Abstract
Roughly one third of all human cancers are attributable to the functional inhibition of the tumor suppressor protein p53 by its two negative regulators MDM2 and MDMX, making dual-specificity peptide antagonists of MDM2 and MDMX highly attractive drug candidates for anticancer therapy. Two pharmacological barriers, however, remain a major obstacle to the development of peptide therapeutics: susceptibility to proteolytic degradation in vivo and inability to traverse the cell membrane. Here we report the design of a fluorescent lanthanide oxyfluoride nanoparticle (LONp)-based multifunctional peptide drug delivery system for potential treatment of acute myeloid leukemia (AML) that commonly harbors wild type p53, high levels of MDM2 and/or MDMX, and an overexpressed cell surface receptor, CD33. We conjugated to LONp via metal-thiolate bonds a dodecameric peptide antagonist of both MDM2 and MDMX, termed PMI, and a CD33-targeted, humanized monoclonal antibody to allow for AML-specific intracellular delivery of a stabilized PMI. The resultant nanoparticle antiCD33-LONp-PMI, while nontoxic to normal cells, induced apoptosis of AML cell lines and primary leukemic cells isolated from AML patients by antagonizing MDM2 and/or MDMX to activate the p53 pathway. Fluorescent antiCD33-LONp-PMI also enabled real-time visualization of a series of apoptotic events in AML cells, proving a useful tool for possible disease tracking and treatment response monitoring. Our studies shed light on the development of antiCD33-LONp-PMI as a novel class of antitumor agents, which, if further validated, may help targeted molecular therapy of AML.
Introduction
Peptide inhibitors of intracellular protein-protein interactions (PPIs) involved in disease initiation and progression are increasingly recognized as potential leads for the development of new classes of therapeutics[1]. Due to their relatively specific and often high-affinity mode of action against protein targets, peptide therapeutics can in theory work at low doses in vivo with a more favorable toxicity profile than that of small molecule drugs[2, 3]. This ideal scenario could certainly play out against the backdrop that small molecule inhibitors are in general ineffective against PPIs[4]. In reality, however, the efficacy of peptide therapeutics is frequently limited by their poor proteolytic stability and inability to traverse the cell membrane [5, 6]. To alleviate these technical hurdles, various elaborate chemistries for peptide modification and delivery vehicles for peptide cargo have been developed, and significant progress has been made in ways of improving the pharmacological properties of peptide therapeutics for clinical use [6–8].
Among the commonly used peptide delivery systems are lipids[9] and biodegradable polymers[10] fabricated often in the forms of liposome, micelle, and dendrimer. However, lipid-based delivery vehicles are rapidly removed by the liver and spleen[11]; polymer-based ones, often highly positively charged, exhibit toxicity and non-specific cellular uptake[12]. Such drawbacks necessarily impede the development of peptide therapeutics for widespread clinical applications. Conceptually novel and clinically viable delivery systems are therefore needed in order to advance peptide drug discovery and development. Toward this end, nanoparticle-mediated peptide drug delivery holds great promise in overcoming many limitations of lipid- and polymer-based delivery systems [13, 14]. Conjugation of peptide cargos to nanoparticles substantially improves peptide resistance to proteolysis, membrane permeability, and bioavailability [6, 15]. In fact, nanoparticle-based drug delivery systems are particularly attractive in the treatment of solid tumors as nanoparticles are capable of actively accumulating through leaky blood vessels in diseased tissues – a phenomenon known as the enhanced permeability and retention (EPR) effect [16, 17]. Importantly, when appropriate materials are used to construct nanoparticles, they can be endowed with a powerful imaging capability for both therapeutic and diagnostic applications [18, 19], yielding multi-functional “theranostics” that combine disease tracking, drug delivery and treatment response monitoring [20].
Recent studies have demonstrated the superiority of lanthanide-doped nanoparticles (LDNp) as an imaging tool in biomedicine due to their excellent photoluminescence property and biocompatibility [15, 21]. LDNp loaded with appropriate therapeutic peptide cargos and disease-targeting molecules may be developed as a novel class of theranostics for clinical use [22–24]. In this work, we interrogated lanthanide oxyfluoride nanoparticles (LONp) as a drug delivery vehicle by conjugating a p53-activating dodecameric peptide termed PMI (TSFAEYWALLSP) [25] for potential treatment of acute myeloid leukemia (AML). Intracellular PMI kills AML cells by antagonizing MDM2 and/or MDMX – the two functional inhibitors of the tumor suppressor protein p53 [26–28]. To endow LONp-PMI with tumor targeting specificity, we also conjugated to the nanoparticle a monoclonal antibody against CD33, a receptor expressed at high levels on leukemic myeloid cells but not on normal hematopoietic pluripotent stem cells in the vast majority of AML patients [29, 30]. Our in vitro and ex vivo data as well as mechanistic studies fully validate the design of antiCD33-LONp-PMI as a novel class of peptide-based antitumor agents with therapeutic potential in the treatment of AML.
Results
Preparation and physicochemical properties of antiCD33-LONp-PMI
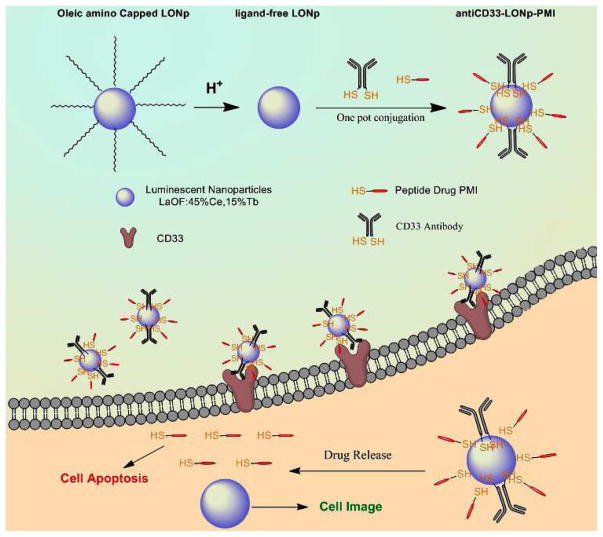
The strategy for the preparation of antiCD33-LONp-PMI is outlined in Fig. 1. Luminescent LONp nanoparticles composed of LaOF (40%), Ce (45%) and Tb (15%) were synthesized as described [22, 31]. Our anti-CD33 monoclonal antibody was derived from a humanized M195 clone [32], combined with four mutations in the Fc region to reduce its effector functions [33], and C-terminally extended by an extra Cys residue in the heavy chain for antibody conjugation. An extra Cys residue was also introduced to the C-terminus of PMI for peptide attachment. AntiCD33-LONp-PMI nanoparticles were obtained by a thiol-induced molecular assembly of PMI-Cys and AntiCD33-Cys with biocompatible LONp (Fig. 1). For comparison, we also prepared antiCD33-LONp and PMI-LONp. UV-Vis and FTIR spectroscopy confirmed conjugation of PMI and/or antiCD33 to LONp as evidenced by characteristic UV and IR absorbance given by peptide and protein (Fig. 2A–B).
Fig. 1.
Schematic Diagram of antiCD33-LONp-PMI for targeted molecular therapy of acute myeloid leukemia. The antiCD33-LONp-PMI were constructed by a thiol-induced molecular assembly of PMI-Cys and AntiCD33-Cys with biocompatible LONp. These nanoparticles can protect the peptides from enzymolysis, AML cells specifically, and get internalized by AML cells. In reductive cytosol, antiCD33-LONp-PMI can be reduced to release potent peptide drugs to kill AML cells.
