Abstract
Cancer cells use glucose and glutamine to facilitate cell growth and proliferation, a process coined “metabolic reprograming” – an emerging hallmark of cancer. Inside the cell, these nutrients synergize to produce metabolic building blocks, such as nucleic acids, lipids and proteins, as well as energy (ATP), glutathione and reducing equivalents (NADPH), required for survival, growth and proliferation. Intense research aimed at understanding the underlying cause of the metabolic rewiring has revealed that established oncogenes and tumor suppressors involved in signaling alter cellular metabolism to contribute to the transition from a normal quiescent cell to a rapidly proliferating cancer cell. Likewise, bona fide metabolic sensors are emerging as regulators of tumorigenesis. This review will focus on one such family of sensors, sirtuins, which utilize NAD+ as a cofactor to catalyze deacetylation, deacylation and ADP-ribosylation of their protein substrates. In this review, we will enumerate how cancer cell metabolism is different from a normal quiescent cell and highlight the emerging role of mitochondrial sirtuin signaling in the regulation of tumor metabolism.
Keywords: Antioxidants, glutamine metabolism, metabolic reprograming, signaling pathways, Warburg effect
Introduction
A major goal in cancer biology is to identify molecular mechanisms that contribute to cancer growth and survival in order to provide insight into new biomarkers for precision medicine, as well as novel targeted therapies. Toward this end, recent studies have elucidated a new promising area of cancer cell biology – metabolic rewiring in tumor cells. Tumor cells rewire the metabolism of glucose, amino acids and fats in order to provide growth and survival advantages (Figure 1). Additionally, it is evident that oncogenic signaling can control the rewiring of fuels. This review will focus on the role of the sirtuin family of proteins in tumor metabolism. We will emphasize the role of sirtuins that have been shown to regulate tumor metabolism by targeting metabolic enzymes, as well as signaling cascades. This review is not meant to provide a comprehensive overview of sirtuins or cancer biology. For a more comprehensive overview of sirtuin biology or tumor metabolism, see Garrett & Grisham (2012); Cantor & Sabatini (2012); Vander Heiden et al. (2012); Ward & Thompson (2012); DeBerardinis et al. (2008); Roth & Chen (2014); Houtkooper et al. (2012); and Sebastian et al. (2012a).
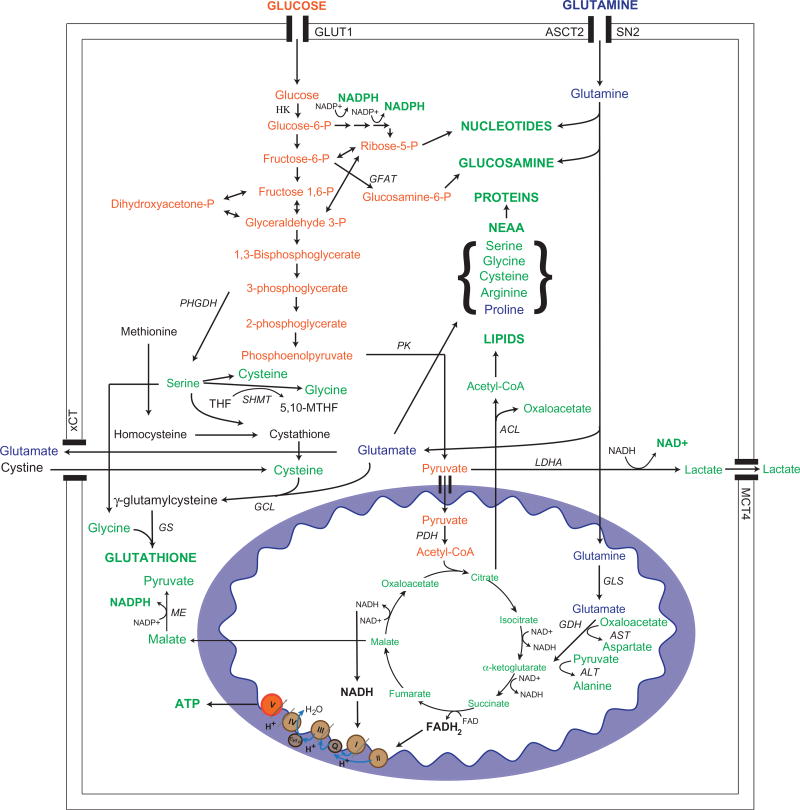
Figure 1.
Alterations in glucose and glutamine metabolism contribute to tumor growth and proliferation. Rewiring of glucose and glutamine metabolism contribute to synthesis of macromolecules, antioxidants and reducing equivalents. Major enzymes involved in each pathway are highlighted above, and expression or activity of these enzymes tends to be altered in tumor cells to contribute to metabolic reprograming, as described in the text. NEAA, non-essential amino acids; HK, hexokinase; GFAT, glutamine fructose-6-phosphate amidotransferase; PHGDH, phosphoglycerate dehydrogenase; SHMT, serine hydroxymethyltransferase; PK, pyruvate kinase; PDH, pyruvate dehydrogenase; GCL, glutamate-cysteine ligase; GS, glutathione synthase; ME, malic enzyme; ACL, ATP citrate lyase; LDHA, lactate dehydrogenase; GLS, glutaminase; GDH, glutamate dehydrogenase; AST, aspartate aminotransferase; ALT, alanine aminotransferase; GLUT1, glucose transporter type 1; ASCT2, sodium-dependent neutral amino acid transporter type 2; SN2, system N transporter 2; MCT4, monocarboxylate transporter type 4; xCT, cystine/glutamate transporter. (see colour version of this figure at www.informahealthcare.com/bmg).
Sirtuins
The sirtuin family of proteins has been implicated in numerous biological processes, as well as longevity and diseases of aging, such as diabetes and cancer (Haigis & Guarente, 2006; Houtkooper et al., 2012; Roth & Chen, 2014; Sebastian et al., 2012a). Mammals express seven sirtuins (SIRT1-7), first identified by their homology to the yeast Sir2 protein (Frye, 1999). Several protein targets through which this family of proteins mediates their function have been identified. The first two enzymatic activities identified for this group of proteins were ADP-ribosylation and deacetylation; however, more recently other enzymatic activities, such as demalonylation, desuccinylation and decrotonylation have been identified (Bao et al., 2014; Park et al., 2013). Sirtuins depend on NAD+ as a cofactor for enzymatic activity. As these proteins catalyze their reactions, they convert NAD+ to nicotinamide and O-acetyl-ADP-ribose, of which nicotinamide functions as a competitive inhibitor of sirtuins (Haigis & Guarente, 2006; Houtkooper et al., 2012; Roth & Chen, 2014; Sebastian et al., 2012a). Levels of NAD+ vary depending on the metabolic state of the cell. Under conditions, such as caloric restriction, fasting or exercise, NAD+ levels increase resulting in sirtuin activation and post-translational modification of their target proteins. Thus, sirtuins sense the metabolic status of the cell allowing them to adjust to stress conditions, such as a low energetic state (Easlon et al., 2008; Kincaid & Bossy-Wetzel, 2013).
Sirtuins localize to various cellular compartments, such as the nucleus (SIRT1, SIRT6, SIRT7), cytosol (SIRT2) and mitochondria (SIRT3, SIRT4, SIRT5), where several targets and the effect of their function have been identified. This localization endows this family of proteins the potential to coordinate cancer metabolism in various ways, from direct regulation of metabolic enzymes in the mitochondria or cytosol to transcriptional regulation of metabolic gene expression in the nucleus. Indeed, recent studies reveal sirtuins to play critical roles in tumor metabolism. Of the seven sirtuins, the best studied sirtuins in tumor metabolism are two mitochondrial sirtuins (SIRT3 and SIRT4), as well as a nuclear sirtuin (SIRT6). This review first highlights metabolic reprograming in cancer and then discusses the roles of sirtuins in regulating this process with emphasis placed on mitochondrial sirtuins.
Cancer cell metabolism and metabolic reprograming
Glucose and glutamine metabolism provide precursors for the production of energy, synthesis of macromolecules and generation of reducing equivalents for survival, growth and proliferation (Figure 1). Recent studies have identified a new facet of oncogenic signaling that controls the rewiring of fuels, providing growth and survival advantages to tumors. How oncogenic signaling contributes to metabolic rewiring in cancer is the topic of the following sections. This section does not cover every aspect of tumor metabolism, but will focus primarily on signaling pathways and metabolism shown to be regulated by sirtuins. For a more comprehensive overview of tumor metabolism, see Garrett & Grisham (2012); Cantor & Sabatini (2012); Vander Heiden et al. (2012); Ward & Thompson (2012); and DeBerardinis et al. (2008).
Glucose metabolism in a quiescent/resting cell
A textbook lesson in glucose metabolism is that glucose oxidation is important for energy production. Indeed, metabolism of glucose through glycolysis and the tricarboxylic acid cycle (TCA cycle), coupled with the electron transport chain (ETC), is necessary for generation of energy, in the form of adenosine triphosphate (ATP). The complete oxidation of glucose yields ~34–36 molecules of ATP, which provides the currency used by the cell to catalyze several reactions, especially biosynthetic reactions (Garrett & Grisham, 2012; Vander Heiden et al., 2009; Lunt & Vander Heiden, 2011).
