Abstract
Although several previous studies have reported the implication of various microRNAs (miRNAs) in regulation of human bladder cancer (BC) development, alterations and function of many miRNAs in bladder cancer growth are not explored yet at present. Here, we screened 1,900 known miRNAs and first discovered that miR-411 was one of the major miRNAs, which was down-regulated in n-butyl-N-(4-hydroxybutyl)-nitrosamine (BBN)-induced BCs. This miR-411 down-regulation was also observed in human BC tissues and cell lines. The results from evaluating the relationship between miR-411 and patient survival in BC using the TCGA (The Cancer Genome Atlas) database indicated that miR-411 was positively correlated with DFS (disease-free survival). Our studies also showed that miR-411 inhibited tumor growth of human BC cells in a xenograft animal model. Mechanistic studies revealed that overexpression of miR-411 repressed the expression of ALL1-fused gene from the chromosome 1q (AF1q) (MLLT11) by binding to the 3′ untranslated region (UTR) of mllt11 mRNA and in turn induced p21 expression and caused cell cycle arrest at the G2/M phase, further inhibiting BC tumor growth. Collectively, our results improve our understanding of the role of miR-411 in BC tumor growth and suggest miR-411 and MLLT11 as potential new targets for the treatment of BC patients.
Keywords: miR-411, MLLT11, bladder cancer, tumor growth
Introduction
Bladder cancer (BC) is one of the most common cancers in the western world; it is the fourth most common cancer in men and seventh most common cancer in women worldwide.1, 2, 3 According to the American Cancer Society, 79,030 new cases of BC are expected to be diagnosed and 16,870 patients will die from this disease in the United States in 2017.4 The incidence and mortality rates of BC have increased from developing countries in Asia.5 In China, the age-standardized incidence rate is 7.68 per 100,000, and the age-standardized mortality rate is 3.03 per 100,000.6, 7, 8 The tumorigenesis of BC involves genetic alterations, epigenetic alterations, and environmental factors.9, 10 Currently, the primary therapeutic strategy for patients with early BC is surgery, followed by chemo-therapeutic approaches, radiotherapy, and biological therapies. In patients with advanced disease, it remains a prevalent and life-threatening malignancy, underscoring the need to identify new targets for diagnosis and treatment.11 Therefore, it is urgent to develop novel diagnostic and therapeutic targets as well as elucidate the molecular mechanisms of these genes and clinical significance for BC to improve the diagnostic accuracy and clinic treatment.
Recent high-throughput transcriptome analyses show that more than 90% of the transcriptome is transcribed for non-coding RNAs, among which microRNAs (miRNAs) are involved in multiple biological processes, such as differentiation, proliferation, cell cycle arrest, apoptosis, and metastasis. miRNAs are a class of small (17–25 nucleotides) endogenous non-coding RNAs.12, 13 miRNAs are involved in the regulation of genes associated with cancer development and progression and can act as oncogenes, contributing to tumor formation14, 15 or as tumor suppressors by inhibiting the expression of oncogenes in different types of cancer.16, 17, 18 In BCs, although alterations in the expression of certain miRNAs have been demonstrated,19 the underlying mechanisms of miRNAs and their clinic significance of BC remains poorly understood. We screened 1,900 known miRNAs by using a microRNA microarray chip, confirmed by real-time PCR subsequently, and first discovered that miR-411 was one of major miRNAs that was downregulated in a n-butyl-N-(4-hydroxybutyl)-nitrosamine (BBN)-induced BC mouse model. We also found the down-regulation of miR-411 expression in human BCs through checking TCGA database and determining the fresh human clinical BC tissues. By utilization of gain- (microRNA overexpression) or loss (microRNA inhibition)-of-function assay, we showed that miR-411 inhibited BC cell proliferation in vitro and in vivo. Therefore, we focused on the roles and related molecular mechanisms of miR-411 in human BC in our current study.
MLLT11, also named ALL1-fused gene from the chromosome 1q (AF1q), which is located in chromosome l band 1q21. The MLLT11 produces a small 9-kDa protein that has no clear function structural domains and no similarities with other known proteins.20 MLLT11 was originally defined as an oncogenic factor implicated in t(1;11) (q21;q23) translocation, which is associated with certain cases of leukemia.21 High expression levels of MLLT11 gene are associated with poor outcomes in pediatric acute myeloid leukemia (AML), adult normal cytogenetic AML, and adult myelodysplastic syndrome.22 The proposed oncogenic role of MLLT11 involves the regulation of the BAD apoptotic pathway via NFκB.21 However, the role and regulatory mechanism of MLLT11 in BCs have never been explored. In the present study, we examined the biological functions and molecular mechanisms of miR-411 in human BC and its role in the regulation of MLLT11 protein expression.
Results
miR-411 Was Downregulated in BC Tissues and Cell Lines
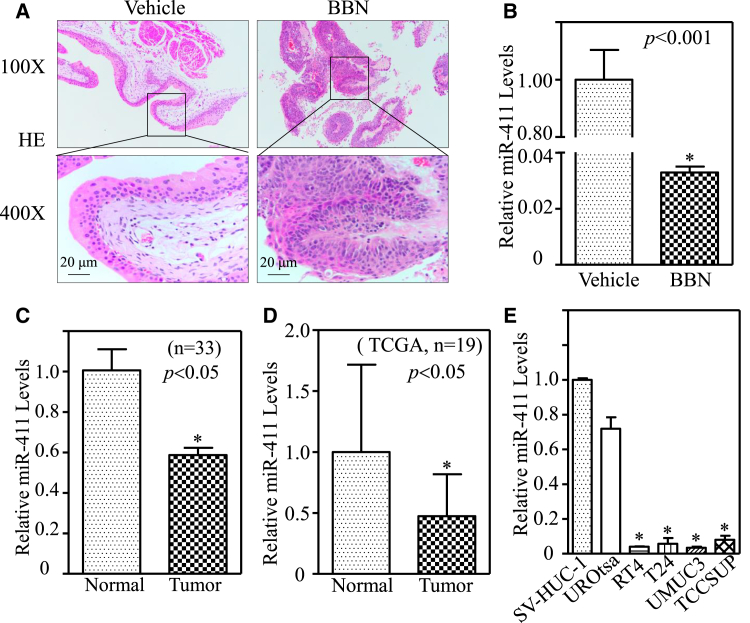
The experimental model of mouse BC induced by BBN is an appropriate and validated model to study human BC development and evaluate the efficacy of therapeutic strategies.23, 24, 25 Mouse bladder tissues were collected from vehicle control and BBN-treated (0.05% in drinking water) mice; the BBN-treated group was diagnosed with high invasive BCs, whereas the vehicle control mice were shown to be normal (Figure 1A). To explore the possible role of miR-411 in BC development, we first utilized miRNA microarray chip (1,900 known miRNAs) to analyze expression levels of miRNAs in 10 mouse BC tissues collected from BBN-treated mice in comparison to vehicle-treated mouse bladder tissues, and the results showed that miR-411 was one of the miRNAs that were dramatically downregulated in BBN-induced BCs (data not shown). The downregulation of miR-411 in BBN-induced mouse BCs was further verified by real-time qPCR in comparison to that in mouse urothelial cells collected from vehicle control mice (Figure 1B; p < 0.05). The miR-411 downregulation was also consistently observed in human BC tissues as compared with the paired adjacent non-tumor bladder tissues (n = 33; Figure 1C; p < 0.05). Because of the limited samples, the TCGA database was also employed to analyze miR-411 expression in all available 19 pairs (BC versus normal bladder tissues; Table S1) of BC samples. The results showed that miR-411 was downregulated in BC tissues (Figure 1D; p < 0.05). The levels of miR-411 were also assessed in human BC cell lines (RT4, T24, UMUC3, and TCCSUP) and normal urothelial cell lines (SV-HUC-1 and UROtsa). As shown in Figure 1E, a similar expression trend of miR-411 was observed in BC cell lines in comparison to normal urothelial cell lines (p < 0.05).