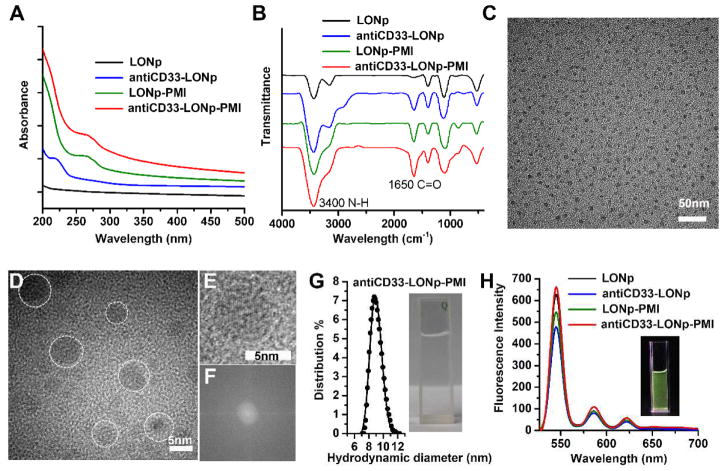
Fig. 2.
Morphology and spectroscopy characterization. (A) UV-VIS absorption spectra of the nano-particles measured in PBS solution (pH7.4). At 214 and 280 nm, the characteristic absorption peaks were for amide bonds in antibody and aromatic structures in PMI respectively. (B) Fluorescence spectra measured after lyophilization. Two bands at 3400 and 1650 cm –1 in FTIR spectra were attributed to the stretching vibration of N–H and C=O groups from peptides and proteins. (C) TEM images of anti-CD33-PMI-LONp, showing the uniformly dispersed particles in PBS buffer. HRTEM images (D), enlarge image (E) and corresponding FFT pattern (F) of an individual particle of anti-CD33-PMI-LONp, exhibiting their single crystal structure. (G) hydrodynamic diameter of anti-CD33-PMI-LONp measured in PBS buffer, pH 7.4, at 25°C by dynamic light scattering. (H) fluorescent photographs of LONp, antiCD33-LONp, LONp-PMI and antiCD33 -LONp-PMI measured in Milli-Q water and excited at 260nm.
We characterized various forms of LONp nanoparticles, “bare” or loaded with PMI, anti-CD33 or both, using high-resolution transmission electron microscopy (TEM), dynamic light scattering and fluorescence spectroscopy techniques. As shown in Fig 2C–D, AntiCD33-LONp-PMI nanoparticles displayed on TEM a uniformly monodispersed nanostructure with a diameter of 5.6 ± 0.4 nm calculated from 100 randomly sampled particles. While well-resolved lattice fringes gave rise to an inter-planar spacing value of 0.37 nm (Fig. 2E), fast Fourier transform patterns confirmed its single crystalline structure (Fig. 2F). A clear solution ensued when antiCD33-LONp-PMI was suspended in PBS buffer, pH 7.4, which yielded a hydrodynamic diameter of the nanoparticle of 8.9 nm as measured by dynamic light scattering (Fig. 2G). When suspended in water, antiCD33-LONp-PMI was well-dispersed and emitted intense green fluorescence (Fig. 2H). Fluorescence spectroscopy confirmed that all four forms of LONp nanoparticles emitted fluorescence primarily at 545 nm with two minor red emission peaks at 585 and 620 nm (Fig. 2H). The fact that peptide and/or antibody conjugation did not alter the fluorescence characteristics of LONp suggests that antiCD33-LONp-PMI nanoparticles are suitable as an imaging tool for tracking drug and monitoring treatment response.
AntiCD33 antibody promotes LONp recognition of and internalization to CD33+ AML cells
Our recombinant antiCD33-Cys was produced with an extra disulfide bond oxidatively formed by the two proximal Cys residues at the C-termini of the heavy chains (Fig. S1). This disulfide-tethered antiCD33-Cys antibody exhibited identical properties to a commercial antiCD33 antibody (ABcam) with respect to binding to CD33+ U937 and OCI-AML2 cell lines (Fig. 3A), suggesting that the engineered antiCD33-Cys antibody was fully functional. For site-specific conjugation to LONp via the metal-thiolate bond, however, controlled partial reduction in situ of tethered antiCD33-Cys in the presence of a low concentration of Tris(2-carboxyethyl)phosphine (TCEP) was necessary to break up the terminal disulfide bond (Fig. S1).
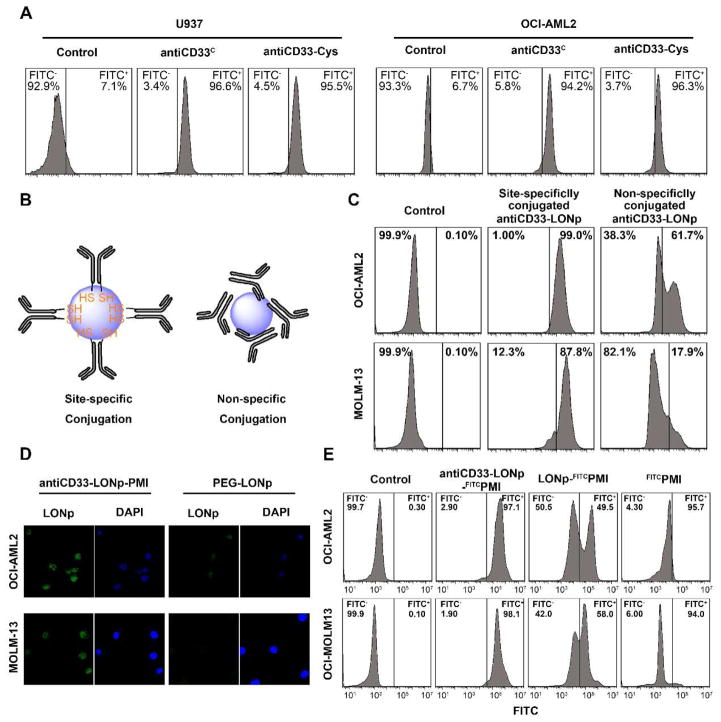
Fig. 3.
antiCD33-Cys promotes AML cells recognition and internalization. (A) The recognition test of Anti-CD33-Cys compared to the commercialized CD33 antibody by flow cytometric analysis. FITC-labeled second antibody was used as signal source (B) Schematic diagram of different conjugations between antiCD33-Cys and LONp. (C) Flow cytometric analysis of site-specifically conjugated antiCD33-LONps uptake, compared to non-specifically conjugated antiCD33-LONps. The fluorescence signal come from the excitation of LONp. (D) Confocal microscopy image of antiCD33-LONps uptake. (E) Flow cytometric analysis of antiCD33-LONp-FITCPMI uptake (detection signal from PMI-FITC. ×600)
Site-specific conjugation of antiCD33-Cys to LONp via two free (reduced) C-terminal Cys residues in the heavy chains ensures a conformationally homogeneous assembly of the antibody on the surface of the nanoparticle for productive binding to CD33+ AML cells (Fig. 3B). This strategy is in stark contrast to the conventional approach [34], where antibody molecules are non-specifically absorbed via electrostatic and hydrophobic forces to the surface of a nanoparticle (Fig. 3B). The latter approach necessarily impedes antibody interactions with an antigen due to its conformationally heterogeneous and functionally unproductive assembly. For comparison, we prepared two forms of antibody-conjugated LONp both site-specifically, using TECP-reduced antiCD33-Cys, and non-specifically, using disulfide-tethered antiCD33-Cys. As shown in Fig. 3C, flow cytometry studies with CD33+ OCI-AML2 and MOLM-13 cell lines showed a significantly more efficient uptake of the site-specifically conjugated antiCD33-LONp than the non-specifically conjugated counterpart. Confocal microscopy studies using PEG-loaded LONp as a control confirmed that the enhanced cellular uptake by and intracellular distribution in OCI-AML2 and MOLM-13 of antiCD33-LONp was specifically mediated by the antibody (Fig. 3D). These results also demonstrated that controlled partial reduction of tethered antiCD33-Cys did not alter the antibody functionally.
