Abstract
Repurposing metformin for cancer therapy is attractive due to its safety profile, epidemiological evidence, and encouraging data from human clinical trials. Although it is known to systemically affect glucose metabolism in liver, muscle, gut, and other tissues, the molecular determinants that predict a patient response in cancer remain unknown. Here we carry out an integrative metabolomics analysis of metformin action in ovarian cancer. Metformin accumulated in patient biopsies and pathways involving nucleotide metabolism, redox and energy status, all related to mitochondrial metabolism, were affected in treated tumors. Strikingly a metabolic signature obtained in a patient with an exceptional clinical outcome mirrored that of a responsive animal tumor. Mechanistically, we demonstrate with stable isotope tracing that these metabolic signatures are due to an inability to adapt nutrient utilization in the mitochondria. This analysis provides new insights into mitochondrial metabolism and may lead to more precise indications of metformin in cancer.
Graphical abstract
INTRODUCTION
Metformin, a biguanide, is a commonly prescribed agent for the management of type II diabetes. It is widely used clinically because of its desirable safety profile and reproducible actions on systemic glucose homeostasis that lead to the reduction glucose levels during hyperglycemia (Knowler et al., 2002). Its effects are attributed to suppression of hepatic gluconeogenesis, reduction of glucose absorption in the intestine, alterations in the composition of the gut microbiota, and direct control of glucose metabolism in muscle and subcutaneous and visceral adipose tissue (Forslund et al., 2015; Fullerton et al., 2013; Madiraju et al., 2014; Zhou et al., 2001). The precise mechanism of action in diabetes however remains controversial (Pernicova and Korbonits, 2014). In epidemiological studies, metformin has been associated with reduced incidence in cancers such as breast, prostate, colorectal, endometrial, and ovarian (Decensi et al., 2010; Evans et al., 2005; Jiralerspong et al., 2009; Romero et al., 2012). Furthermore, a prospective clinical trial in non-diabetic patients showed its efficacy in patients presenting with adenomatous polyps when administered after surgical resection (Higurashi et al., 2016). Moreover, multiple clinical trials using metformin as a treatment in non-diabetic cancer patients are ongoing (Camacho et al., 2015).Thus there is significant interest in using metformin as a cancer therapeutic, especially in cancers with limited treatment options such as ovarian cancer (OvCa) (Bowtell et al., 2015; Febbraro et al., 2014). The mechanism of its anti-cancer properties has also been controversial and it remains poorly understood whether metformin acts through altering host metabolism or through direct action on tumor cells (Chandel et al., 2016; Dowling et al., 2016). Further clinical advances are limited by controversies surrounding the biology of metformin and the poorly understood determinants of a response that lead to biomarkers that can be used to predict which patients may benefit.
At the cellular level, metformin is believed to disrupt mitochondrial function by partially inhibiting NADH dehydrogenase (Wheaton et al., 2014) in general or by inhibiting glycerol phosphate dehydrogenase in liver cells that also results in alterations to the electron transport chain (ETC) (Madiraju et al., 2014). As a result, electrons contained in NADH and FADH2 are not as effectively transported through the ETC. Since each of these processes is central to numerous aspects of cell physiology, the cellular effects of metformin are pleiotropic. For example, it has been documented to affect AMPK signaling, protein kinase A signaling, folate metabolism, and anabolic metabolism (Birsoy et al., 2014; Cabreiro et al., 2013; Griss et al., 2015; Janzer et al., 2014; Madiraju et al., 2014; Miller et al., 2013; Shaw et al., 2005; Wheaton et al., 2014; Zhou et al., 2001). Most of these mechanisms point to mitochondrial biology as its mechanistic target.
Mitochondria utilize glucose, amino acids and fatty acids as substrates to allow for ATP production, redox balance, and biomass precursor production. Thus metformin results in alterations to the tricarboxylic acid (TCA) cycle, the generation of reactive oxygen species, alterations of the mitochondrial phosphate to oxygen ratio that affects ATP production, and other essential mitochondrial functions. For example it has recently been reported that one such essential function is to provide aspartate, which is used for the synthesis of nucleotides and protein (Birsoy et al., 2015; Cardaci et al., 2015; Sullivan et al., 2015). Other studies have found that metformin alters lipid synthesis from the mitochondria by affecting reductive carboxylation (Fendt et al., 2013). Consistently, metformin has also been found to suppress nucleotide levels (Janzer et al., 2014). Each of these mechanisms occurs through changes in the capacity of the ETC.
In light of this extensive body of literature, there is a lack of analysis of metabolism in physiological environments where clinically relevant metabolic signatures of metformin-associated cytotoxicity can be observed. This limits many of these findings and it is thus not understood how mitochondrial metabolism is altered to produce these changes and what compensatory pathways induced by disruption of the ETC must be overcome for metformin-induced cytotoxicity. There is furthermore a lack of direct study of metformin action on tumor metabolism in patients and comparisons to mechanisms observed in cell culture and in animal models. With the advent of recent metabolomics approaches, multiple aspects of tumor cell metabolism can be profiled in patient samples (Liu et al., 2014). We therefore hypothesized that such experiments could shed light on the anti-cancer mechanisms of metformin when integrated with studies on experimental models.