Figure 1.
miR-411 Was Downregulated in BBN-Treated Mouse BCs, Human BCs, and Cell Lines
(A) H&E staining was performed to show mouse high-invasive BC that was collected from C57BL/6J mouse that was with drinking water containing BBN (0.05%; v/v) for 20 weeks. (B) miR-411 levels in urothelial cells collected from BBN-treated mice (n = 10) versus vehicle control mice (n = 10) were evaluated by real-time PCR and down-regulation was observed in BBN-treated mice as compared with the control group (*p < 0.05). (C) Total RNA was extracted from human BC tissues (tumor) and the paired adjacent normal tissues (normal) of 33 patients and then subjected to real-time PCR analyses to determine miR-411 expression levels. Data represent mean ± SD (*p < 0.05). (D) miR-411 expression levels in 19 BC tissues were compared with the paired normal tissues obtained from the TCGA BC database and miR-411 was down-regulated in BC patients (n = 19; *p < 0.05). (E) miR-411 levels in human BC cell lines (RT4, T24, UMUC3, and TCCSUP) were determined and compared with two normal urothelial cell lines (SV-HUC-1 and UROtsa) by real-time PCR. miR-411 expression was normalized to U6 expression; bars represent the mean ± SD, and the asterisk (*) indicates a significant change relative to the control group (p < 0.05).
To assess the clinical significance of miR-411 in BCs, the potential association of miR-411 expression levels with BC patient disease-free survival (DFS) was analyzed by using available data from the TCGA database. Kaplan-Meier survival analysis revealed that miR-411 downregulation was associated with poor prognosis in patients with BC (higher levels of miR-411 improve DFS; Figure S1). Taken together, above results indicate that miR-411 downregulation might play a role in BC development and has a potential to serve as a new prognostic biomarker for clinic BC patients.
Ectopic Expression of miR-411 Suppressed BC Cell Proliferation and Induced G2/M Cell Cycle Arrest in Human BC Cells
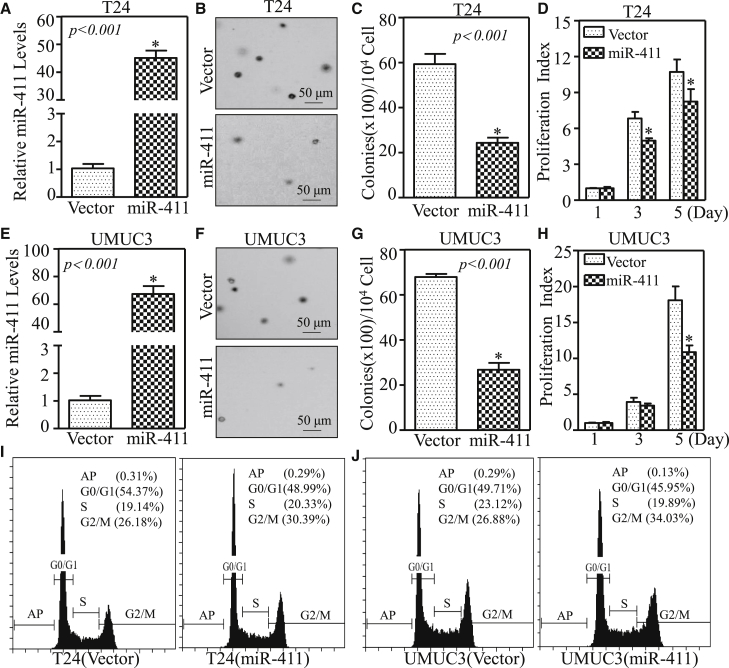
To evaluate the effects of miR-411 on the regulation of human BC, miR-411 was stably transfected into T24 and UMUC3 cells. The stable transfectants T24(miR-411) and UMUC3(miR-411) and their corresponding control vector transfectants were established by using puromycin selection and identified by real-time qPCR (Figures 2A and 2E; p < 0.05). The results from soft-agar assays indicated that miR-411 overexpression led to a dramatic decrease of anchorage-independent growth of BC cells (Figures 2B, 2C, 2F, and 2G; p < 0.001). Moreover, the assessment of cell proliferation using the ATP assay showed that the monolayer BC cell growth ability was also lower in T24(miR-411) and UMUC3(miR-411) cells than in control vector transfectants, especially at the later time point (5 days; Figures 2D and 2H; p < 0.05). These results suggest that miR-411 inhibits BC cell growth in vitro.
Figure 2.
Overexpression of miR-411 Suppressed Anchorage-Independent Growth and Monolayer Proliferation and Induced Cell Cycle Arrest in G2/M Phase in Human BC Cells
(A and E) miR-411 and control vector plasmids were stably transfected into T24 (A) and UMUC3 (E) cells, and stable transfectants were identified by real-time PCR. Bars represent the mean ± SD, Student’s t test was used to determine the p value, and the asterisk (*) indicates a significant increase relative to control vector cells (*p < 0.05). (B and F) A soft-agar assay was performed to determine the effect of miR-411 overexpression on T24 (B) and UMUC3 (F) anchorage-independent growth. Representative images of colonies in the indicated cells were captured by microscopy after 3 weeks of incubation. (C and G) Colonies with >32 cells in T24 (vector) versus T24 (miR-411) cells (C) and UMUC3 (vector) versus UMUC3 (miR-411) (G) were counted, and the results are presented as colonies per 10,000 cells from three independent experiments. The asterisk (*) indicates a significant decrease relative to the vector control cells (p < 0.05). (D and H) The effect of miR-411 on the monolayer proliferative rates of T24 (D) and UMUC3 (H) cells were evaluated by ATP assays using a CellTiter-Glo Luminescent Cell Viability Assay kit. The results are presented as the mean ± SD, and the asterisk (*) indicates a significant inhibition of proliferation relative to vector control cells (p < 0.05). (I and J) The indicated cells were seeded into 6-well plates and cultured to 70%–80% confluence; after synchronization in 0.1% FBS medium for 24 hr, cells were cultured in complete medium for another 24 hr and then subjected to cell cycle analysis by flow cytometry as described in Materials and Methods, and G2/M arrest was induced by miR-411 in T24 (I) and UMUC3 (J) cells.
To explore the molecular mechanisms underlying the anticancer activity of miR-411, the effect of miR-411 on the regulation of cell cycles progression was determined by flow cytometry in T24 and UMUC3 cells. As shown in Figures 2I and 2J, miR-411 induced a significant G2/M cell growth arrest, suggesting that the induction to G2/M phase growth arrests might be associated with the inhibitory effect of miR-411 in human BC cells.
miR-411 Targeted the 3′ UTR of mllt11 mRNA and Downregulated Its Protein Expression in Human BC Cells
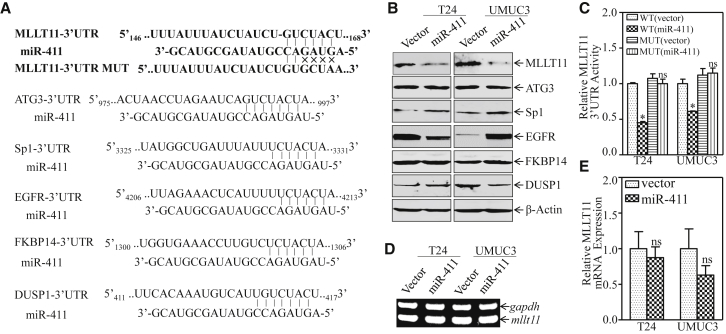
miRNAs play biological roles by modulating target gene expression via its binding to the 3′ UTR of target genes to cause mRNA stability alteration or protein translation suppression.26, 27 A bioinformatics tool (TargetScan, http://www.targetscan.org) was used to identify candidate targets of miR-411. As shown in Figure 3A, the mRNA 3′ UTR regions of MLLT11, ATG3, epidermal growth factor receptor (EGFR), dual-specificity phosphatase 1 (DUSP1), FKBP14, and Sp1 were predicted to contain potential miR-411-binding sites. The effect of miR-411 on regulating these genes was determined by comparing the protein expressions of miR-411 overexpressed cells and the vector controls transfectants. As shown in Figure 3B, MLLT11 was significantly down-regulated in both T24(miR-411) and UMUC3(miR-411) cells as compared with their levels in control vector transfectants, suggesting that MLLT11 could be a direct target of miR-411. Other genes were excluded to be targeted by miR-411 because ATG3 and Sp1 were up-regulated in miR-411-overexpressing cells, and EGFR was down-regulated in T24(miR-411) cells but upregulated in UMUC3(miR-411) cells; DUSP1 and FKBP14 showed no obvious changes (Figure 3B).