To ascertain the ability of antiCD33-LONp to efficiently deliver peptides into AML cells, we labelled the N terminus of PMI with fluorescein isothiocyanate (FITC) and conjugated FITCPMI to LONp and antiCD33-LONp, respectively, via a C-terminally extended Cys residue. FITC-based quantitative FACS analysis showed that free FITCPMI peptide failed, as expected, to penetrate into AML cells (Fig. 3E). While FITCPMI-LONp was able to traverse the cell membrane, the uptake of antiCD33-LONp-FITCPMI by OCI-AML2 and MOLM-13 cell lines was substantially more efficient (Fig. 3E). Taken together, these findings indicate that our antiCD33-Cys antibody, when conjugated along with PMI to LONp, can significantly enhance in a target-specific fashion the delivery of the peptide cargo into CD33+ AML cells.
Addition of a C-terminal Cys residue to PMI for LONp conjugation is functionally inconsequential
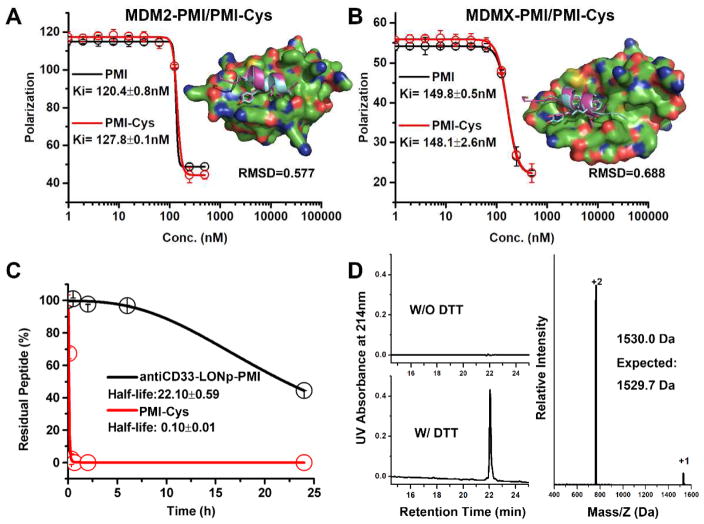
To form a redox-responsive metal-thiolate bond for PMI conjugation to LONp, an extra Cys residue was introduced to the C-terminus of the peptide. Prior structural studies of PMI in complex with MDM2 or MDMX as well as mutational analysis suggested that Cys addition to the C-terminus of PMI would be functionally neutral [35]. In fact, the C-terminal residue Pro12 of PMI made little or no contact with MDM2/MDMX, and the P12A mutation was largely inconsequential at the functional level [35]. To verify, however, we comparatively characterized PMI and PMI-Cys with respect to their interactions with MDM2 and MDMX using a previously described fluorescence polarization-based competitive binding assay [36], where increasing concentrations of PMI or PMI-Cys were added to a preformed complex of either FITC-(17–28)p53-25–109MDM2 or FITC-(17–28)p53-24–108MDMX to displace FITC-(17–28)p53. This titration process was accompanied by a dose-dependent reduction in fluorescence polarization, from which the Ki value of PMI or PMI-Cys could be derived through a non-linear regression analysis [37]. As shown in Fig. 4A–B, PMI and PMI-Cys displayed nearly identical Ki values for either MDM2 or MDMX, supporting the premise that Cys addition to the C-terminus of PMI is functionally inconsequential. Molecular dynamics simulation studies of PMI-Cys in complex with MDM2 or MDMX showed that PMI-Cys nicely overlapped with PMI complexed with MDM2 or MDMX [25] (PDB codes: 3EQS and 3EQY) with a respective RMSD value of 0.577 Å or 0.688 Å (Fig. 4A–B). As expected, however, the extra Cys residue at the C-terminus of PMI made no contact with either protein, structurally validating the above functional findings.
Fig. 4.
C-terminal Cys residue to PMI for LONp conjugation is functionally inconsequential; antiCD33-LONp-PMI resist PMI proteolytic extracellular and redox-responsibly release PMI intracellular. (A) fluorescence polarization-based competitive binding assay of PMI or PMI-Cys to MDM2, and the simulated structure of MDM2/PMI-Cys complex compared to MDM2/PMI complex (PDB code: 3EQS). (B) fluorescence polarization assay and structural simulation (compared to the MDMX/PMI complex, PDB code 3EQY) of MDMX/PMI-Cys. For fluorescence polarization measurements at room temperature on a Tecan Infinite M2000 plate reader, FITC was covalently conjugated to the N terminal of (15–29)p53. Readings were taken at λex = 495 nm,λem = 520 nm. Non-linear regression analyses were performed to give rise to Ki values (mean ± SEM, n=3); each curve is the mean of four independent measurements with the error bars denoting SEM. (C) antiCD33-LONp-PMI protecting peptides from proteolytic of chymotrypsin. RP-HPLC was used to monitor and quantify time-dependent peptide hydrolysis. (D) Redox-dependent release profiles of peptides from antiCD33 -LONp-PMI in PBS with 50mg/ml DTT at pH 7.4.
LONp-conjugated PMI is more resistant to proteolysis and can be efficiently cleaved off LONp by reducing thiols
Small peptides are generally unstructured or conformationally disordered in solution on their own and, thus, highly susceptible to proteolytic degradation in vitro and in vivo. This important pharmacological obstacle impedes the development of peptide therapeutics for clinical use [5, 6]. Conjugation of small peptides to nanoparticles is expected to enhance their resistance to proteolysis due to steric hindrance to the action of proteolytic enzymes [6, 15, 38]. We quantitatively examined the susceptibility of antiCD33-LONp-PMI and free PMI peptide to chymotrypsin using analytical HPLC coupled with mass spectrometry. Not surprisingly, while free PMI peptide was quickly digested at room temperature by the enzyme with a half-life under ~6 min, LONp-conjugated PMI was significantly more resistant to chymotrypsin-mediated degradation, with a half-life over 20 hours (Fig. 4C).
Therapeutic efficacy of antiCD33-LONp-PMI hinges upon an efficient release of PMI into the cytosol, a process controlled by intracellular reducing thiols such as reduced glutathione to break the metal-thiolate bond. On the other hand, a stable metal-thiolate bond is highly desirable for circulating antiCD33-LONp-PMI for obvious reasons. We incubated antiCD33-LONp-PMI in PBS (~0.5mmol/ml, PMI) for one week and, following centrifugation, injected the supernatant for analysis on analytical HPLC. As shown in Fig. 4D, no PMI peptide was detected, suggesting that the metal-thiolate bond was indeed stable. By sharp contrast, 30 min after DTT at 50 mg/ml was added to the nanoparticle solution, PMI-Cys peptide was quantitatively recovered from the supernatant (Fig. 4D). Taken together, these results demonstrate that conjugation to LONp greatly enhances PMI resistance to proteolysis and that conjugated PMI can be efficiently cleaved off LONp by reducing thiols.