Glucose oxidation occurs in multiple steps, offering many opportunities to the cell to regulate the metabolism of this fuel. Glucose is internalized by cells via glucose transporters, after which it is phosphorylated by hexokinase or glucokinase to trap it within the cell (Garrett & Grisham, 2012; Warburg et al., 1927). Glucose is then oxidized to pyruvate in nine steps via glycolysis, a process that consumes two ATP molecules to generate four ATP molecules. Cytosolic pyruvate undergoes several changes, and it can be converted to alanine, lactate or enter the mitochondria to generate acetyl CoA (Lunt & Vander Heiden, 2011; Vander Heiden et al., 2012; Warburg et al., 1927; Weinhouse, 1976). In a normal, quiescent cell and in the presence of oxygen, pyruvate is transported into the mitochondria via pyruvate carriers where it is converted into acetyl CoA by pyruvate dehydrogenase (PDH), resulting in the production of one molecule of NADH. Acetyl CoA enters the TCA cycle as it is combined with oxaloacetate (OAA) to generate citrate, which is oxidatively decarboxylated to produce the electron donors, NADH and FADH2. Under oxygen-rich conditions, electrons from both NADH and FADH2 pass through complexes I to III in the ETC and combine with oxygen, the final electron acceptor in complex IV to produce water (Cantor & Sabatini, 2012; Garrett & Grisham, 2012; Locasale et al., 2011; Vander Heiden et al., 2012). In this process, protons are pumped from the mitochondrial membrane to the intermembrane space generating a proton gradient across the inner mitochondrial membrane. The proton gradient is used by the F0F1 ATP synthase, also known as complex V, to generate ATP from ADP and inorganic phosphate, a process known as oxidative phosphorylation (Cantor & Sabatini, 2012; DeBerardinis et al., 2008; Garrett & Grisham, 2012; Vander Heiden et al., 2012; Ward & Thompson, 2012). Approximately two and three molecules of ATP are generated for every FADH2 or NADH molecule, respectively. In total, glycolysis generates a net of two ATP molecules, while oxidative phosphorylation generates an additional 32–34 ATP molecules – an efficient process for a resting cell to generate energy to support necessary cellular processes and maintain homeostasis. During low oxygen availability, anaerobic glycolysis is the main source of energy production since pyruvate is shifted away from the mitochondria and is converted by lactate dehydrogenase (LDHA) to lactate, which is secreted from cells (Garrett & Grisham, 2012; Lunt & Vander Heiden, 2011; Vander Heiden et al., 2009). This repartitioning of pyruvate metabolism provides a major way for the cell to adapt to stress and maintain homeostasis.
Cancer cells and the Warburg effect
In the 1920s, Otto Warburg made the first observation that cancer cells display aberrant metabolism compared to normal cells. Warburg noted that cancer cells utilized glucose and secreted lactate at higher rates than a normal cell in the presence of oxygen – a phenomenon termed the Warburg effect (Garrett & Grisham, 2012; Warburg et al., 1927). He hypothesized that this process was a result of a rapidly dividing tumor cell’s requirement for energy in the presence of defective mitochondria; however, this conclusion has proven to be untrue for many tumors (Lunt & Vander Heiden, 2011; Vander Heiden et al., 2012; Warburg et al., 1927; Weinhouse, 1976). Why would a cell increase glycolysis, if only two ATP molecules are generated in this process? Recent studies have revealed that glucose oxidation is rewired by tumors to generate macromolecules to fuel increased biomass, which is necessary for rapid proliferation (Figure 1) (Garrett & Grisham, 2012; Locasale et al., 2011; Vander Heiden et al., 2012). For example, 3-phosphoglycerate (3PG) is diverted from glycolysis and is utilized for synthesis of non-essential amino acids. In a series of three steps, 3PG is converted to serine, which is further converted to glycine and cysteine (Greenberg & Ichihara, 1957; Lunt & Vander Heiden, 2011; Possemato et al., 2011). The first enzyme involved in redirecting glucose to serine is phosphoglycerate dehydrogenase (PHGDH), and expression of this enzyme is high in a number of cancers, including breast and melanoma cancers (Locasale et al., 2011; Possemato et al., 2011).
Glucose also contributes to biosynthesis via the pentose phosphate pathway (PPP), which provides reducing equivalents for anabolic reactions and 5-carbon sugars for nucleotide synthesis (Figure 1) (Andrés et al., 1980; Boros et al., 2000; DeBerardinis et al., 2007). The PPP is composed of two phases – the oxidative and non-oxidative phase (DeBerardinis, 2008; Jones & Thompson, 2009). Glucose-6-phosphate (G6P) dehydrogenase (G6PD) utilizes G6P in the first step of the PPP to divert it from glycolysis and support production of NADPH from NADP+ in the oxidative phase of the PPP. G6PD expression is elevated in tumors and expression of this protein has also been shown to increase NADPH levels (Polat et al., 2002; Rao et al., 1997; Van Driel et al., 1999). In the non-oxidative phase of the PPP, fructose-6-phosphate and glyceraldehyde-3-phosphate (G3P) contribute to generation of ribose-5-phosphate, which is used for synthesis of nucleotides. In addition, G3P is converted to dihydroxyacetone phosphate, which is utilized for triglyceride synthesis (Vander Heiden et al., 2012). Another major regulatory node for glycolysis is the last step of glycolysis catalyzed by pyruvate kinase (PK). Cancer cells and proliferating cells express the PKM2 isoform, which has low enzymatic activity in its dimeric form, and thus, contributes to decreased entry of pyruvate to the mitochondria to allow diversion of glycolytic metabolites to other pathways to support biomass production (Christofk et al., 2008). In sum, tumors adjust glucose metabolism through coordinated rewiring of multiple nodes to maximize production of metabolites used for nucleotide, amino acid and fatty acid synthesis.
Glutamine metabolism
For many years, cancer metabolism research focused on understanding the Warburg effect and the importance of glucose metabolism to a cancer cell. However, more recently, the importance of glutamine in cancer was unveiled, as reprograming of glutamine metabolism has been found in various types of cancers (Dang, 2012a; Daye & Wellen, 2012; DeBerardinis & Cheng, 2009; Lu et al., 2010; Ward & Thompson, 2012; Wise et al., 2008). Glutamine is the most abundant amino acid in human plasma, and is usually categorized as a non-essential amino acid for many resting cells (Lu et al., 2010). However, glutamine becomes an essential amino acid in a rapidly dividing cell as it complements glucose usage by providing its nitrogen and carbon atoms for synthesis of macromolecules, such as nucleic acids, proteins and lipids to support cellular growth and proliferation, as well as for regulation of redox homeostasis (Figure 1) (Dang, 2012a; Daye & Wellen, 2012; DeBerardinis & Cheng, 2009; Lu et al., 2010; Ward & Thompson, 2012; Wise et al., 2008).
The metabolic reprograming of glutamine can be observed by increased glutamine uptake and glutaminolysis in some cancer cells. Cells take up glutamine via high affinity glutamine transporters, such as ASCT2 or SN2, which are overexpressed in various types of tumors (Cairns et al., 2011; Wise et al., 2008). Once, inside the cell, glutamine has many fates (Figure 1) (Dang, 2012a; DeBerardinis & Cheng, 2009). Glutaminolysis takes place in the mitochondria, where glutamine is converted to glutamate by glutaminase (GLS), and glutamate is deaminated by glutamate dehydrogenase (GDH) to produce α-ketoglutarate (Kovacević, 1971; Lukey et al., 2013; Matsuno, 1989). Ammonia is produced in both of these steps and high levels of ammonia are observed in cancer cells that utilize glutamine (Matés et al., 2009). However, the fate of this ammonia is still not clear. Glutamine-derived α-ketoglutarate is then oxidized to produce TCA cycle intermediates, such as malate and citrate, which are important for NADPH and lipid synthesis, respectively (Le et al., 2012; Sabine et al., 1973). Malate can be exported from the mitochondria and is converted to pyruvate, by malic enzyme, resulting in the generation of NADPH, which is utilized in anabolic reactions as well as to maintain redox balance (Dang, 2012a; DeBerardinis et al., 2007; Ward & Thompson, 2012). In the TCA cycle, malate is converted to OAA, which is condensed with acetyl-coA to form citrate. Citrate can be exported from the mitochondria. In the cytosol, citrate is converted to OAA and acetyl CoA by ATP citrate lyase (Wellen & Thompson, 2012). The acetyl CoA derived from citrate is used for fatty acid and cholesterol synthesis, and may be used for epigenetic and post-translational modification. Additional rewiring of glutamine metabolism has been observed in cancer cells under hypoxic conditions (Metallo et al., 2012; Wise et al., 2011). Under oxygen-limiting conditions, glutamine-derived alpha-ketoglutarate is reductively carboxylated to contribute to the citrate pool. Thus, mitochondrial glutamine metabolism is essential for production of ATP, NADPH, and lipids necessary for proliferation.
In addition to contributing to lipid synthesis, glutamine provides the nitrogen for nucleotide, hexosamine, and amino acid synthesis (Figure 1) (DeBerardinis & Thompson, 2012; Tong et al., 2009; Wellen et al., 2010). Glutamine donates its amide group in three steps of purine synthesis and two steps of pyrimidine synthesis to support nucleotide production for DNA synthesis, which is essential to generate the genetic material of a new cell. Hexosamine synthesis also relies on nitrogen from glutamine in the conversion of fructose-6-phosphate to glucosamine-6-phosphate by glutamine fructose-6-phosphate amidotransferase (GFAT) (Marshall et al., 1991). Synthesis of hexosamines, such as uridine diphosphate N-acetylglucosamine, plays a crucial role in cellular growth and proliferation, as glycosylation of cell surface receptors is necessary for nutrient uptake (Wellen et al., 2010). Amino acids are important for synthesis of proteins to carry out reactions necessary for cellular division, and glutamine contributes to synthesis of non-essential amino acids. For example, alanine transaminase (ALT) or aspartate transaminase (AST) transfer nitrogen from glutamine-derived glutamate to pyruvate or oxaloacetate to produce alanine and aspartate, respectively (Matés et al., 2009). In addition, glutamine contributes to synthesis of other amino acids, such as serine, cysteine, glycine, threonine, proline and arginine (Matés et al., 2009).