Figure 3.
MLLT11 Was the Direct Target of miR-411 in Human BC Cells
(A) Potential miR-411 targeting sequences included the 3′ UTR of MLLT11, ATG3, EGFR, DUSP1, Sp1, and FKBP14 mRNAs that were analyzed through TargetScan software. The predicted miR-411 binding site existed in the 3′-UTR of mllt11 mRNA and its mutants (MUTs) were generated in the binding site. (B) Cell lysates from the indicated cells were subjected to western blot to determine MLLT11, ATG3, EGFR, DUSP1, Sp1, and FKBP14 protein expression. β-actin was used as a loading control. (C) The pMIR-MLLT11 3′ UTR reporters were co-transfected with pRL-TK into the indicated cells. Twenty-four hours post-transfection, the transfectants were extracted for determination of the luciferase activity, and TK was used as the internal control. The results are shown as MLLT11 3′ UTR activity relative to vector control transfectant, and each bar indicates the mean ± SD from three independent experiments. The symbol (*) indicates a significant difference (p < 0.05). (D and E) Total RNAs were extracted from the indicated cells, and RT-PCR (D) and real-time PCR (E) were performed to determine mllt11 mRNA expression levels. gapdh was used as an internal control. Bars represent mean ±SD from three independent experiments; ns represents not significant.
To determine whether the effect of miR-411 on MLLT11 protein translation was due to its specific binding to a potential binding site in the mllt11 mRNA 3′ UTR, mllt11 mRNA 3′ UTR-driven luciferase reporter (wild-type [WT]), and mllt11 mRNA 3′ UTR mutant luciferase reporter (MUT), constructs were generated using the pMIR-report luciferase vector as shown in Figure 3A. WT and mutant mllt11 3′ UTR luciferase reporter vectors were transiently transfected into T24(vector), T24(miR-411), UMUC3(vector), and UMUC3(miR-411) cells with pRL-TK. As shown in Figure 3C, miR-411 significantly reduced mllt11 3′ UTR WT luciferase reporter activity in both cells, whereas mutation of the miR-411-binding site at the mllt11 mRNA 3′ UTR-impaired miR-411-mediated inhibition of mllt11 3′ UTR luciferase reporter activity. This result indicates that miR-411 binds to the mllt11 3′ UTR directly and regulates MLLT11 protein expression. To further investigate the molecular mechanism underlying the regulation of MLLT11 by miR-411, RT-PCR and real-time PCR were performed to examine mRNA expression (Figures 3D and 3E). The results showed no significant differences in the expression levels of mllt11 mRNA, suggesting that miR-411 had no effect on mllt11 mRNA expression. This excluded the possibility that miR-411 affected mllt11 expression at the RNA degradation level. Combined with the results of MLLT11 protein expression in T24(miR-411) and UMUC3(miR-411) cells, we anticipated that miR-411 downregulated MLLT11 by inhibiting protein translation. Taken together, these results demonstrate that MLLT11 is a direct target gene of miR-411 in BC in vitro.
MLLT11 as a miR-411 Downstream-Regulated Gene Mediated Its Effect on Inhibiting BC In Vitro
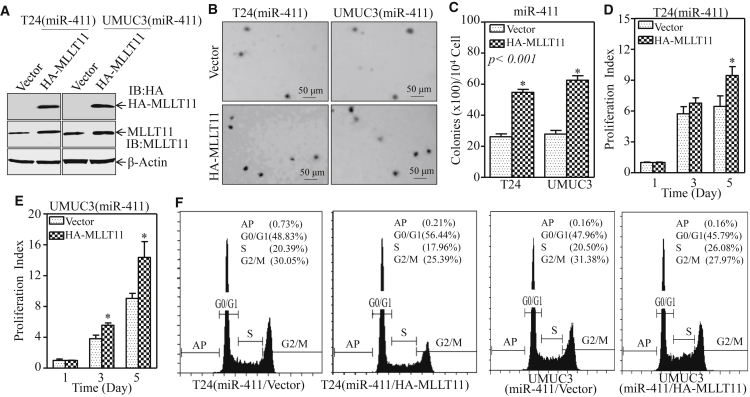
Our results showed that miR-411 inhibited BC cell proliferation and anchorage-independent growth and MLLT11 was a direct target of miR-411. To determine whether MLLT11 was responsible for the inhibitory effect of miR-411 on BC cell proliferation and anchorage-independent growth, T24(miR-411) and UMUC3(miR-411) cells were stably transfected with a hemagglutinin (HA)-tagged MLLT11 plasmid to restore MLLT11 expression (Figure 4A). As shown in Figures 4B–4F, ectopic expression of HA-MLLT11 markedly increased cell proliferation and anchorage-independent growth and reversed the G2/M growth arrest induced by miR-411 compared with those in scramble control vector cells. These results suggested that overexpression of HA-MLLT11 reversed the inhibitory effect of miR-411 on BC cells. Collectively, above data provide evidence that down-regulation of the MLLT11 protein is an important event associated with the cancer inhibitory activities of miR-411 in BCs.
Figure 4.
MLLT11 Mediated miR-411 Promoting BC Cell Growth
(A) HA-MLLT11 and its vector control plasmids were transfected into T24(miR-411) and UMUC3(miR-411) cells, and cell lysates from the indicated cells were subjected to western blot to analyze HA-MLLT11 and MLLT11 expression. (B) A soft-agar assay was performed to determine the effect of HA-MLLT11 on anchorage-independent growth of T24 (miR-411) and UMUC3 (miR-411) cells. Representative images of colonies from the indicated cells were captured under microscopy after 3 weeks of incubation. (C) The number of colonies was scored and presented as colonies per 104 seeded cells. Bars represent the mean ± SD, Student’s t test was used to determine the p value, and the asterisk (*) indicates a significant increase (p < 0.05). (D and E) The effect of HA-MLLT11 on the monolayer proliferative rates of T24(miR-411) (D) and UMUC3(miR-411) (E) cells was evaluated by ATP assay using the CellTiter-Glo Luminescent Cell Viability Assay kit. The results are presented as the mean ± SD, and the asterisk (*) indicates a significant increase of the proliferation index compared with vector control cells (p < 0.05). (F) The indicated cells were seeded into 6-well plates and cultured to 70%–80% confluence; after synchronization in 0.1% FBS medium for 24 hr, cells were cultured in complete medium for another 24 hr and then subjected to cell cycle analysis by flow cytometry.