Anti-CD33-LONp-PMI induces apoptosis of MDM2/MDMX-overexpressed AML cells harboring wild type p53
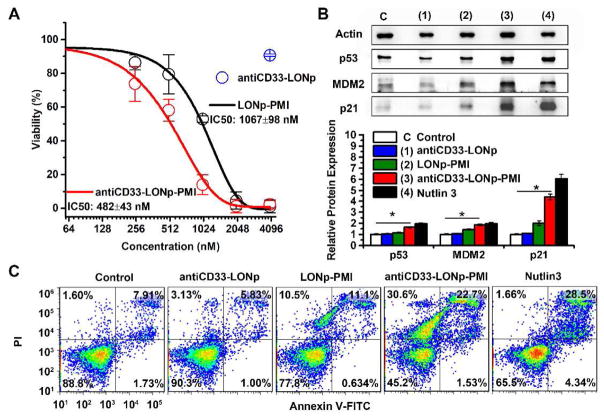
Most AML cells harbor wild type p53 and elevated levels of MDM2 and/or MDMX [39–41]; MDM2 and MDMX antagonists such as PMI are expected to activate p53, which, in turn, induces apoptotic cell death [25]. We treated the AML cell line MOLM-13 with LONp-PMI and antiCD33-LONp-PMI at concentrations from 0.25 to 4.0 μM (based on quantification of thiol-cleaved PMI-Cys) in a cell proliferation assay. AntiCD33-LONp at a concentration of 4 μg/ml, equivalent in weight to antiCD33-LONp-PMI at 4.0 μM, was used as a negative control. As shown in Fig. 5A, a dose-dependent inhibition of cell growth was observed for LONp-PMI (IC50 = 1070 nM) and antiCD33-LONp-PMI (IC50 = 480 nM), whereas antiCD33-LONp was, as expected, non-inhibitory at the highest concentration tested. These data suggest that MOLM-13 growth was inhibited solely by PMI and that the antiCD33 antibody mediated an improved cellular uptake or internalization of nanoparticles and, thus, enhanced killing of tumor cells.
Fig. 5.
antiCD33-LONp-PMI killed MDM2 overexpressed AML cell line - MOLM-13. (A) Dose-dependent growth inhibition of MOLM-13 cells upon various treatments as determined by the CCK-8 cell viability assay. AntiCD33-LONp at a concentration of 40 μg/ml, equivalent in weight to antiCD33-LONp-PMI at 4.0 μM. (B) Protein expression analysis of p53, p21, MDM2, expression in MOLM13 cells after 48 h treatment with 500 nM antiCD33-LONp-PMI, antiCD33-LONp, LONp-PMI or 5uM Nutlin3 by western blot, normalized by actin. Protein levels of p53, p21, MDM2, were quantified using Image J. Data are shown as the mean ± standard error (n = 3), and a t-test between control and antiCD33-LONp-PMI shows statistically significant difference (p<0.005). (C) Effects of antiCD33-LONp-PMI, antiCD33- LONp, LONp-PMI and Nutlin3 on MOLM-13 cells as determined by flow cytometric analysis. Viable cells are Annexin V and PI negative; cells in early apoptosis are Annexin V positive and PI negative; and cells that are in late apoptosis or already dead are both Annexin V and PI positive.
To investigate into the mechanisms of action of intracellular PMI, we analyzed the expression of p53, MDM2 and p21 in MOLM-13 cells by Western blotting. As shown in Fig. 5B, 24 h after treatment, antiCD33-LONp-PMIl and Nutlin-3 – a small molecule antagonist of MDM2 [42] significantly induced the expression of p53 and its responsive gene p21 – a cyclin-dependent kinase inhibitor that promotes cell cycle arrest [43, 44]. MDM2 expression also increased in response to the treatment as it is transcriptionally inducible by p53 in a positive feedback loop essential for maintaining low levels of p53 during normal development [45]. While LONp-PMI was less effective than either antiCD33-LONp-PMI or Nutlin-3, antiCD33-LONp was inactive, as expected. Consistent with these results, FACS analysis confirmed that MOLM-13 cells underwent different degrees of apoptosis 24 h after treatment with LONp-PMI, antiCD33-LONp-PMI, or Nutlin-3 (Figs. 5C, S4). In fact, the largest increase in percent apoptotic cells (from a basal level of ~10% to 55%) was seen with antiCD33-LONp-PMI. These findings mechanistically validate the mode of action of PMI, that is, PMI induces apoptotic cell death of p53+ AML cells by antagonizing MDM2 to activate the p53 signaling pathway.
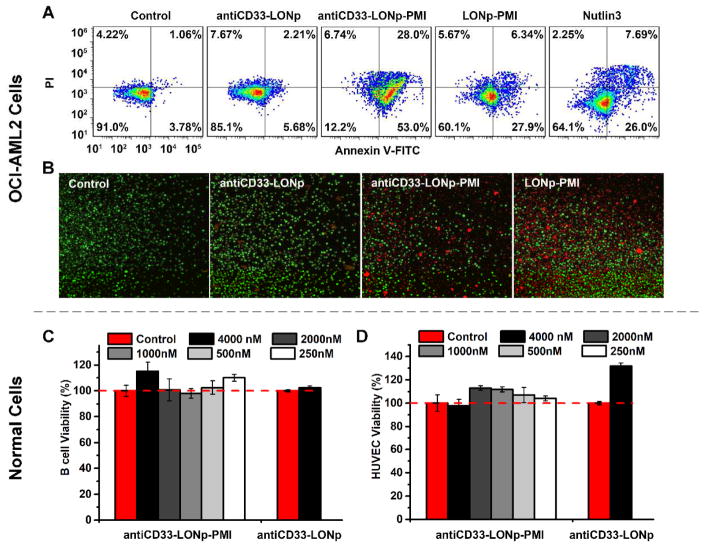
Similar results were found with the cell line OCI-ALM2, which harbors wild type p53 and was originally established from the peripheral blood of AML patients [46]. Unlike MOLM-13 where p53 is sequestered primarily by MDM2, however, OCI-AML2’s p53 function and stability is predominantly regulated MDMX [47]. Since small molecule antagonists of MDM2 such as Nutlin-3 are generally ineffective against MDMX [47–49], PMI as a dual-specificity antagonist of both MDM2 and MDMX is particularly suitable for killing MDMX-overexpressed tumor cells. As shown in Figs. 6A–B and S4 flow cytometry and live-dead cell staining studies demonstrated that OCI-AML2 was extremely sensitive to antiCD33-LONp-PMI (1 μM) with most cells undergoing apoptosis after 48 h, but significantly less sensitive to treatment with 1 μM PMI-LONp or 6 μM Nutlin-3. As expected, antiCD33-LONp had little effect on OCI-AM2 cells, indicative of a PMI-dependent apoptotic cell death of OCI-AML2 treated with anti-CD33-LONp-PMI or PMI-LONp.
Fig. 6.
