Abstract
Serpins are a broadly distributed superfamily of protease inhibitors that are present in all kingdoms of life. The acronym, serpin, is derived from their function as potent serine proteases inhibitors. Early studies of serpins focused on their functions in haemostasis since modulating serine proteases activities are essential for coagulation. Additional research has revealed that serpins function in infection and inflammation, by modulating serine and cysteine proteases activities. The aim of this review is to summarize the accumulating findings and current understanding of the functions of serpins in host-pathogen interactions, serving as host defense proteins as well as pathogenic factors. We also discuss the potential crosstalk between host and pathogen serpins. We anticipate that future research will elucidate the therapeutic value of this novel target.
Keywords: Serpin, Serine protease, Host-pathogen interaction, Infection, Inflammation
Introduction
Serpins are a superfamily of proteins. The family name is derived from the functional attributes of the members as they are serine protease inhibitors (serpin). They are the most broadly distributed protease inhibitors, and are present in all kingdoms of life including plants, animals, bacteria, archaea and viruses (Silverman et al., 2001). While the majority of serpins function as serine protease inhibitors, some serpins function as “cross-class” inhibitors of other kinds of proteases. For example, viral serpin CrmA inhibits a cysteine protease, interleukin-1 beta converting enzyme (Irving et al., 2002; Ray et al., 1992). In addition, there are a few serpins that exhibit no inhibitory functions but participate in biological processes in other ways. For examples, serpin HSP47 serves as a chaperone, and ovalbumin (another serpin), functions as a storage protein (Law et al., 2006).
As potent protease inhibitors, serpins modulate a wide variety of proteolytic cascades thus controlling many physiological and pathological reactions. For instance, human serpins are found to regulate the proteolytic cascade that is central to blood clotting. Antithrombin, a serpin superfamily member, can inhibit multiple key enzymes in blood coagulation such as thrombin, activated factor X (FXa), FIXa and FXIa (Aguila et al., 2017; Hepner & Karlaftis, 2013). In addition to blood coagulation, serpins also participate in a wide variety of other biological processes. These processes include thrombosis (Van de Water et al., 2004), immune-regulation (Pemberton et al., 1988), tumour-suppression (Dzinic et al., 2017), chromatin condensation (Grigoryev & Woodcock, 1998) and apoptosis regulation (Ray et al., 1992). Furthermore, studies reveal serpins have clinical relevance. For example, patients with papillary thyroid cancer have high-concentrations of SERPINE2 and SLPI (secretory leukocyte protease inhibitor) (Stein & Chothia, 1991). The serum concentrations of both anticoagulant proteins are considered markers for the development of this disease. Kallistatin, another serpin family member, has been shown to regulate cardiovascular function and blood vessel development. Its levels are elevated in patients with type 1 and type 2 diabetes with chronic diabetic complications (Gateva et al., 2017).
Recently, the study of serpin functions in infection and inflammation has been of particular interest, especially as more serpins from pathogens are identified and characterized. One example is crmA, a cowpox viral serpin, one of the smallest members of the serpin superfamily. CrmA is thought to be important in allowing viruses to avoid host inflammatory and apoptotic responses (Ekert, Silke & Vaux, 1999; Renatus et al., 2000). Usually, viral genomes are compact to fit their unique life style. The presence of viral serpins indicates their essential functions for the survival of the pathogen and the infection of hosts. Since serpins are present in both pathogen and host organisms, we will discuss the functions of serpins on each side of these processes, and their potential interactions, in a variety of organisms during infection and inflammation.
Survey Methodology
In this study, we reviewed articles related to the functions of serpins in different organisms that either serve as hosts or pathogens. All references in this review were retrieved using search engines such as PubMed and Google Scholar. Keywords such as serpin, serine protease, host-pathogen interaction, infection and inflammation were used to search for the references. Figures related to protein structures were searched and modified from the Protein Data Bank.
Mechanism of serpin inhibition
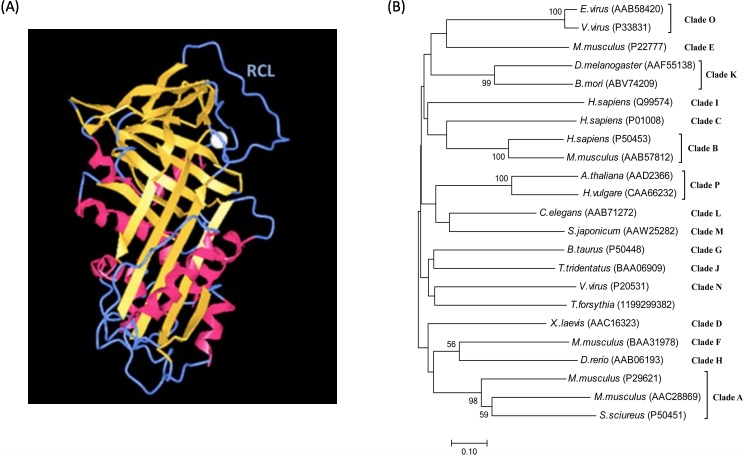
The mechanism of serpin-inhibiting proteases relies on a reactive center loop (RCL) that interacts with the target enzyme. Structural studies reveal that serpins are characterized by a common core domain consisting of three β-sheets and 8–9 α-helices and a reactive center loop, as shown in Fig. 1A (Moreira et al., 2014; Patschull et al., 2011; Pearce et al., 2008). Although this core region is present in all serpins, sequence homology among members in the family could be as low as 25%. A phylogenetic study of the superfamily divided serpins into 16 ‘clades’, designated clade A-P (Gettins, 2002), and includes serpins from vertebrates, invertebrates, plants and viruses. A phylogenetic tree of representative serpins from each clade is shown in Fig. 1B.
Figure 1. Serpin structure and phylogenetic tree.
(A) Structure of the serpin alpha-1 antitrypsin. Human alpha-1 antitrypsin is representative of serpin structures. It contains α-helices (red), β-sheets (golden) and a reactive center loop (RCL, the upright blue region) (PDB: 3NE4). (B) Phylogenetic tree of serpin superfamily. The neighbour-joining tree is based on serpin protein sequences and different clades are represented by a single identifier (e.g., Antithrombin III, P01008), where possible. The phylogenetic analysis was performed using MEGA version 7.0. Analysis was done on 1,000 bootstrapped datasets and values of >50% are shown.