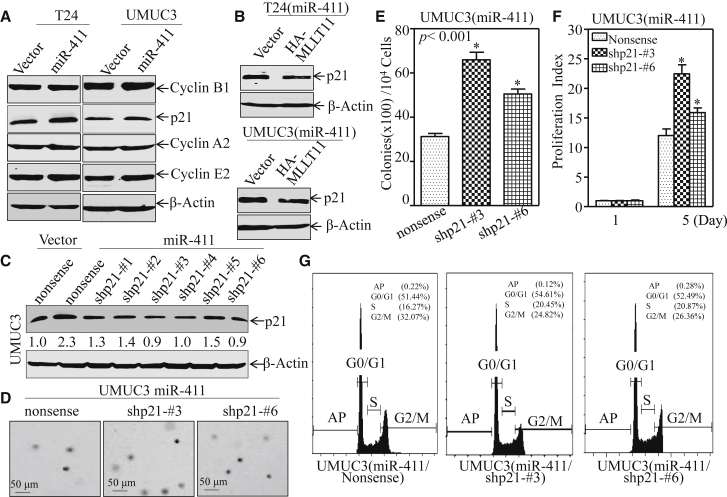
The miR-411-MLLT11 Pathway Inhibited BC Cell Growth by Upregulating the G2/M Cell Cycle Inhibitor p21
To elucidate the molecular mechanism leading to G2/M growth arrests induced by miR-411 in BC, the effect of miR-411 on major cell cycle regulators associated with the G2/M phase was examined by western blot. The results showed that ectopic expression of miR-411 did not affect the expression of the cell cycle regulatory proteins cyclin B1, cyclin A2, and cyclin E2, whereas it affected p21, which was remarkably increased in both T24(miR-411) and UMUC3(miR-411) transfectants in comparison to their corresponding scramble vector transfectants (Figure 5A). Consistently, introduction of MLLT11 into miR-411-overexpressing cells could completely reverse the p21 expression (Figure 5B). Because p21 is a well-known cell cycle inhibitor involved in cell cycle regulation,28, 29 we anticipate that p21 acts as a MLLT11 downstream effector being responsible for the miR-411 inhibition of BC growth. To test this notion, p21 expression was knocked down by short hairpin RNA (shRNA) in miR-411-overexpressing UMUC3 cells; western blot analysis showed that shp21 No. 3 and shp21 No. 6 were able to knock down p21 successfully (Figure 5C). The results obtained showed that knockdown of p21 reversed the inhibitory effect of miR-411 on both monolayer growth and anchorage-independent growth in UMUC3 cells (Figures 5D–5F). Flow cytometry also indicated that UMUC3 (miR-411/shp21 No. 3) and UMUC3 (miR-411/shp21 No. 6) exhibited a reversed effect of miR-411 on inducing G2/M cell cycle arrest (Figure 5G). These results further demonstrate that p21 contributes to miR-411-mediated G2/M cell cycle arrest and anchorage-independent growth in human BCs.
Figure 5.
Ectopic Expression of miR-411 Induced Cell Cycles Arrest in G2/M Phase by Up-regulating p21 Protein in Human BC Cells
(A) Cell lysates from the indicated cells were subjected to western blot to analyze expression of p21, cyclin B1, cyclin A2, and cyclin E2. β-actin was used as an internal control. (B) Western blot was used to determine the expression of p21 in T24 (miR-411/HA-MLLT11) and UMUC3 (miR-411/HA-MLLT11) cells in comparison to their vector control transfectants. β-actin was used as an internal control. (C) UMUC3 (miR-411) cells stably transfected with shRNA targeting human p21 or its nonsense control plasmid. The cell lysates from the indicated transfectants were subjected to western blot to assess the knockdown efficiency of the p21 protein. (D) UMUC3 (miR-411/nonsense), UMUC3 (miR-411/shp21 No. 3), and UMUC3 (miR-411/shp21 No. 6) cells were subjected to soft-agar assay to determine the effect of p21 on miR-411-induced inhibition of anchorage-independent growth. The images of colonies from the indicated cells were captured under microscopy after 3 weeks of incubation. (E) The number of colonies was scored and presented as colonies per 10,000 cells. The symbol (*) indicates a significant increase in comparison with UMUC3 (miR-411/Nonsense) transfectants (*p < 0.05). (F) The ATP assay was used to determine the monolayer proliferative rates of UMUC3(miR-411/nonsense), UMUC3 (miR-411/shp21 No. 3), and UMUC3 (miR-411/shp21 No. 6) cells. The asterisk (*) indicates a significant increase compared with UMUC3 (miR-411/nonsense) cells. (G) Cell cycle distribution in UMUC3 (miR-411/nonsense), UMUC3 (miR-411/shp21 No. 3), and UMUC3 (miR-411/shp21 No. 6) cells was determined by flow cytometry after cells were synchronization in 0.1% FBS medium for 24 hr and then cultured in 10% FBS medium for another 24 hr.
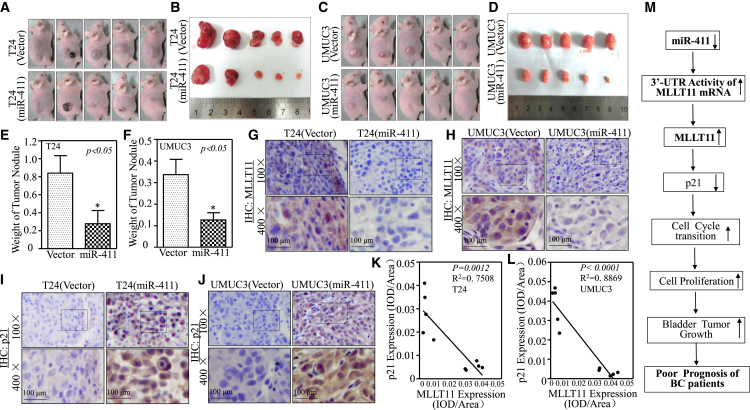
miR-411 Repressed Tumor Growth In Vivo in a Xenograft Nude Mouse Model
To extend our in vitro finding that miR-411 inhibits BC cell growth in vivo, a xenograft tumorigenic nude mouse model was employed to examine the effect of miR-411 on tumorigenicity in both T24 and UMUC3 cells. The results showed that miR-411 overexpression markedly attenuated xenograft tumor growth compared with that in scramble vector control group (p < 0.05; n = 5; Figures 6A–6F and S2), revealing the strong inhibitory effect of miR-411 on bladder tumor growth in vivo. Immunohistochemistry (IHC) staining of MLLT11 expression showed that MLLT11 expression was decreased in tumor tissues obtained from tumor-bearing mice injected with miR-411-overexpressed cells as compared with that in the mice injected with the corresponding vector cells. In contrast to MLLT11 expression, p21 expression was upregulated in tumor tissues obtained from tumor-bearing mice injected within miR-411-overexpressed cells (Figures 6G–6J). Quantitative analysis of the relative expression of MLLT11 and p21 in tumor tissues obtained from xenograft nude mice revealed that MLLT11 and p21 were negatively correlated in response to miR-411 overexpression (Figures 6K and 6L), which was consistent with our in vitro results and supports our conclusion that miR-411 acted as a strong tumor suppressor in human BCs and its downregulation plays a critical role in BC development.
Figure 6.
miR-411 Inhibited Tumor Growth Accompanied with Attenuation of MLLT11 In Vivo and a Negatively Correlative Expression of MLLT11 with p21 in Tumors
(A and C) Female BALB/c athymic nude mice were divided into four groups and subcutaneously injected with T24(vector), T24(miR-411) (A) and UMUC3(vector), UMUC3 (miR-411) (C) cells. The mice were photographed at 5 weeks after cell injection. (B and D) The mice injected with T24(vector), T24(miR-411) cells (B) and UMUC3(vector), UMUC3(miR-411) cells (D) were sacrificed and the tumor were surgically removed and imaged. (E & F) The tumor removed from T24(miR-411) group (E) and UMUC3(miR-411) group (F) were weighed. Symbol “*” indicates a significant difference between vector group and miR-411 overexpression group (P < 0.05). (G and I) Representative IHC images showing the expression of MLLT11 (G) and p21 (I) in T24(vector) and T24(miR-411) tumor tissues. (H & J) Representative IHC images showing the expression of MLLT11 (H) and p21 (J) in UMUC3(vector) and UMUC3(miR-411) tumor tissues. (K) The negative correlative expression of MLLT11 with p21 was analyzed in T24(vector) and T24(miR-411) tumor tissues. (L) The negative correlative expression of MLLT11 with p21 was analyzed in UMUC3(vector) vs. UMUC3(miR-411) tumor tissues. (M) The proposed schematic for the mechanism underlying miR-411 inhibition of human BC growth.