AntiCD33-LONp-PMI killed MDMX overexpressed AML cell line - OCI-AML2; in vitro cytotoxicity of antiCD33-LONp-PMI to B cells and HUVEC cells. (A) Effects of antiCD33-LONp-PMI, antiCD33-LONp, LONp-PMI and Nutlin3 on OCI-AML2 cells as determined by flow cytometric analysis of apoptosis. (B) LIVE/DEAD staining (×600) of AML2 cells after treatment with 500 nM Anti-CD33-PMI-LONp, PMI-LONp or 5uM Nutlin3. Viable cells are green, and cells that are in late apoptosis or already dead are red. (C) B cells and (D) HUVEC cells viability after incubation with different dosages of antiCD33-LONp-PMI and 4 μg/ml antiCD33-LONp (equivalent in weight to antiCD33-LONp-PMI at 4.0 μM) for 48 h using standard CCK-8 assay (n=5).
Importantly, anti-CD33-LON-PMI is not toxic to normal cells. Systemic toxicity has always been a major challenge in current chemotherapy [50, 51]. Despite the specificity of antiCD33 and PMI for their respective cellular and protein targets, it remained unclear whether or not antiCD33-LONp-PMI would be selectively cytotoxic against AML cells but not normal cells. We evaluated the cytotoxicity of antiCD33-LONp-PMI at concentrations from 0.25 to 4.0 μM (based on quantification of thiol-cleaved PMI-Cys) toward B lymphoblastoid cell line HMy2.CIR and human vascular endothelial cells (HUVEC) in a cell proliferation assay. As shown in Fig. 6C–D, neither antiCD33-LONp-PMI nor antiCD33-LONp had any effect on cell viability at the highest concentration tested, suggesting that they are not toxic to normal cells. Taken together, our in vitro models demonstrate the therapeutic efficacy and safety profile of antiCD33-LONp-PMI as a novel class of antitumor agents for potential treatment of AML.
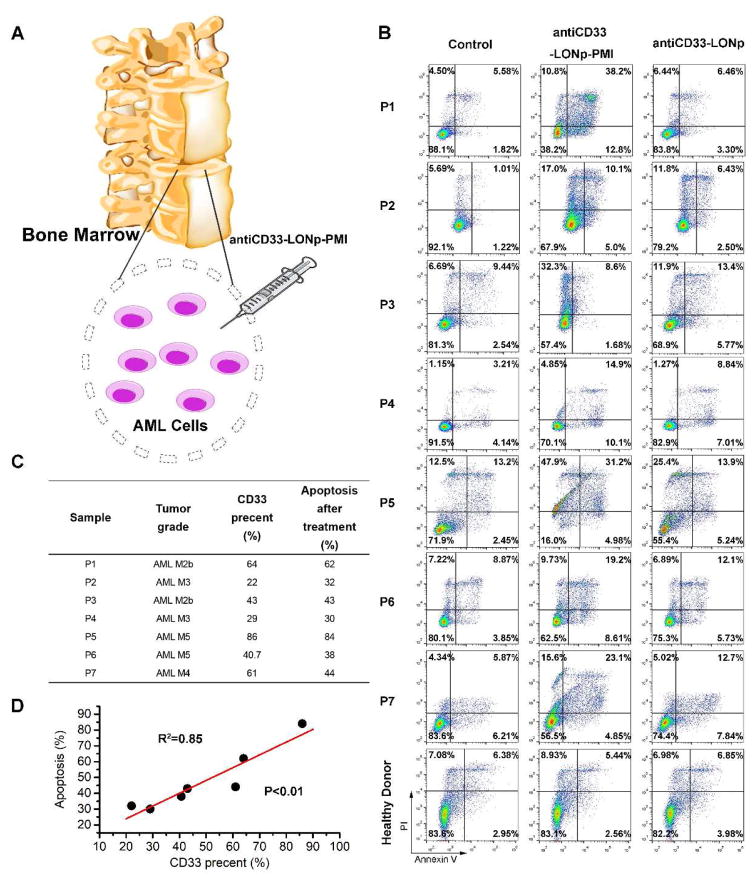
Anti-CD33-LONp-PMI induces apoptosis of primary AML cells ex vivo in a CD33-dependent fashion
Ex vivo cytotoxicity assays using freshly isolated cancer cells from patients can be a powerful tool for evaluating the therapeutic efficacy of antitumor agents against hematological malignancies [52, 53]. We obtained primary leukemic cells from 7 newly diagnosed AML patients and subjected them to treatment with antiCD33-LONp-PMI and antiCD33-LONp (Fig. 7A); PMBCs from a healthy donor were used as a control. As shown in Fig. 7B, all AML blasts were sensitive to the killing induced by antiCD33-LONp-PMI, but significantly less so with respect to antiCD33-LONp, suggesting that PMI specifically induced apoptotic cell death of AML cells ex vivo. Of note, neither antiCD33-LONp-PMI nor antiCD33-LONp had any effect on the growth of PBMCs. Importantly, the amount of CD33 expressed on cell surface strongly correlated to the sensitivity of primary AML cells to the killing by antiCD33-LONp-PMI (Fig. 7C–D), underscoring the functional importance of a CD33-mediated uptake of nanoparticles and subsequent intracellular effects of PMI. These findings with fresh patient samples amply demonstrate that a favorable clinical response could potentially be achieved to the treatment of AML patients with antiCD33-LONp-PMI.
Fig. 7.
The ex vivo sensitivity of primary cells from patients with AML to antiCD33-LONp-PMI. (A) Schemata of the treatment for AML specimens. (B) Apoptosis of AML and healthy specimens after antiCD33-LONp-PMI treatment detected by flow cytometric analysis of apoptosis. (C) The CD33 positive percentage of AML specimens, measured by flow cytometric analysis via the commercial antiCD33 antibody purchased from Miltenyi Biotech (Percent CD33 positive cells in healthy donor is 1.59%) and their apoptosis percent after antiCD33-LONp-PMI treatment (measured by flow cytometric analysis of apoptosis). (D) Linear regression between the CD33 positive percent and the corresponding apoptosis percent, in which the amount of CD33 expressed on cell surface strongly correlated to the sensitivity of primary AML cells to the killing by antiCD33-LONp-PMI.
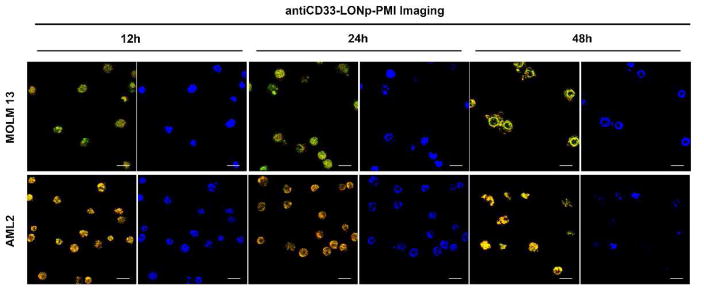
Imaging power of anti-CD33-LONp-PMI in real-time monitoring of PMI-induced apoptosis
One important application of LONp as a carrier of theranostics lies in its intrinsic luminescence, which endows antiCD33-LONp-PMI with a remarkable imaging capability useful for real-time monitoring of its pharmacological effects on tumor cells [22, 31]. Apoptosis is accompanied by a series of morphological changes of the nucleus: shrinkage, dramatic reorganization, membrane blebbing and, ultimately, fragmentation into membrane enclosed vesicles [54]. This process can be visualized with AML cells and antiCD33-LONp-PMI due to the ability of LONp to stain the nucleus [22]. As shown in Fig. 8, the morphology of the nucleus of MOLM-13 and OCI-AML2 cells stained in yellow by antiCD33-LONp-PMI, confirmed by Hochest staining in situ, progressively changed over time, entirely consistent with PMI-induced apoptotic events.