RESULTS
Altered mitochondrial metabolism in human tumors from patients taking metformin
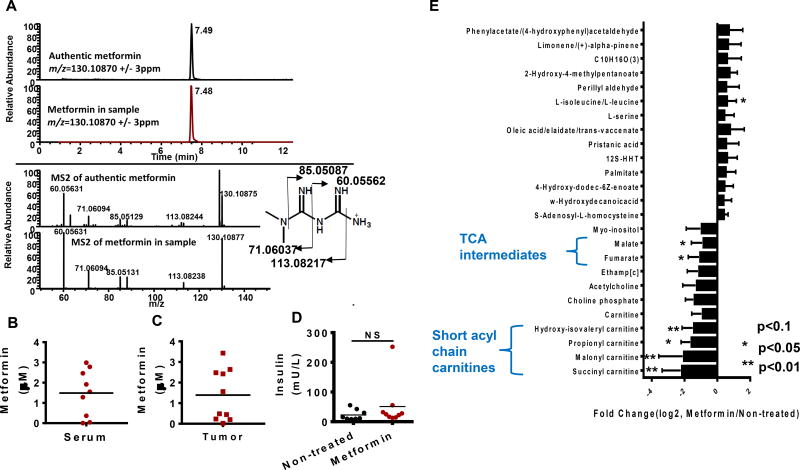
There has been a paucity of data surrounding the concentration of metformin that can be achieved in human tumors. We first developed a sensitive and quantitative method using liquid chromatography coupled to high resolution mass spectrometry (LC-HRMS) to quantify metformin concentrations in tissue and serum. With an authentic reference compound and fragmentation pattern from tandem mass spectrometry (Fig 1a), we were able to unequivocally identify and quantify metformin in the sera (Fig 1b) and tumors (Fig 1c) of patients taking metformin prior to surgery and found concentrations reaching micromolar values. Interestingly, no differences in the insulin levels in these patients were observed (Fig 1d). We then generated a metabolite profile (Liu et al., 2014) of the tumors from ten patients taking metformin presenting with stage III/IV high grade papillary serous OvCa (Fig S1a) compared to ten patients not taking metformin with similar diagnoses, ages and medication profiles. An inspection of the profile (Fig 1e) showed that most of the over three hundred metabolites measured did not show a difference indicating extensive heterogeneity as expected (Hensley et al., 2016). Interestingly, TCA intermediates and short chain acyl carnitines (succinyl carnitine and branched chain acyl carnitines), both related to mitochondrial metabolism (Koves et al., 2008; Liu et al., 2015), were the classes of compounds most frequently suppressed in the tumors of metformin-treated patients. Three of the known functions of mitochondrial metabolism are to maintain nucleotide levels, energy status, and oxidation status (Shadel and Horvath, 2015; Weinberg and Chandel, 2015; Zong et al., 2016). In each case, and as is expected since cancer is a heterogeneous disease, there was large variation in metabolites involved in each of these processes with some patients exhibiting alterations in the metabolites involved in each of these functions (Fig 1e, Fig S1b,c and d). Long chain acyl carnitines, markers of fatty acid oxidation (Koves et al., 2008), were variable in the patient tumors indicating that a defect in fatty acid oxidation although possibly present in certain tumors was not commonly observed (Fig S1e). Together these results indicate that metformin accumulates in human tumors and affects known mitochondrial biology.
Figure 1. Altered mitochondrial metabolism in human ovarian tumors from patients taking metformin.
A. Extracted ion chromatogram of authentic standard metformin (top) and metformin extracted from mouse serum (bottom) with an m/z window of 6 ppm (+/− 3ppm). MS2 spectrum of authentic metformin (top) and metformin extracted from mouse serum (bottom). B. Metformin levels in serum of ovarian cancer (OvCa) patients. C. Metformin concentration in ovarian tumors from patients. D. Insulin levels in the serum of OvCa patients after overnight fasting. E. Metabolites with trending changes (p<0.1, * p<0.05, ** p<0.01, student’s t-test two tailed) in tumors from patients on metformin compared to tumor not exposed to metformin treatment (N=10). The fold change is calculated by dividing MS intensity (integrated peak area of metabolites) values of each metabolite in metformin treated patient tumors by those with no metformin treatment. Error bars obtained from SEM of n=10 independent measurements.
Therapeutic doses of metformin directly suppress tumor mitochondrial metabolism
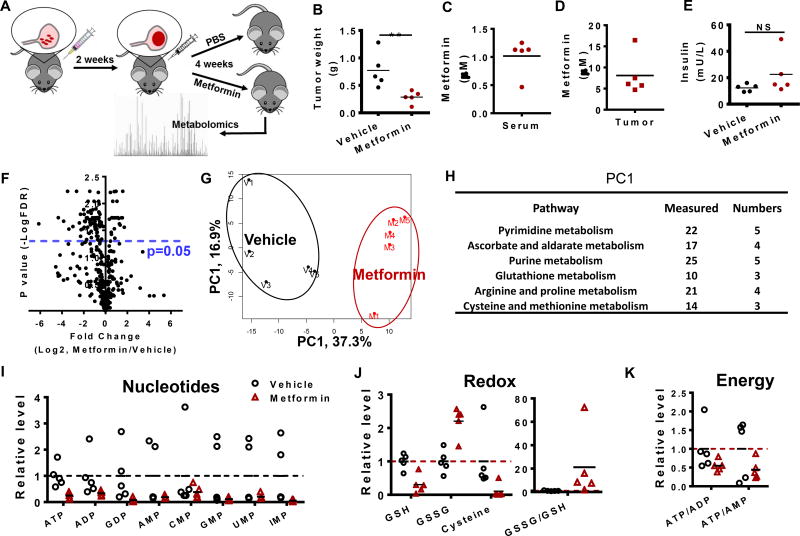
The metabolomics of human tumors (Fig 1) appear consistent with the conclusion that metformin affects cell intrinsic mitochondrial metabolism. Nevertheless, these studies involved a limited set of patient samples. We therefore considered laboratory models of ovarian cancer. HeyA8 cells were injected in the intraperitoneal cavity of female mice and tumors formed in the omentum resembling a serous-papillary cystadenocarcinoma (Lengyel et al., 2014) (Fig 2a). Treatment of mice two weeks after initiation of OvCa showed that metformin effectively reduced tumor weight (p = 0.01, student’s T-test, one tailed) (Fig 2b) and the concentration of metformin reached micromolar concentrations in serum (Fig 2c) and tumor (Fig 2d). Both serum and intratumoral levels of metformin correlated (p = 0.07, p = 0.03, respectively, Spearman’s correlation coefficient) with treatment response. As observed in the patients, insulin levels in mouse serum were unaffected (Fig 2e). Metabolite profiles from metformin- and vehicle-treated animal tumors were generated using LC-HRMS (Fig 2f), and first analyzed by a principle component (PC) analysis (Fig 2g), revealing that the largest source variation in the cohort was treatment. Metabolites enriched in the pyrimidine/purine metabolism pathways, and the glutathione pathway (Fig 2h), contributed to the first PC. Metabolites involved in nucleotide synthesis (Fig 2i), redox status indicated by increased oxidized glutathione (GSSG) levels (Fig 2j) and energy status (Fig 2k) were generally reduced. Together, these findings indicate that metformin affects several features related to mitochondrial metabolism in animal tumors.
Figure 2. Metformin directly suppress tumor mitochondrial metabolism in mouse ovarian tumors.