Discussion
The diverse functions of miRNAs are mediated by their role in regulating the expression of various genes involved in mammalian development and carcinogenesis. Identification of novel miRNA in tumor biology and understanding the mechanisms underlying the action of individual miRNAs would be advantageous for the diagnosis, prognosis, and treatment of many cancers. Despite accumulating evidence showing the function of various miRNAs in cancers, available information about the miRNAs and their function in BC remains to be limited. Here, we first discovered that miR-411 was downregulated in a BBN-induced BC mouse model, and this was validated in BC tissues and cell lines. Our data suggest a potential diagnostic/prognostic role of miR-411 in predicting survival and indicate that miR-411 is a tumor suppressor in BC. Downregulation of miR-411 up-regulated the downstream target MLLT11 and in turn attenuated p21 expression, resulting in inhibiting G2/M cell cycle phase transition, consequently increasing BC cell proliferation and tumor growth. These findings not only identify miR-411 as a new miRNA for inhibiting BC growth but also provide a novel scenario linking miR-411 to favorable prognosis (Figure 6M).
Previous reports suggested that miR-411 is a multi-functional molecule that plays critical roles in a variety of biological processes of various human cancers. The expression of miR-411 in cancer is controversial, as it is up-regulated and plays an oncogenic role in hepatocellular carcinoma30 and is up-regulated in facioscapulohumeral muscular dystrophy (FSHD) myoblasts in comparison with control myoblasts,31 whereas it acts as a tumor suppressor in breast cancer.32 In the present study, we evaluated the relationship between miR-411 and patient survival in BC using the TCGA database. The results indicated that miR-411 was positively correlated with disease-free survival. Although studies reported that miR-411 could regulate MMP13 or Sp1 in relation to cancer invasion, we found that miR-411 had no effect on BC cell migration in our wound-healing assay (Figure S3). Taken together, these results indicate that miR-411 is an important anticancer molecule in BCs and may serve as a new target to improve prognosis of BC patients.
MLLT11 was identified with AML patients with chromosomal abnormalities.33 MLLT11 promotes the emergence and migration of bone marrow pro-thymocytes to the thymus during T cell development through interaction with the Notch signaling pathway.34 MLLT11 is an oncogenic factor involved in leukemia development, thyroid tumorigenesis, and metastasis.33 However, the clinical significance of MLLT11 in solid tumors has not been systematically explored to date. In accordance with the classic mechanism of miRNA, our team first found and confirmed that MLLT11 was the direct target of miR-411 in BC by binding to 3′ UTR and assessing its activity. Sp1 was another target candidate of miR-411 screened from TargetScan. Sp1 is a transcription factor involved in the regulation of many genes; it plays an important role in cancer cell proliferation and survival by regulating the expression of oncogenes.35 Studies suggest that Sp1 is a direct target of miR-411 in breast cancer and is involved in the inhibition of cell growth, migration, and invasion.36 However, in the present study, miR-411 had no obvious effect on Sp1 protein expression in BC. DUSP1 is involved in various processes, including proliferation and differentiation, and the role of DUSP1 in different tumors is also controversial.37 DUSP1 plays a tumor suppressor role in prostate, ovarian, colon, and gastric cancers. In prostatic cancer, DUSP1 promotes cancer progression.38 In the present study, although DUSP1 was screened as a potential target of miR-411 in BC, its protein expression was not suppressed by miR-411, suggesting that it is not a target of miR-411 in BC. EGFR is overexpressed in many solid malignancies. It acts as an oncogene and affects tumor cell proliferation and transformation.39 A previous study showed that p85α enhances EGFR mRNA stability and EGF-induced malignant cellular transformation.40 However, the effect of miR-411 on EGFR expression differed between T24(miR-411) and UMUC3(miR-411) cells. Therefore, there is little evidence that EGFR is regulated by miR-411 in BC.
The present study showed that miR-411 up-regulated p21 expression and induced G2/M cell cycle arrest, and knockdown of p21 in UMUC3(miR-411) cells reversed the inhibitory effect of miR-411 on cell monolayer and anchorage-independent growth, as well as G2/M arrest. These results indicate that the anti-cancer effect of miR-411 may be attributed to its induction of p21 expression. Our results define p21 acting as a downstream molecule of miR-411 in BC tumor biology. In addition, we showed a negative relationship between p21 and MLLT11 in response to miR-411 in BC cells and a negative correlation in tumor tissues. The association of MLLT11 with p21 provide novel information insight into understanding the anticancer mechanism of miR-411 in BCs. However, the mechanism by which MLLT11 regulates the expression of p21 needs to be further investigated.
In summary, the present study defines that miR-411 is down-regulated in both human and mouse BCs, whereas miR-411 directly inhibits oncogenic MLLT11 translation, revealing the suppressive effect of miR-411 in BCs. miR-411 overexpression suppressed BC cell monolayer and anchorage-independent growth and induced G2/M cell cycle arrest. The expression of miR-411 was associated with disease-free survival; the patients with higher miR-411 have better DFS in contrast to the patients with lower miR-411 in TCGA database. Elevation of miR-411 level in BCs may serve as a therapeutic biomarker for the treatment of BC patients.
Materials and Methods
Plasmids, Antibodies, and Reagents
miR-411 and its control vector plasmids were purchased from GenePharma (No. C5619; Shanghai, China); human MLLT11 cDNA was cloned into the pcDNA3.1+ HA vector through the KpnI and EcoRI sites. The human mllt11 3′ UTR was cloned into the pMIR-report luciferase vector through the KpnI and HindIII sites. mllt11 3′ UTR point mutation was amplified from the WT template by overlap PCR using the following primers: forward: 5′-ATTTATCTATCTGTCCTAACCATTTCTCTCAAAA-3′; reverse: 5′-TTTTGAGAGAAATGGTTAGGACAGATAGATAAT-3′. The shRNA for p21 was purchased from Open Biosystems (No. RHS4531-EG1026; Lafayette, CO, USA). The antibodies against cyclin A2 (No. 4656), cyclin B1 (No. 12231), cyclin E2 (No. 4132), EGFR (No. 4267S), and ATG3 (No. 3415) were obtained from Cell Signaling Technology (Boston, MA, USA). The antibodies against β-actin (No. ab8224), MLLT11 (No. ab109016), and DUSP1 (No. ab195261) were bought from Abcam (Cambridge, UK), and FKBP14 (No. GTX81835) was obtained from GeneTex (Irvine, CA, USA). Anti-Sp1 antibody was bought from Santa Cruz Biotechnology (No. sc-59; Santa Cruz, CA, USA).
