, we detected a modest but consistent reduction of m6A levels in xio RNAi cells and in xioC mutant pupae (Fig. 5G). The amount of reduction was comparable with that seen in nito mutants (12), and the modest reduction was likely due to the contamination of rRNA in the sample and/or incomplete loss of function of xio in these conditions. Nevertheless, these results validate that Xio is required for proper m6A levels and, together with our phenotypic analysis and biochemical interaction data, strongly support the model that Xio is a new bona fide subunit of the m6A methyltransferase complex (Fig. 5H).
Xio Regulates a Broad Spectrum of Gene Expression and Alternative Splicing Events.
To gain a global view of Xio-mediated gene expression, we performed RNA-Seq experiments in control and xio mutant animals. As xioA hemizygous males are sterile, we were unable to generate xioA homozygous females and thus performed RNA-Seq in xioA males. Since Sxl regulates the expression of numerous genes in females, using males has the advantage of dissecting Sxl independent events. We used the pharate stage, just before eclosure, as this is the period with very high m6A levels and most xioA mutants cannot develop beyond this stage. Differential gene expression analysis revealed 2,002 down-regulated genes and 842 up-regulated genes (fold change ≥ 1.5 and P value < 0.05) (Fig. S7A and Dataset S2). Kyoto Encyclopedia of Genes and Genomes (KEGG)-pathway analysis indicated that metabolic pathways, including fatty acid, carbohydrate, and amino acid metabolism genes, are strongly enriched (Fig. S7C and Dataset S2). Consistent with the adult phenotypes, Gene Ontology (GO) analysis found a significant enrichment of neuron-related categories, such as sleep, neuron projection, circadian rhythm, and motor neuron axon guidance (Fig. S7B). In addition, we also analyzed the alternative splicing changes in these mutants and found that 105 alternative splicing events in 96 genes were significantly different (Bayes factor > 10, ΔPSI (difference in percentage spliced in) > 0.2) (Dataset S3). GO term enrichment revealed similar neuron-related categories, such as synaptic growth, gravitaxis, axon guidance, and neuromuscular synaptic transmission, as well as other developmental processes (Fig. S7D and Dataset S3). A few examples of differentially alternative spliced genes are shown in Fig. S8. In summary, our data revealed a broad range of genes and splicing events regulated by Xio and provide an important dataset for further mechanistic studies.
Discussion
The Drosophila sex determination pathway, comprised of a hierarchy of alternative splicing events, remains a textbook paradigm for sex determination mechanisms and alternative splicing. Here, we describe the characterization of Xio as a component of the Drosophila sex determination pathway. Xio loss of function results in female-specific lethality and striking sexual transformation phenotypes. We further show that Xio regulates the master sex determination gene Sxl by controlling its alternative splicing. Such function is reminiscent of previously reported genes, such as snf (26, 28), U2AF (27), U1-70K (29), SPF45 (30, 31), PPS (24), fl(2)d (8, 9), vir (5, 7), and nito (6), that play similar roles in Sxl splicing regulation. While several of these genes mainly act as splicing factors, Fl(2)d, Vir, Nito, and Xio are now known as the core subunits of the RNA m6A methyltransferase complex.
m6A modification has been known for more than 40 y (32) but recently gained attention due to the emergence of new technologies to map m6A sites throughout the transcriptome (33, 34), as well as the identification of the writers, erasers, and readers of this pathway (35–37). The key methyltransferase METTL3 was first discovered in 1994 (38); then, other subunits of the writer complex were identified mainly through biochemical interaction experiments and genetic screens (39, 40). For example, a proteomic study to identify WTAP interacting proteins in human cells revealed Vir/KIAA1429, RBM15, and the ortholog of Xio, ZC3H13 (or KIAA0853) (41). Drosophila sex determination provides an unambiguous phenotype to screen and characterize m6A pathway components. Fl(2)d and Vir have been known to be involved in sex determination for more than two decades, and recently we identified Nito as a new component of the sex determination pathway in an RNAi screen (6). Finally, METTL3, METTL14, and the reader YT521-B were shown to be also required for sex determination (10–12).