Fig. 8.
antiCD33-LONp-PMI real-time monitored the pharmacological effects of PMI. CLSM images of MOLM13 and OCI-AML2 at 12 h, 24 h and 48 h after 500 nM antiCD33 -LONp-PMI treatment, scale bars: 60 μm. After first 12 h treatment, except for the intact cells and nuclei, some of the yellow nuclear of MOLM13 or OCI-AML2 showed some special morphologies of early apoptosis with the collapse of the chromatin against the nuclear periphery. After 24 h incubation, the counts of intact cells and nuclei revealed significant losses, meanwhile some entire nucleuses were condensed into a single dense ball, both of which were the typical hallmarks of apoptosis. After 48 h, nearly all the MOLM13 and OCI-AML2 cells were dead or in terminal apoptosis. In accordance with the theoretical morphology in this time, the nucleus of two cell lines showed diffuse and granular substructure with divisions by fine septate-like structures, which were called the apoptotic bodies or ring-like structures.
Discussion
The tumor suppressor protein p53 induces powerful growth inhibitory and apoptotic responses to cellular stress and plays a pivotal role in preventing damaged cells from becoming cancerous [26, 55, 56]. Not surprisingly, impairment of the p53 pathway is a hallmark of almost all human cancers, where either the TP53 gene is mutated or wild-type p53 is functionally inactivated by the E3 ubiquitin ligase MDM2 and its homolog MDMX [48, 57]. In many tumor cells harboring wild-type p53, MDM2 and/or MDMX are elevated and often cooperate to inhibit p53 transactivation activity and target p53 for degradation, conferring tumor development and progression [48]. Numerous studies have validated MDM2 and/or MDMX antagonism as a viable therapeutic paradigm for cancer treatment, and several small molecule antagonists specific for MDM2 are in various phases of clinical trials [58, 59].
Growing evidence suggests that the interplay between MDM2 and MDMX confers robust p53 inactivation in tumorigenesis and that antagonizing both MDM2 and MDMX affords a powerful, synergistic and sustained inhibition of tumor growth.[48] [60] as amply demonstrated by us and others [61, 62], representing powerful p53 activators for anticancer therapy. However, major pharmacological hurdles still impede the development of anticancer peptide therapeutics with optimal therapeutic efficacy, including: (1) short circulation half-life due to proteolytic degradation and renal excretion (<20 KDa), (2) poor cellular uptake, and (3) lack of tumor-targeting specificity. To overcome these technical obstacles, we resorted to a clinically validated therapeutic strategy by conjugating both a therapeutic peptide and a tumor-targeting monoclonal antibody to fluorescent lanthanide oxyfluoride nanoparticles (LONp). In this proof of concept study with AML, we used a dual-specificity peptide antagonist of MDM2 and MDMX, termed PMI [25, 35], and a humanized monoclonal antibody against CD33.
The rationales for our experimental design of this multifunctional peptide drug delivery system are several-fold. First, AML, characterized by accumulation in the bone marrow of myeloblasts, remains a difficult cancer to treat as evidenced by a high rate of remission and a low overall survival rate [63]. In AML patients the TP53 gene is rarely mutated, and MDM2 and/or MDMX levels are significantly elevated [39, 64, 65], making PMI an ideal drug candidate for AML treatment. Second, the vast majority of AML patients have elevated levels of CD33 expression on the surface of malignant blast cells, but not on normal hematopoietic pluripotent stem cells, making the antigen an attractive candidate for targeted therapy of AML [66, 67]. Third, anti-CD33 mAbs rapidly internalize upon binding to AML cells and, thus, are an ideal vehicle for intracellular delivery of PMI. Fourth, peptide conjugation to nanoparticles can significantly enhance its resistance to proteolysis, improving the bioavailability of PMI. Fifth, nanoparticle conjugation also increases the apparent size of a peptide cargo to minimize its renal excretion. Finally, LONp emits intense green fluorescence, endowing the antiCD33-LONp-PMI nanoparticle with a powerful imaging capability, which may be useful for tracking disease and monitoring treatment response.
The experimental findings reported here fully validate our design strategy aims at improving peptide drugs’ bioavailability, cellular deliverability, and tumor-targeting specificity. We also demonstrated lanthanide oxyfluoride-derived nanoparticles as superior carriers of peptide and protein therapeutics that can be readily and reversibly released in the reducing intracellular environment. Although nanoparticles are capable of actively accumulating in solid tumors through “EPR” effects [16, 17], they are non-specific for hematologic malignancies such as AML. Conjugation of antiCD33 to LONp largely alleviated this problem and significantly improved LONp recognition of and internalization to CD33+ AML cells. Consequently, PMI efficiently induced apoptosis of CD33+ AML cell lines and primary leukemic cells isolated from AML patients by activating the p53 pathway, showcasing its potential as a novel class of peptide therapeutics for the treatment of AML. Of note, our very recent acute toxicity study suggests that antiCD33-LONp-PMI nanoparticles injected IP and IV at 50 μM (in 200 μl) are well tolerated in NSG mice, paving the way for an in vivo efficacy study in the future using appropriate mouse models.
Conclusion
Redox-responsive peptide delivery may become a robust technological platform in nanomedicine for the development of anticancer peptide therapeutics. We here developed lanthanide oxyfluoride nanoparticles (LONp) functionalized with an anti-CD33 mAb and a p53-activating antitumor peptide, PMI, for targeted molecular therapy of acute myeloid leukemia. Our fluorescent antiCD33-LONp-PMI nanoparticles can be efficiently taken up by AML cells overexpressing CD33 and kill p53+ tumor cells in vitro and ex vivo by antagonizing MDM2 and/or MDMX, promising a novel class of peptide therapeutics not only for potential treatment of AML but also for possible AML tracking and treatment response monitoring. Several different endocytotic pathways such as phagocytosis, macropinocytosis, clathrin- and caveolin-mediated endocytosis are involved in cellular uptake of nanoparticles [68], depending on a number of factors such as the cell type and cellular environment, particle size, shape, material, surface coating, and how nanoparticles interact with the cell, among others. Future studies are warranted to better understand the endocytotic uptake mechanisms of LONp-derived nanoparticles in order to exploit this nanomedicine strategy for possible applications to other human diseases.
