Abstract
Helminth infections of the gastrointestinal tract and lungs can lead to devastating economical losses to the pastoral based animal production. Farm animals can suffer from malnutrition, tissue damage, and blood loss resulting in impaired production traits and reproduction parameters. In Austria, pastures grazed by sheep, goats, and cattle overlap with the habitats of several species of wild cervids (roe deer, red deer, sika deer, and fallow deer) and bovids (mouflon, chamois, and ibex), and transmission of parasites between different ruminant species seems likely. A complete and updated overview on the occurrence of helminths of domestic and wild ruminants in Austria is presented. Based on these data, intersections of the host spectrum of the determined parasites were depicted. The “liability index” was applied to identify the ruminant species, which most likely transmit parasites between each other. A degree for host specificity was calculated for each parasite species based on the average taxonomic distance of their host species. Of the 73 identified helminth species 42 were identified as generalists, and 14 transmission experiments supported the assumed broad host specificity for 14 generalists and 1 specialist helminth species. Overall, 61 helminths were found to infect more than one host species, and 4 were found in all 10 ruminant species investigated. From these analyses, it can be concluded that a number of helminth parasites of the gastrointestinal tract and the lungs are potentially transmitted between domestic and wild ruminants in Austria. For some parasites and host species, experimental evidence is in support for possible transmission, while for other such studies are lacking. Host preference of different genotypes of the same parasite species may have a confounding effect on the evaluation of cross-transmission, but so far this has not been evaluated systematically in helminths in Austria. Further studies focusing on experimental cross-transmission and genetic characterization are needed to define the potential consequences for the epidemiology of those parasites, animal welfare, and economic impact.
Keywords: cervidae, bovidae, trematodes, cestodes, nematodes, liability index, biodiversity index, ABOL
Introduction
Transmission of helminths between different host species is a biological feature that is a key element for the understanding of parasite epidemiology and can play a major role for the design of strategies for the control of parasites of veterinary as well as medical importance. Transmission requires a common environment, where host species share the same resources like pasture or watering holes. In some cases, further requirements are necessary, e.g., the presence of a suitable intermediate host. In this study, we analyzed the occurrence of helminths parasitizing ruminants in Austria and evaluated the extent of parasite cross-transmission between different species of hosts. In Austria, 634.071 ha of land are used as grassland. This includes, as a specific characteristic of the alpine country Austria, 330.545 ha of mountain pasture (excluding unused areas like forests and abandoned land). Almost 2 million cattle, 380,000 sheep and 83,000 goats are reared in Austria, with around 250,000 cattle, 113,000 sheep, and 10,000 goats kept on mountain pasture.1 Seven species of wild ruminants form free-living populations in Austria, which include four species of cervids: red deer, roe deer, sika deer, and fallow deer with annual harvests (2015/2016) of 52,024, 276,222, 1,053, and 805 individuals, respectively; and three species of bovids: chamois, mouflon, and ibex with annual (2015/2016) harvests of 20,371, 2,450 and 549 individuals, respectively.2
As the habitats of red deer (1), roe deer (2), chamois (3), mouflon (4), and ibex (5) overlap with pasture of livestock (3) and protective fences are rare, contact of a particular ruminant species with the local helminth populations of the abovementioned other species on mountain pasture is possible. Fallow deer prefer habitats below 800 m above sea level (6). Sika deer often occurs in the same habitats as roe deer and fallow deer (7). As the habitat of ibex ranges on average between 1,600 and 3,200 m above sea level (5), contacts between the helminth population of fallow deer and sika deer and the helminth population of ibex are highly unlikely.
To evaluate theoretical transmission scenarios for Austria, we developed a model of likeliness of transmission of helminths between different species of ruminants and compared these findings with the results from transmission experiments described in the international literature.
Materials and Methods
Species of Ruminants Included in the Analyses
Our study included cattle (Bos primigenius taurus), sheep (Ovis gmelini aries), and goat (Capra aegagrus hircus) as domestic species and red deer (Cervus elaphus), roe deer (Capreolus capreolus), sika deer (Cervus nippon), fallow deer (Dama dama), mouflon (Ovis gmelini musimon), ibex (Capra ibex), and chamois (Rupicapra rupicapra) as wild species.