The sexual phenotype associated with xio and the biochemical interactions between Xio and other m6A factors indicate that Xio is a new component of the m6A writer complex. We further show that xio mutants phenocopy Mettl3 mutant adult defects and that the m6A level is reduced in xio mutant cells and fly. As ZC3H13 was found in the WTAP-associating protein complex (41), Xio/ZC3H13 is likely an evolutionarily conserved m6A factor (Fig. 5H). Our study also shed light on how the m6A modification is involved in Sxl splicing regulation. Similar to Fl(2)d and Nito (6, 8), we show that Xio can interact with Sxl in a co-IP experiment. Furthermore, we tested whether the m6A writer complex physically interacts with the spliceosome and found that Nito is the major component that strongly interacts with SNF in the absence of Sxl. This is not particularly surprising since Nito has three RRM domains, compared with two RRMs in the Sxl protein. These results suggest a new mechanism for m6A-mediated splicing in which the m6A writer complex can interact with the spliceosome independently of Sxl and Nito may serve as a bridge between the spliceosome and the m6A catalytic core (Fig. 4A).
Besides Sxl-mediated sex determination, m6A mutants in Drosophila exhibit several characteristic phenotypes. Mettl3 and Mettl14 mutants are homozygous viable and show held-out wing and flightless phenotypes (10–12). fl(2)d, vir, and nito mutants die during larval stages, preventing the analysis of their adult phenotypes. A strong allele of xio also causes lethality during the pupal stage; however, by using a slightly weaker allele of xio, we were able to examine its adult phenotype. xioA mutants resemble Mettl3 held-out wing and flightless phenotypes, strongly arguing that Xio is a core subunit of the m6A writer complex. These phenotypes are likely due to the function of m6A modifications in the nervous system where its level is highest in both fly and mammals. Finally, RNA-Seq analysis revealed that many neuronal genes are differentially expressed and/or alternatively spliced in xio mutants, suggesting that it will be important to pinpoint the critical mRNA species that are m6A-modified in the nervous system. Another group of genes that were found from the RNA-Seq analysis are metabolic genes, which are also significantly enriched in Mettl3 mutant flies (10). These genes are of particular interest since overexpression of the m6A demethylase Fto in mice leads to increased food intake and obesity (42). In vertebrates, m6A has been shown to regulate embryonic stem cell differentiation, somatic cell differentiation, maternal-to-zygotic transition, circadian rhythm, and spermatogenesis (17); whether m6A plays similar roles in Drosophila remains to be determined.
Methods
Details on the fly strains and antibodies used in this study, as well as how xio clones were generated and how Xio antibody was generated, can be found in SI Methods. Protocols used for antibody staining, cell culture and RNA interference, coimmunoprecipitation, RT-PCR, analyzing m6A levels by LC-MS, and RNA-Seq can be found in SI Methods.
Supplementary Material
Acknowledgments
We thank Eric Lai, Jean-Yves Roignant, Helen Salz, and Matthias Soller for fly stocks. This work was supported by National Natural Science Foundation of China Grant 31771586, the Shanghai Pujiang Program, and a start-up fund from the Chinese Academy of Sciences (to D.Y.). N.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE110047).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720945115/-/DCSupplemental.