Supplementary Material
Acknowledgments
This work was partially supported by the Natural Science Foundation of Shaanxi Province of China (2017ZDJC-02), 985 Program of XJTU, and the National Institutes of Health Grants CA167296 and CA219150 (to W. L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Milroy LG, Grossmann TN, Hennig S, Brunsveld L, Ottmann C. Modulators of protein–protein interactions. Chemical reviews. 2014;114(9):4695–4748. doi: 10.1021/cr400698c. [DOI] [PubMed] [Google Scholar]
- 2.Azzarito V, Long K, Murphy NS, Wilson AJ. Inhibition of α-helix-mediated protein–protein interactions using designed molecules. Nature chemistry. 2013;5(3):161. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- 3.Mullard A. Protein–protein interaction inhibitors get into the groove. Nature reviews Drug discovery. 2012;11(3):173–175. doi: 10.1038/nrd3680. [DOI] [PubMed] [Google Scholar]
- 4.Scott DE, Bayly AR, Abell C, Skidmore J. Small molecules, big targets: drug discovery faces the protein–protein interaction challenge. Nature Reviews Drug Discovery. 2016;15(8):533. doi: 10.1038/nrd.2016.29. [DOI] [PubMed] [Google Scholar]
- 5.Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discovery Today. 2015;20(1):122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Acar H, Ting JM, Srivastava S, LaBelle JL, Tirrell MV. Molecular engineering solutions for therapeutic peptide delivery. Chemical Society Reviews. 2017;46(21):6553–6569. doi: 10.1039/c7cs00536a. [DOI] [PubMed] [Google Scholar]
- 7.Almeida AJ, Souto E. Solid lipid nanoparticles as a drug delivery system for peptides and proteins. Advanced Drug Delivery Reviews. 2007;59(6):478–490. doi: 10.1016/j.addr.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Lu Y, Sun W, Gu Z. Stimuli-responsive nanomaterials for therapeutic protein delivery. Journal of controlled release. 2014;194:1–19. doi: 10.1016/j.jconrel.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu Z, Conejos-Sanchez I, Griffin BT, O’Driscoll CM, Alonso MJ. Lipid-based nanocarriers for oral peptide delivery. Advanced drug delivery reviews. 2016;106:337–354. doi: 10.1016/j.addr.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Nicolas J, Mura S, Brambilla D, Mackiewicz N, Couvreur P. Design, functionalization strategies and biomedical applications of targeted biodegradable/biocompatible polymer-based nanocarriers for drug delivery. Chemical Society Reviews. 2013;42(3):1147–1235. doi: 10.1039/c2cs35265f. [DOI] [PubMed] [Google Scholar]
- 11.Allen TM, Cullis PR. Liposomal drug delivery systems: from concept to clinical applications. Advanced drug delivery reviews. 2013;65(1):36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 12.Ulbrich K, Hola K, Subr V, Bakandritsos A, Tucek J, Zboril R. Targeted drug delivery with polymers and magnetic nanoparticles: covalent and noncovalent approaches, release control, and clinical studies. Chemical reviews. 2016;116(9):5338–5431. doi: 10.1021/acs.chemrev.5b00589. [DOI] [PubMed] [Google Scholar]
- 13.Jain RK, Stylianopoulos T. Delivering nanomedicine to solid tumors. Nature reviews Clinical oncology. 2010;7(11):653. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nature biotechnology. 2015;33(9):941. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan J, He W, Yan S, Niu F, Liu T, Ma B, Shao Y, Yan Y, Yang G, Lu W, Du Y, Lei B, Ma PX. Self-Assembled Peptide–Lanthanide Nanoclusters for Safe Tumor Therapy: Overcoming and Utilizing Biological Barriers to Peptide Drug Delivery. ACS Nano. 2018 doi: 10.1021/acsnano.8b00081. [DOI] [PubMed] [Google Scholar]
- 16.Maeda H, Nakamura H, Fang J. The EPR effect for macromolecular drug delivery to solid tumors: Improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Advanced drug delivery reviews. 2013;65(1):71–79. doi: 10.1016/j.addr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Torchilin V. Tumor delivery of macromolecular drugs based on the EPR effect. Advanced drug delivery reviews. 2011;63(3):131–135. doi: 10.1016/j.addr.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Tao Y, Li M, Ren J, Qu X. Metal nanoclusters: novel probes for diagnostic and therapeutic applications. Chemical Society Reviews. 2015;44(23):8636–8663. doi: 10.1039/c5cs00607d. [DOI] [PubMed] [Google Scholar]
- 19.Savla R, Minko T. Nanoparticle design considerations for molecular imaging of apoptosis: Diagnostic, prognostic, and therapeutic value. Advanced drug delivery reviews. 2017;113:122–140. doi: 10.1016/j.addr.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Kumar CSSR, Mohammad F. Magnetic nanomaterials for hyperthermia-based therapy and controlled drug delivery. Advanced Drug Delivery Reviews. 2011;63(9):789–808. doi: 10.1016/j.addr.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouzigues C, Gacoin T, Alexandrou A. Biological applications of rare-earth based nanoparticles. ACS nano. 2011;5(11):8488–8505. doi: 10.1021/nn202378b. [DOI] [PubMed] [Google Scholar]
- 22.Yan J, He W, Li N, Yu M, Du Y, Lei B, Ma PX. Simultaneously targeted imaging cytoplasm and nucleus in living cell by biomolecules capped ultra-small GdOF nanocrystals. Biomaterials. 2015;59:21–29. doi: 10.1016/j.biomaterials.2015.04.041. [DOI] [PubMed] [Google Scholar]
- 23.Wu J, Tian Q, Hu H, Xia Q, Zou Y, Li F, Yi T, Huang C. Self-assembly of peptide-based multi-colour gels triggered by up-conversion rare earth nanoparticles. Chemical Communications. 2009;(27):4100–4102. doi: 10.1039/b907517h. [DOI] [PubMed] [Google Scholar]
- 24.Liu C, Hou Y, Gao M. Are Rare-Earth Nanoparticles Suitable for In Vivo Applications? Advanced Materials. 2014;26(40):6922–6932. doi: 10.1002/adma.201305535. [DOI] [PubMed] [Google Scholar]
- 25.Pazgier M, Liu M, Zou G, Yuan W, Li C, Li C, Li J, Monbo J, Zella D, Tarasov SG. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proceedings of the National Academy of Sciences. 2009;106(12):4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 27.Shvarts A, Steegenga W, Riteco N, Laar Tv, Dekker P, Bazuine M, Ham Rv, Houven van Oordt W, Hateboer G, Eb A. MDMX: a novel p53-binding protein with some functional properties of MDM2. The EMBO journal. 1996;15(19):5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 28.Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nature reviews Clinical oncology. 2011;8(1):25. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- 29.Legrand O, Perrot JY, Baudard M, Cordier A, Lautier R, Simonin G, Zittoun R, Casadevall N, Marie JP. The immunophenotype of 177 adults with acute myeloid leukemia: proposal of a prognostic score. Blood. 2000;96(3):870–877. [PubMed] [Google Scholar]
- 30.Linenberger M. CD33-directed therapy with gemtuzumab ozogamicin in acute myeloid leukemia: progress in understanding cytotoxicity and potential mechanisms of drug resistance. Leukemia. 2005;19(2):176. doi: 10.1038/sj.leu.2403598. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Yan J, Yang Q, Yan Y, Li S, Wang L, Li C, Lei B, Yang G, He W. Arginine-modified dual emission photoluminescent nanocrystals for bioimaging at subcellular resolution. Journal of biomaterials applications. 2017;32(4):533–542. doi: 10.1177/0885328217725203. [DOI] [PubMed] [Google Scholar]
- 32.Caron PC, Bull MK, Avdalovic NM, Queen C, Scheinberg DA. Biological and immunological features of humanized M195 (anti-CD33) monoclonal antibodies. Cancer Research. 1992;52(24):6761–6767. [PubMed] [Google Scholar]