A. Description of timeline and treatment of the ovarian cancer model where HeyA8 cells are injected intraperitoneally. B. Mouse tumor weight (N=5). C. Metformin concentration in mouse serum. D. Metformin concentration in mouse tumor. E. Insulin levels in mouse serum. F. Volcano plot of metabolites in mouse tumor. G. PCA analysis of metabolites in mouse tumor. H. Pathway analysis of metabolites in mouse tumor. “Measured” is the total number of metabolites detected in mouse tumor, while “Number” is the number of metabolites with significant changes (p<0.05) in the metformin treatment group. I to K. Representative metabolites among nucleotides, redox and energy state. * p<0.05, ** p<0.01
A metformin-treated patient with an exceptional outcome shares the metabolic profile of the animal tumors responding to metformin
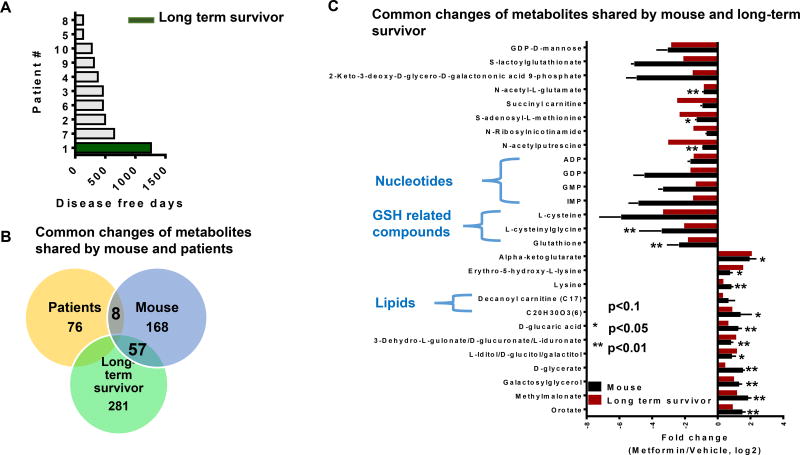
While the metabolic profile of the metformin response in human tumors was variable, metformin treatment in controlled settings using laboratory animals appeared reproducible. Of note, the time to recurrence was also variable in the patients taking metformin and one patient exhibited an exceptionally long disease free interval (long term survivor) with no recurrence to date (Fig 3a). Although single patient responses have long been considered merely anecdotal in nature and lack statistical rigor, there have recently been considerable advances made by using these observations to understand the mechanism underlying these exceptional outcomes (Grisham et al., 2015; Marx, 2015; Wagle et al., 2014). Utilizing this long term survivor, we sought to compare the metabolic features of this patient, the cohort of patients, and the mouse responding to metformin. Overall a small amount of overlap in the profiles of the animal tumors and the patients was observed. Remarkably, when comparing this long term survivor, a substantial amount of the metabolome overlapped with the signature in the mouse ovarian tumor responding to metformin (Fig 3b). An analysis of overlapping features (Fig 3c) revealed decreases in acyl-carnitines, nucleotides, and glutathione-related metabolites and increases in nucleotide intermediates suggesting a buildup in these pathways. These intriguing results indicate a possible a cell-intrinsic human signature of a response to metformin in ovarian cancer.
Figure 3. Metabolite changes are shared by patient and mouse in response to metformin treatment.
A. Disease free days of ten patients on metformin. The long term survivor patient is the one with longest disease free days shown in green. B. Number of metabolites overlapped in patients and mouse tumor with larger effects in the metformin treatment group (fold change >2 or <0.6). This number indicates the number of metabolites with changes in response to metformin treatment. C. Representative metabolites sharing similar changes in response to metformin treatment in mouse (p<0.1, * p<0.05, ** p<0.01) and a long term survivor patient tumor. Error bars obtained from SEM of n=5 independent measurements.
In vivo metabolite profiles of metformin response can be modeled in a glucose-limited environment
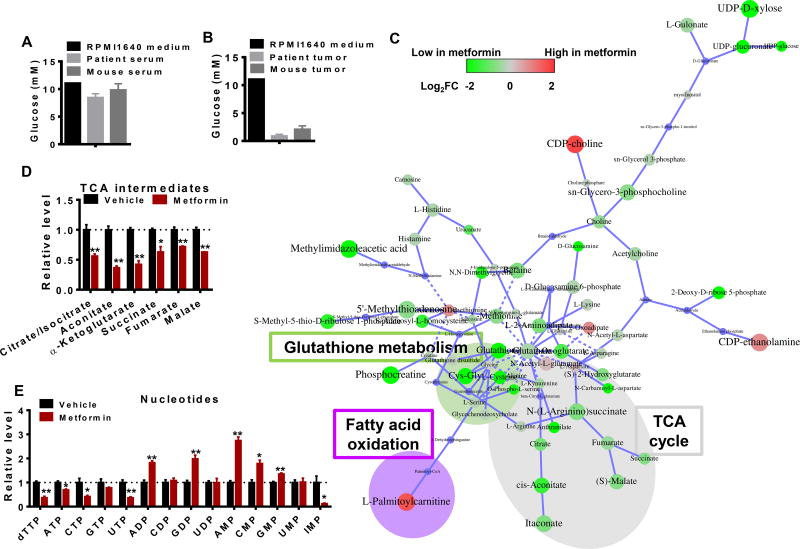
We next questioned the specific biochemical events and environmental conditions that directly give rise to these correlative signatures seen in human and mouse xenograft tumors. Although serum levels of glucose in the mice and humans were in the range of 5–10mM (Fig 4a), the values measured in human and mouse tumors were markedly lower, ranging from 0–1mM (Fig 4b). We thus reasoned that the metformin responses observed in vivo may require limiting mitochondrial substrate availability in the glucose-deprived tumor microenvironment. To test this hypothesis, we generated a metabolite profile in HeyA8 cells treated with metformin or vehicle in glucose-limited (1mM) conditions that mimic the tumor microenvironment (Fig 4c). Notably, intracellular metformin concentration could reach values comparable to that in the culture media (Fig 2c). An increase in NADH indicating disruption of ETC (Fig S2b) was observed with no changes glucose uptake and lactate secretion fluxes (Fig S2c,d). Moreover, decreases in glutathione-related metabolites were also observed (Fig 4c) in addition to depletions in the TCA cycle (Fig 4d) and nucleotide levels (Fig 4e). An additional feature associated with ETC inhibition involving a decrease in short chain acyl carnitines metabolites was observed (Fig S2e), consistent with observations in patient (Fig 1e) and mouse tumors (Fig S2f). Notably, metformin also induced an increase of NADH in glucose rich medium (Fig S2g), indicating metformin also targets on the ETC when there is sufficient glucose. However, no changes in glutathione (GSH) related metabolites (Fig S2f), or TCA intermediates (Fig S2h), or nucleotides were observed (Fig S2i), indicating that global metabolic reprogramming is not induced upon metformin treatment in high glucose conditions. Thus most of the metabolic alterations found in metformin-responsive mouse and in human tumor tissues can be reproduced in glucose-limited but not in the high-glucose conditions observed in typical cell culture media.
Figure 4. In vivo metabolite profiles of metformin treatment can be modeled in a glucoselimited environment.