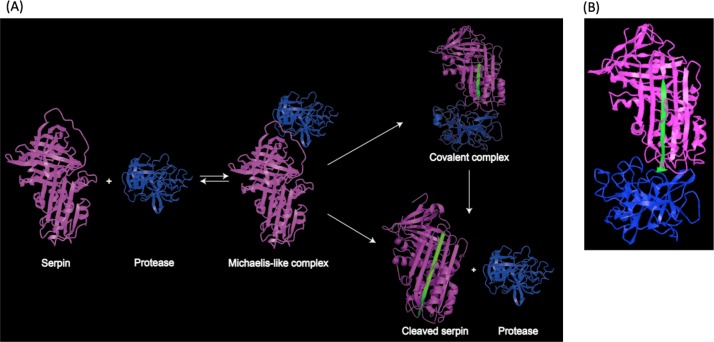
Structural studies demonstrated that the RCL region contains a scissile bond between residues P1 and P1′, that interacts with and can be cleaved by the target protease (Li et al., 1999). Upon cleavage, the reactive center loop of serpin inserts into the β-sheet A. This conformational change makes serpins thermodynamically more stable (Dementiev, Dobo & Gettins, 2006; Gong et al., 2015). In addition, a fluorescence study demonstrated that the protease in the complex was also conformationally distorted (Elliott et al., 1996). As a result, the target protease is trapped in a covalent and irreversible complex with the serpin, and thus is inhibited irreversibly (Irving et al., 2000; Stein & Chothia, 1991). This process is illustrated in Fig. 2A, and a structure model showing covalent serpin-protease complex is shown in Fig. 2B.
Figure 2. Representation of serpin-protease interaction.
(A) Proposed process of serpin-protease interaction. A serpin (magenta) interacts with targeted protease (blue), and the Michaelis-like complex of serpin and protease is formed. The complex either undergo peptide bond hydrolysis resulting in a kinetically trapped loop-inserted covalent complex (inhibitory pathway), or a cleaved serpin and free protease (non-inhibitory/substrate pathway). The cleaved and inserted RCL is highlighted in green. Serpin-protease complex is stable. Possibility of transition from covalent complex to cleaved form exists yet slim, since complex in vivo would be cleared long before complex decay could occur. (B) Structure of stable serpin- protease complex (PDB: 2D26). The complex is formed by serpin α1PI (magenta) and protease elastase (blue). The inserted RCL is highlighted in green.
It is worth noting that co-factors are sometimes needed or can enhance serpins’ inhibitory functions. For example, the glycosaminoglycan heparin, a known anti-coagulant, enhances inhibition of cathepsin L by serpin B3 and B4 (Higgins et al., 2010). It is also interesting to know that serpins can be secreted or intracellular, thus may also impact their targeted proteases and ways of functions. For instances, the secreted serpins such as SERPINA1 and SEPRINA3 can inhibit inflammatory response molecules; while the intracellular serpins such as SERPINB9 acts on cytosolic proteases thus participate in cellular events (Law et al., 2006; Lomas, 2005; Sun et al., 1996). This does not mean that different forms of serpins have distinct functions, in fact many intracellular serpins participate in inflammatory responses, or vice versa; it is just something we shall keep in mind when discuss the underlying mechanisms of serpin functions as defense factors and pathogenic agents.
Serpins serve as host defense factors
The defense strategies of serpins derived from the host are variable, including direct inhibition of pathogen proteases, inhibition of pathogen binding, and enhancement of host immune cell functions. Here we discuss mechanisms of how serpins function as host defense factors in a few representative organisms, such as humans, insects and plants.
Serpins in vertebrates
In humans and other vertebrates, neutrophil extracellular traps (NETs) are web-like DNA structures extruded into the extracellular environment by activated neutrophils. NETs are thought to represent a unique defense strategy against microbial infection. A serpin superfamily member expressed by macrophages and neutrophils is SerpinB1. It is capable of restricting NET production. Studies indicate that SerpinB1 inhibits neutrophil elastase, cathepsin G, and proteinase 3 (Farley et al., 2012). More recently, serpins expressed at the mucosal surface have been linked to inhibition of HIV binding, replication and reduction of inflammation of susceptible cells. These serpins, together with other protease inhibitors, are found to be expressed at the epithelial layer of the female genital tract, and thus are considered as essential in the frontline defense against infection. In addition, their potential applications in disease treatment have also been explored (Aboud et al., 2014).
Serpins are also found to regulate defense reactions in other mammalian species, such as mouse. A serpin superfamily member α1-antitrypsin promotes lung defense against Pseudomonas aeruginosa (PA) infection in mice. A study by Jiang et al. (2013) demonstrated that the underlying mechanism by which α1-antitrypsin reduces lung bacterial infection is through inhibiting neutrophil elastase-mediated host defense protein degradation. Potential therapeutic application of α1-antitrypsin to both humans and mice during PA infection has been proposed.
Serpins in invertebrates
Serpins have been described in invertebrates, particularly insects. Insects utilize innate immunity as the major defense system against pathogen invasion. The immune responses include hemocyte clotting, melanization and antimicrobial peptide expression (Meekins, Kanost & Michel, 2017). To set off these innate responses, cascades of serine proteases activation must be triggered, and these proteolytic cascades are tightly regulated by serpins. Many serpin genes have been identified in species of insects including 34 in Bombyx mori, 32 in Manduca sexta, 31 in the beetle Tribolium castaneum and 29 in Drosophila melanogaster. Insect serpins are found expressed in various organs including fat bodies, midgut and hemocytes (Meekins, Kanost & Michel, 2017). The majority of these insect serpins are believed to be related to innate immunity. For example, when Bombyx mori was challenged by pathogens such as Micrococcus luteus and Serratia marcescens, the expression of serpin6 (BmSerpin6) was increased significantly (Li et al., 2017). It was found that BmSerpin6 directly inhibited the expression of the antimicrobial proteins drosomycin and gloverin2, and the prophenoloxidase (PPO) activity in the melanization cascade.