Red deer, sika deer, fallow deer, and roe deer belong to the ungulate family Cervidae (8). Red deer, sika deer, and fallow deer belong to the tribe Cervini or Old World deer (Plesiometacarpalia) (9, 10). Red deer and sika deer belong to the same genus Cervus; they can interbreed and produce fertile offspring (7, 11). Red deer is widespread in Austria from the alpine areas of the west to the eastern Danube flood lands. Sika deer and fallow deer are not native to Austria. Sika deer was introduced to Europe in the mid-nineteenth century. Its distribution in Austria is largely restricted to the lowlands of Lower Austria (12).2 Fallow deer was present in Europe until the last ice age and has been reintroduced in the sixteenth to seventeenth century; it also prefers lowland areas (13, 14). Roe deer is the most numerous and widespread wild ruminant species in Austria.2 It belongs to the tribe Capreolini or New World deer (Telemetacarpalia) (8).
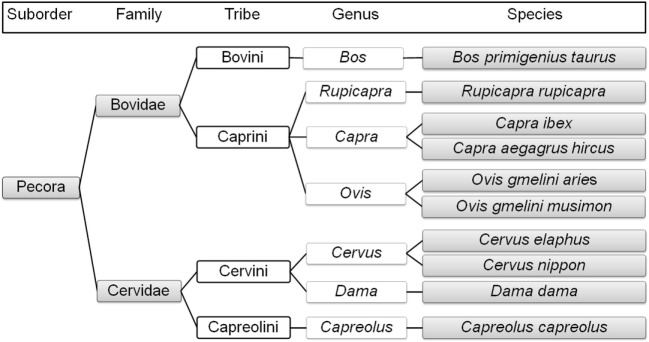
Sheep, mouflon, ibex, goat, chamois, and domestic cattle belong to the family Bovidae. Free-living mouflon, ibex, and chamois are essentially restricted to the higher-altitude regions of Austria. The tribe Caprini includes sheep, mouflon, ibex, goat, and chamois. Sheep and mouflon both belong to the genus Ovis and are close relatives, as are ibex and goat (both belonging to the genus Capra), while chamois belong to the genus Rupicapra (8). Domestic cattle belong to the genus Bos within the tribe Bovini (8, 15) (Figure 1).
Figure 1.
Phylogenetic relationships of the selected ruminant species in Austria (1–8, 11, 14, 15) created with TimeTree; www.timetree.org.
Literature Search for Helminths of the Gastrointestinal Tract and Lungs of Ruminants in Austria
Helminth species recorded for the gastrointestinal tract and lungs of ruminants in Austria were summarized from published post mortem records, prevalence surveys (only including those with records of worms determined to species) and experimental transmission studies (Table S1 in Supplementary Material). Reclassification of parasites (synonymization of names and acceptance of the concept of polymorphism) was taken into account. Literature research at the Veterinary Library of the Veterinary University Vienna and using the search engines “PubMed,”3 “Scopus,”4 and “VetmedSeeker”5 provided 27 publications, which ranged from 1969 (16) to 2014 (17).
The animals included in these records were derived from different parts of Austria, both the (pre-)alpine west and the central and eastern lowlands.
Host Specificity of the Listed Helminths
Inspired by Poulin and Mouillot (18), the biodiversity index (BI) (19) was calculated in its general form and used as a measure for host specificity of each parasite (Eq. 1). Its value increases with greater taxonomic distance (genus, tribe, family, and infraorder) which the host species, infected by the parasite in question, have to a common ancestor host and is inversely proportional to host specificity. The BI for two host species which share a common ancestor in the same genus will be 1; it will be 2 for species in the same tribe, 3 for those from the same family, and 4 for those in the same infraorder. Its value will be 0 if there is only one infected host,
| (1) |
BI—this formula (19) factors the number of host species used by a parasite (s), the average taxonomic distance (ωij), which is the mean number of steps up the Linnaean hierarchy (genus, tribe, family, and infraorder) between the host species i and j {i: 1, … s; j: 1, …, s, i ≠ j}, that must be taken to reach a taxon, which is common to both host species.
Taxonomic heterogeneity of the parasitized host species is reflected by the standard variance of the BI Λ+ (19) (Eq. 2). Its value increases proportionally to the variability in taxonomic distinctness of all infected host species. The variance is by nature not assessable if there is only one infected host species and is 0 if there are only two infected host species or under rare particular taxonomic distance combinations of infected host species.
A generalist parasite was defined to have the ability to infect host species which have common ancestors in the same host family or infraorder. A minimum of three infected host species was set to increase the informative value. Thus, the generalist parasite BI is >2 and its standard variance >0. Specific parasites infect host species with a common ancestor within the same tribe. Their BI value is ≤2, and its standard variance >0,
| (2) |
Standard variance of the BI: s: number of host species that are infected by the parasite in question, i and j: taxonomic distance between host species {i: 1, … 4; j: 1, …, 4, i ≠ j}, ωij: average taxonomic distance, : average taxonomic distinctness (≜BI).
