Eukaryotic voltage gated sodium-selective channels (VGSCs) enable influx of Na+ into excitable cells in response to a change in the transmembrane potential. This movement of ions causes the membrane depolarization occurring during the rising phase of the action potential and, as such, underlies propagation of electrical signals in neurons. The transmembrane region of VGSCs is characterized by a fourfold pseudosymmetrical architecture. In particular, the channel is constituted of four homologous repeats (referred to as domains, DI through DIV), each comprising six helical segments (S1 through S6). The first four helices (S1–S4) of each domain assemble into a separate helix bundle, the so-called voltage sensor domain, which undergoes a conformational transition in response to membrane depolarization. The remaining S5 and S6 helices from all of the domains form a tetrameric assembly, the pore domain, containing a lumen in its center. The latter constitutes a pathway connecting the extracellular and intracellular compartments, enabling diffusion of water molecules and ions across the membrane. Crucial milestones along this pathway are the selectivity filter, a section permeable to Na+ but not K+, and the activation gate, a hydrophobic plug that hinders the passage of waters and ions when the channel is in the closed state.
The major features of this biological nanomachine are remarkably conserved along evolution: Voltage-gated ion channels from all kingdoms of life share a common “blueprint” with the same architecture and basic rules of functioning. In particular, VGSCs are members of a large phylogenetic family, the six-transmembrane family, also containing VGSCs from bacteria. Despite the large degree of sequence similarity, the structure of prokaryotic VGSCs is less complex than that of eukaryotic ones. While the latter are constituted of a single polypeptide chain containing four homologous repeats, the former are genuine homotetramers. Moreover, prokaryotic VGSCs lack almost completely the large intracellular and extracellular domains characterizing eukaryotic VGSCs. In other words, bacteria possess a minimalist version of VGSCs. This inherent simplicity enabled a wealth of structural and functional studies that resulted in a detailed microscopic picture of the VGSC activation mechanism (1–3).
Understanding the molecular details of VGSCs sheds light not only on fundamental aspects of electrical signaling but also on the causes of many diseases associated with disorders of excitable cell function. VGSC malfunctioning is involved, for instance, in cardiac arrhythmias, epilepsy, and pain syndromes (4–7). Accordingly, small-molecule modulation of VGSCs is one of the major therapeutic strategies to treat these diseases. Often, however, the safety, and thus the viability, of these drugs is limited by their lack of selectivity. The human genome contains nine VGSC genes with distinct expression profiles between the heart, central nervous system, and peripheral nervous system. Simultaneous inhibition of several VGSC subtypes, as in the case of local anesthetics, increases the risk of life-threatening side effects, and thus severely limits the possible routes of administration. Developing selective inhibitors is thus a necessary strategy to discover effective yet safe drugs. However, success in this endeavor is still episodic and has not yet resulted in approved drugs (8–10). Part of the problem is the lack of a detailed understanding of VGSC inhibitor mechanism of action.
Since the first pioneering studies, VGSC inhibition appeared as a complex process with several puzzling aspects. For instance, VGSCs are inhibited in two distinct ways: through a “tonic” block, in which the drug binds to the closed channel, and through a “use-dependent” block, which requires prior opening of the channel (11, 12). Intriguingly, these modes of action entail the existence of distinct drug-binding pathways: While the drug molecule binds the channel pore in both cases, the route to access cannot be the same in the closed and open states (13). The drug crosses the open activation gate in use-dependent inhibition, while it follows an alternative hydrophobic pathway in tonic block. Several simulation studies have shown that the so-called fenestrations, cavities connecting the pore to the hydrophobic section of the lipid bilayer, provide a viable route for the drug to reach the binding site (14–16). However, no microscopic picture was available to rationalize the difference between tonic and use-dependent block. In particular, the observation that mutation of specific amino acids can significantly affect use-dependent block without appreciably affecting tonic block suggests that different interactions are established between the drug and the channel in the two modes of action.