A. Glucose concentrations in RPMI medium, patient and mouse serum. B. Glucose concentration in RPMI medium, patient and mouse ovarian tumors. C. Network of significant metabolic changes in ovarian cancer cells in response to Metformin treatment. The node size represents p value while the color represents the fold change. The size of the node indicates magnitude of the change. Red represents metabolites that are higher in metformin treated cells, while green denotes lower in metformin treated cells. D to E. Representative metabolites among TCA and nucleotides. Relative level was calculated by dividing MS intensity of each metabolite by corresponding MS intensity in Vehicle. * p<0.05, ** p<0.01 Error bars obtained from SEM of n=3 independent measurements.
Metabolite profiles of metformin are the result of altered substrate utilization in the mitochondria
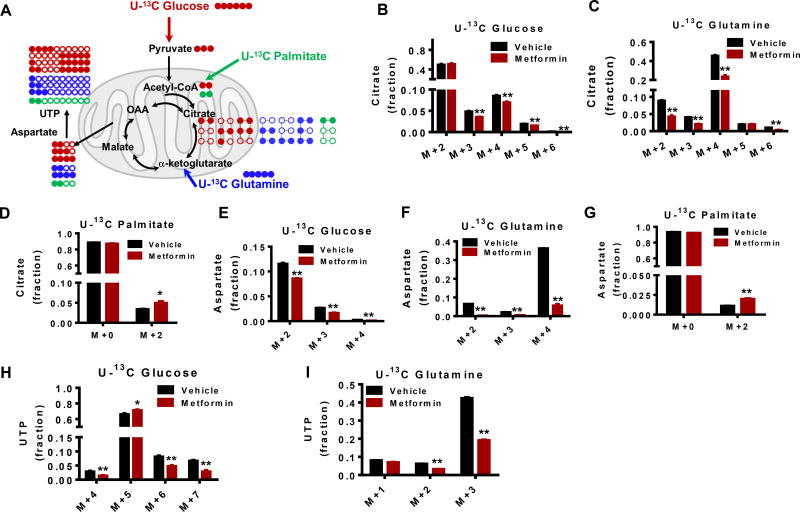
Although metabolite levels are direct reflections of physiological status, their interpretation is confounded by the dynamic nature of metabolism that involves the flow of nutrients or flux (Zamboni et al., 2015). Therefore, the utilization of each key macronutrient that is catabolized in the mitochondria was investigated. Stable isotopes of carbohydrate (U-13C glucose), lipid (U-13C palmitate), and amino acid (U-13C glutamine) were used to trace the fate of mitochondrial substrate utilization in response to metformin treatment (Fig 5a). Glucose entry into the mitochondria, as measured by the mass isotopologue patterns of citrate (Fig 5b), glutamine (Fig 5c), and palmitate (Fig 5d) in the TCA cycle were each affected by metformin treatment. In addition glucose-derived (Fig 5e), glutamine-derived (Fig 5f), and palmitate-derived (Fig 5g) aspartate, an intermediate for pyrimidine synthesis were decreased, as were corresponding labeling patterns of glucose- (Fig 5h) and glutamine- (Fig 5i) derived pyrimidine that originates from the TCA cycle. Moreover, glutathione biosynthesis, which uses glutamate generated from the TCA cycle, was reduced (Fig S3a–d). However, addition of the antioxidant N-acetylcysteine (NAC) didn’t increase GSH levels in the absence of metformin (Fig S3e), but decreased GSSG levels (Fig S3f). Surprisingly, intracellular aspartate levels remained constant in the presence of metformin (Fig S4a), and meanwhile, aspartate was actively exported out of the cell indicating its excess rather than limitation (Fig S4b). Indeed compared to cells grown in high glucose, there was also a decreased uptake of non-essential amino acids, such as lysine and leucine/isoleucine (Fig S4c,d), which further demonstrates the decreased consumption of these amino acids in low glucose environments. Together these findings suggest that metformin alters substrate utilization and anabolic metabolism in the mitochondria with anabolic metabolism being affected by a previously uncharacterized aspartate overflow mechanism.
Figure 5. Metabolite profiles of metformin are the result of altered substrate utilization in the mitochondria.
A. 13C enrichment patterns that are derived from different 13C labeled fuel sources. Solid circle denotes 13C carbon while open circle denotes 12C carbon. B to D. 13C isotopologues distribution of citrate from cells treated with 13C labeled glucose, glutamine or palmitate for 6 hrs. E to G. 13C enrichment in aspartate from 13C labelled glucose, glutamine or palmitate. H to I. UTP 13C distribution in the presence of 13C labelled glucose or glutamine. 13C labelled UTP was not detected from 13C labeled palmitate. * p<0.05, ** p<0.01 Error bars obtained from SEM of n=3 independent measurements.
Metformin response and resistance is determined by the availability of specific nutrients
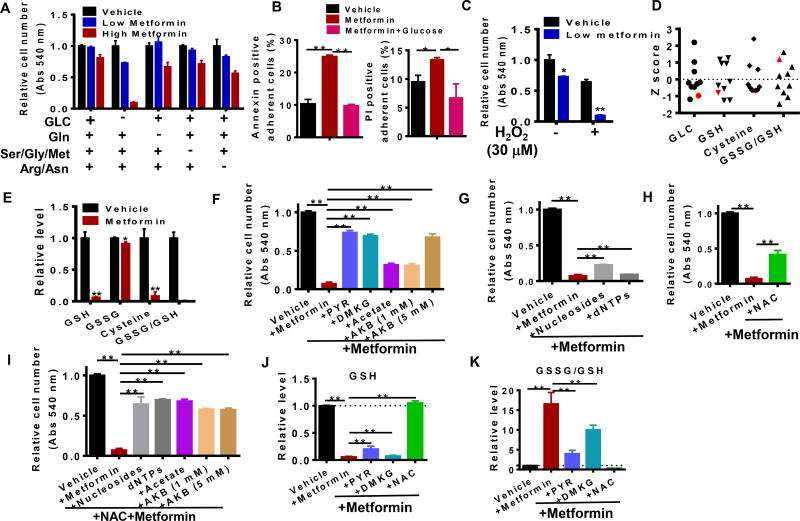
Next, we asked whether the nutrients utilized and metabolic outputs generated by mitochondria determine the cytotoxic effects of metformin. Consistent with previous observations (Birsoy et al., 2014; Litchfield et al., 2015; Wheaton et al., 2014), we found that HeyA8 OvCa cells are resistant to metformin in nutrient rich conditions containing typical concentrations of glucose in culture media. When subjected to nutrient limiting conditions including glucose and amino acid deprivation, these cells become sensitive to metformin implying that these environments impose sensitivity to metformin (Fig 6a). Annexin V and propidium iodide staining of HeyA8 cells in glucose limited (Vehicle) and glucose complete media (+Glucose) demonstrated that glucose availability affects metformin-induced apoptosis and cell death in this model (Fig 6b). Furthermore, oxidative stress involving sub-lethal concentrations (30 µM) of hydrogen peroxide (Fig 6c) sensitizes cells to metformin-induced cell death. Interestingly, the tumor from the long term survivor (Fig 6d) exhibited low levels of glucose and a higher oxidative state as measured by glutathione potential and other related metabolites. Similarly, increased oxidative stress was also observed in metformin-treated cells (Fig 6e). To understand the metabolic features that underlie this sensitivity, we attempted to rescue metformin-cytotoxicity using a series of nutrients and quantified cell number using both a colorimetric tetrazolium-based assay and direct cell counting. We found that pyruvate, an esterified version of alpha-ketoglutarate (DMKG), acetate, and alpha-ketobutyrate (Sullivan et al., 2015), all immediate mitochondrial substrates, were able to rescue metformin-induced cytotoxicity to various degrees (Fig 6f). Furthermore, we investigated