Glutamine regulates redox homeostasis by contributing to production of glutathione and NADPH (Cairns et al., 2011; Dang, 2012b; Lora et al., 2004). Glutathione is the most abundant antioxidant, which is synthesized in two steps and is composed of three amino acids: glutamate, cysteine and glycine – all of which can be generated from glutamine (Lu, 2009, 2013; Ortega et al., 2011). Glutamine is the main contributor to the pool of glutamate, which is incorporated into glutathione, but is also important for import of the cysteine, the rate-limiting amino acid, for glutathione synthesis (Lo et al., 2008; Sato et al., 1999; Wu et al., 2004). Glutathione protects cells from oxidative stress by reducing ROS and in the process glutathione becomes oxidized (Lu, 2009). Regeneration of glutathione is catalyzed by glutathione reductase, a reaction requiring NADPH (Bause & Haigis, 2013). As already mentioned glutaminolysis generates NADPH, as malate is exported from the mitochondria and converted to pyruvate. Thus, glutamine plays a crucial role in protecting cells from oxidative stress.
Transcription factors mediate metabolic reprograming in cancer
Transcription factors have the ability to rewire metabolism by affecting expression of various genes, including metabolic genes (Figure 2). Although various transcription factors may be capable of altering gene expression to alter tumor metabolism, it is well established that the transcription factors: hypoxia inducible factor (HIF) 1 and c-Myc play key roles in metabolic reprograming of cancer cells (Dang, 2013; Denko, 2008; Jones & Schulze, 2012; Jones & Thompson, 2009; Levine & Puzio-Kuter, 2010; Semenza, 2010).
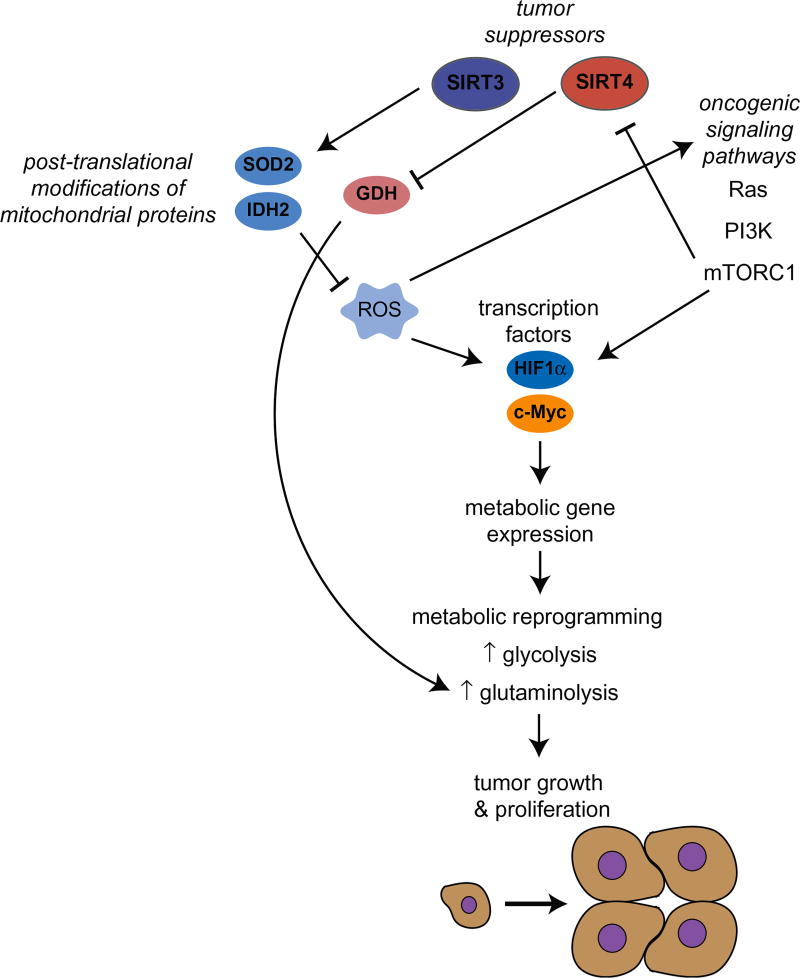
Figure 2.
Oncogenic signaling pathways and tumor suppressors mediate metabolic reprograming in cancer to increase tumor growth and proliferation. Signaling cascades, such as Ras, PI3K and mTORC1, regulate metabolism and overexpression or mutations in components of these pathways contribute to altered metabolism in cancer. Additionally, signaling cascades stabilize transcription factors, such as HIF1 and c-Myc, which control metabolic gene expression to increase glycolysis or glutaminolysis. Mitochondrial sirtuins, SIRT3 and SIRT4, repress aberrant metabolism in cancer. SIRT3 represses reactive oxygen species (ROS) by deacetylating direct targets, such as manganese superoxide dismutase (SOD2) and isocitrate dehydrogenase (IDH2), to destabilize HIF1 and repress the Warburg effect. SIRT4 ADP-ribosylates glutamate dehydrogenase (GDH), and thus, represses glutaminolysis. (see colour version of this figure at www.informahealthcare.com/bmg).
HIF1 supports the Warburg effect
The transcription factor HIF1 is composed of two subunits: HIF1α and HIF1β (also known as ARNT) (Schulze & Harris, 2012; Semenza, 2000). The HIF1α subunit is stabilized under low oxygen conditions (hypoxia). In the presence of oxygen, HIF1α is hydroxylated by prolyl hydroxylases (PHD), a family of dioxygenases, activated by α-ketoglutarate and inhibited by succinate (Kaelin & Thompson, 2010; Selak et al., 2005). Hydroxylation results in recruitment of the von Hippel-Lindau (VHL) tumor suppressor protein, an E3-ubiquitin ligase, which ubiquitinates HIF1α and targets it for destruction by the proteasome (Kaelin & Thompson, 2010; Marxsen et al., 2004). Increased HIF1 protein levels are found in human tumors, such as pancreatic, breast, colon and lung cancer (Zhong et al., 1999). Loss of VHL is observed in cancers, leading to HIF1 stability (Kaelin, 2002). In addition, under normoxic conditions, mutations in other tumor suppressors, such as succinate dehydrogenase or fumarate hydratase also stabilize HIF1 potentially via increased α-ketoglutarate levels (Baysal et al., 2004; Tomlinson et al., 2002).
HIF1 regulates cellular metabolism by inducing the expression of several glycolytic genes (Semenza, 2010). HIF1 contributes to increased glucose uptake by increasing the expression of the glucose transporter GLUT1. In addition, HIF1 induces glycolysis by stimulating expression of hexokinase (HK2), and an isoform of phosphofructokinase (PFK1), one of the three key regulatory enzymes in glycolysis (Semenza, 2010). HIF1 also upregulates expression of pyruvate dehydrogenase kinase (PDK1), which inhibits PDH, and thus, HIF1 represses oxidative phosphorylation and promotes reliance on glycolysis (Kim et al., 2006; Papandreou et al., 2006; Semenza, 2010). HIF1 supports regeneration of NAD+ (required to maintain glycolysis) by increasing LDHA expression. Lastly, HIF1 stimulates expression of the monocarboxylate transporter MCT4 to allow excretion of the high levels of lactate generated by high rates of glycolysis (Denko, 2008; Jones & Schulze, 2012).
c-Myc promotes glycolysis and glutaminolysis
Increased levels of the c-Myc (Myc) transcription factor are observed in many tumors, such as Burkitt’s lymphoma, breast cancer and neuroblastoma (Dang, 2013; Lin et al., 2012; Liu et al., 2008). Myc regulates a wide variety of biological processes, such as mitochondrial biogenesis, protein synthesis and metabolism (Levine & Puzio-Kuter, 2010; Miller et al., 2012). Recent studies show that Myc coordinates this wide range of cellular processes by amplifying transcription of already active genes in the cell (Lin et al., 2012; Nie et al., 2012). This transcription factor activates glucose and glutamine metabolism by directly binding to the promoters and increasing the expression of glycolytic and glutaminolytic genes. Like HIF1, Myc has been shown to upregulate transcription of GLUT1, HK2 and LDHA to increase glycolysis (Dang, 2013; Miller et al., 2012; Son et al., 2013; Ying et al., 2012). Additionally, Myc increases expression of PKM2 to enhance glycolysis (Sun et al., 2011).
Myc is a major driver for glutamine metabolism, and many cancer cells expressing high levels of Myc rely on glutamine for survival (Wise et al., 2008; Yuneva et al., 2007). Myc upregulates expression of the glutamine transporter ASCT2 contributing to an increased glutamine uptake in cancers. Moreover, Myc stimulates glutaminolysis by increasing GLS1 expression in a mechanism that involves inhibition of a miRNA that targets a sequence in the GLS 3′UTR, miR23a/b (Gao et al., 2009). Lastly, Myc targets other metabolic enzymes, such as serine hydroxymethyltransferase (SHMT), which converts serine to glycine and is important for nucleotide synthesis (Miller et al., 2012). Thus, Myc may also play a key role in regulating nucleic acid synthesis for a rapidly proliferating cell (Liu et al., 2008).
Metabolic reprograming can be attributed to oncogenic signaling
For many years, cancer research focused on identifying oncogenes and tumor suppressors. In recent years, metabolism research experienced a renaissance as studies revealed that oncogenes and tumor suppressors mediate metabolic reprograming in cancer cells (Jones & Thompson, 2009; Levine & Puzio-Kuter, 2010; Liang et al., 2013). Oncogenic signaling by Ras, PI3K and mTORC, discussed below, contributes to an altered tumor metabolism, partly by regulating HIF1 or c-Myc (Figure 2) (Csibi et al., 2013; DeBerardinis et al., 2008; Düvel et al., 2010; Son et al., 2013; Ying et al., 2012). Additionally, tumor suppressors, such as p53 and Nrf2, not discussed in this review, also rewire the metabolic landscape of a cancer cell (Liang et al., 2013; Mitsuishi et al., 2012). Interestingly, all of these signaling pathways have been linked to sirtuins, which function to rewire metabolism in response to nutritional status.