Cell Culture and Transfection
The human BC cell lines RT4, T24, UMUC3, and TCCSUP and the normal urinary epithelial cell lines SV-HUC-1 and UROtsa were used in the study. All cancer cell lines and SV-HUC-1 were subjected to DNA tests and authenticated before use for the studies. UROtsa was generously provided by Dr. Scott H. Garrent (University of North Dakota, Grand Forks, ND, USA).23, 41 UMUC3 cells were maintained at 37°C in a 5% CO2 incubator in DMEM (No. 11995-065; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (No. 10437-028; Gibco). T24 cells were cultured with a 1:1 mixture of DMEM/Ham’s F12 medium (No. 10565-018; Gibco) supplemented with 5% FBS. UROtsa and RT4 cells were cultured with 1640 (No. 11875-093; Gibco) supplemented with 10% FBS, and TCCSUP cells were cultured with MEM (No. 11095-080; Gibco) supplemented with 10% FBS. SV-HUC-1 cells were cultured with F-12K medium supplemented with 10% FBS (No. 21127-022; Gibco) medium. Stable transfections were performed with constructs using PolyJet DNA In Vitro Transfection Reagent (SignaGen Laboratories, Gaithersburg, MD, USA) according to the manufacturer’s instructions, and stable transfectants were selected with puromycin (0.2–0.3 μg/mL) or hygromycin B (200–400 μg/mL) for 3 or 4 weeks according to the different antibiotic resistance plasmids transfected.23, 42
Lentivirus Packaging and Infection
Stable p21 knockdown cells were established by lentivirus infection. Briefly, 293T cells were seeded into 6-well plates until reaching 60%–70% confluence. The transfection complex consisted of 2 μg DNA plasmid and two packaging vectors (1.2 μg pMD2.G and 1.2 μg psPAX2) diluted in 100 μL serum-free DMEM and 4 μL PolyJet DNA diluted in 100 μL serum-free DMEM. The diluted PolyJet reagent was immediately added to the diluted DNA solution and incubated for 15 min at room temperature to allow transfection complexes to form after shaking and mixing. The complex was added to 293T cells, and viral supernatant fractions produced at 48 hr were purified by centrifugation at 2,500 rpm for 30 min and filtration through a 0.45-μm-pore-size membrane. The viral supernatant fractions of 2.0 mL were used to transfect target cells at a ratio of 100 μL:1 mL (viral supernatant fractions versus medium). Cells were cultured in 6-well plates, and after 48 hr, the medium was replaced with fresh complete medium containing 3 μg/mL puromycin. Similar experiments were performed with all cells until the control cells (without infections) completely died (2 or 3 days) in the puromycin medium.43
Anchorage-Independent Growth
The potential miR-411 inhibitory effect on anchorage-independent growth (soft-agar assay) in human BC cells was determined in the UMUC3 and T24 cell lines. In brief, 1 × 104 of UMUC3(miR-411) or T24(miR-411) stable transfectants and control vector transfectants were exposed to basal medium eagle (BME) containing 0.33% agar and seeded on the bottom layer of 0.5% agar in 10% FBS BME in each well of 6-well plates. The cultures were maintained at 37°C in a 5% CO2 incubator for 3 or 4 weeks, and the cell colonies with >32 cells were scored. Colonies were observed and counted under a microscope (DMi1; Leica Microsystems, Germany). The results were presented as the mean ± SD of colony number per 10,000 seeded cells in soft agar.44
Monolayer Cell Proliferation
Confluent monolayers of cells were trypsinized with 0.25% trypsin (No. 25200-072; Gibco), and 0.4 × 103 viable cells suspended in 200 μL complete medium were added to each well of 96-well plates. After adherence, cells were synchronized by replacing with 0.1% FBS medium for 24 hr and then cultured with complete medium for the indicated days. The proliferation index of the cells was determined using a Cell Titer-Glo Luminescent Cell Viability Assay kit (Promega, Madison, WI, USA), and luminescence was measured with a luminometer (Centro LB 960; Berthold, Germany).
Cell Cycle Analyses
The cells were trypsinized and washed with ice-cold PBS (No. 8116531; Gibco) two times and fixed in ice-cold 70% ethanol at 4°C overnight. Then, the cells were washed with PBS two times, incubated with RNase A (No. KGA511; KeyGEN BioTECH, Nanjing, China) at 37°C for 30 min to remove RNA, and stained with propidium iodide (PI) (No. KGA512, KeyGEN BioTECH, Nanjing, China) at 4°C for 30 min. The DNA content was determined by flow cytometry using a CytoFLEX (Beckman Coulter, San Diego, CA, USA), and CytExpert software was used to analyze the results.
Western Blot
Cells were lysed with boiling buffer (1% SDS, 1 mM Na3VO4, and 10 mM Tris-HCl [pH 7.4]) on ice, heated at 100°C for 5–10 min, and exposed to ultrasound to break the nucleic acids. Protein concentration was determined using a BCA Protein Assay kit (No. 23228; Thermo Scientific, USA). The cell extracts were subjected to SDS-PAGE, transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA), and probed with the indicated primary antibodies (rabbit anti-MLLT11, 1:3,000; rabbit anti-ATG3, 1:800; rabbit anti-SP1, 1:500; rabbit anti-EGFR, 1:800; rabbit anti-FKBP14, 1:500; rabbit anti-DUSP1, 1:1,000; mouse anti-cyclin A2, 1:1,000; rabbit anti-cyclin B1, 1:1,000; rabbit anti-cyclin E2, 1:1,000; rabbit anti-p21, 1:800; and mouse anti-β-actin, 1:5,000) at 4°C overnight, followed by incubation with alkaline phosphatase (AP)-conjugated second antibodies (1:2,000) for 3 hr at 4°C. Signals were detected using the ECF (No. RPN5787; GE Healthcare, Pittsburgh, PA, USA) western blotting system and the Typhoon FLA 7000 imager (GE Healthcare).45, 46
RT-PCR
Total RNA was extracted with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and cDNAs were synthesized with oligodT primers using the SuperScript First-Strand Synthesis system (Invitrogen, Grand Island, NY, USA). The target mRNA amount present in the cells was measured by semiquantitative RT-PCR. The specific primers were 5′-GACTCATGACCACAGTCCATGC-3′ and 5′-CAGGTCAGGTCCACCACTGA-3′ for human GAPDH (223 bp) as a loading control, and the MLLT11 cDNA fragment (159 bp) was amplified using human-MLLT11-specific PCR primers (forward: 5′-GGGACCCTGTGAGTAGCCAG-3′ and reverse: 5′-CTGCAGTTGCTTGCCCGA-3′). The PCR products were separated on 2% agarose gels and stained with gold view (No. G8140; Solarbio, Beijing, China). The results were imaged with a Doc XRS+ system and analyzed with Lab Software (Bio-Rad).
Real-Time PCR
Total miRNAs were extracted using the miRNeasy Mini Kit (QIAGEN, Valencia, CA, USA). Total RNA (1.0 μg) was used for reverse transcription following the manufacturer’s instructions, and miRNA expression was determined by the Q6 real-time PCR system (Applied Biosystems, Carlsbad, CA, USA) using the miScript PCR Kit (QIAGEN). The primer for miR-411 (5′-TAGTAGACCGTATAGCGTACG-3′) was synthesized by Sunny Biotechnology (Shanghai, China), and U6 was used as an internal loading control. Cycle threshold (CT) values were determined, and the relative expression of miRNAs was calculated using the values of 2−ΔΔCT.
Dual-Luciferase Reporter Assay
UMUC3 (vector), UMUC3 (miR-411), T24 (vector), and T24 (miR-411) cells were transiently co-transfected with the mllt11 3′ UTR WT or mllt11 3′ UTR mutant luciferase reporter and pRL-TK for 24 hr. The transfectants were washed with PBS and extracted with passive lysis buffer on a rocking platform at room temperature for 15 min to ensure complete lysis of the cells. The lysates were transferred to a 96-well plate, and the Luciferase Assay Reagent II was added to examine luciferase activity. The Stop&Glo Reagent was added into the wells to detect pRL-TK activity using a luminometer (Centro LB 960; Berthold, Germany) using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).47
Xenograft Model in Nude Mice In Vivo
All animal studies were performed in the animal institute of Wenzhou Medical University according to the protocols approved by the Laboratory Animal Ethics Committee of Wenzhou Medical University and Laboratory Animal Centre of Wenzhou Medical University. The female BALB/c athymic nude mice (3 or 4 weeks old) were purchased from Shanghai Silaike Experimental Animal Company (license No. SCXK; Shanghai 2010-0002). Mice aged 5 or 6 weeks were randomly divided into four groups and subcutaneously injected with 0.1 mL T24 (miR-411) and T24 (vector) or UMUC3 (miR-411) and UMUC3 (vector) cells (2 × 106 suspended in 100 μL PBS) at the right side of the back region. After 4 or 5 weeks, the mice were sacrificed and the tumor was surgically removed, imaged, and weighed. One-third of the tumor was fixed in 4% paraformaldehyde for IHC, one-third was frozen at −80°C, and the remaining third was used to extract RNA if necessary.