downstream pathways altered by metformin and observed that nucleosides, deoxynucleotide triphosphates (dNTPs) (Fig 6g), or N-acetyl cysteine (NAC) (Fig 6h), exhibited modest but significant effects on rescuing cytotoxicity. The combinations of NAC with nucleotides, nucleosides, acetate, or alpha-ketobutyrate, could to a larger extent rescue cells from metformin-induced cell death (Fig 6i). Importantly, results from the tetrazolium colorimetric assay were consistent with those obtained both from cell counting and cell imaging (Fig S5a–e). The supply of aspartate has been proposed to be an essential role of electron transfer chain (Birsoy et al., 2015; Cardaci et al., 2015; Sullivan et al., 2015), but aspartate alone failed to rescue metformin-induced cell death, while the combination with NAC exerted only a modest effect (Fig S5f). Taken together, these results demonstrate that consistent with signatures of metformin response in mice and humans, metformin cytotoxicity is caused by limited mitochondrial substrate availability and that redox and nucleotide biosynthesis are the critical functions required for cell viability in the presence of metformin. In agreement, redox status was preserved with the addition of mitochondrial substrates or NAC (Fig 6j) and oxidative stress was diminished (Fig 6k). Furthermore, we found that metformin also decreased energy status (ATP/ADP and ATP/AMP) and this effect was partially recovered in all four conditions that rescued cell proliferation (Fig S6a). Increased NADH/NAD+ due to NADH accumulation however was not rescued by these nutrients (Fig S6b). This finding is in contrast to previous observations where pyruvate preserved the NAD+/NADH ratio in the presence of electron chain transport inhibitors (Sullivan et al., 2015). The discrepancy highlights the heterogeneity of metabolic requirements and likely depends on other fluxes that metabolize NAD+. TCA intermediates were dramatically elevated by PYR and DMKG, but not by NAC or nucleosides (Fig S6c). Similarly, nucleotides levels are also differentially regulated by these nutrients (Fig S6d). Therefore, these findings additionally confirm that redox, energy homeostasis, and nucleotide biosynthesis in combination appear to be determining factors to the cytotoxic response in these environments.
Figure 6. Metformin response and resistance depends on the availability of specific nutrients.
A. Response to metformin treatment in different medium conditions. Abs: Absorbance (at 540 nm from MTT assay). GLC: Glucose (11 mM), Gln: Glutamine, Ser/Gly/Met: Serine, glycine and methionine, Arg/Asn: Arginine and Asparagine. Low metformin: 0.5 mM metformin, High metformin: 1.5 mM metformin. B. Metformin effect on cell proliferation in the presence of low glucose (1mM) and/or hydrogen peroxide (H2O2, 30 µM). C. Annexin and PI positive populations of adherent cells after 40 hours of metformin treatment. D. Z score distribution of glucose (GLC) and redox metabolites (GSH, cysteine and GSSG/GSH) in patients after metformin treatment. The long term survivor is highlighted in red color. E. Relative levels of redox metabolites in ovarian cancer cells with or without metformin treatment for 24 hrs. F. Cell viability in the presence of metformin and precursors of the TCA cycle (PYR: pyruvate; DMKG: dimethyl α-ketoglutarate; Acetate: sodium acetate; AKB: α-ketobutyrate). G. Rescue of metformin-induced loss of cell viability by nucleosides or deoxynucleotide triphosphates (dNTPs). H to I. Cell viability in various medium with or without N-acetylcysteine (NAC). J to K. Redox status in different treatment conditions. * p<0.05, ** p<0.01 Error bars obtained from SEM of n=3 independent measurements.
The response to metformin depends on whether substrate utilization in the mitochondria is flexible
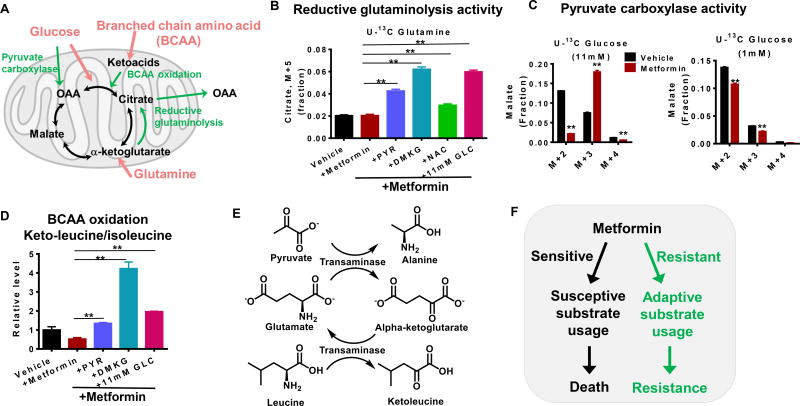
Altered nutrient utilization in cells with defective mitochondria is well documented with numerous pathway fluxes involved (Fendt et al., 2013; Worth et al., 2014). These include pyruvate carboxylase, reductive glutaminolysis, and branched chain amino acid oxidation, which are used as compensatory pathways when cells with functioning mitochondria are under mitochondrial stress (Fig 7a). In glucose-limited conditions, cells were unable to increase reductive glutaminolysis activity when oxidative glutamine metabolism is inhibited by metformin, while the presence of PYR, DMKG, NAC, or glucose causes upregulation of the activity of this pathway upon treatment with metformin (Fig 7b). Moreover, the addition of excess glucose (11.1 mM) increased pyruvate carboxylase activity in response to metformin and maintained a stable carbon supply into the mitochondria (Fig 7c). In contrast, when glucose-limited, these cells were not able to use this pathway. Moreover, branched chain amino acid oxidation (BCAA), also a pathway used as a fuel source, was significantly diminished by metformin treatment, and recovered with addition of PYR, DMKG or glucose (Fig 7d). Consistently, pyruvate was found to increase the concentration of TCA cycle intermediates (Fig S6c). Indeed, upon adding U-13C pyruvate for 6 hours, citrate (M+2) and malate (M+3) representing flux into the TCA cycle through citrate synthase and pyruvate carboxylase were both increased (Fig 7e) as was alanine (M+3) indicating transamination and generation of alpha-ketoglutarate (Fig 7f). Thus, the inability to use these adaptive pathways in the TCA cycle in response to metformin treatment contributes to the sensitivity in glucose-limited conditions (Fig 7g).