Signaling pathways regulate nutrient uptake
Many of the metabolic alterations in cancer cells mirror metabolism in rapidly proliferating cells (such as immune cells; see Pearce & Pearce, 2013; Wang et al., 2011). However, rapidly proliferating cells adjust metabolism in response to growth signals, whereas cancer cells have evolved mechanisms that allow them to evade checkpoints to inhibit proliferation. Aberrant activation of cellular signaling is a hallmark of cancers, resulting in deregulated growth and proliferation, as well as directly leading to metabolic reprograming (Hanahan & Weinberg, 2011). Here, we focus on signaling pathways important to cancer regulated by mitochondrial sirtuins (Figure 2).
Ras signaling
The Ras family of proteins controls differentiation, growth and proliferation. Ras is a small GTPase, active in its GTP-bound form and inactive when bound to GDP (Downward, 2003). Hyperactivation of this pathway, due to oncogenic mutations in the Ras protein, is common in cancer. The most common mutation is a point mutation in the Ras gene, resulting in a constitutively active Ras protein. Ras activates Raf, which phosphorylates and activates mitogen-activated protein kinases, kinases 1 and 2 (MEK1 and MEK2). MEK1 and MEK2 phosphorylate ERK1 and ERK2, which activate PI3K and c-Myc to alter cancer metabolism (Downward, 2003; Sears et al., 2000).
Ras contributes to metabolic reprograming in cancer. Cells transformed with oncogenic KRAS depend on glucose and glutamine for survival and proliferation (Gaglio et al., 2011; Son et al., 2013; Ying et al., 2012). Initially, transformation of cells with oncogenic KRAS resulted in an increased glucose uptake. Upon further analysis tracing the fate of glucose in the cell, oncogenic KRAS was demonstrated to increase glycolysis, but decrease glucose utilization in the TCA cycle (Gaglio et al., 2011). Moreover, glutamine-tracing experiments showed an increased anaplerosis from glutamine with KRAS transformation, highlighting how these fuels complement each other to promote cellular growth and proliferation (Gaglio et al., 2011; Son et al., 2013). Mechanistically, microarray data comparing non-transformed and KRAS transformed cells showed an increased expression of genes in glycolysis, nucleotide synthesis and glutamine metabolism further supporting these findings (Gaglio et al., 2011). A mouse model of oncogenic KRAS-driven pancreatic ductal adenocarcinoma supported the role of KRAS in metabolic reprograming (Ying et al., 2012). In this mouse model, inactivation of KRAS resulted in tumor regression, which was accompanied by a decrease in glucose uptake and lactate secretion, as well as a decrease in glycolytic intermediates known to be precursors for various biosynthetic pathways. Indeed, other anabolic pathways, such as the hexosamine biosynthesis and the non-oxidative arm of the PPP, are also activated by oncogenic KRAS to promote protein glycosylation and ribose production for DNA and RNA synthesis. Knockdown of enzymes in the hexosamine and PPP pathway resulted in a decreased xenograft tumor growth, supporting the regulation of these pathways by KRAS to promote tumorigenesis. Lastly, metabolic gene expression decreases with KRAS inactivation, which is also the case with MAPK inhibition and Myc knockdown, suggesting that regulation of these pathways by KRAS contributes to metabolic reprograming in cancer (Ying et al., 2012).
Regulation of metabolism by PI3K/AKT
In a normal cell, growth factors stimulate receptor tyrosine kinases to activate the phosphatidylinositol 3-kinase (PI3K) signaling cascade (Courtney et al., 2010). PI3K is recruited to the phosphorylated receptors and is phosphorylated and activated. In addition, Ras can directly activate PI3K (Downward, 2003). Activation of PI3K results in its localization to the plasma membrane where it converts phosphatidylinositol-4,5-biphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3). Phosphoinositide-dependent kinase 1 (PDK1) and protein kinase B (also known as AKT) are recruited to the plasma membrane by PIP3, which results in phosphorylation and activation of AKT (Cairns et al., 2011; Hers et al., 2011). As a result, AKT promotes various processes important to a cancer cell, such as survival, growth, proliferation, angiogenesis, metastasis and metabolism (Bellacosa et al., 2005; Martini et al., 2014). Deregulation of the PI3K/AKT signaling pathway is common across various cancers. Activation of this pathway in cancer is often due to an activating mutation in the subunits of the PI3K complex, or inactivation/loss of PTEN, a tumor suppressor (Courtney et al., 2010). Hyperactivation of this pathway increases glucose metabolism in cancer. PI3K regulates glucose metabolism via its effector AKT. In addition to supporting the localization of the glucose transporter GLUT1 to the plasma membrane, AKT increases the expression and activates HK2 (Vander Heiden et al., 2009). Moreover, AKT phosphorylates PFK2 resulting in production of fructose-2,6-bisphosphate, which allosterically activates PFK1 to increase glycolysis (Dang, 2012a). AKT also stimulates de novo fatty acid synthesis by activating SREBP (Chang, 2005). Lastly, AKT promotes c-Myc stabilization by repressing GSK-3, which phosphorylates c-Myc thereby targeting it for degradation (Sears et al., 2000).
The mTORC1 signaling pathway regulates metabolism
The mammalian target of rapamycin (mTOR) signaling pathway regulates cellular growth, proliferation and metabolism (Yecies & Manning, 2011a). Like Ras and PI3K/AKT, mTOR signaling is deregulated in cancer (Guertin & Sabatini, 2005; Menon & Manning, 2008). mTOR is a highly conserved serine/threonine kinase found in two complexes in the cell, mTOR complex 1 and mTOR complex 2 (mTORC1 and mTORC2), and of the two complexes, more is known about mTORC1 (Laplante & Sabatini, 2009). mTORC1 is regulated by signals from growth factors, energy status, amino acids and oxygen. Most of these signals are integrated by the tuberous sclerosis complex (TSC), composed of TSC1 and TSC2, which functions as a GTPase activating protein (GAP) for the GTPase Rheb. In turn, Rheb activates mTORC1 in its GTP-bound state. mTORC is activated in the presence of growth factors and under nutrient-rich conditions, and in the presence of amino acids. As already mentioned, growth factors activate the PI3K and Ras signaling cascades, and signals from these pathways are sensed by the TSC complex to regulate mTORC1 activity (Laplante & Sabatini, 2009). Activated AKT phosphorylates and inhibits TSC2 to activate mTORC1. Likewise, ERK1/2 and RSK1, activated by the Ras signaling cascade, activate mTORC1 in the same manner (Laplante & Sabatini, 2009). In addition, amino acids, such as leucine and glutamine, are required for mTORC1 activation (Nicklin et al., 2009). Import of leucine relies on export of glutamine via the Slc7a5 (or LAT1) transporter, and inhibition of the glutamine transporter (ASCT2) or LAT1 suppresses mTORC1 signaling. Lastly, under nutrient-rich conditions, ATP is produced at a level necessary to support cell growth and proliferation. However, under nutrient-poor conditions, or under hypoxia, energy levels decline resulting in an increase in the AMP:ATP ratio. AMP activated kinase (AMPK), the energy status sensor of the cell, is activated when energy levels decrease, and AMPK negatively regulates mTORC1 by phosphorylating and activating TSC2 (Dang, 2012b; Laplante & Sabatini, 2009).
mTORC stimulation supports anabolic metabolism and represses catabolic processes to promote growth and proliferation (Düvel et al., 2010; Yecies & Manning, 2011b). mTORC1 regulates transcription, ribosome biogenesis, translation and autophagy by phosphorylating and thus activating or repressing downstream targets. Two well-known downstream effectors of mTORC1 are p70 ribosomal S6 kinase (S6K) and eukaryotic initiation factor 4E binding protein 1 (4E-BP1), and phosphorylation of these targets promotes protein synthesis by activating mRNA and ribosome synthesis, as well as translation elongation. Moreover, mTORC1 phosphorylates and inactivates ULK1 and ATG13 to inhibit autophagy (Jung et al., 2009). In addition, mTORC1 has been shown to regulate several metabolic pathways. Through S6K, mTORC1 stabilizes HIF1α and sterol regulatory element-binding protein (SREBP1 and SREBP2) to increase their metabolic target gene expression and promote glucose uptake, glycolysis, de novo lipid synthesis, and the oxidative arm of the PPP. Recently, mTORC1 was shown to promote de novo pyrimidine synthesis by activating the enzyme that catalyzes the first three steps of pyrimidine synthesis, CAD (carbamoylphosphate synthetase 2, aspartate transcarbamylase and dihydroorotase), via S6K (Ben-Sahra et al., 2013).
Regulation of tumor metabolism by mitochondrial sirtuins
Three of the seven sirtuins, SIRT3, SIRT4 and SIRT5, reside in the mitochondria where they regulate metabolism via posttranslational modifications on various types of proteins, including metabolic enzymes (Haigis & Guarente, 2006). Mitochondria are recognized as the powerhouse of the cell. In addition to producing energy in the form of ATP, this organelle is a metabolic hub for several metabolic pathways, such as fatty acid oxidation, the TCA cycle, and glutaminolysis. As discussed above, these metabolic pathways play an important role in tumors by generating (1) reducing equivalents utilized in the ETC for energy production, as well as in other anabolic reactions; and (2) metabolic intermediates that can contribute to synthesis of macromolecules.