In studies of Manduca sexta, researchers demonstrated that serpin-1 could form a complex with the serine protease hemolymph protease 8 (HP8), to inhibit the activation of the Toll pathway (An, Ragan & Kanost, 2011) during bacterial infection. Serpin-7 was found to inhibit prophenoloxidase-activating protease-3 (PAP3) in the melanization pathway to down-regulate innate immune responses (Suwanchaichinda et al., 2013). In addition, more recent studies showed that Manduca sexta serpin-1, 4, 9, 13 and serpin-3, 5, 6 were all able to complex with pro-hemolymph protease 1 (ProHP1), which is a key proteinase in innate immunity of insects (He et al., 2017).
In Drosophila, necrotic protein is one of the many serpins that have been related to innate immunity. Necrotic protein inhibits the clip domain of serine protease persephone, and Drosophila with necrotic protein mutations constitutively express anti-microbial peptide drosomycin in the Toll inflammatory signaling pathway (Robertson et al., 2003).
Thus, most insect serpins negatively regulate innate immunity by inhibiting serine proteases that are essential for immune responses. In addition, several studies revealed that insect serpins could also possess direct anti-pathogen activity upon infection (Levashina et al., 1999). For example, serpin protein SRPN6 from Anopheles gambiae was highly up-regulated in epithelia immediately after bacterial and parasitic exposures. The AgSRPN6 acts directly on parasite clearance by inhibiting melanization and promoting parasite lysis (Abraham et al., 2005).
Serpins in plants
Serpins are present in almost all land plants (Roberts et al., 2011). However, the functions of plant serpins remain to be characterized. In vitro studies have demonstrated the protease-inhibiting activities of plant serpins, but the lack of target chymotrypsin-like proteases within plants suggests that plant serpins may target digestive proteases from invaded pathogens or parasites. In addition, there is abundant accumulation of serpins in plant seeds endosperm, phloem of coleoptiles and leaves (Roberts et al., 2003). These localizations of plant serpins further imply the defensive roles of serpins in plants against exogenous proteases and pathogens. Although plant serpins may have distinct characteristics from their insect and animal counterparts, they have been shown to have a role in the pathways regulating the host immune responses. For example, Yoo et al. (2000) demonstrated a serpin protein, Cucurbita maxima phloem serpin-1 (CmPS), had effective elastase-inhibiting activity. They showed that increased expression of CmPS-1 within the phloem sap was associated with reduced ability of sap-sucking insect aphids to survive and reproduce.
Plants serpins are also found to participate in plant immunity as negative regulators of stress-induced cell death, or so-called hypersensitive response (HR). For instance, Arabidopsis thaliana serpins AtSRP4 and AtSRP5 negatively regulate stress-induced cell death induced by bacteria (Bhattacharjee et al., 2017). This kind of cell death or response usually occurs at sites where pathogens attempt to invade. Thus the activities of serpins may have a protective role for plants when facing pathogen attack.
In addition to the above mechanisms of serpins acting on proteases to function as host defense factors, there are also other ways that serpins participate in host defense. For example, serpins can induce the expression of host antimicrobial peptides and cytokines (Kausar et al., 2017; Zhao et al., 2014). Serpins can also directly bind to bacterial pathogens and cause membrane disruptions (Malmstrom et al., 2009). Interestingly, even non-inhibitory serpin can exert antibacterial activity. For example, non-inhibitory serpin ovalbumin-related protein X possesses antibacterial activity through heparin-binding ability (Rehault-Godbert et al., 2013). All these findings broaden our understanding of the mechanisms of serpin functions as host defense factors.
Pathogen-derived serpins in infection and inflammation
As mentioned above, serpins are present in almost all kingdoms of life including microbes and other pathogenic organisms. Pathogen-derived serpins may facilitate infection or survival of pathogens, but the mechanisms remain to be fully elucidated. The data indicate that pathogen-derived serpins are capable of inhibiting host inflammatory proteins or cells, and abrogating host cell apoptosis. Studies are on-going to find additional strategies used by pathogen-derived serpins to facilitate infection. In addition, the potential of pathogen-derived serpins as novel candidates of clinical therapies or vaccination has drawn great interest from scientists and physicians.
Serpins from viruses
Viral genomes are usually kept at minimum scales to fit their unique life style. Thus the presence of viral serpins must be essential for the survival and/or function of the virus. In fact, researchers have identified several serpins that are required for virulence and infectivity (Nathaniel et al., 2004). In the myxoma viruses, three serpins, SERP1, SERP2 and SERP3 have been identified (MacNeill, Turner & Moyer, 2006). Similarly, there are three serpins in orthopox viruses, designated SPI-1, SPI-2 and SPI-3 (Macen et al., 1996). In addition, the P1 positions of SPI-2-like and SERP2 serpins contain an aspartyl residue, which indicates their potential targets are mammalian caspases and the serine protease granzyme B (Turner et al., 1999). By inhibiting these host proteases, the virus may be able to restrain apoptosis in host cells thus down-regulatinghost immune responses. Other viruses have also been shown to contain one or more serpins, including swinepox, lumpy skin disease virus, fowlpox, and members of the rhadinovirus genus. Recently, a baculovirus serpin Hesp018 has been identified in the Hemileuca species nuclear polyhedrosis virus (NPV). This serpin protein has been suggested to abrogate host cell apoptosis, resulting in accelerated production of virus in Sf9 insect cells (Ardisson-Araujo et al., 2015).
The ability of viral serpins to abrogate host immune systems has been proposed as a strategy to treat certain diseases. One such example is the proposed application of the viral serpin Serp-1 to treat acute unstable coronary syndromes (Lucas et al., 2009). Several other viral serpins are being studied for their potential applications as novel anti-inflammatory therapeutics as well (Mangan et al., 2017; Zheng et al., 2012).
Serpins from bacteria
Tannerella forsythia is an anaerobic, gram-negative bacteria species that usually resides in the human mouth and contributes to chronic periodontitis (Pereira et al., 2017). To inhibit host endopeptidases, T. forsythia secretes a serpin-type protease inhibitor called miropin, which irreversibly inhibits serine and cysteine endopeptidases of the host (Goulas et al., 2017). Phylogenetic analysis of this serpin protein shows that it does not follow a vertical descent model, indicating micropin may arise from the host by horizontal gene transfer. In fact, the studies of serpins from human commensal bacteria and their therapeutic applications have become exciting research areas. For instance, Mkaouar et al. (2016) characterized two novel serpins from the human gastrointestinal tract commensal bacteria. These two serpins are called siropin-1 and siropin-2. These two serpins are found to preferentially inhibit two human serine proteases, neutrophil elastase and proteinase 3, that are associated with human inflammatory bowel disease (Mkaouar et al., 2016). Thus, siropins or other serpins from human commensal bacteria have been suggested as novel therapeutics against human inflammatory diseases.