Figure 7. The response to metformin depends on whether substrate utilization in the mitochondria is flexible.
A. Schematic of substrate utilization. The adaptive pathways under mitochondrial stress are highlighted in green color. OAA: oxaloacetate, Keto acids: ketoleucine/ketoisoleucine. B. Reductive glutaminolysis activity as represented by the fraction of 13C enriched citrate isotopologue (M+5) from cells treated with 13C-glutamine in different medium conditions. C. Pyruvate carboxylase activity as estimated by the fraction of malate (M+3) from cells treated with 13C-glucose in different medium conditions. D. Ketoleucine/ketoisoleucine levels in different media conditions, representing BCAA oxidation. E. Representative metabolite enrichment pattern from U-13C pyruvate treated cells. F. Scheme of coupled transamination reactions demonstrating stoichiometric alpha-ketoglutarate generation from alanine labeling. G. Model of resistant and sensitive response to metformin treatment. * p<0.05, ** p<0.01 Error bars obtained from SEM of n=3 independent measurements.
DISCUSSION
Cell intrinsic metabolic reprogramming by metformin in tumors
Epidemiological data have strongly associated metformin usage with improved cancer outcomes in multiple cohorts in numerous different cancers including OvCa (Decensi et al., 2010; Evans et al., 2005; Ezewuiro et al., 2016; Jiralerspong et al., 2009; Libby et al., 2009; Romero et al., 2012; Wright and Stanford, 2009). However, understanding how metformin might act to protect against or treat cancer has been complicated by controversies concerning its mechanism of action and the organ site on which it acts. By quantifying metformin concentrations in patient and mouse tumors, we were able to show it accumulates in tumors at concentrations that are sufficient to induce metabolic alterations consistent with changes in the activity of the electron transport chain. Importantly, the extent of the anti-tumor effect correlated with the amount of the compound that was measured in the tumor. Accordingly, critical aspects of mitochondrial metabolism were altered after metformin treatment, such as the levels of TCA intermediates, the accumulation of NADH and the increase of ROS. The accumulation of ROS could be attributed either to decreased glutathione (GSH) biosynthesis or electron leakage from electron transport chain, whose efficiency is inhibited by metformin. Aspartate biosynthesis was recently identified as the essential function of the respiratory chain (Birsoy et al., 2015; Cardaci et al., 2015; Sullivan et al., 2015). Although metformin inhibited nucleotide synthesis in several settings, aspartate surprisingly did not appear to be limiting in these conditions.
As mitochondria function as a centralized hub in the metabolic network, we observed global metabolic reprogramming in response to metformin treatment in human, mouse and cell culture. Nevertheless most of these metabolic alterations can be rationalized from a perturbation in mitochondrial metabolism and rescue of metformin-induced cell death was achieved either by supplementing cells with mitochondrial substrates or with combinations of nutrients that produce metabolites involved in downstream processes such as nucleotides and antioxidants.
Metformin and nutrient limitations in the mitochondria
Glucose concentrations were found to be less than 1mM in tumors from OvCa patients. Consistently, cytotoxicity at physiologically meaningful doses of metformin is achieved when glucose availability is limited (Birsoy et al., 2014; Litchfield et al., 2015; Wheaton et al., 2014). Interestingly in conditions with sufficient nutrient availability such as standard tissue culture conditions, cells readily resist metformin treatment by bringing other nutrients into the mitochondria to compensate for partially impaired ETC activity. Accordingly, metformin treatment enhanced pyruvate carboxylase activity and increased the reverse exchange flux in the TCA cycle, which resulted in increased production of malate, fumarate and oxaloacetate and decreased flux along the TCA cycle. Such effects may also result in suppression of hepatic gluconeogenesis that is thought to be a determinant of its mechanism of action in diabetes. Each of these features is thought to confer requirements for tumor growth in different contexts (Davidson et al., 2016; Hensley et al., 2016). However, in contrast to recent findings on biguanides and aspartate synthesis (Birsoy et al., 2015; Cardaci et al., 2015; Sullivan et al., 2015), our data show that under limiting conditions of nutrient availability, inhibition of the respiratory chain by metformin can actually cause secretion of aspartate as a waste product (Fig S4b). This finding exemplifies the heterogeneity of tumor metabolism and may extend to other cases when cells have certain respiratory chain defects, such as the case of succinate dehydrogenase mutations that increase dependence on pyruvate carboxylase (Cardaci et al., 2015). Thus, inhibition of the respiratory chain in proliferating cells does not always cause a defect in biosynthesis derived from aspartate metabolism.
We have reported that OvCa cells obtain fatty acids from adjacent adipocytes in the tumor microenvironment (Nieman et al., 2011). Here we observed that catabolism of lipids by fatty acid oxidation (FAO) could be used to fuel the TCA cycle in environments of limited glucose availability. However, when increased consumption of fatty acid was observed, cells were sensitive to metformin likely due to the fact that FAO is much slower in fueling the TCA cycle relative to the rate of glucose or glutamine oxidation. Therefore, in glucose-limited conditions, adaptive pathways such as FAO that are upregulated and available in response to metformin are unable to fully compensate.
Finally, recent studies have suggested an enhanced dependency on mitochondrial metabolism during tumor dormancy or latency, a poorly characterized state of cancer cells that may underlie how tumors recur (Viale et al., 2014). Since nearly complete inhibition of the respiratory chain that can be achieved with compounds such as rotenone and cyanide is limited by toxicity, a speculative suggestion is that the safety profile of metformin may lend itself to a treatment for this cancer cell state of dormancy as a means to prevent recurrence.
Molecular determinants of metformin action and a long term survivor
In addition to an overall signature pointing to alterations in mitochondrial metabolism across a set of human tumors, we further provide evidence, albeit an “n of 1”, that a late stage ovarian cancer patient taking metformin had an exceptionally long disease free interval (long term survivor) after surgical resection and exhibited numerous intratumoral features of what defined a response to metformin in both a mouse model of OvCa and in cultured cells. It is understood that this long term survivor provides only anecdotal evidence of a patient response to metformin; nevertheless, examples of defining a mechanistic basis for an exceptional patient response have led to numerous new and important insights(Grisham et al., 2015; Marx, 2015; Wagle et al., 2014). Typically these responses are characterized by further validation of a predictive model for why a patient with a defined genetic lesion responded to a particular therapy. In this case, we show that a mechanistic basis of a long term survivor can be understood through analysis of the specific metabolism occurring in the patient’s tumor. Thus knowledge of the metabolic profile of the tumor can be predictive of therapeutic response independent of cancer genetics.