Mitochondria are also the main source of ROS, which is generated in the ETC as electrons can leak at complexes I or III during oxidative phosphorylation to produce superoxide (Bause & Haigis, 2013; McBride et al., 2006; Ray et al., 2012). ROS has been implicated in cancer, but the connection between ROS and cancer is complex as ROS can function as a double-edged sword (Pelicano et al., 2004; Schumacker, 2006). At very high levels, ROS can halt tumor proliferation and result in cancer cell death (Pelicano et al., 2004; Trachootham et al., 2009). However, moderate elevations in ROS levels can activate signaling pathways to promote cellular adaptation. For example, increased ROS inhibits PHD activity to promote HIF1α stability and induce glycolytic gene expression to promote the Warburg effect in tumors (Ray et al., 2012; Thannickal & Fanburg, 2000). Altogether, the mitochondria seem indispensable to many cancer cells as the metabolism taking place in this important cellular compartment is necessary for the synthesis of macromolecules to support tumor growth.
Recent studies have begun to elucidate the role of mitochondrial sirtuins in the regulation of cancer cell metabolism, providing insight into new biomarkers for tumor metabolism, as well as new therapeutic targets. SIRT3 and SIRT4 directly regulate metabolic enzymes or affect oncogenic signaling known to orchestrate metabolic reprogramming (Finley et al., 2011a; Jeong et al., 2013). SIRT5 biology remains the least studied and has not been reported to regulate cancer cell metabolism, although this possibility cannot be completely ruled out.
SIRT3 represses the Warburg effect
SIRT3 activates a program in the mitochondria to promote energy production, oxidative metabolism, and redox homeostasis (Finley & Haigis, 2012). SIRT3 has a multi-faceted control of metabolism. First, SIRT3 mediates this metabolic control by deacetylating a number of mitochondrial proteins to boost their activity. Secondly, SIRT3 can affect cellular signaling pathways to control metabolism. The direct role of SIRT3 on cellular metabolism is clear from a wide-range of studies. Several SIRT3 substrates have been identified and validated, and these enzymes are involved in mitochondrial metabolic pathways, such as the electron transport chain, fatty acid oxidation, amino acid metabolism and in the maintenance of cellular redox homeostasis (Hebert et al., 2013). For example, SIRT3 activates the electron transport chain by deacetylating NDUFA9 and succinate dehydrogenase, components of electron transport complexes I and II, respectively, which supports the observation that tissues with low SIRT3 levels also have low ATP levels (Ahn et al., 2008; Finley et al., 2011b). Additionally, SIRT3 deacetylates long-chain acyl coenzyme A dehydrogenase (LCAD) to promote fatty acid oxidation in the liver (Bharathi et al., 2013; Hirschey et al., 2010). SIRT3 is also involved in the regulation of amino acid catabolism by activating GDH and promotes ammonia detoxification by deacetylating ornithine transcarbamoylase (OTC) and inducing the urea cycle (Hallows et al., 2011; Lombard et al., 2007; Schlicker et al., 2008). Moreover, SIRT3 promotes reactive oxygen species (ROS) detoxification by activating manganese superoxide dismutase 2 (SOD2) and isocitrate dehydrogenase 2 (IDH2) (Chen et al., 2011; Jacobs et al., 2008; Qiu et al., 2010; Someya et al., 2010; Tao et al., 2010). SOD2 scavenges superoxide and converts it to hydrogen peroxide, a substrate for catalase, which generates water and oxygen from hydrogen peroxide (Chen et al., 2011). IDH2, a TCA cycle enzyme, decarboxylates isocitrate to produce alpha-ketoglutarate, under normal conditions (Someya et al., 2010). This reaction generates reducing equivalents as it uses NADP+ to generate NADPH, which is essential for anabolic reactions and is used by glutathione reductase for regeneration of a major antioxidant, glutathione (Someya et al., 2010). In line with these findings, tissues from knockout (KO) mice display an increased acetylation of mitochondrial proteins (Lombard et al., 2007). A recent study describes a new role of SIRT3 as a decrotonylase (Bao et al., 2014), but it remains to be seen how this other sirtuin activity impacts tumor metabolism. Taken together, numerous studies suggest that SIRT3 coordinates a program to protect a cell from oxidative stress and cellular damage.
The importance of this pathway in human cancers is beginning to crystallize. First, decreased SIRT3 levels are observed across a wide range of human tumors, and particularly in breast cancer where SIRT3 gene deletions are also observed (Finley et al., 2011a; Kim et al., 2010). Compared to normal tissue, breast cancers have decreased SIRT3 protein expression (Finley et al., 2011a). Lastly, SIRT3 is undetectable in metastatic samples, suggesting that SIRT3 may play a key role in repressing metastasis (Kim et al., 2010). It is important to note that many tumors also display increased SIRT3 expression, which may indicate a pro-survival role for SIRT3 in these cancers (Alhazzazi et al., 2011). It will be critical for future studies to elucidate the importance of SIRT3 deletion versus amplification in human cancers. Moreover, in tumors lacking SIRT3, it will also be critical to determine whether SIRT3 loss plays an active role in initiating tumorigenesis or promoting tumor growth.
Insights may arise from studies of cellular and mouse models. Indeed, using allograft studies and SIRT3 KO mouse models, our lab and others have demonstrated that SIRT3 possesses tumor suppressive activity, in part by rewiring metabolism (Bell et al., 2011; Finley et al., 2011a; Kim et al., 2010). First, SIRT3 null MEFs compared to WT MEFS (when both are transformed by E1A/Ras) display features of cancer cells, such as rapid proliferation, aneuploidy, anchorage-independent colony formation and allograft tumor formation in nude mice (Finley et al., 2011a; Kim et al., 2010). Moreover, SIRT3 null MEFs have higher superoxide levels under stress conditions – consistent with the role of SIRT3 in deacetylating and activating SOD2 (Finley et al., 2011a; Qiu et al., 2010). Second, SIRT3 null mice spontaneously develop tumors, particularly in breast, at 12 months. Mammary tumors from SIRT3 null mice have increased protein damage, suggesting that these mice have increased levels of ROS (Kim et al., 2010). Additionally, SIRT3 overexpression can inhibit the in vitro formation of breast and pancreatic cell lines in mice (Finley et al., 2011a; Jeong et al., 2014b). Together, these studies indicate that SIRT3 may inhibit tumorigenesis of certain cancers.
How does SIRT3 regulate tumor growth? One mechanism is that SIRT3 deacetylates a specific target with strong tumor suppressive activity. One such target has not been identified. Based on published data, thus far, it is more likely that SIRT3 deacetylates an array of targets that either changes the metabolic status or redox of the cell. For example, SIRT3 plays a pivotal role in diminishing oxidative stress, in part, by regulating SOD2 (Qiu et al., 2010; Tao et al., 2010) and IDH2 (Yu et al., 2012), but it may also control ROS production by regulating the efficiency of the ETC (Bause & Haigis, 2013). This particular arm of SIRT3 activity may have important roles in cancer, as many cancer cells have increased ROS levels in comparison with non-tumorigenic cells (Schumacker, 2006). Paradoxically, chronic elevation of ROS in tumors may trigger important signaling cascades that promote cellular proliferation and survival (Figure 2) (Schieber & Chandel, 2014). Indeed, numerous studies have observed that SIRT3 suppresses ROS (Finley et al., 2011a; Kim et al., 2010; Qiu et al., 2010; Tao et al., 2010; Yu et al., 2012). It was proposed that elevated ROS in SIRT3 null cells and mice contributes to an increased genomic instability, leading to an increased tumorigenesis. We and others have shown that SIRT3 regulates signaling pathways via ROS to repress metabolic reprograming in cancer (Figure 2). It will be interesting for future studies to elucidate the roles of SIRT3 substrates in control of cellular signaling.
How does SIRT3 regulate signaling? Our lab and the Guarente Lab independently discovered that increased ROS in SIRT3 null cells results in HIF1α stabilization. HIF1α stability is regulated by prolyl hydroxylases (PHDs), which hydroxylate HIF1α under normoxia to allow the von Hippel-Lindau protein (vHL) to ubiquitinate HIF1α, leading to proteosomal degradation (Kaelin & Thompson, 2010). SIRT3 null MEFs demonstrated decreased HIF1α hydroxylation (Finley et al., 2011a), and overexpression of SIRT3 was able to repress in numerous cell types. Importantly, although PHDs function as oxygen sensors, the regulation of PHD activity by SIRT3 occurred during normoxia.
As discussed above, elevated HIF activity has important consequences in the metabolism of human cancers and this fact is supported by studies of SIRT3. First, SIRT3 null MEFs display elevated glycolytic metabolism, which can be inhibited with anti-oxidants or by knockdown of HIF1α (Figure 2) (Bell et al., 2011; Finley et al., 2011a). Steady-state metabolomics analysis shows that the absence of SIRT3 alters glucose metabolism by increasing several intermediates in the glycolytic pathway and the pentose phosphate pathway (PPP), suggesting rewiring of glucose metabolism and the potential for glucose to contribute to other pathways, such as the PPP, to support synthesis of macromolecules for cellular proliferation (Finley et al., 2011a). Flux analysis of glucose metabolism yielded similar results (Hebert et al., 2013). Cells and tissues from SIRT3 KO mice demonstrate a signature of elevated HIF1α gene expression. Finally, expression of SIRT3 decreases HIF1α levels, represses the Warburg effect, and decreases cell growth in breast cancer cells (Finley et al., 2011a). While these studies highlight HIF1α as a signaling pathway regulated by SIRT3, many other signaling pathways are controlled by mitochondria or ROS. Thus, it is important to examine: (1) whether SIRT3 affects other mitochondrial-rooted signaling pathways, and (2) if this control of other signaling pathways contribute to the tumor suppressive activity of SIRT3.