Susceptibility of Host Species to Be Infected with Generalist Parasites of Other Hosts
The liability index L (20) was applied to measure the degree to which each ruminant species in Austria is susceptible to infection with generalist helminths from another ruminant species (Eq. 3). The value of the L index ranges from −1 to 1. Hosts with an entirely unique parasite spectrum have a liability index of −1, hosts with half-unique and half-shared parasites with another host have an index of 0, and those hosts which share all their parasites with another host have an index of 1 (20). The liability index was calculated for all ruminant species in Austria. Calculations were done with RStudio (21),
| (3) |
Liability index (20)—this formula calculates the index by dividing the difference of the number of shared parasites (SAB) by one selected host species (A) with another host species (B), and the number of parasites unique to that selected host species (UA) by the total number of parasites of that selected host species (dA).
Likelihood of Parasites to Occur in Selected Ruminant Species
Liability index, BI, and standard variance of the BI were combined to assess which ruminants are likely to be hosts for the same helminths.
It was assumed that generalist parasites of a selected host species will occur in other ruminant species if their liability index is >0. Specific parasites of a selected host species are expected to occur in other ruminants if the liability index is >0 and the host species are members of the same tribe/genus. An adapted liability index L* (Eq. 4) was developed which takes this assumed situation into account. Numbers of shared generalist, shared specialist, unique generalist and unique specialist (US) parasites were calculated between pairs of ruminant species, sharing a liability index ≥0. Parasites which could not be clearly attributed to generalist and specialist parasites were excluded from the calculation,
| (4) |
Adapted liability index for ruminants of different tribes with a liability index ≥0, A, B: compared ruminant species, SGAB: shared generalist parasites, SSAB: shared specialist parasites, UGA: unique generalist parasites, and USA: unique specialist parasites.
As US parasites are seen to be occurring in the same tribe/genus, the adapted liability index will be 1 for a pair of ruminant species of the same tribe/genus.
Furthermore, an adapted ratio for shared generalist parasites (RSGAB) and shared specialist parasites (RSSAB) was calculated to determine which of the two groups of helminths are mainly shared (Eq. 5). The results give additional indication which parasites are likely to be shared between two ruminant species,
| (5) |