Serpins from parasites
Parasites are organisms that live in another organism and may cause major public health problems such as zoonotic diseases. Usually the parasites need to evade host defense system in order to survive. Serpins are found to be important during host-parasite interaction, and parasites utilize their serpins to facilitate infection and survival in the host. For example, cattle tick Rhipicephalus microplus encodes at least 24 serpins, of which RmS-3, RmS-6, and RmS-17 were identified in the saliva and later confirmed to inhibit pro-inflammatory and pro-coagulatory proteases of the host (Tirloni et al., 2014). In a follow-up study, rRmS-3 was found to inhibit chymotrypsin, cathepsin G, and pancreatic elastase. Among these serpins, rRmS-6 was found to inhibit trypsin, chymotrypsin, factor Xa, factor XIa and plasmin; while rRmS-17 inhibited trypsin, cathepsin G, chymotrypsin, plasmin and factor XIa (Tirloni et al., 2016). This study also claimed that polyclonal antibodies to saliva proteins of Amblyomma americanum, Ixodes scapularis and Rhipicephalus sanguineus were able to cross-react with these three R. microplus saliva serpins. These findings suggest serpins from pathogens could be applied as novel candidates of vaccination (De la Fuente et al., 2007).
Researchers have identified several serpins associated with Trichinella spiralis including Tsp03044 and TspAd5 (Knox, 2007; Zhang et al., 2016). Both of them inhibit trypsin, α-chymotrypsinand pepsin of mammals. These data support the inference that serpins from parasites facilitate invasion into host tissues (Zhang et al., 2016).
Similarly, Schistosoma mansoni has at least eight serpins. Among those, Smpi56 and SmSPI have been characterized. Smpi56 was purified from extracts of adult S. mansoni, and is able to inhibit neutrophil elastase, pancreatic elastase and an endogenous cercarial protease (Ghendler, Arnon & Fishelson, 1994; Quezada & McKerrow, 2011). SmSPI, also known as S. mansoni serpin isoform 3, was found to inhibit chymotrypsin, neutrophil elastase and porcine pancreatic elastase (Pakchotanon et al., 2016). In addition, SmSPI was found predominantly expressed in the head gland of the parasite, as well as on its spines. These findings suggest that serpins from S. mansoni facilitate intradermal and intravenous survival of this pathogen.
Potential crosstalk between host and pathogen serpins
As we discussed above, under most circumstances serpins target proteases to modulate inflammatory responses during host-pathogen interactions. Yet it is still of great interest to know whether serpins function through serpin-serpin interactions. In fact, there is evidence that serpins can be inactivated through polymerization (Gettins & Olson, 2016). Serpin polymerization occurs when the RCL region of one serpin docks into the β-sheet A of another serpin to form an inactive serpin polymer. Actually, this kind of serpin inactivation is not uncommon and many examples are found in human deficiency and diseases. For example, the Z allele of SERPINA1 accumulates in patients’ liver through serpin polymerization (Lomas et al., 1992). Thus, we would not be surprised to find out in the future that pathogen serpins utilize this mechanism to inactivate host serpins, or vice versa, during infection.
Conclusions
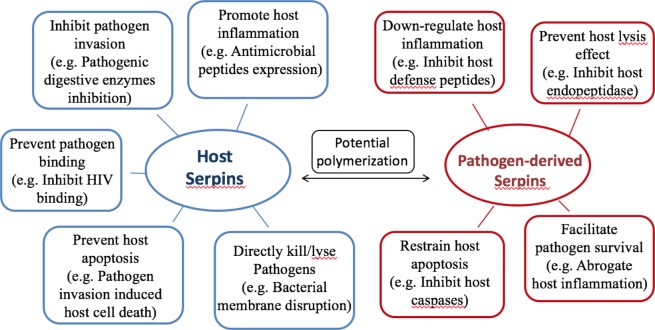
Almost all organisms express serpins, and serpins play critical roles in host-pathogen interactions and regulation of inflammatory responses. The evidence indicates that serpins of the host provide protection and those of the pathogens enhance infectivity. A brief summary of representative serpin functions during host-pathogen interactions is shown in Fig. 3.
Figure 3. Summary of serpin functions in host-pathogen interactions.
Hypothesis of protective mechanisms offered byhost serpins (on the left, blue), and pathogenic mechanisms exerted by pathogen-derived serpins (on the right, red). Host serpins may act directly or indirectly upon pathogen infections. The representative mechanisms include inhibiting pathogenic digestive proteases, promoting host antimicrobial peptide expression and so on. Pathogen-derived serpins also utilize various mechanisms and representative ones are listed.
On the basis of existing studies, we conclude that serpins, either from hosts or pathogens, modulate inflammatory responses by inhibiting target proteases associated with host-pathogen interactions. There is also evidence indicating potential cross interactions between host and pathogen serpins. In addition to all that, there are a number of studies demonstrating that serpin-proteases complexes can lead to downstream signals that result in responses such as inflammation, cytoskeleton rearrangement, proliferation and apoptosis. For example, R1-Anti-chymotrypsin-cathepsin G complexes have been shown to stimulate production of interleukin 6 and activation of NADPH oxidase (Kurdowska & Travis, 1990; Schuster et al., 1992). More recently, serpin α-1 antitrypsin was found to be able to form complexes independently of the inhibitory site with neutrophil-expressedpro-inflammatory leukotriene B4. The resulting complexes modulate downstream signaling events, and augmentation of serpin α-1 antitrypsin was suggested as a potential therapy for inflammatory diseases (O’Dwyer et al., 2015). These signaling properties of serpin-protease complexes shed light on the versatility of serpins modulating inflammatory responses. Another interesting yet poorly explored aspect is the activities of the peptide derivatives resulting from proteolytic cleavage of the reactive center loop (RCL) of serpins. A few studies have demonstrated that the peptides derived from serpin RCL have expanded functions such as anti-inflammatory and antimicrobial activities (Ambadapadi et al., 2016; Andersson et al., 2004). Further explorations are needed for additional activities and potential applications of these peptide derivatives.