Potential modulation of tumor metabolism via regulation of cellular signaling by SIRT3
In normal, primary tissues, SIRT3 has been implicated in the regulation of cellular signaling in heart, skeletal muscle, brown adipose tissue and liver (Finley et al., 2011b; Jing et al., 2011; Shi et al., 2010; Sundaresan et al., 2009). SIRT3 plays a key role in response to cardiac hypertrophy as its expression increases in hearts of mice treated with hypertrophy agonists in part by activating the transcription factor Foxo3a to induce expression of antioxidant genes (Pillai et al., 2010; Sundaresan et al., 2009). Induction of signaling cascades, such as the PI3K/Akt and MAPK/ERK pathways, also may contribute to the pathogenesis of cardiac hypertrophy, as these pathways were repressed in transgenic mice overexpressing SIRT3 (Pillai et al., 2014). In addition, SIRT3 repressed activation of Ras, which is upstream of PI3K and MAPK/ERK signaling. Finally, the mTORC signaling pathway was activated in hearts of SIRT3 null mice and this was abolished by overexpression of SIRT3 (Sundaresan et al., 2009). The mTORC1 signaling pathway is negatively regulated by AMP kinase (AMPK), and its regulator LKB1 (Laplante & Sabatini, 2009). In concordance with increased mTORC signaling, SIRT3 null mouse hearts have decreased phosphorylation and activity of AMPK and LKB1 (Sundaresan et al., 2009). In addition to regulating signaling in heart, SIRT3 plays a key role in insulin signaling in skeletal muscle, which is important as SIRT3 KO mice have impaired insulin signaling, which is an early feature of type 2 diabetes (Jing et al., 2011). However, cell signaling differs between skeletal muscle and heart. Skeletal muscle from SIRT3 null mice displays decreased insulin signaling evidenced by decreased phosphorylation of tyrosine residues in the insulin receptor (IR). In addition, SIRT3 loss decreases phosphorylation of IR targets, such as PI3K and AKT, which is accompanied by decreased glucose uptake (Jing et al., 2011). Likewise, decreased Erk phosphorylation is evident in skeletal muscle from SIRT3 null mice stimulated with insulin. Like hearts from SIRT3 null mice, skeletal muscle from these mice displays increased oxidative stress and increased expression of genes involved in oxidative stress response. Various kinases, such as the Jun N-terminal kinase (JNK), protein kinase C and S6 kinase, are activated by oxidative stress and phosphorylate the IR and IRS-1 to decrease PI3K activation (Benhar et al., 2002; Jing et al., 2011). Upon closer examination, skeletal muscle from SIRT3 null mice have increased phosphorylation of JNK, which is in agreement with observations in mouse embryonic fibroblasts from these mice. The study proposes increased ROS in SIRT3 null mouse tissue as a mechanism for activation of JNK and deregulation of insulin signaling (Jing et al., 2011). As discussed above, these signaling pathways have importance in tumor metabolism. Thus, it will be interesting for future studies to probe whether SIRT3 controls PI3K or mTOR signaling in human cancers.
Although the absence of SIRT3 results in altered signal transduction, the direction in which SIRT3 regulates these pathways is not clear as signaling varies based on tissue or cell type. SIRT3 may also contribute to metabolic reprograming by regulating signaling pathways known to affect metabolism; however, the role of SIRT3 in the regulation of cellular signaling in cancer has not been examined. Further studies could focus on elucidating the mechanism by which SIRT3 regulates cellular signaling, and whether this is the case in cancer.
SIRT4 regulates glutamine metabolism
SIRT4 is a lesser studied mitochondrial sirtuin that connects metabolism to cell growth and survival. Unlike SIRT3, SIRT4 does not possess a robust, promiscuous deacetylase activity. Moreover, activation of SIRT4 seems to repress carbon flow into the TCA cycle via numerous nodes. SIRT4 was initially identified as an inhibitor of glutamate dehydrogenase (GDH) via ADP-ribosylation (Haigis et al., 2006). By inhibiting GDH, SIRT4 suppresses the flow of amino acids into the TCA cycle and inhibited insulin secretion from the pancreatic β cells. Additionally, SIRT4 negatively regulates insulin secretion via insulin degrading enzyme and adenine nucleotide translocases (Ahuja et al., 2007). SIRT4 also reduces fat oxidation while promoting lipogenesis by deacetylating and inhibiting malonyl-CoA decarboxylase (MCD), which converts malonyl-CoA into acetyl-CoA (Laurent et al., 2013). SIRT4 also represses PDH to block entry of pyruvate into central mitochondrial metabolism (Mathias et al., 2014). Given these observations, it is likely that SIRT4 is uniquely poised to regulate whole-body metabolic homeostasis, which can mediate anabolic signaling to promote growth, both in normal cells as well as in cancer cells.
A clue to the physiological relevance of SIRT4 came from studies of its regulation. Unlike other mitochondrial sirtuins, SIRT4 mRNA and protein levels were highly stabilized by genotoxic stress, suggesting that sirtuin may integrate metabolic adaptation with DNA damage responses. SIRT4 expression increased upon DNA damage by camptothecin (topoisomerase I inhibitor) and ultraviolet (UV) light, and this increase occurs concomitantly with a decrease in glutamine uptake by the cells (Jeong et al., 2013; Rardin et al., 2013). These observations suggest that DNA damage leads to decreased glutaminolysis and glutamine anaplerosis, which are supported by the fact that TCA cycle intermediates are decreased after exposure of cells to UV light. Additional metabolic flux analysis of 13C-labeled glutamine demonstrates a clearly decreased contribution of glutamine to TCA cycle intermediates after DNA damage. Moreover, SIRT4 is required to suppress glutamine flux into the TCA cycle after DNA damage, which suggests that SIRT4 plays a critical role in cellular response to DNA damage (Jeong et al., 2013; Lu et al., 2014; Ouaïssi et al., 2008). This hypothesis demonstrates the presence of a connection between mitochondrial metabolism and nuclear/cytosolic stress responses, which remains to be understood.
Consistent with the idea that SIRT4 triggers a metabolic checkpoint, mTOR inhibition induces SIRT4 expression. In this case, mTORC1 repression stabilizes cAMP responsive element binding 2 (CREB2), which is responsible for upregulating SIRT4 transcription (Csibi et al., 2013) (Figure 2). Thus, inhibition of mTORC1 by rapamycin increases the expression of SIRT4, which subsequently decreases GDH activity and glutamine anaplerosis (Figure 2) (Csibi et al., 2013). Importantly, the increase in SIRT4 is required for repression of GDH activity upon mTORC1 inhibition.
The above studies demonstrate that SIRT4 functions as a sensor of cellular stress to trigger a metabolic pause by repressing glutamine metabolism. This ability of SIRT4 to reprogram glutamine metabolism suggests that SIRT4 could also have tumor suppressive activity. Indeed, loss of SIRT4 increases cell proliferation, an effect that disappears with inhibition of glutaminolysis (Jeong et al., 2013). In addition, transformed SIRT4 null MEFs form larger tumors in mice, and SIRT4−/− mice have increased incidence of spontaneous cancers, especially lung cancer. Of relevance to human cancers, analysis of microarray data revealed that many human cancers, including lung, gastric, bladder and breast cancers – as well as leukemias – have decreased SIRT4 expression (Csibi et al., 2013; Jeong et al., 2013, 2014a). In addition, lower SIRT4 levels in lung cancer leads to decreased survival, highlighting the importance of SIRT4 in cancer progression and mortality. It is not yet clear whether SIRT4 represses tumors via control of the DNA damage response, mTOR, or glutamine metabolism. It is unlikely that all of the tumor suppression by SIRT4 is mediated via glutamine inhibition, as SIRT4 has many other metabolic targets. Thus, it will be important for future studies to parse out the mechanisms involved.
Potential role of SIRT5 in regulating tumor metabolism
SIRT5 is a mitochondrial sirtuin with a unique enzymatic activity that does not center around deacetylation–desuccinylation. In addition to desuccinylation, SIRT5 has been shown to possess demalonylation and deglutarylation activity (Lin et al., 2013; Park et al., 2013; Rardin et al., 2013; Tan et al., 2014). It is important to note, that although proteomic studies have been performed identifying succinylated proteins (Hornbeck et al., 2015; Park et al., 2013; Peng et al., 2011; Rardin et al., 2013; Weinert et al., 2013), we still have little understanding of the scope and physiological relevance of these modifications in vivo. An important clue will come from systematic studies comparing levels of acetylation with other modifications. Nearly 80% of TCA cycle and 60% of fatty acid metabolism enzymes are succinylated when SIRT5 is deleted, but future studies will need to assess whether this is due directly to changes in SIRT5 activity or altered succinate metabolism.
SIRT5 substrates range from detoxifying enzymes to mitochondrial metabolic enzymes. The first well-described substrate for this sirtuin was carbamoyl phosphate synthase-1 (CPS1). In the liver, SIRT5 activates CPS1 via deacetylation to promote the urea cycle and increase the clearance of ammonia produced from catabolism of amino acids (Nakagawa et al., 2009; Park et al., 2013). Other well defined substrates include succinate dehydrogenase, pyruvate dehydrogenase and SOD1, and the effect of SIRT5 on their activity is described below (Lin et al., 2013; Park et al., 2013).