References
- 1.Cline TW, Meyer BJ. Vive la différence: Males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 2.Salz HK, Erickson JW. Sex determination in Drosophila: The view from the top. Fly (Austin) 2010;4:60–70. doi: 10.4161/fly.4.1.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camara N, Whitworth C, Van Doren M. The creation of sexual dimorphism in the Drosophila soma. Curr Top Dev Biol. 2008;83:65–107. doi: 10.1016/S0070-2153(08)00403-1. [DOI] [PubMed] [Google Scholar]
- 4.Lucchesi JC, Kuroda MI. Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol. 2015;7:a019398. doi: 10.1101/cshperspect.a019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilfiker A, Amrein H, Dübendorfer A, Schneiter R, Nöthiger R. The gene virilizer is required for female-specific splicing controlled by Sxl, the master gene for sexual development in Drosophila. Development. 1995;121:4017–4026. doi: 10.1242/dev.121.12.4017. [DOI] [PubMed] [Google Scholar]
- 6.Yan D, Perrimon N. spenito is required for sex determination in Drosophila melanogaster. Proc Natl Acad Sci USA. 2015;112:11606–11611. doi: 10.1073/pnas.1515891112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.SchuLtt C, Hilfiker A, Nothiger R. virilizer regulates Sex-lethal in the germline of Drosophila melanogaster. Development. 1998;125:1501–1507. doi: 10.1242/dev.125.8.1501. [DOI] [PubMed] [Google Scholar]
- 8.Penn JK, et al. Functioning of the Drosophila Wilms’-tumor-1-associated protein homolog, Fl(2)d, in Sex-lethal-dependent alternative splicing. Genetics. 2008;178:737–748. doi: 10.1534/genetics.107.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granadino B, Campuzano S, Sánchez L. The Drosophila melanogaster fl(2)d gene is needed for the female-specific splicing of Sex-lethal RNA. EMBO J. 1990;9:2597–2602. doi: 10.1002/j.1460-2075.1990.tb07441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haussmann IU, et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 11.Lence T, et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540:242–247. doi: 10.1038/nature20568. [DOI] [PubMed] [Google Scholar]
- 12.Kan L, et al. The m6A pathway facilitates sex determination in Drosophila. Nat Commun. 2017;8:15737. doi: 10.1038/ncomms15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for notch signaling during oogenesis. Proc Natl Acad Sci USA. 2011;108:14855–14860. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer KD, Jaffrey SR. Rethinking m6A readers, writers, and erasers. Annu Rev Cell Dev Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patil DP, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu PJ, Shi H, He C. Epitranscriptomic influences on development and disease. Genome Biol. 2017;18:197. doi: 10.1186/s13059-017-1336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roignant JY, Soller M. m6A in mRNA: An ancient mechanism for fine-tuning gene expression. Trends Genet. 2017;33:380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Guruharsha KG, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, et al. FlyExpress 7: An integrated discovery platform to study coexpressed genes using in situ hybridization images in Drosophila. G3 (Bethesda) 2017;7:2791–2797. doi: 10.1534/g3.117.040345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto S, et al. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson ML, Nagengast AA, Salz HK. PPS, a large multidomain protein, functions with sex-lethal to regulate alternative splicing in Drosophila. PLoS Genet. 2010;6:e1000872. doi: 10.1371/journal.pgen.1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell LR, Horabin JI, Schedl P, Cline TW. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991;65:229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- 26.Flickinger TW, Salz HK. The Drosophila sex determination gene snf encodes a nuclear protein with sequence and functional similarity to the mammalian U1A snRNP protein. Genes Dev. 1994;8:914–925. doi: 10.1101/gad.8.8.914. [DOI] [PubMed] [Google Scholar]
- 27.Nagengast AA, Stitzinger SM, Tseng CH, Mount SM, Salz HK. Sex-lethal splicing autoregulation in vivo: Interactions between SEX-LETHAL, the U1 snRNP and U2AF underlie male exon skipping. Development. 2003;130:463–471. doi: 10.1242/dev.00274. [DOI] [PubMed] [Google Scholar]
- 28.Oliver B, Perrimon N, Mahowald AP. Genetic evidence that the sans fille locus is involved in Drosophila sex determination. Genetics. 1988;120:159–171. doi: 10.1093/genetics/120.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salz HK, et al. The Drosophila U1-70K protein is required for viability, but its arginine-rich domain is dispensable. Genetics. 2004;168:2059–2065. doi: 10.1534/genetics.104.032532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaouki AS, Salz HK. Drosophila SPF45: A bifunctional protein with roles in both splicing and DNA repair. PLoS Genet. 2006;2:e178. doi: 10.1371/journal.pgen.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lallena MJ, Chalmers KJ, Llamazares S, Lamond AI, Valcárcel J. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell. 2002;109:285–296. doi: 10.1016/s0092-8674(02)00730-4. [DOI] [PubMed] [Google Scholar]
- 32.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 34.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 36.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- 39.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong S, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horiuchi K, et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Church C, et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.