Nevertheless, it is worth nothing there are several confounding factors in the patient study. The patients taking metformin have diabetes that is its major clinical indication and some patients were also taking other medications. Also two other patients of the twenty total have not experienced recurrence at the time of this paper’s submission, albeit these patients are much closer to the time of their surgical resection. Nevertheless we found signatures of mitochondrial inhibition in certain human tumors that we studied more deeply with model systems, and our conclusions were drawn from the integration of three independent systems (human, mouse, cell culture).
SUMMARY
The data presented to our knowledge provide the most comprehensive metabolomics analysis of metformin action in ovarian cancer to date. By integrating data from metabolic features remnant in human patient tumors with idealized responses in pre-clinical models, we were able to provide insight into the nature of metformin’s effect in cancer. In doing so, we were also able to identify new aspects of metabolism that are controlled by mitochondria. With this deeper biochemical and systems-level understanding using a thorough metabolomics platform, we identified many of the pleiotropic features that determine an anti-cancer response to metformin. Hopefully these results will inform the design and patient selection for upcoming clinical investigations of this intriguing agent.
EXPERIMENTAL PROCEDURES
Reagents
A detailed list of reagents is provided in Supplemental Information.
Animal
HeyA8 cells were injected intraperitoneally (Lengyel et al., 2014). Two weeks after injection, mice were treated with metformin (250 mg/kg/day) or placebo (PBS) administered intraperitoneally for additional 4 weeks before mice were sacrificed. The mice were fed food and water ad libitum before sacrifice. Briefly, mice were anesthetized with isoflurane and euthanized with cervical dislocation. The time from sacrifice to flash freezing of the mouse tumors was approximately 2–4 min. The entire section of tumor was then stored at −80 °C before metabolomics analysis, and there was no additional processing performed. All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Chicago (#71951).
Patient samples
Patient samples and clinical data were extracted from the biorepository and ongoing dataset of patients treated for OvCa at the University of Chicago. The last update on patient information was on May 1st, 2016. As previously reported, the ovarian cancer dataset contains information on clinicopathologic parameters, use of metformin, and cancer outcomes (Romero et al., 2012). Recurrence of disease was defined using previously published clinical criteria (Therasse et al., 2000). Disease-free days were calculated from the date of diagnosis until the date of ovarian cancer recurrence or death. For patients without recurrence or death, date of last follow-up was utilized. Patients had been fasting overnight and until after surgery. Patient serum were collected after overnight starvation and before surgery. Patients were administered an IV with 278 mmol/L dextrose in normal saline before or during the surgery Patients were under general anesthesia for a period of 30–90 min before surgery, and stayed under anesthesia at the time of sample collection. The time from removal from the patient to flash freezing was less than 20 min. The entire section of human tumor was then stored at −80 °C freezer metabolomics analysis. There was no additional processing performed prior to metabolomics analysis. The institutional review board at the University of Chicago approved the study (#13372B and 13248A).
Cell culture
The HeyA8 OvCa cell lines were provided by Dr. Gordon Mills. Cell lines were authenticated using CellCheck (IDEXX Bioresearch). HeyA8 cells were first cultured in 10 cm dish with full growth medium containing RPMI 1640, 10 % FBS, 100 U/ml penicillin and 100 µg/ml streptomycin.
Cell viability assays
Cell viability was assessed using both tetrazolium-based MTT assay (Invitrogen) and cell counting (Moxi™ Z Mini Automated Cell Counter, ORFLO Technologies). The detailed procedure is described in Supplemental Information.
Metabolite extraction in tumor and serum
Both patient and mouse tumors were extracted as described previously (Liu et al., 2015) and also in Supplemental Information. For absolute quantitation of metformin and glucose, [2H6]-metformin and 13C labeled standards were added to extraction solvent before metabolite extraction. More detailed information is included in Supplemental Information.
Metabolite extraction in cell culture
HeyA8 cells were seeded at a density of 150000 cells per well in 6 well plates. After overnight incubation in full growth medium, the old medium was removed and cells were washed with 1 ml PBS before the addition of 2 ml of treatment medium. Glucose consumption and lactate secretion measurement are described in Supplemental Information. For intracellular metabolite analysis, after an incubation for 20 hours, metabolites were extracted as described in a previous study (Liu et al., 2014). Detailed information on 13C tracing is provided in Supplemental Information.
Insulin measurement
The concentration of insulin concentration in serum was measured using Ultra Sensitive Mouse Insulin ELISA Kit (Crystal Chem Inc) according to the manufacturer’s protocol. This insulin kit was kindly provided by Drs. Daniel Cooper and David Kirsch.
Apoptotic Cell Death
Phosphatidylserine on the external surface and propidium iodide uptake were measured using the Annexin V Alexa Fluor® 488 & Propidium Iodide (PI) Dead Cell Apoptosis Kit (ThermoFisher Scientific) according to the manufacturer's recommendations. A detailed experimental procedure is described in Supplemental Information.
Statistical analysis and bioinformatics
LC-HRMS data analysis method was described Supplemental Information. Pathway analysis of metabolites using the KEGG pathway database (http://www.genome.jp/kegg/) was carried out with software Metaboanalyst (http://www.metaboanalyst.ca/MetaboAnalyst/faces/Home.jsp). Principle component analysis (PCA) and statistical analysis were done using R programmer (https://www.r-project.org/). Metabolic network was constructed based on HMDB database (http://www.hmdb.ca/) using open source software GAM (https://artyomovlab.wustl.edu/shiny/gam/). All data are represented as mean ± SEM. All p values are obtained from student’s t-test two-tailed unless otherwise noted.
Supplementary Material
Acknowledgments
We acknowledge support from National Institutes of Health awards R01CA193256 (JWL), P50CA136393 (EL and ILR), and K12HD000849 to ILR. We thank Dr. Donald McDonnell, Dr. Deborah Muoio, members of the Locasale Lab, and Dr. Abir Mukherjee for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
ILR and EL initiated the studies by suggesting analysis of the patient and animal samples. JWL, XL designed the metabolism experiments, performed all data analysis and interpretation, and wrote the paper with essential edits from ILR and EL. LL and ILR performed the animal experiments. XL performed all other experiments. EL and ILR contributed the human patient samples.
DISCLOSURES
The authors declare no conflicts of interest at this time.