- 33.Raju TS. Terminal sugars of Fc glycans influence antibody effector functions of IgGs. Current opinion in immunology. 2008;20(4):471–478. doi: 10.1016/j.coi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Chinen AB, Guan CM, Ferrer JR, Barnaby SN, Merkel TJ, Mirkin CA. Nanoparticle probes for the detection of cancer biomarkers, cells, and tissues by fluorescence. Chemical reviews. 2015;115(19):10530–10574. doi: 10.1021/acs.chemrev.5b00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C, Pazgier M, Li C, Yuan W, Liu M, Wei G, Lu WY, Lu W. Systematic mutational analysis of peptide inhibition of the p53–MDM2/MDMX interactions. Journal of molecular biology. 2010;398(2):200–213. doi: 10.1016/j.jmb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Gohain N, Zhan C, Lu W, Pazgier M, Lu W. Structural basis of how stress-induced MDMX phosphorylation activates p53. Oncogene. 2016;35(15):1919. doi: 10.1038/onc.2015.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossi AM, Taylor CW. Analysis of protein-ligand interactions by fluorescence polarization. Nature protocols. 2011;6(3):365. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruno BJ, Miller GD, Lim CS. Basics and recent advances in peptide and protein drug delivery. Therapeutic delivery. 2013;4(11):1443–1467. doi: 10.4155/tde.13.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kojima K, Konopleva M, Samudio IJ, Shikami M, Cabreira-Hansen M, McQueen T, Ruvolo V, Tsao T, Zeng Z, Vassilev LT. MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood. 2005;106(9):3150–3159. doi: 10.1182/blood-2005-02-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seliger B, Papadileris S, Vogel D, Hess G, Brendel C, Storkel S, Kolbe K, Huber C, Huhn D, Neubauer A. Analysis of the p53 and MDM-2 gene in acute myeloid leukemia. European journal of haematology. 1996;57(3):230–240. doi: 10.1111/j.1600-0609.1996.tb01369.x. [DOI] [PubMed] [Google Scholar]
- 41.Quintas-Cardama A, Hu C, Qutub A, Qiu Y, Zhang X, Post S, Zhang N, Coombes K, Kornblau S. p53 pathway dysfunction is highly prevalent in acute myeloid leukemia independent of TP53 mutational status. Leukemia. 2017;31(6):1296. doi: 10.1038/leu.2016.350. [DOI] [PubMed] [Google Scholar]
- 42.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 43.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. nature. 1993;366(6456):701. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 44.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J, Sedivy J, Kinzler K, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 45.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. The EMBO journal. 1993;12(2):461. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutcliffe T, Fu L, Abraham J, Vaziri H, Benchimol S. A functional wild-type p53 gene is expressed in human acute myeloid leukemia cell lines. Blood. 1998;92(8):2977–2979. [PubMed] [Google Scholar]
- 47.Long J, Parkin B, Ouillette P, Bixby D, Shedden K, Erba H, Wang S, Malek SN. Multiple distinct molecular mechanisms influence sensitivity and resistance to MDM2 inhibitors in adult acute myelogenous leukemia. Blood. 2010;116(1):71–80. doi: 10.1182/blood-2010-01-261628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wade M, Li Y-C, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nature Reviews Cancer. 2013;13(2):83. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nature Reviews Cancer. 2009;9(12):862. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 50.Verwaal VJ, van Ruth S, de Bree E, van Slooten GW, van Tinteren H, Boot H, Zoetmulder FA. Randomized trial of cytoreduction and hyperthermic intraperitoneal chemotherapy versus systemic chemotherapy and palliative surgery in patients with peritoneal carcinomatosis of colorectal cancer. Journal of clinical oncology. 2003;21(20):3737–3743. doi: 10.1200/JCO.2003.04.187. [DOI] [PubMed] [Google Scholar]
- 51.Pels H, Schmidt-Wolf IG, Glasmacher A, Schulz H, Engert A, Diehl V, Zellner A, Schackert G, Reichmann H, Kroschinsky F. Primary central nervous system lymphoma: results of a pilot and phase II study of systemic and intraventricular chemotherapy with deferred radiotherapy. Journal of Clinical Oncology. 2003;21(24):4489–4495. doi: 10.1200/JCO.2003.04.056. [DOI] [PubMed] [Google Scholar]
- 52.Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nature reviews Cancer. 2015;15(12):747. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gordon MJ, Tardi P, Loriaux MM, Spurgeon SE, Traer E, Kovacsovics T, Mayer LD, Tyner JW. CPX-351 exhibits potent and direct ex vivo cytotoxicity against AML blasts with enhanced efficacy for cells harboring the FLT3-ITD mutation. Leukemia Res. 2017;53:39–49. doi: 10.1016/j.leukres.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 54.Arends M, Morris R, Wyllie A. Apoptosis. The role of the endonuclease. The American journal of pathology. 1990;136(3):593. [PMC free article] [PubMed] [Google Scholar]
- 55.Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nature reviews Drug discovery. 2014;13(3):217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 56.Kirsch DG, Kastan MB. Tumor-suppressor p53: implications for tumor development and prognosis. Journal of clinical oncology. 1998;16(9):3158–3168. doi: 10.1200/JCO.1998.16.9.3158. [DOI] [PubMed] [Google Scholar]
- 57.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 58.Burgess A, Chia KM, Haupt S, Thomas D, Haupt Y, Lim E. Clinical overview of MDM2/X-targeted therapies. Frontiers in oncology. 2016;6:7. doi: 10.3389/fonc.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y, Aguilar A, Bernard D, Wang S. Small-molecule inhibitors of the MDM2–p53 protein– protein interaction (MDM2 Inhibitors) in clinical trials for cancer treatment: miniperspective. Journal of medicinal chemistry. 2014;58(3):1038–1052. doi: 10.1021/jm501092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan BX, Liew HP, Chua JS, Ghadessy FJ, Tan YS, Lane DP, Coffill CR. Anatomy of Mdm2 and Mdm4 in evolution. Journal of molecular cell biology. 2017;9(1):3–15. doi: 10.1093/jmcb/mjx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhan C, Lu W. Peptide activators of the p53 tumor suppressor. Current pharmaceutical design. 2011;17(6):603–609. doi: 10.2174/138161211795222577. [DOI] [PubMed] [Google Scholar]
- 62.Brown CJ, Quah ST, Jong J, Goh AM, Chiam PC, Khoo KH, Choong ML, Lee MA, Yurlova L, Zolghadr K. Stapled peptides with improved potency and specificity that activate p53. ACS chemical biology. 2012;8(3):506–512. doi: 10.1021/cb3005148. [DOI] [PubMed] [Google Scholar]
- 63.Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. Journal of Clinical Oncology. 2011;29(5):487–494. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- 64.Prokocimer M, Molchadsky A, Rotter V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: projections on diagnostic workup and therapy. Blood. 2017;130(6):699–712. doi: 10.1182/blood-2017-02-763086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lepelley P, Preudhomme C, Vanrumbeke M, Quesnel B, Cosson A, Fenaux P. Detection of p53 mutations in hematological malignancies: comparison between immunocytochemistry and DNA analysis. Leukemia. 1994;8(8):1342–1349. [PubMed] [Google Scholar]
- 66.Bernstein I. Monoclonal antibodies to the myeloid stem cells: therapeutic implications of CMA-676, a humanized anti-CD33 antibody calicheamicin conjugate. Leukemia. 2000;14(3):474. doi: 10.1038/sj.leu.2401663. [DOI] [PubMed] [Google Scholar]
- 67.Sievers E, Appelbaum F, Spielberger R, Forman S, Flowers D, Smith F, Shannon-Dorcy K, Berger M, Bernstein I. Selective Ablation of Acute Myeloid Leukemia Using Antibody-Targeted Chemotherapy: A Phase I Study of an Anti-CD33 Calicheamicin Immunoconjugate. Blood; Presented in part at the 1997 Annual Meeting of the American Society of Clinical Oncology, Denver, CO; the 1997 European Cancer Conference, Hamburg, Germany; and the 1997 Annual Meeting of the American Society of Hematology; San Diego, CA. 1999. pp. 3678–3684. [PubMed] [Google Scholar]
- 68.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annual review of biochemistry. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.