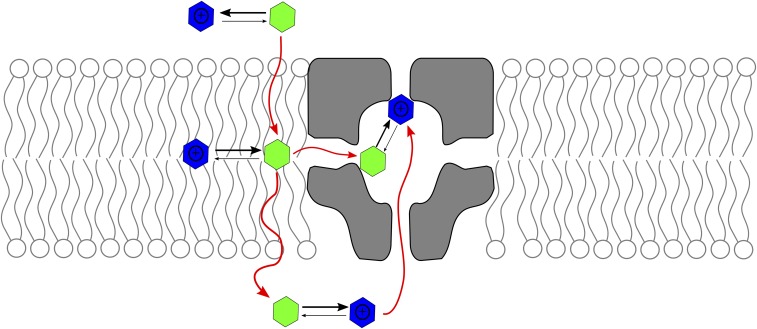
In PNAS, Buyan et al. (17) provide a rigorous and systematic exploration of the binding of six molecules known to act as sodium channel pore blockers. The authors use molecular dynamics simulations in connection with an enhanced sampling technique to exhaustively explore the conformational space of the drug molecule. The resulting equilibrium probability density functions provide information about the precise location and relative affinity of each binding site. Three of the six molecules (benzocaine, lamotrigine, and carbamazepine) are electrically neutral, while the other three (PF-5215786, PF-6305591, and lidocaine) exist in two forms at pH 7.0: neutral and positively charged. It is found that both charged and neutral forms partition into the bilayer even though neutral molecules show a smaller free energy barrier to cross the membrane. Importantly, it is shown that there are two possible binding sites: While neutral molecules bind to an already known site on S6, charged inhibitors bind to a so far unreported binding site, with the positively charged group in direct contact with the selectivity filter (Fig. 1).
Fig. 1.
Protonation state and binding sites of sodium channel pore blockers. Many use-dependent drugs have a pKa near 7.0; therefore, they exist in solution in equilibrium between charged (blue) and neutral (green) states. The neutral form crosses the lipid bilayer more easily than the charged one; thus, it is the species that likely reaches the channel pore through the fenestrations. Buyan et al. (17) find that inside the pore, the neutral and charged species bind in different locations. In particular, the charged form binds the selectivity filter, thereby hindering diffusion of ions to the central cavity.
Buyan et al. (17) show that the results are robust with respect to the choice of the molecule (they consider three perpetually neutral and three titrable molecules) and are consistent for two VGSCs: one prokaryotic (NavMs) and the other eukaryotic (NavPas). Importantly, the novel binding site can explain the halogen electronic density measured by Bagnéris et al. (18) by cocrystallizing NavMs with PF-5215786. A crucial aspect of these results is that they provide a viable model for inhibition of Na+ conduction. Indeed, the authors show that the positively charged groups of the drug molecules compete with Na+ ions for the binding site in the selectivity filter. As such, the drug molecule constitutes an obstruction along the permeation pathway, which hinders diffusion of ions. The most interesting observation, however, concerns the different pattern of interactions between the drug and the channel observed for the two binding sites, which can help to rationalize the difference between tonic and use-dependent blockers.
Overall, the results by Buyan et al. (17) provide accurate predictions about the binding mode of pore blockers and, for the first time, mechanistic insight about the differences between tonic and use-dependent blockers. Taken together, these predictions will help to design novel pore binders and to predict their behavior in future experiments. Importantly, in providing solid ground to proceed in the investigation of VGSC mechanism of inhibition, this study will necessarily prompt further investigations. For instance, the work by Buyan et al. (17) does not consider the issue of state-dependent affinities, whereby the drug molecules bind selectively a conformational state of the channel. While most of the pore blockers bind the inactivated state strongly, the authors used an open state in their simulations. Future studies will, no doubt, fill this gap and bring us even closer to a quantitative model of VGSC inhibition.
Acknowledgments
My research is supported by the National Institute of General Medical Sciences of the NIH (Award R01GM093290) and by the National Science Foundation (Award ACI-1614804).