As with SIRT3 and SIRT4, control of metabolic enzymes and detoxifying enzymes by SIRT5 may alter cancer metabolism. In contrast to SIRT3 and SIRT4, which show tumor suppressive activity in animal studies, SIRT5 has been implicated as a promoter of tumorigenesis. Elevated SIRT5 mRNA levels have been reported in pancreatic cancer and non-small cell lung cancer (Lu et al., 2014; Matsushita et al., 2010; Ouaïssi et al., 2008; Pfister et al., 2008). SIRT5 inhibition decreases lung cancer cell proliferation in vitro and in vivo (Lu et al., 2014). SIRT5 also contributes to drug resistance by increasing expression of antioxidant response genes, and silencing of SIRT5 results in decreased xenograft tumor size and enhanced susceptibility to various anticancer drugs (Lu et al., 2014). Additionally, elevated SIRT5 levels correlate with increased incidence of metastasis, advanced tumor stage, and overall, poor prognosis. How SIRT5 regulates tumor metabolism has not been examined; however, given the clear role of mitochondrial metabolism in cancer, it is possible that SIRT5 rewires metabolism to have a pro-survival role. For example, SIRT5 inhibits succinate dehydrogenase and pyruvate dehydrogenase, which also suggests that SIRT5 may potentially play a key role in cancer by stabilizing tumorigenic pathways dependent on (proto)oncometabolites (Park et al., 2013). Finally, SIRT5 desuccinylates and activates Cu/Zn superoxide dismutase, thereby promoting ROS scavenging, which may repress ROS signaling and tumorigenesis in a manner similar to SIRT3 (Lin et al., 2013). It is important to note that SIRT5 is present both inside the mitochondria as well as in the cytosol and the nucleus, suggesting the potential for a much broader effect on cancer metabolism than other mitochondrial sirtuins (Matsushita et al., 2010; Pfister et al., 2008).
Potential regulation of cancer metabolism by other sirtuins
In addition to mitochondrial sirtuins, mammalian cells contain four other sirtuins (SIRT1, SIRT2, SIRT6 and SIRT7), which have been well studied regarding cellular signaling and in some cases tumorigenesis. For example, SIRT1 deacetylates HIF1α thereby repressing HIF1 activity (Lim et al., 2010), and SIRT1 also deacetylates Myc to destabilize it (Yuan et al., 2009). Additionally, SIRT1 activation promotes mitochondrial biogenesis (Gerhart-Hines et al., 2007; Lagouge et al., 2006; Rodgers et al., 2005). Like SIRT1, SIRT7 has been shown to bind to and inhibit Myc (Shin et al., 2013). Although the roles of SIRT1 or SIRT7 have not been directly assessed in tumor metabolism, it is tempting to speculate that SIRT1 and SIRT7 may inhibit some tumors, in part via metabolic reprograming. Additionally, SIRT2 deacetylates and activates phosphoglycerate mutase, a glycolytic protein whose inhibition decreases proliferation and tumor growth (Xu et al., 2014). In addition, SIRT2 deacetylates K-Ras and decreases its transformation activity (Yang et al., 2013). In conclusion, SIRT1, SIRT2 and SIRT7 have the potential to regulate metabolic reprograming by modulating key players known to regulate this process.
SIRT6 has been described to coordinate metabolic reprograming in cancer by regulating HIF1 and Myc. More specifically, SIRT6 regulates glucose metabolism via deacetylation of H3K9 at the promoters of glycolytic genes, as well as by destabilizing HIF1α (Zhong et al., 1999). Additionally, SIRT6 co-represses Myc and inhibits expression of ribosomal genes (Sebastian et al., 2012b). Thus, SIRT6 seems to function as a tumor suppressor and its loss results in a cellular metabolic phenotype similar to that described by Warburg. It will be interesting for future studies to assess whether SIRT6 also promotes mitochondrial biogenesis or metabolism through transcriptional control.
Conclusions
Despite intense biomedical research, cancer continues to be one of the leading causes of death in the United States. A recent burst in research focusing on tumor metabolism has shed light on the importance of metabolic reprograming in cancer cells. The connections between oncogenes, tumor supressors, signaling pathways and metabolic reprograming are emerging, and this knowledge may provide insights into identifying new targets for the development of cancer treatment. As highlighted in this review, several key players contributing to altered tumor metabolism are regulated by sirtuins, suggesting this family of proteins may pose a new venue to modulate aberrant signaling and metabolic reprograming in cancer. For example, as SIRT3 plays an important role in maintaining redox homeostasis, it will be interesting for future studies to identify whether SIRT3 activation rewires metabolism of tumors exhibiting a proliferative advantage as a result of increased oxidative stress. Alternatively, inhibition of SIRT3 may be useful in cancers with increased oxidative stress to increased ROS levels high enough to cause tumor cell death. Further investigation is necessary to determine the therapeutic benefit of sirtuin modulation in cancer.
Acknowledgments
Declaration of interest
Karina N. Gonzalez was supported by the Paul & Daisy Soros Fellowship for New Americans. Jaewon Lee was supported by HHMI. Marcia C. Haigis, PhD, was supported by an American Cancer Society Research Scholar Grant, the National Institutes of Health (NIH) Grant AG032375, and the Glenn Foundation for Medical Research.
References
- Ahn B-H, Kim H-S, Song S, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA. 2008;105:14447–52. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja N, Schwer B, Carobbio S, et al. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–92. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- Alhazzazi TY, Kamarajan P, Joo N, et al. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117:1670–8. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés A, Satrústegui J, Machado A. Development of NADPH-producing pathways in rat heart. Biochem J. 1980;186:799–803. doi: 10.1042/bj1860799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Wang Y, Li X, et al. Identification of “erasers’” for lysine crotonylated histone marks using a chemical proteomics approach. Elife. 2014;3:e02999. doi: 10.7554/eLife.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bause AS, Haigis MC. SIRT3 regulation of mitochondrial oxidative stress. Exp Gerontol. 2013;48:634–9. doi: 10.1016/j.exger.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Baysal BE, Willett-Brozick JE, Filho PAA, et al. An Alumediated partial SDHC deletion causes familial and sporadic paraganglioma. J Med Genet. 2004;41:703–9. doi: 10.1136/jmg.2004.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult, et al. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–96. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellacosa A, Kumar CC, Di Cristofano A, et al. Activation of AKT kinases in cancer: implications for therapeutic targeting. Adv Cancer Res. 2005;94:29–86. doi: 10.1016/S0065-230X(05)94002-5. [DOI] [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–8. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, Engelberg D, Levitzki A. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep. 2002;3:420–5. doi: 10.1093/embo-reports/kvf094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathi SS, Zhan Y, Mohsen A-W, et al. Sirtuin 3 (SIRT3) protein regulates long-chain acyl-CoA dehydrogenase by deacetylating conserved lysines near the active site. J Biol Chem. 2013;288:33837–47. doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros LG, Torday JS, Lim S, et al. Transforming growth factor beta2 promotes glucose carbon incorporation into nucleic acid ribose through the nonoxidative pentose cycle in lung epithelial carcinoma cells. Cancer Res. 2000;60:1183–5. [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–98. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. KGF induces lipogenic genes through a PI3K and JNK/SREBP-1 pathway in H292 cells. J Lipid Res. 2005;46:2624–35. doi: 10.1194/jlr.M500154-JLR200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang J, Lin Y, et al. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. 2011;12:534–41. doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–83. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibi A, Fendt S-M, Li C, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–54. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. Cancer cell metabolism: there is no ROS for the weary. Cancer Discov. 2012a;2:304–7. doi: 10.1158/2159-8290.CD-12-0069. [DOI] [PubMed] [Google Scholar]
- Dang CV. Links between metabolism and cancer. Genes Dev. 2012b;26:877–90. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013;3:a014217. doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daye D, Wellen KE. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Sem Cell Dev Biol. 2012;23:362–9. doi: 10.1016/j.semcdb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ. Is cancer a disease of abnormal cellular metabolism? New angles on an old idea. Genet Med. 2008;10:767–77. doi: 10.1097/GIM.0b013e31818b0d9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2009;29:313–24. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–44. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–50. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Denko NC. Hypoxia, HIF1α and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–13. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- Düvel K, Yecies JL, Menon S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–83. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easlon E, Tsang F, Skinner C, et al. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 2008;22:931–44. doi: 10.1101/gad.1648308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LWS, Haigis MC. Metabolic regulation by SIRT3: implications for tumorigenesis. Trends Mol Med. 2012;18:516–23. doi: 10.1016/j.molmed.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LWS, Carracedo A, Lee J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011a;19:416–28. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LWS, Haas W, Desquiret-Dumas V, et al. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS One. 2011b;6:e23295. doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–9. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- Gaglio D, Metallo CM, Gameiro PA, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang T-C, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett R, Grisham C. Biochemistry. Belmont (CA): Cengage Learning; 2012. [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–23. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DM, Ichihara A. Further studies on the pathway of serine formation from carbohydrate. J Biol Chem. 1957;224:331–40. [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. An expanding role for mTOR in cancer. Trends Mol Med. 2005;11:353–61. doi: 10.1016/j.molmed.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins – emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–21. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell. 2006;126:941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Yu W, Smith BC, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–49. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–99. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hers I, Vincent EE, Tavaré JM. Akt signalling in health and disease. Cell Signal. 2011;23:1515–27. doi: 10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–5. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–20. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–38. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs KM, Pennington JD, Bisht KS, et al. SIRT3 interacts with the daf-16 homolog FOXO3a in the mitochondria, as well as increases FOXO3a dependent gene expression. Int J Biol Sci. 2008;4:291–9. doi: 10.7150/ijbs.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SM, Lee A, Lee J, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23:450–63. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SM, Lee A, Lee J, Haigis MC. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J Biol Chem. 2014a;289:4135–44. doi: 10.1074/jbc.M113.525949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SM, Lee J, Finley LW, et al. SIRT3 regulates cellular iron metabolism and cancer growth by repressing iron regulatory protein 1. Oncogene [Epub ahead of print] 2014b doi: 10.1038/onc.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing E, Emanuelli B, Hirschey MD, et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci USA. 2011;108:14608–1. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NP, Schulze A. Targeting cancer metabolism – aiming at a tumour’s sweet-spot. Drug Discov Today. 2012;17:232–41. doi: 10.1016/j.drudis.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–48. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro S-H, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG. Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2:673–82. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Thompson CB. Q&A: cancer: clues from cell metabolism. Nature. 2010;465:562–4. doi: 10.1038/465562a. [DOI] [PubMed] [Google Scholar]
