PLANT BIOLOGY Correction for “AtsPLA2-α nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response,” by Solène Froidure, Joanne Canonne, Xavier Daniel, Alain Jauneau, Christian Brière, Dominique Roby, and Susana Rivas, which was first published August 9, 2010; 10.1073/pnas.1009056107 (Proc Natl Acad Sci USA 107:15281–15286).
The undersigned authors wish to note, “We acknowledge inappropriate reuse of two Ponceau images from Fig. 2B and Fig. S4 of the PNAS paper in later publications of our group (see doi.org/10.1105/tpc.17.00567 and doi.org/10.1371/journal.pone.0190773). We hereby confirm that the original images are those first published in PNAS. When preparing the figures for submission to PNAS, the raw images were cropped and/or stretched to match the other blots and saved in the format for submission. Unfortunately, we did not systematically archive an independent copy of each raw image, and only the final version of the figures was stored. Fig. 3 appears to have areas of unmarked splicing and background inconsistencies, but we are confident, however, in the scientific accuracy of the data despite being unable to provide the original images.
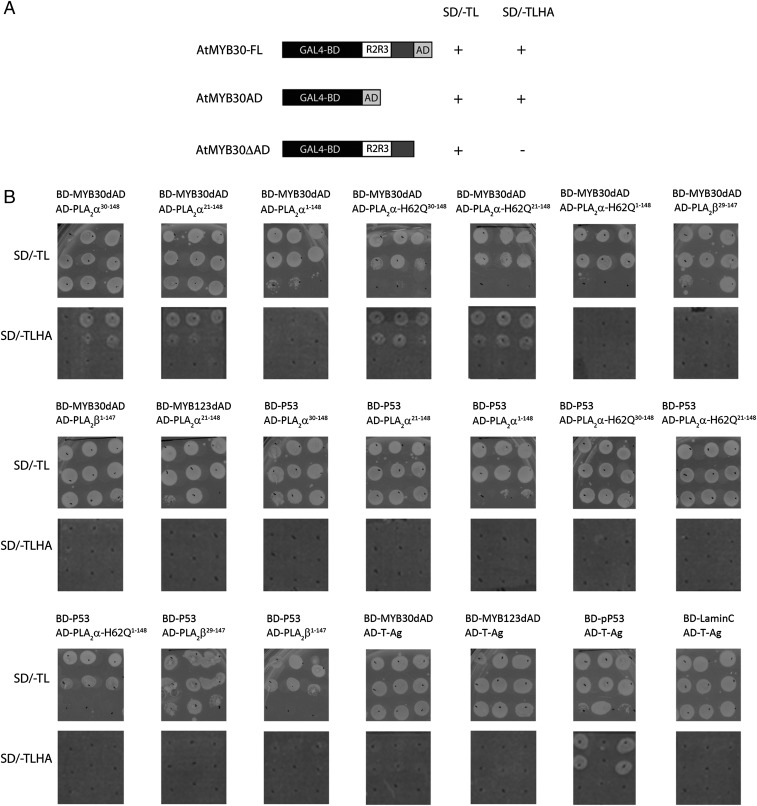
“Further, we acknowledge that images of yeast colonies in Fig. S1B are indeed duplicated. Our intention was to represent the presence (or absence) of yeast growth observed with the different protein combinations. We recognize that this should have been clearly indicated in the figure legend and apologize for this omission. We have been able to retrieve the original results and prepared the revised figure below showing growth of yeast colonies expressing the different protein combinations. We apologize for any inconvenience the publication of these figures may have caused.” The corrected Fig. S1 and its corrected legend appear below. The SI has been corrected online.
Fig. S1.
Specific interaction between MYB30 and AtsPLA2-α in yeast. (A) AtMYB30 putative transcriptional transactivation domain is functional in yeast. To identify AtMYB30-interacting proteins, an AtMYB30 version deleted from its C-terminal potential transcriptional activation domain was used as bait to screen the generate yeast two-hybrid Arabidopsis cDNA library. The choice of the bait was due to the fact that the presence of AtMYB30 activation domain (AtMYB30AD) led to autoactivation of yeast reporter genes. AtMYB30-FL (Top), AtMYB30AD (Middle), and AtMYB30ΔAD (Bottom) were used as N-terminal fusions to the DNA binding domain of the GAL4 protein (GAL4-BD) to transform the AH109 yeast strain. Yeast grow on nonselective (SD/-TL) and high stringency medium (SD/-TLHA), 3 d after transformation, is indicated. (B) Yeast two-hybrid assay of the interaction between AtMYB30 and AtsPLA2-α. Growth of three independent yeast colonies for each protein combination is shown after 5 d on low stringency (SD/-TL) or high stringency medium (SD/-TLHA). Three individual colonies were disposed horizontally on top and three dilutions (1/10 serial dilutions) were vertically spotted for each individual colony. AtsPLA2-α30-148, AtsPLA2-α clone identified in the yeast two-hybrid screen; AtsPLA2-α21-148, AtsPLA2-α after cleavage of its signal peptide; AtsPLA2-α1-148, noncleaved AtsPLA2-α; AtsPLA2-α-H62Q, inactive catalytic mutant version of AtsPLA2-α; AtsPLA2-β29-147, AtsPLA2-β after cleavage of its signal peptide; AtsPLA2-β1-147, noncleaved AtsPLA2-β. BD-p53, BD-laminC, and AD-T-antigen are controls provided by Clontech. BD, GAL4 DNA-binding domain; AD, GAL4 activation domain. Coexpression of full length AtsPLA2-α (AtsPLA2-α1-148) and AtMYB30ΔAD did not result in yeast growth on selective medium. The lack of interaction between the two proteins in yeast is probably due to the presence of an HA tag at the N terminus of AtsPLA2-α, which blocks cleavage of its signal peptide. Indeed, AtsPLA2-α deleted from its signal peptide (AtsPLA2-α21-148) was able to interact with AtMYB30ΔAD in yeast. In a control experiment, yeast cells expressing both AtsPLA2-α and the nonrelated MYB transcription factor AtMYB123ΔAD were not able to grow on selective medium. Furthermore, no protein interaction could be detected when yeasts were cotransformed with AtMYB30ΔAD and the closest AtsPLA2-α ortholog, AtsPLA2-β, containing or not containing its signal peptide. Taken together, these data demonstrate the specificity of the interaction between AtsPLA2-α and AtMYB30 in yeast.
Solène Froidure, Joanne Canonne, Alain Jauneau, Christian Brière, Susana Rivas