Footnotes
The author declares no conflict of interest.
See companion article on page E3135.
References
- 1.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yarov-Yarovoy V, et al. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci USA. 2012;109:E93–E102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sula A, et al. The complete structure of an activated open sodium channel. Nat Commun. 2017;8:14205. doi: 10.1038/ncomms14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meisler MH, Kearney JA. Sodium channel mutations in epilepsy and other neurological disorders. J Clin Invest. 2005;115:2010–2017. doi: 10.1172/JCI25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roden DM, George AL., Jr The cardiac ion channels: Relevance to management of arrhythmias. Annu Rev Med. 1996;47:135–148. doi: 10.1146/annurev.med.47.1.135. [DOI] [PubMed] [Google Scholar]
- 6.Ruan Y, Liu N, Priori SG. Sodium channel mutations and arrhythmias. Nat Rev Cardiol. 2009;6:337–348. doi: 10.1038/nrcardio.2009.44. [DOI] [PubMed] [Google Scholar]
- 7.Dib-Hajj SD, Cummins TR, Black JA, Waxman SG. Sodium channels in normal and pathological pain. Annu Rev Neurosci. 2010;33:325–347. doi: 10.1146/annurev-neuro-060909-153234. [DOI] [PubMed] [Google Scholar]
- 8.Jarvis MF, et al. A-803467, a potent and selective Nav1.8 sodium channel blocker, attenuates neuropathic and inflammatory pain in the rat. Proc Natl Acad Sci USA. 2007;104:8520–8525. doi: 10.1073/pnas.0611364104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kort ME, et al. Subtype-selective Na(v)1.8 sodium channel blockers: Identification of potent, orally active nicotinamide derivatives. Bioorg Med Chem Lett. 2010;20:6812–6815. doi: 10.1016/j.bmcl.2010.08.121. [DOI] [PubMed] [Google Scholar]
- 10.Ahuja S, et al. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science. 2015;350:aac5464. doi: 10.1126/science.aac5464. [DOI] [PubMed] [Google Scholar]
- 11.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Molecular determinants of state-dependent block of Na+ channels by local anesthetics. Science. 1994;265:1724–1728. doi: 10.1126/science.8085162. [DOI] [PubMed] [Google Scholar]
- 12.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Common molecular determinants of local anesthetic, antiarrhythmic, and anticonvulsant block of voltage-gated Na+ channels. Proc Natl Acad Sci USA. 1996;93:9270–9275. doi: 10.1073/pnas.93.17.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hille B. Local anesthetics: Hydrophilic and hydrophobic pathways for the drug-receptor reaction. J Gen Physiol. 1977;69:497–515. doi: 10.1085/jgp.69.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boiteux C, et al. Local anesthetic and antiepileptic drug access and binding to a bacterial voltage-gated sodium channel. Proc Natl Acad Sci USA. 2014;111:13057–13062. doi: 10.1073/pnas.1408710111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin LJ, Corry B. Locating the route of entry and binding sites of benzocaine and phenytoin in a bacterial voltage gated sodium channel. PLOS Comput Biol. 2014;10:e1003688. doi: 10.1371/journal.pcbi.1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raju SG, Barber AF, LeBard DN, Klein ML, Carnevale V. Exploring volatile general anesthetic binding to a closed membrane-bound bacterial voltage-gated sodium channel via computation. PLOS Comput Biol. 2013;9:e1003090. doi: 10.1371/journal.pcbi.1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buyan A, Sun D, Corry B. Protonation state of inhibitors determines interaction sites within voltage-gated sodium channels. Proc Natl Acad Sci USA. 2018;115:E3135–E3144. doi: 10.1073/pnas.1714131115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagnéris C, et al. Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism. Proc Natl Acad Sci USA. 2014;111:8428–8433. doi: 10.1073/pnas.1406855111. [DOI] [PMC free article] [PubMed] [Google Scholar]