There is no doubt that further studies of serpins will identify many more biological targets and underlying molecular mechanisms. The study of serpins will remain an important area for basic research, as well as for clinical applications.
Acknowledgments
We appreciate Dr. Judith S. Bond, Evan Pugh Professor Emeritus, Department of Biochemistry and Molecular Biology Penn State University College of Medicine at Hershey; Adjunct Professor, Department of Biochemistry & Biophysics, University of North Carolina School of Medicine at Chapel Hill, for her great support for correcting the language in this manuscript.
Funding Statement
This work was supported by the Fundamental Research Funds for the Central Universities (No. XDJK2017B002) and the National Natural Science Foundation of China (No. 31470250, No. 31472151). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Jialing Bao conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Guoqing Pan, Junhong Wei and Zeyang Zhou authored or reviewed drafts of the paper, approved the final draft.
Mortimer Poncz and Maoshuang Ran prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The research in this review article did not generate any data or code.
References
- Aboud et al. (2014).Aboud L, Ball TB, Tjernlund A, Burgener A. The role of serpin and cystatin antiproteases in mucosal innate immunity and their defense against HIV. American Journal of Reproductive Immunology. 2014;71:12–23. doi: 10.1111/aji.12166. [DOI] [PubMed] [Google Scholar]
- Abraham et al. (2005).Abraham EG, Pinto SB, Ghosh A, Vanlandingham DL, Budd A, Higgs S, Kafatos FC, Jacobs-Lorena M, Michel K. An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16327–16332. doi: 10.1073/pnas.0508335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguila et al. (2017).Aguila S, Izaguirre G, Martinez-Martinez I, Vicente V, Olson ST, Corral J. Disease-causing mutations in the serpin antithrombin reveal a key domain critical for inhibiting protease activities. Journal of Biological Chemistry. 2017 doi: 10.1074/jbc.M117.787325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambadapadi et al. (2016).Ambadapadi S, Munuswamy-Ramanujam G, Zheng D, Sullivan C, Dai E, Morshed S, McFadden B, Feldman E, Pinard M, McKenna R, Tibbetts S, Lucas A. Reactive Center Loop (RCL) peptides derived from serpins display independent coagulation and immune modulating activities. Journal of Biological Chemistry. 2016;291:2874–2887. doi: 10.1074/jbc.M115.704841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, Ragan & Kanost (2011).An C, Ragan EJ, Kanost MR. Serpin-1 splicing isoform J inhibits the proSpatzle-activating proteinase HP8 to regulate expression of antimicrobial hemolymph proteins in Manduca sexta. Developmental and Comparative Immunology. 2011;35:135–141. doi: 10.1016/j.dci.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson et al. (2004).Andersson E, Rydengard V, Sonesson A, Morgelin M, Bjorck L, Schmidtchen A. Antimicrobial activities of heparin-binding peptides. European Journal of Biochemistry. 2004;271:1219–1226. doi: 10.1111/j.1432-1033.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- Ardisson-Araujo et al. (2015).Ardisson-Araujo DM, Rohrmann GF, Ribeiro BM, Clem RJ. Functional characterization of hesp018, a baculovirus-encoded serpin gene. Journal of General Virology. 2015;96:1150–1160. doi: 10.1099/vir.0.000041. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee et al. (2017).Bhattacharjee L, Singh D, Gautam JK, Nandi AK. Arabidopsis thaliana serpins AtSRP4 and AtSRP5 negatively regulate stress-induced cell death and effector-triggered immunity induced by bacterial effector AvrRpt2. Physiologia Plantarum. 2017;159:329–339. doi: 10.1111/ppl.12516. [DOI] [PubMed] [Google Scholar]
- De la Fuente et al. (2007).De la Fuente J, Almazan C, Canales M, Perez de la Lastra JM, Kocan KM, Willadsen P. A ten-year review of commercial vaccine performance for control of tick infestations on cattle. Animal Health Research Reviews. 2007;8:23–28. doi: 10.1017/S1466252307001193. [DOI] [PubMed] [Google Scholar]
- Dementiev, Dobo & Gettins (2006).Dementiev A, Dobo J, Gettins PG. Active site distortion is sufficient for proteinase inhibition by serpins: structure of the covalent complex of alpha1-proteinase inhibitor with porcine pancreatic elastase. Journal of Biological Chemistry. 2006;281:3452–3457. doi: 10.1074/jbc.M510564200. [DOI] [PubMed] [Google Scholar]
- Dzinic et al. (2017).Dzinic SH, Bernardo MM, Li X, Fernandez-Valdivia R, Ho YS, Mi QS, Bandyopadhyay S, Lonardo F, Vranic S, Oliveira DS, Bonfil RD, Dyson G, Chen K, Omerovic A, Sheng X, Han X, Wu D, Bi X, Cabaravdic D, Jakupovic U, Wahba M, Pang A, Harajli D, Sakr WA, Sheng S. An essential role of maspin in embryogenesis and tumor suppression. Cancer Research. 2017;77:886–896. doi: 10.1158/0008-5472.CAN-16-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekert, Silke & Vaux (1999).Ekert PG, Silke J, Vaux DL. Inhibition of apoptosis and clonogenic survival of cells expressing crmA variants: optimal caspase substrates are not necessarily optimal inhibitors. EMBO Journal. 1999;18:330–338. doi: 10.1093/emboj/18.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott et al. (1996).Elliott PR, Lomas DA, Carrell RW, Abrahams JP. Inhibitory conformation of the reactive loop of alpha 1-antitrypsin. Nature Structural Biology. 1996;3:676–681. doi: 10.1038/nsb0896-676. [DOI] [PubMed] [Google Scholar]