Ratio of shared generalist (RSGAB) and ratio of shared specialist (RSSAB): A, B: compared ruminant species, SGAB: shared generalist parasites, SSAB: shared specialist parasites, and UGA: unique generalist parasites.
Results
Helminth Parasites of the Gastrointestinal Tract and Lungs of Ruminants in Austria
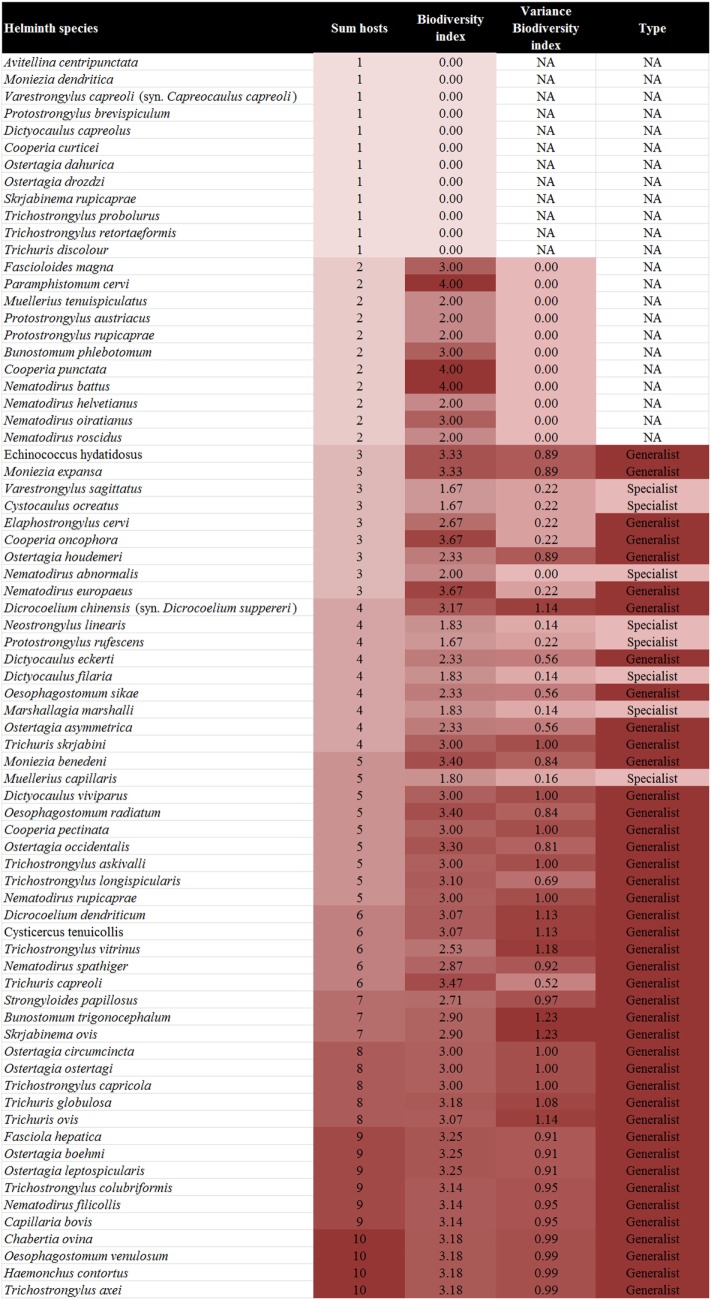
In total, 73 helminth species of the gastrointestinal tract and lung of ruminants in Austria were recorded from the literature, belonging to the families Fasciolidae (2 species), Dicrocoeliidae (2), Paramphistomidae (1), Taeniidae (2; only larval stages), Anoplocephalidae (4), Strongyloididae (1), Protostrongylidae (11), Dictyocaulidae (4), Chabertiidae (4), Ancylostomatidae (2), Trichostrongylidae (25), Molineidae (9), and Trichuridae (6) (Figure 2).
Figure 2.
Biodiversity index, variance of the biodiversity index, and type of the different helminth species by ascending number of host species.
Host Range of the Helminth Parasites Recorded
Of the 73 helminths species recorded in 10 ruminant species in Austria 21 (28.8%; sika deer) to 45 (61.6%; roe deer) have been described per ruminant host. The individual parasite species were recorded in 1–10 host species (Figure 2).
Host Specificity of the Helminth Parasites Recorded
The BI of the helminths ranged from 0 (n = 12) for species with only one host listed to four (n = 3) for helminths occurring in two hosts belonging to different families. The standard variance of the BI ranged from 0 (n = 12), for helminths infecting only two hosts (and in one case three hosts, Nematodirus abnormalis, due to the combination of hosts) to 1.23 for helminths (Bunostomum trigonocephalum and Skrjabinema ovis) infecting a range of distantly related hosts (Figure 2).
In total, 42 of 73 helminths (54.5%) had a BI > 2 with a variance of >0 and were therefore defined as generalist parasites, while seven helminth species (9.1%) had a BI ≤ 2 with a variance of >0 and therefore were defined as specialist parasites. Out of these, six species occur in the tribe Caprini and one in the tribe Cervini. One helminth species, N. abnormalis, had a BI ≤ 2 with a variance of 0 but was described from three different hosts and was therefore regarded as a specialist parasite. No clear attribution was possible for 23 (31.5%) species (Figure 2).
For a number of helminths cross-infection between ungulate species was confirmed in experimental studies; this concerned 17 generalist species, 1 specialist (Muellerius capillaris), and 4 species without clear host-type specificity (Table 1).
Table 1.
Parasites that have been successfully transmitted between ruminants in comparison with the calculated biodiversity index (BI), variance of the biodiversity index (VBI), and host specificity type (type—G, generalist; S, specialist; NA, not applicable).