References
- Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B, Wang T, Chen WW, Clish CB, Sabatini DM. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508:108–112. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, Weinkove D, Schuster E, Greene ND, Gems D. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho L, Dasgupta A, Jiralerspong S. Metformin in breast cancer - an evolving mystery. Breast cancer research : BCR. 2015;17:88. doi: 10.1186/s13058-015-0598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardaci S, Zheng L, MacKay G, van den Broek NJ, MacKenzie ED, Nixon C, Stevenson D, Tumanov S, Bulusu V, Kamphorst JJ, et al. Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis. Nat Cell Biol. 2015;17:1317–1326. doi: 10.1038/ncb3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS, Avizonis D, Reczek CR, Weinberg SE, Menz S, Neuhaus R, Christian S, Haegebarth A, Algire C, Pollak M. Are Metformin Doses Used in Murine Cancer Models Clinically Relevant? Cell Metab. 2016;23:569–570. doi: 10.1016/j.cmet.2016.03.010. [DOI] [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O'Brien JP, Pierce KA, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell metabolism. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decensi A, Puntoni M, Goodwin P, Cazzaniga M, Gennari A, Bonanni B, Gandini S. Metformin and cancer risk in diabetic patients: a systematic review and meta-analysis. Cancer Prev Res (Phila) 2010;3:1451–1461. doi: 10.1158/1940-6207.CAPR-10-0157. [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Lam S, Bassi C, Mouaaz S, Aman A, Kiyota T, Al-Awar R, Goodwin PJ, Stambolic V. Metformin Pharmacokinetics in Mouse Tumors: Implications for Human Therapy. Cell Metab. 2016;23:567–568. doi: 10.1016/j.cmet.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt SM, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, Vokes NI, Guarente L, Vander Heiden MG, Stephanopoulos G. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nature communications. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Krogh Pedersen H, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton MD, Galic S, Marcinko K, Sikkema S, Pulinilkunnil T, Chen ZP, O'Neill HM, Ford RJ, Palanivel R, O'Brien M, et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat Med. 2013;19:1649–1654. doi: 10.1038/nm.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisham RN, Sylvester BE, Won H, McDermott G, DeLair D, Ramirez R, Yao Z, Shen R, Dao F, Bogomolniy F, et al. Extreme Outlier Analysis Identifies Occult Mitogen-Activated Protein Kinase Pathway Mutations in Patients With Low-Grade Serous Ovarian Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:4099–4105. doi: 10.1200/JCO.2015.62.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griss T, Vincent EE, Egnatchik R, Chen J, Ma EH, Faubert B, Viollet B, DeBerardinis RJ, Jones RG. Metformin Antagonizes Cancer Cell Proliferation by Suppressing Mitochondrial-Dependent Biosynthesis. PLoS Biol. 2015;13:e1002309. doi: 10.1371/journal.pbio.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi T, Hosono K, Takahashi H, Komiya Y, Umezawa S, Sakai E, Uchiyama T, Taniguchi L, Hata Y, Uchiyama S, et al. Metformin for chemoprevention of metachronous colorectal adenoma or polyps in post-polypectomy patients without diabetes: a multicentre double-blind, placebo-controlled, randomised phase 3 trial. Lancet Oncol. 2016 doi: 10.1016/S1470-2045(15)00565-3. [DOI] [PubMed] [Google Scholar]
- Janzer A, German NJ, Gonzalez-Herrera KN, Asara JM, Haigis MC, Struhl K. Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10574–10579. doi: 10.1073/pnas.1409844111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiralerspong S, Palla SL, Giordano SH, Meric-Bernstam F, Liedtke C, Barnett CM, Hsu L, Hung MC, Hortobagyi GN, Gonzalez-Angulo AM. Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:3297–3302. doi: 10.1200/JCO.2009.19.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM Diabetes Prevention Program Research, G. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell metabolism. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Lengyel E, Burdette JE, Kenny HA, Matei D, Pilrose J, Haluska P, Nephew KP, Hales DB, Stack MS. Epithelial ovarian cancer experimental models. Oncogene. 2014;33:3619–3633. doi: 10.1038/onc.2013.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield LM, Mukherjee A, Eckert MA, Johnson A, Mills KA, Pan S, Shridhar V, Lengyel E, Romero IL. Hyperglycemia-induced metabolic compensation inhibits metformin sensitivity in ovarian cancer. Oncotarget. 2015;6:23548–23560. doi: 10.18632/oncotarget.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sadhukhan S, Sun S, Wagner GR, Hirschey MD, Qi L, Lin H, Locasale JW. High-Resolution Metabolomics with Acyl-CoA Profiling Reveals Widespread Remodeling in Response to Diet. Mol Cell Proteomics. 2015;14:1489–1500. doi: 10.1074/mcp.M114.044859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ser Z, Locasale JW. Development and quantitative evaluation of a high-resolution metabolomics technology. Analytical chemistry. 2014;86:2175–2184. doi: 10.1021/ac403845u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, Prigaro BJ, Wood JL, Bhanot S, MacDonald MJ, et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2014;510:542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. Cancer: A most exceptional response. Nature. 2015;520:389–393. doi: 10.1038/520389a. [DOI] [PubMed] [Google Scholar]
- Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494:256–260. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nature medicine. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernicova I, Korbonits M. Metformin--mode of action and clinical implications for diabetes and cancer. Nat Rev Endocrinol. 2014;10:143–156. doi: 10.1038/nrendo.2013.256. [DOI] [PubMed] [Google Scholar]
- Romero IL, McCormick A, McEwen KA, Park S, Karrison T, Yamada SD, Pannain S, Lengyel E. Relationship of type II diabetes and metformin use to ovarian cancer progression, survival, and chemosensitivity. Obstet Gynecol. 2012;119:61–67. doi: 10.1097/AOG.0b013e3182393ab3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Horvath TL. Mitochondrial ROS signaling in organismal homeostasis. Cell. 2015;163:560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, Montminy M, Cantley LC. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Grabiner BC, Van Allen EM, Hodis E, Jacobus S, Supko JG, Stewart M, Choueiri TK, Gandhi L, Cleary JM, et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 2014;4:546–553. doi: 10.1158/2159-8290.CD-13-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nature chemical biology. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth AJ, Basu SS, Snyder NW, Mesaros C, Blair IA. Inhibition of neuronal cell mitochondrial complex I with rotenone increases lipid beta-oxidation, supporting acetylcoenzyme A levels. The Journal of biological chemistry. 2014;289:26895–26903. doi: 10.1074/jbc.M114.591354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni N, Saghatelian A, Patti GJ. Defining the metabolome: size, flux, and regulation. Molecular cell. 2015;58:699–706. doi: 10.1016/j.molcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, et al. Role of AMP-activated protein kinase in mechanism of metformin action. The Journal of clinical investigation. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong WX, Rabinowitz JD, White E. Mitochondria and Cancer. Molecular cell. 2016;61:667–676. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.