- Farley et al. (2012).Farley K, Stolley JM, Zhao P, Cooley J, Remold-O’Donnell E. A serpinB1 regulatory mechanism is essential for restricting neutrophil extracellular trap generation. Journal of Immunology. 2012;189:4574–4581. doi: 10.4049/jimmunol.1201167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateva et al. (2017).Gateva A, Assyov Y, Velikova T, Kamenov Z. Increased kallistatin levels in patients with obesity and prediabetes compared to normal glucose tolerance. Endocrine Research. 2017;42:163–168. doi: 10.1080/07435800.2017.1286671. [DOI] [PubMed] [Google Scholar]
- Gettins (2002).Gettins PG. Serpin structure, mechanism, and function. Chemical Reviews. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- Gettins & Olson (2016).Gettins PGW, Olson ST. Inhibitory serpins. New insights into their folding, polymerization, regulation and clearance. Biochemical Journal. 2016;473:2273–2293. doi: 10.1042/Bcj20160014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghendler, Arnon & Fishelson (1994).Ghendler Y, Arnon R, Fishelson Z. Schistosoma mansoni: isolation and characterization of Smpi56, a novel serine protease inhibitor. Experimental Parasitology. 1994;78:121–131. doi: 10.1006/expr.1994.1013. [DOI] [PubMed] [Google Scholar]
- Gong et al. (2015).Gong L, Liu M, Zeng T, Shi X, Yuan C, Andreasen PA, Huang M. Crystal structure of the michaelis complex between tissue-type plasminogen activator and plasminogen activators inhibitor-1. Journal of Biological Chemistry. 2015;290:25795–25804. doi: 10.1074/jbc.M115.677567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas et al. (2017).Goulas T, Ksiazek M, Garcia-Ferrer I, Sochaj-Gregorczyk AM, Waligorska I, Wasylewski M, Potempa J, Gomis-Ruth FX. A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome. Journal of Biological Chemistry. 2017;292:10883–10898. doi: 10.1074/jbc.M117.786533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev & Woodcock (1998).Grigoryev SA, Woodcock CL. Chromatin structure in granulocytes. A link between tight compaction and accumulation of a heterochromatin-associated protein (MENT) Journal of Biological Chemistry. 1998;273:3082–3089. doi: 10.1074/jbc.273.5.3082. [DOI] [PubMed] [Google Scholar]
- He et al. (2017).He Y, Wang Y, Yang F, Jiang H. Manduca sexta hemolymph protease-1, activated by an unconventional non-proteolytic mechanism, mediates immune responses. Insect Biochemistry and Molecular Biology. 2017;84:23–31. doi: 10.1016/j.ibmb.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepner & Karlaftis (2013).Hepner M, Karlaftis V. Antithrombin. Methods in Molecular Biology. 2013;992:355–364. doi: 10.1007/978-1-62703-339-8_28. [DOI] [PubMed] [Google Scholar]
- Higgins et al. (2010).Higgins WJ, Fox DM, Kowalski PS, Nielsen JE, Worrall DM. Heparin enhances serpin inhibition of the cysteine protease cathepsin L. Journal of Biological Chemistry. 2010;285:3722–3729. doi: 10.1074/jbc.M109.037358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving et al. (2002).Irving JA, Pike RN, Dai W, Bromme D, Worrall DM, Silverman GA, Coetzer TH, Dennison C, Bottomley SP, Whisstock JC. Evidence that serpin architecture intrinsically supports papain-like cysteine protease inhibition: engineering alpha(1)-antitrypsin to inhibit cathepsin proteases. Biochemistry. 2002;41:4998–5004. doi: 10.1021/bi0159985. [DOI] [PubMed] [Google Scholar]
- Irving et al. (2000).Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Research. 2000;10:1845–1864. doi: 10.1101/gr.GR-1478R. [DOI] [PubMed] [Google Scholar]
- Jiang et al. (2013).Jiang D, Persinger R, Wu Q, Gross A, Chu HW. alpha1-Antitrypsin promotes SPLUNC1-mediated lung defense against Pseudomonas aeruginosa infection in mice. Respiratory Research. 2013;14:122. doi: 10.1186/1465-9921-14-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kausar et al. (2017).Kausar S, Abbas MN, Qian C, Zhu B, Sun Y, Sun Y, Wang L, Wei G, Maqsood I, Liu CL. Serpin-14 negatively regulates prophenoloxidase activation and expression of antimicrobial peptides in Chinese oak silkworm Antheraea pernyi. Developmental and Comparative Immunology. 2017;76:45–55. doi: 10.1016/j.dci.2017.05.017. [DOI] [PubMed] [Google Scholar]
- Knox (2007).Knox DP. Proteinase inhibitors and helminth parasite infection. Parasite Immunology. 2007;29:57–71. doi: 10.1111/j.1365-3024.2006.00913.x. [DOI] [PubMed] [Google Scholar]
- Kurdowska & Travis (1990).Kurdowska A, Travis J. Acute phase protein stimulation by alpha 1-antichymotrypsin-cathepsin G complexes. Evidence for the involvement of interleukin-6. Journal of Biological Chemistry. 1990;265:21023–21026. [PubMed] [Google Scholar]
- Law et al. (2006).Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC. An overview of the serpin superfamily. Genome Biology. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levashina et al. (1999).Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffmann JA, Reichhart JM. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- Li et al. (2017).Li B, Yu HZ, Ye CJ, Ma Y, Li X, Fan T, Chen FS, Xu JP. Bombyx mori Serpin6 regulates prophenoloxidase activity and the expression of antimicrobial proteins. Gene. 2017;610:64–70. doi: 10.1016/j.gene.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Li et al. (1999).Li J, Wang Z, Canagarajah B, Jiang H, Kanost M, Goldsmith EJ. The structure of active serpin 1K from Manduca sexta. Structure. 1999;7:103–109. doi: 10.1016/S0969-2126(99)80013-6. [DOI] [PubMed] [Google Scholar]