IHC
Tumor tissues obtained from the sacrificed mice were formalin fixed and paraffin embedded. For IHC staining, antibodies specific against MLLT11 (1:80; No. ab109016; Abcam, Cambridge, UK) and p21 (No. sc-397; 1:30; Santa Cruz, CA, USA) were used. The staining was performed using a kit from Boster Bio-Engineering Company (No. SA1022; Wuhan, China) according to the manufacturer’s instructions. The resultant immunostaining images were captured using the Nikon Eclipse Ni microsystem (Nikon DS-Ri2, Japan). Protein expression levels were analyzed by calculating the integrated optical density per stained area (IOD/area) using Image-Pro Plus version 6.0 (Media Cybernetics).48
Clinical Specimens
The present study was approved by the Protection of Human Subjects Committee of The First Affiliated Hospital of Wenzhou Medical University (Wenzhou, Zhejiang, China). In total, 33 pairs of human BC tissues and their corresponding adjacent normal tissues were obtained with patient approval (Table S2). The tissue samples were snap frozen in liquid nitrogen at the time of surgery, RNA was extracted, and synthesized cDNA was stored at −80°C.
Statistical Analysis
The Student’s t test was used to determine the significance of differences between different groups. The differences were considered to be significant at p < 0.05.
Author Contributions
H.H. and C.H. conceived and designed the study. H.J., W.S., C.C., J.G., and X.H. detected the cells’ biological function; conducted the RT-PCR assays; carried out the soft agar, ATP assay, cell flow cytometry, western blot, and luciferase reporter assays; and performed the statistical analysis. S.W., H.Y., H.L., and Y.Z. carried out the animal studies and the IHC staining assays. L.Z. diagnosed and classified the BBN-induced mouse bladder tissues and human bladder cancer tissue pathologic types. G.J. and D.R. provided the human bladder cancer tissue specimens. H.H., H.J., Q.X., and C.H. drafted the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The results published here are in whole based upon data generated by the TCGA Research Network (https://cancergenome.nih.gov/). We also thank the participants, specimen donors, and research groups who developed the TCGA bladder cancer dataset resource for their contributions in database construction. This work was partially supported by the Natural Science Foundation of China (NSFC81601849, NSFC81702530, and NSFC81773391), the Wenzhou Science and Technology Bureau (Y20170028 and Y20160075), Wenzhou Medical University (89216021), and grants from the NIH/NCI (CA177665 and CA165980) and NIH/NIEHS (ES000260).
Footnotes
Supplemental Information includes three figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.03.003.
Contributor Information
Qipeng Xie, Email: pandon2002@163.com.
Chuanshu Huang, Email: chuanshu.huang@nyumc.org.
Supplemental Information
References
- 1.El-Arabey A.A. New insight for metformin against bladder cancer. Genes Environ. 2017;39:13. doi: 10.1186/s41021-017-0074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinney C.P., McConkey D.J., Millikan R.E., Wu X., Bar-Eli M., Adam L., Kamat A.M., Siefker-Radtke A.O., Tuziak T., Sabichi A.L. Focus on bladder cancer. Cancer Cell. 2004;6:111–116. doi: 10.1016/j.ccr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Kamat A.M., Hahn N.M., Efstathiou J.A., Lerner S.P., Malmström P.-U., Choi W., Guo C.C., Lotan Y., Kassouf W. Bladder cancer. Lancet. 2016;388:2796–2810. doi: 10.1016/S0140-6736(16)30512-8. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 5.Chen W., Zheng R., Zhang S., Zeng H., Xia C., Zuo T., Yang Z., Zou X., He J. Cancer incidence and mortality in China, 2013. Cancer Lett. 2017;401:63–71. doi: 10.1016/j.canlet.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 6.Chen W., Zheng R., Zhang S., Zhao P., Zeng H., Zou X., He J. Annual report on status of cancer in China, 2010. Chin. J. Cancer Res. 2014;26:48–58. doi: 10.3978/j.issn.1000-9604.2014.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W., Zheng R., Zeng H., Zhang S., He J. Annual report on status of cancer in China, 2011. Chin. J. Cancer Res. 2015;27:2–12. doi: 10.3978/j.issn.1000-9604.2015.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 9.Mahdavifar N., Ghoncheh M., Pakzad R., Momenimovahed Z., Salehiniya H. Epidemiology, incidence and mortality of bladder cancer and their relationship with the development index in the world. Asian Pac. J. Cancer Prev. 2016;17:381–386. doi: 10.7314/apjcp.2016.17.1.381. [DOI] [PubMed] [Google Scholar]
- 10.Luke C., Tracey E., Stapleton A., Roder D. Exploring contrary trends in bladder cancer incidence, mortality and survival: implications for research and cancer control. Intern. Med. J. 2010;40:357–362. doi: 10.1111/j.1445-5994.2009.01980.x. [DOI] [PubMed] [Google Scholar]
- 11.Ploeg M., Aben K.K., Kiemeney L.A. The present and future burden of urinary bladder cancer in the world. World J. Urol. 2009;27:289–293. doi: 10.1007/s00345-009-0383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L., Cui Z., Liu Y., Bai Y., Lan F. MicroRNAs as biomarkers for the diagnostics of bladder cancer: a meta-analysis. Clin. Lab. 2015;61:1101–1108. doi: 10.7754/clin.lab.2015.150204. [DOI] [PubMed] [Google Scholar]
- 13.Yoshino H., Seki N., Itesako T., Chiyomaru T., Nakagawa M., Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat. Rev. Urol. 2013;10:396–404. doi: 10.1038/nrurol.2013.113. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B., Pan X., Cobb G.P., Anderson T.A. MicroRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 15.Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 16.Shenouda S.K., Alahari S.K. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 17.Zhou M., Wang S., Hu L., Liu F., Zhang Q., Zhang D. miR-199a-5p suppresses human bladder cancer cell metastasis by targeting CCR7. BMC Urol. 2016;16:64. doi: 10.1186/s12894-016-0181-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo J., Cao R., Yu X., Xiao Z., Chen Z. MicroRNA-223-3p inhibits human bladder cancer cell migration and invasion. Tumour Biol. 2017;39 doi: 10.1177/1010428317691678. 1010428317691678. [DOI] [PubMed] [Google Scholar]
- 19.Trang P., Weidhaas J.B., Slack F.J. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27(Suppl 2):S52–S57. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiberio P., Cavadini E., Callari M., Daidone M.G., Appierto V. AF1q: a novel mediator of basal and 4-HPR-induced apoptosis in ovarian cancer cells. PloS One. 2012;7:e39968. doi: 10.1371/journal.pone.0039968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Co N.N., Tsang W.P., Tsang T.Y., Yeung C.L.A., Yau P.L., Kong S.K., Kwok T.T. AF1q enhancement of γ irradiation-induced apoptosis by up-regulation of BAD expression via NF-кB in human squamous carcinoma A431 cells. Oncol. Rep. 2010;24:547–554. [PubMed] [Google Scholar]
- 22.Tse W., Meshinchi S., Alonzo T.A., Stirewalt D.L., Gerbing R.B., Woods W.G., Appelbaum F.R., Radich J.P. Elevated expression of the AF1q gene, an MLL fusion partner, is an independent adverse prognostic factor in pediatric acute myeloid leukemia. Blood. 2004;104:3058–3063. doi: 10.1182/blood-2003-12-4347. [DOI] [PubMed] [Google Scholar]