- Kim H-S, Patel K, Muldoon-Jacobs K, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Kincaid B, Bossy-Wetzel E. Forever young: SIRT3 a shield against mitochondrial meltdown, aging, and neurodegeneration. Front Aging Neurosci. 2013;5:48. doi: 10.3389/fnagi.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacević Z. The pathway of glutamine and glutamate oxidation in isolated mitochondria from mammalian cells. Biochem J. 1971;125:757–63. doi: 10.1042/bj1250757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–94. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent G, German NJ, Saha AK, et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell. 2013;50:686–98. doi: 10.1016/j.molcel.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–21. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–4. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- Liang Y, Liu J, Feng Z. The regulation of cellular metabolism by tumor suppressor p53. Cell Biosci. 2013;3:9. doi: 10.1186/2045-3701-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Lee Y-M, Chun Y-S, et al. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1α. Mol Cell. 2010;38:864–78. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z-F, Xu H-B, Wang JY, et al. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem Biophys Res Commun. 2013;441:191–5. doi: 10.1016/j.bbrc.2013.10.033. [DOI] [PubMed] [Google Scholar]
- Liu Y-C, Li F, Handler J, et al. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS One. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M, Wang YZ, Gout PW. The xc–cystine/glutamate antiporter: a potential target for therapy of cancer and other diseases. J Cell Physiol. 2008;215:593–602. doi: 10.1002/jcp.21366. [DOI] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–74. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, et al. Mammalian Sir2 Homolog SIRT3 Regulates Global Mitochondrial Lysine Acetylation. Mol Cell Biol. 2007;27:8807–14. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lora J, Alonso FJ, Segura JA, et al. Antisense glutaminase inhibition decreases glutathione antioxidant capacity and increases apoptosis in Ehrlich ascitic tumour cells. Eur J Biochem/FEBS. 2004;271:4298–306. doi: 10.1111/j.1432-1033.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- Lu SC. Regulation of glutathione synthesis. Mol Aspects Med. 2009;30:42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC. Glutathione synthesis. Biochim Biophys Acta. 2013;1830:3143–53. doi: 10.1016/j.bbagen.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Zuo Y, Feng Y, Zhang M. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumor Biol. 2014;35:10699–705. doi: 10.1007/s13277-014-2372-4. [DOI] [PubMed] [Google Scholar]
- Lu W, Pelicano H, Huang P. Cancer metabolism: is glutamine sweeter than glucose? Cancer Cell. 2010;18:199–200. doi: 10.1016/j.ccr.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukey MJ, Wilson KF, Cerione RA. Therapeutic strategies impacting cancer cell glutamine metabolism. Future Med Chem. 2013;5:1685–700. doi: 10.4155/fmc.13.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–64. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–12. [PubMed] [Google Scholar]
- Martini M, De Santis MC, Braccini M, et al. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med. 2014;46:372–83. doi: 10.3109/07853890.2014.912836. [DOI] [PubMed] [Google Scholar]
- Marxsen JH, Stengel P, Doege K, et al. Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-alpha-prolyl-4-hydroxylases. Biochem J. 2004;381:761–7. doi: 10.1042/BJ20040620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matés JM, Segura JA, Campos-Sandoval JA, et al. Glutamine homeostasis and mitochondrial dynamics. Int J Biochem Cell Biol. 2009;41:2051–61. doi: 10.1016/j.biocel.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Mathias RA, Greco TM, Oberstein A, et al. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159:1615–25. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno T. End products of glutamine oxidation in MC-29 virus-induced chicken hepatoma mitochondria. Biochem Med Metab Biol. 1989;42:125–31. doi: 10.1016/0885-4505(89)90048-0. [DOI] [PubMed] [Google Scholar]
- Matsushita N, Yonashiro R, Ogata Y, et al. Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes Cells. 2010;16:190–202. doi: 10.1111/j.1365-2443.2010.01475.x. [DOI] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–60. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2008;27:S43–51. doi: 10.1038/onc.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Thomas SD, Islam A, et al. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546–53. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, et al. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–70. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega AL, Mena S, Estrela JM. Glutathione in cancer cell death. Cancer. 2011;3:1285–310. doi: 10.3390/cancers3011285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouaïssi M, Sieleznef I, Silvester R, et al. High histone deacetylase 7 (HDAC7) expression is significantly associated with adenocarcinomas of the pancreas. Ann Surg Oncol. 2008;15:2318–28. doi: 10.1245/s10434-008-9940-z. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, et al. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–97. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Park J, Chen Y, Tishkoff DX, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–30. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–43. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicano H, Carney D, Huang P. ROS stress in cancer cells and therapeutic implications. Drug Resist Updates. 2004;7:97–110. doi: 10.1016/j.drup.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.012658. M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister JA, Ma C, Morrison BE, et al. Opposing effects of sirtuins on neuronal survival: SIRT1-mediated neuroprotection is independent of its deacetylase activity. PLoS One. 2008;3:e4090. doi: 10.1371/journal.pone.0004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai VB, Sundaresan NR, Kim G, et al. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol Chem. 2010;285:3133–44. doi: 10.1074/jbc.M109.077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai VB, Sundaresan NR, Gupta MP. Regulation of Akt signaling by sirtuins: its implication in cardiac hypertrophy and aging. Circ Res. 2014;114:368–78. doi: 10.1161/CIRCRESAHA.113.300536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polat MF, Taysi S, Gul M, et al. Oxidant/antioxidant status in blood of patients with malignant breast tumour and benign breast disease. Cell Biochem Funct. 2002;20:327–31. doi: 10.1002/cbf.980. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, et al. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–7. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Rao KN, Elm MS, Kelly RH, et al. Hepatic hyperplasia and cancer in rats: metabolic alterations associated with cell growth. Gastroenterology. 1997;113:238–48. doi: 10.1016/s0016-5085(97)70101-x. [DOI] [PubMed] [Google Scholar]
- Rardin MJ, He W, Nishida Y, et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–33. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PD, Huang B-W, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–90. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–18. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene. 2014;33:1609–20. doi: 10.1038/onc.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabine JR, Kopelovich L, Abraham S, et al. Control of lipid metabolism in hepatomas: conversion of glutamate carbon to fatty acid carbon via citrate in several transplantable hepatomas. Biochim Biophys Acta. 1973;296:493–8. doi: 10.1016/0005-2760(73)90109-4. [DOI] [PubMed] [Google Scholar]
- Sato H, Tamba M, Ishii T, et al. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J Biol Chem. 1999;274:11455–8. doi: 10.1074/jbc.274.17.11455. [DOI] [PubMed] [Google Scholar]
- Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453–62. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, et al. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–73. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- Schumacker PT. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Cancer Cell. 2006;10:175–6. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian C, Satterstrom FK, Haigis MC, et al. From sirtuin biology to human diseases: an update. J Biol Chem. 2012a;287:42444–52. doi: 10.1074/jbc.R112.402768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian C, Zwaans BMM, Silberman DM, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012b;151:1185–99. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J App Physiol. 2000;88:1474–80. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–6. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Fan GQ, Xiao SD. SIRT3 reduces lipid accumulation via AMPK activation in human hepatic cells. J Dig Dis. 2010;11:55–62. doi: 10.1111/j.1751-2980.2009.00416.x. [DOI] [PubMed] [Google Scholar]
- Shin J, He M, Liu Y, et al. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep. 2013;5:654–65. doi: 10.1016/j.celrep.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya S, Yu W, Hallows WC, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–12. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–5. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Chen X, Ma J, et al. Mammalian target of rapamycin upregulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc Natl Acad Sci USA. 2011;108:4129–34. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan NR, Gupta M, Kim G, et al. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–71. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Peng C, Anderson KA, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–17. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Coleman MC, Pennington JD, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol. 2000;279:L1005–28. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- Tomlinson IPM, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–10. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009;19:32–7. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–91. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- Van Driel BE, Valet GK, Lyon H, et al. Prognostic estimation of survival of colorectal cancer patients with the quantitative histochemical assay of G6PDH activity and the multiparameter classification program CLASSIF1. Cytometry. 1999;38:176–83. doi: 10.1002/(sici)1097-0320(19990815)38:4<176::aid-cyto4>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Lunt SY, Dayton TL, et al. Metabolic pathway alterations that support cell proliferation. Cold Spring Harb Symp Quant Biol. 2012;76:325–34. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–82. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–30. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Schölz C, Wagner SA, et al. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–51. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Weinhouse S. The Warburg hypothesis fifty years later. Zeitschrift für Krebsforschung und klinische Onkologie. Cancer Res Clin Oncol. 1976;87:115–26. doi: 10.1007/BF00284370. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–6. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, et al. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010;24:2784–99. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JES, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci USA. 2011;108:19611–16. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Fang Y-Z, Yang S, et al. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–92. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- Xu Y, Li F, Lv L, et al. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Res. 2014;74:3630–42. doi: 10.1158/0008-5472.CAN-13-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Laurent G, Bause AS, et al. HDAC6 and SIRT2 regulate the acetylation state and oncogenic activity of mutant K-RAS. Mol Cancer Res. 2013;11:1072–7. doi: 10.1158/1541-7786.MCR-13-0040-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med. 2011a;89:221–8. doi: 10.1007/s00109-011-0726-6. [DOI] [PubMed] [Google Scholar]
- Yecies JL, Manning BD. Transcriptional control of cellular metabolism by mTOR signaling. Cancer Res. 2011b;71:2815–20. doi: 10.1158/0008-5472.CAN-10-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–70. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. 2012;287:14078–86. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc–SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185:203–11. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, et al. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, De Marzo AM, Laughner E, et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5. [PubMed] [Google Scholar]