- Lomas (2005).Lomas DA. Molecular mousetraps, alpha1-antitrypsin deficiency and the serpinopathies. Clinical Medicine. 2005;5:249–257. doi: 10.7861/clinmedicine.5-3-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas et al. (1992).Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanism of Z-Alpha-1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- Lucas et al. (2009).Lucas A, Liu L, Dai E, Bot I, Viswanathan K, Munuswamy-Ramunujam G, Davids JA, Bartee MY, Richardson J, Christov A, Wang H, Macaulay C, Poznansky M, Zhong R, Miller L, Biessen E, Richardson M, Sullivan C, Moyer R, Hatton M, Lomas DA, McFadden G. The serpin saga development of a new class of virus derived anti-inflammatory protein immunotherapeutics. Advances in Experimental Medicine and Biology. 2009;666:132–156. doi: 10.1007/978-1-4419-1601-3_11. [DOI] [PubMed] [Google Scholar]
- Macen et al. (1996).Macen JL, Garner RS, Musy PY, Brooks MA, Turner PC, Moyer RW, McFadden G, Bleackley RC. Differential inhibition of the Fas- and granule-mediated cytolysis pathways by the orthopoxvirus cytokine response modifier A/SPI-2 and SPI-1 protein. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9108–9113. doi: 10.1073/pnas.93.17.9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeill, Turner & Moyer (2006).MacNeill AL, Turner PC, Moyer RW. Mutation of the Myxoma virus SERP2 P1-site to prevent proteinase inhibition causes apoptosis in cultured RK-13 cells and attenuates disease in rabbits, but mutation to alter specificity causes apoptosis without reducing virulence. Virology. 2006;356:12–22. doi: 10.1016/j.virol.2006.07.049. [DOI] [PubMed] [Google Scholar]
- Malmstrom et al. (2009).Malmstrom E, Morgelin M, Malmsten M, Johansson L, Norrby-Teglund A, Shannon O, Schmidtchen A, Meijers JC, Herwald H. Protein C inhibitor—a novel antimicrobial agent. PLOS Pathogens. 2009;5:e1000698. doi: 10.1371/journal.ppat.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan et al. (2017).Mangan MS, Melo-Silva CR, Luu J, Bird CH, Koskinen A, Rizzitelli A, Prakash M, Scarff KL, Mullbacher A, Regner M, Bird PI. A pro-survival role for the intracellular granzyme B inhibitor Serpinb9 in natural killer cells during poxvirus infection. Immunology and Cell Biology. 2017;95(10):884–894. doi: 10.1038/icb.2017.59. [DOI] [PubMed] [Google Scholar]
- Meekins, Kanost & Michel (2017).Meekins DA, Kanost MR, Michel K. Serpins in arthropod biology. Seminars in Cell & Developmental Biology. 2017;62:105–119. doi: 10.1016/j.semcdb.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mkaouar et al. (2016).Mkaouar H, Akermi N, Mariaule V, Boudebbouze S, Gaci N, Szukala F, Pons N, Marquez J, Gargouri A, Maguin E, Rhimi M. Siropins, novel serine protease inhibitors from gut microbiota acting on human proteases involved in inflammatory bowel diseases. Microbial Cell Factories. 2016;15 doi: 10.1186/S12934-016-0596-2. Article 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira et al. (2014).Moreira CJ, Waniek PJ, Valente RH, Carvalho PC, Perales J, Feder D, Geraldo RB, Castro HC, Azambuja P, Ratcliffe NA, Mello CB. Isolation and molecular characterization of a major hemolymph serpin from the triatomine, Panstrongylus megistus. Parasites & Vectors. 2014;7:23. doi: 10.1186/1756-3305-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathaniel et al. (2004).Nathaniel R, MacNeill AL, Wang YX, Turner PC, Moyer RW. Cowpox virus CrmA, Myxoma virus SERP2 and baculovirus P35 are not functionally interchangeable caspase inhibitors in poxvirus infections. Journal of General Virology. 2004;85:1267–1278. doi: 10.1099/vir.0.79905-0. [DOI] [PubMed] [Google Scholar]
- O’Dwyer et al. (2015).O’Dwyer CA, O’Brien ME, Wormald MR, White MM, Banville N, Hurley K, McCarthy C, McElvaney NG, Reeves EP. The BLT1 inhibitory function of alpha-1 antitrypsin augmentation therapy disrupts leukotriene B4 neutrophil signaling. Journal of Immunology. 2015;195:3628–3641. doi: 10.4049/jimmunol.1500038. [DOI] [PubMed] [Google Scholar]
- Pakchotanon et al. (2016).Pakchotanon P, Molee P, Nuamtanong S, Limpanont Y, Chusongsang P, Limsomboon J, Chusongsang Y, Maneewatchararangsri S, Chaisri U, Adisakwattana P. Molecular characterization of serine protease inhibitor isoform 3, SmSPI, from Schistosoma mansoni. Parasitology Research. 2016;115:2981–2994. doi: 10.1007/s00436-016-5053-y. [DOI] [PubMed] [Google Scholar]
- Patschull et al. (2011).Patschull AO, Segu L, Nyon MP, Lomas DA, Nobeli I, Barrett TE, Gooptu B. Therapeutic target-site variability in alpha1-antitrypsin characterized at high resolution. Acta Crystallographica Section F: Structural Biology Communications. 2011;67:1492–1497. doi: 10.1107/S1744309111040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce et al. (2008).Pearce MC, Morton CJ, Feil SC, Hansen G, Adams JJ, Parker MW, Bottomley SP. Preventing serpin aggregation: the molecular mechanism of citrate action upon antitrypsin unfolding. Protein Science. 2008;17:2127–2133. doi: 10.1110/ps.037234.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton et al. (1988).Pemberton PA, Stein PE, Pepys MB, Potter JM, Carrell RW. Hormone binding globulins undergo serpin conformational change in inflammation. Nature. 1988;336:257–258. doi: 10.1038/336257a0. [DOI] [PubMed] [Google Scholar]
- Pereira et al. (2017).Pereira RS, Rodrigues VAA, Furtado WT, Gueiros S, Pereira GS, Avila-Campos MJ. Microbial analysis of root canal and periradicular lesion associated to teeth with endodontic failure. Anaerobe. 2017;48:12–18. doi: 10.1016/j.anaerobe.2017.06.016. [DOI] [PubMed] [Google Scholar]
- Quezada & McKerrow (2011).Quezada LA, McKerrow JH. Schistosome serine protease inhibitors: parasite defense or homeostasis? Anais da Academia Brasileira de Ciencias. 2011;83:663–672. doi: 10.1590/S0001-37652011000200025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray et al. (1992).Ray CA, Black RA, Kronheim SR, Greenstreet TA, Sleath PR, Salvesen GS, Pickup DJ. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-Y. [DOI] [PubMed] [Google Scholar]