- 23.Huang H., Jin H., Zhao H., Wang J., Li X., Yan H., Wang S., Guo X., Xue L., Li J. RhoGDIβ promotes Sp1/MMP-2 expression and bladder cancer invasion through perturbing miR-200c-targeted JNK2 protein translation. Mol. Oncol. 2017;11:1579–1594. doi: 10.1002/1878-0261.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu J., Li Y., Chen C., Ma J., Sun W., Tian Z., Li J., Xu J., Liu C.S., Zhang D. NF-κB p65 overexpression promotes bladder cancer cell migration via FBW7-mediated degradation of RhoGDIα protein. Neoplasia. 2017;19:672–683. doi: 10.1016/j.neo.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iida K., Itoh K., Maher J.M., Kumagai Y., Oyasu R., Mori Y., Shimazui T., Akaza H., Yamamoto M. Nrf2 and p53 cooperatively protect against BBN-induced urinary bladder carcinogenesis. Carcinogenesis. 2007;28:2398–2403. doi: 10.1093/carcin/bgm146. [DOI] [PubMed] [Google Scholar]
- 26.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 27.Kim D.H., Saetrom P., Snøve O., Jr., Rossi J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C., Ge Q., Chen Z., Hu J., Li F., Song X., Xu H., Ye Z. A new double stranded RNA suppresses bladder cancer development by upregulating p21 (Waf1/CIP1) expression. BioMed Res. Int. 2015;2015:304753. doi: 10.1155/2015/304753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbas T., Dutta A. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia K., Zhang Y., Cao S., Wu Y., Guo W., Yuan W., Zhang S. miR-411 regulated ITCH expression and promoted cell proliferation in human hepatocellular carcinoma cells. Biomed. Pharmacother. 2015;70:158–163. doi: 10.1016/j.biopha.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Harafuji N., Schneiderat P., Walter M.C., Chen Y.W. miR-411 is up-regulated in FSHD myoblasts and suppresses myogenic factors. Orphanet J. Rare Dis. 2013;8:55. doi: 10.1186/1750-1172-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G., Zhang Y., Zhao X., Meng C., Ma L., Kong Y. MicroRNA-411 inhibited matrix metalloproteinase 13 expression in human chondrocytes. Am. J. Transl. Res. 2015;7:2000–2006. [PMC free article] [PubMed] [Google Scholar]
- 33.Park J., Schlederer M., Schreiber M., Ice R., Merkel O., Bilban M., Hofbauer S., Kim S., Addison J., Zou J. AF1q is a novel TCF7 co-factor which activates CD44 and promotes breast cancer metastasis. Oncotarget. 2015;6:20697–20710. doi: 10.18632/oncotarget.4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong Y., Li Z., Ji M., Tan A.C., Bemis J., Tse J.V., Huang G., Park J., Ji C., Chen J. MIR29B regulates expression of MLLT11 (AF1Q), an MLL fusion partner, and low MIR29B expression associates with adverse cytogenetics and poor overall survival in AML. Br. J. Haematol. 2011;153:753–757. doi: 10.1111/j.1365-2141.2011.08662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng X., Xu Z., Gu J., Huang H., Gao G., Zhang X., Li J., Jin H., Jiang G., Sun H., Huang C. Induction of miR-137 by isorhapontigenin (ISO) directly targets Sp1 protein translation and mediates its anticancer activity both in vitro and in vivo. Mol. Cancer Ther. 2016;15:512–522. doi: 10.1158/1535-7163.MCT-15-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo L., Yuan J., Xie N., Wu H., Chen W., Song S., Wang X. miRNA-411 acts as a potential tumor suppressor miRNA via the downregulation of specificity protein 1 in breast cancer. Mol. Med. Rep. 2016;14:2975–2982. doi: 10.3892/mmr.2016.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen J., Zhang Y., Yu H., Shen B., Liang Y., Jin R., Liu X., Shi L., Cai X. Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med. 2016;5:2061–2068. doi: 10.1002/cam4.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chintharlapalli S., Papineni S., Ramaiah S.K., Safe S. Betulinic acid inhibits prostate cancer growth through inhibition of specificity protein transcription factors. Cancer Res. 2007;67:2816–2823. doi: 10.1158/0008-5472.CAN-06-3735. [DOI] [PubMed] [Google Scholar]
- 39.Mooso B.A., Vinall R.L., Mudryj M., Yap S.A., deVere White R.W., Ghosh P.M. The role of EGFR family inhibitors in muscle invasive bladder cancer: a review of clinical data and molecular evidence. J. Urol. 2015;193:19–29. doi: 10.1016/j.juro.2014.07.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie Q., Guo X., Gu J., Zhang L., Jin H., Huang H., Li J., Huang C. p85α promotes nucleolin transcription and subsequently enhances EGFR mRNA stability and EGF-induced malignant cellular transformation. Oncotarget. 2016;7:16636–16649. doi: 10.18632/oncotarget.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin H., Xu J., Guo X., Huang H., Li J., Peng M., Zhu J., Tian Z., Wu X.R., Tang M.S., Huang C. XIAP RING domain mediates miR-4295 expression and subsequently inhibiting p63α protein translation and promoting transformation of bladder epithelial cells. Oncotarget. 2016;7:56540–56557. doi: 10.18632/oncotarget.10645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang Y., Cao Z., Hou Q., Ma C., Yao C., Li J., Wu X.R., Huang C. Cyclin d1 downregulation contributes to anticancer effect of isorhapontigenin on human bladder cancer cells. Mol. Cancer Ther. 2013;12:1492–1503. doi: 10.1158/1535-7163.MCT-12-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horn P.A., Topp M.S., Morris J.C., Riddell S.R., Kiem H.P. Highly efficient gene transfer into baboon marrow repopulating cells using GALV-pseudotype oncoretroviral vectors produced by human packaging cells. Blood. 2002;100:3960–3967. doi: 10.1182/blood-2002-05-1359. [DOI] [PubMed] [Google Scholar]
- 44.Luo W., Liu J., Li J., Zhang D., Liu M., Addo J.K., Patil S., Zhang L., Yu J., Buolamwini J.K. Anti-cancer effects of JKA97 are associated with its induction of cell apoptosis via a Bax-dependent and p53-independent pathway. J. Biol. Chem. 2008;283:8624–8633. doi: 10.1074/jbc.M707860200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin H., Yu Y., Hu Y., Lu C., Li J., Gu J., Zhang L., Huang H., Zhang D., Wu X.R. Divergent behaviors and underlying mechanisms of cell migration and invasion in non-metastatic T24 and its metastatic derivative T24T bladder cancer cell lines. Oncotarget. 2015;6:522–536. doi: 10.18632/oncotarget.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Y., Zhang D., Huang H., Li J., Zhang M., Wan Y., Gao J., Huang C. NF-κB1 p50 promotes p53 protein translation through miR-190 downregulation of PHLPP1. Oncogene. 2014;33:996–1005. doi: 10.1038/onc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding J., Li J., Chen J., Chen H., Ouyang W., Zhang R., Xue C., Zhang D., Amin S., Desai D., Huang C. Effects of polycyclic aromatic hydrocarbons (PAHs) on vascular endothelial growth factor induction through phosphatidylinositol 3-kinase/AP-1-dependent, HIF-1alpha-independent pathway. J. Biol. Chem. 2006;281:9093–9100. doi: 10.1074/jbc.M510537200. [DOI] [PubMed] [Google Scholar]
- 48.Jiang G., Wu A.D., Huang C., Gu J., Zhang L., Huang H., Liao X., Li J., Zhang D., Zeng X. Isorhapontigenin (ISO) inhibits invasive bladder cancer formation in vivo and human bladder cancer invasion in vitro by targeting STAT1/FOXO1 axis. Cancer Prev. Res. (Phila.) 2016;9:567–580. doi: 10.1158/1940-6207.CAPR-15-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.