- Rehault-Godbert et al. (2013).Rehault-Godbert S, Labas V, Helloin E, Herve-Grepinet V, Slugocki C, Berges M, Bourin MC, Brionne A, Poirier JC, Gautron J, Coste F, Nys Y. Ovalbumin-related protein X is a heparin-binding ov-serpin exhibiting antimicrobial activities. Journal of Biological Chemistry. 2013;288:17285–17295. doi: 10.1074/jbc.M113.469759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renatus et al. (2000).Renatus M, Zhou Q, Stennicke HR, Snipas SJ, Turk D, Bankston LA, Liddington RC, Salvesen GS. Crystal structure of the apoptotic suppressor CrmA in its cleaved form. Structure. 2000;8:789–797. doi: 10.1016/S0969-2126(00)00165-9. [DOI] [PubMed] [Google Scholar]
- Roberts et al. (2011).Roberts TH, Ahn JW, Lampl N, Fluhr R. Plants and the study of serpin biology. Methods in Enzymology. 2011;499:347–366. doi: 10.1016/B978-0-12-386471-0.00017-1. [DOI] [PubMed] [Google Scholar]
- Roberts et al. (2003).Roberts TH, Marttila S, Rasmussen SK, Hejgaard J. Differential gene expression for suicide-substrate serine proteinase inhibitors (serpins) in vegetative and grain tissues of barley. Journal of Experimental Botany. 2003;54:2251–2263. doi: 10.1093/jxb/erg248. [DOI] [PubMed] [Google Scholar]
- Robertson et al. (2003).Robertson AS, Belorgey D, Lilley KS, Lomas DA, Gubb D, Dafforn TR. Characterization of the necrotic protein that regulates the Toll-mediated immune response in Drosophila. Journal of Biological Chemistry. 2003;278:6175–6180. doi: 10.1074/jbc.M209277200. [DOI] [PubMed] [Google Scholar]
- Schuster et al. (1992).Schuster MG, Enriquez PM, Curran P, Cooperman BS, Rubin H. Regulation of neutrophil superoxide by antichymotrypsin-chymotrypsin complexes. Journal of Biological Chemistry. 1992;267:5056–5059. [PubMed] [Google Scholar]
- Silverman et al. (2001).Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O’Donnell E, Salvesen GS, Travis J, Whisstock JC. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. Journal of Biological Chemistry. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- Stein & Chothia (1991).Stein P, Chothia C. Serpin tertiary structure transformation. Journal of Molecular Biology. 1991;221:615–621. doi: 10.1016/0022-2836(91)80076-7. [DOI] [PubMed] [Google Scholar]
- Sun et al. (1996).Sun J, Bird CH, Sutton V, McDonald L, Coughlin PB, De Jong TA, Trapani JA, Bird PI. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. Journal of Biological Chemistry. 1996;271:27802–27809. doi: 10.1074/jbc.271.44.27802. [DOI] [PubMed] [Google Scholar]
- Suwanchaichinda et al. (2013).Suwanchaichinda C, Ochieng R, Zhuang S, Kanost MR. Manduca sexta serpin-7, a putative regulator of hemolymph prophenoloxidase activation. Insect Biochemistry and Molecular Biology. 2013;43:555–561. doi: 10.1016/j.ibmb.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirloni et al. (2016).Tirloni L, Kim TK, Coutinho ML, Ali A, Seixas A, Termignoni C, Mulenga A, Da Silva Vaz Jr I. The putative role of Rhipicephalus microplus salivary serpins in the tick-host relationship. Insect Biochemistry and Molecular Biology. 2016;71:12–28. doi: 10.1016/j.ibmb.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirloni et al. (2014).Tirloni L, Reck J, Terra RM, Martins JR, Mulenga A, Sherman NE, Fox JW, Yates 3rd JR, Termignoni C, Pinto AF, Vaz Ida Jr S. Proteomic analysis of cattle tick Rhipicephalus (Boophilus) microplus saliva: a comparison between partially and fully engorged females. PLOS ONE. 2014;9:e94831. doi: 10.1371/journal.pone.0094831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner et al. (1999).Turner PC, Sancho MC, Thoennes SR, Caputo A, Bleackley RC, Moyer RW. Myxoma virus Serp2 is a weak inhibitor of granzyme B and interleukin-1beta-converting enzyme in vitro and unlike CrmA cannot block apoptosis in cowpox virus-infected cells. Journal of Virology. 1999;73:6394–6404. doi: 10.1128/jvi.73.8.6394-6404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Water et al. (2004).Van de Water N, Tan T, Ashton F, O’Grady A, Day T, Browett P, Ockelford P, Harper P. Mutations within the protein Z-dependent protease inhibitor gene are associated with venous thromboembolic disease: a new form of thrombophilia. British Journal of Haematology. 2004;127:190–194. doi: 10.1111/j.1365-2141.2004.05189.x. [DOI] [PubMed] [Google Scholar]
- Yoo et al. (2000).Yoo BC, Aoki K, Xiang Y, Campbell LR, Hull RJ, Xoconostle-Cazares B, Monzer J, Lee JY, Ullman DE, Lucas WJ. Characterization of cucurbita maxima phloem serpin-1 (CmPS-1). A developmentally regulated elastase inhibitor. Journal of Biological Chemistry. 2000;275:35122–35128. doi: 10.1074/jbc.M006060200. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2016).Zhang Z, Mao Y, Li D, Zhang Y, Li W, Jia H, Zheng J, Li L, Lu Y. High-level expression and characterization of two serine protease inhibitors from Trichinella spiralis. Veterinary Parasitology. 2016;219:34–39. doi: 10.1016/j.vetpar.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Zhao et al. (2014).Zhao YR, Xu YH, Jiang HS, Xu S, Zhao XF, Wang JX. Antibacterial activity of serine protease inhibitor 1 from kuruma shrimp Marsupenaeus japonicus. Developmental and Comparative Immunology. 2014;44:261–269. doi: 10.1016/j.dci.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Zheng et al. (2012).Zheng D, Chen H, Bartee MY, Williams J, Davids JA, Huang E, Moreb J, Lucas A. Virus-derived anti-inflammatory proteins: potential therapeutics for cancer. Trends in Molecular Medicine. 2012;18:304–310. doi: 10.1016/j.molmed.2012.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following information was supplied regarding data availability:
The research in this review article did not generate any data or code.