| Helminth species | Cattle | Sheep | Mouflon | Ibex | Goat | Chamois | Roe deer | Red deer | Fallow deer | Sika deer | BI | VBI | Type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cestoda | |||||||||||||
| Anoplocephalidae | |||||||||||||
| Moniezia benedeni | 9 | 9 | 3.40 | 0.84 | G | ||||||||
| Nematoda | |||||||||||||
| Strongyloididae | |||||||||||||
| Strongyloides papillosus | 3, 8 | 3, 8, 13 | 13 | 13 | 2.71 | 0.97 | G | ||||||
| Protostrongylidae | |||||||||||||
| Muellerius capillaris | 4 | 4 | 1.80 | 0.16 | S | ||||||||
| Dictyocaulidae | |||||||||||||
| Dictyocaulus eckerti | 2 | 2 | 2.33 | 0.56 | G | ||||||||
| Dictyocaulus viviparus | 2, 5, 7 | 7 | 5, 7 | 2, 5 | 3.00 | 1.00 | G | ||||||
| Chabertiidae | |||||||||||||
| Oesophagostomum venulosum | 3, 7, 12 | 1, 3 | 1, 7, 12 | 3.18 | 0.99 | G | |||||||
| Trichostrongylidae | |||||||||||||
| Cooperia curticei | 3, 6, 8, 12 | 3, 6, 8, 11 | 11, 12 | 0.00 | NA | NA | |||||||
| Cooperia oncophora | 3, 6, 9, 12 | 3, 6, 9 | 1 | 12 | 3.67 | 0.22 | G | ||||||
| Cooperia pectinata | 8 | 8 | 3.00 | 1.00 | G | ||||||||
| Cooperia punctata | 3, 8, 12 | 3, 8 | 12 | 4.00 | 0.00 | NA | |||||||
| Haemonchus contortus | 3, 8, 10, 12 | 1, 3, 8, 10, 11, 13 | 7, 13 | 11, 13 | 1, 7, 13 | 7, 11, 12 | 7 | 3.18 | 0.99 | G | |||
| Ostertagia leptospicularis | 1, 7 | 1, 13 | 1, 13 | 1, 7, 13 | 7 | 3.25 | 0.91 | G | |||||
| Ostertagia boehmi | 7 | 1 | 13 | 1, 7, 13 | 7 | 3.25 | 0.91 | G | |||||
| Ostertagia circumcincta | 3 | 3, 11, 13 | 13 | 13 | 11 | 3.00 | 1.00 | G | |||||
| Ostertagia ostertagi | 3, 12 | 3 | 12 | 3.00 | 1.00 | G | |||||||
| Trichostrongylus axei | 8, 10, 12, 13 | 8, 10, 13 | 13 | 13 | 13 | 12 | 3.18 | 0.99 | G | ||||
| Trichostrongylus colubriformis | 1, 12 | 1, 13 | 13 | 1, 13 | 1, 13 | 12 | 3.14 | 0.95 | G | ||||
| Trichostrongylus vitrinus | 3 | 1, 3 | 1, 13 | 13 | 2.53 | 1.18 | G | ||||||
| Molineidae | |||||||||||||
| Nematodirus helvetianus | 8, 10 | 8, 10 | 2.00 | 0.00 | NA | ||||||||
| Nematodirus roscidus | 7 | 7 | 2.00 | 0.00 | NA | ||||||||
| Nematodirus rupicaprae | 3.00 | 1.00 | G | ||||||||||
| Trichuridae | |||||||||||||
| Trichuris capreoli | 7 | 7 | 3.47 | 0.52 | G | ||||||||
| Trichuris ovis | 8, 9 | 8, 9 | 3.07 | 1.14 | G | ||||||||
|
Number 1 2 3 4 5 6 7 |
Citation Barth and Dollinger (22) Bienioschek et al. (24) Borgsteede (26) Ten Doesschate et al. (28) Foreyt et al. (30) Johnson et al. (32) Koutz and Rebrassier (34) |
Number 8 9 10 11 12 13 |
Citation Kutzer (23) Porter (25) Smith and Archibald (27) Stoll (29) Tapia-Escárate et al. (31) Wetzel and Fortmeyer (33) |
||||||||||
Susceptibility of Host Species to Helminth Parasites
The liability index for the one-to-one comparison between individual ruminant species ranged from −0.55 (goat–sika deer) to 1.000 (goat–sheep and fallow deer–roe deer) with a mean of 0.19 and a median of 0.13. The mean number of helminth species shared between two ruminant species was 19 (range 5–34, median: 19), the mean number of unique helminth species was 14 (range 0–33, median: 13) (Table 2).
Table 2.
Shared and unique helminths species and liability indices for the pairs of ruminant species included in the present study.
| Shared | Shared G | Shared S | S, no clear attribution | Unique | Unique G | Unique S | Unique, no clear attribution | D | Ratio shared G:S | Ratio shared S:G | L | L* | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cattle–sheep | 19 | 18 | 0 | 1 | 7 | 4 | 0 | 3 | 26 | 1 | 0 | 0.46 | 1 |
| Cattle–mouflon | 16 | 16 | 0 | 0 | 10 | 6 | 0 | 4 | 26 | 1 | 0 | 0.23 | 1 |
| Cattle–ibex | 14 | 14 | 0 | 0 | 12 | 8 | 0 | 4 | 26 | 1 | 0 | 0.08 | 1 |
| Cattle–goat | 13 | 13 | 0 | 0 | 13 | 9 | 0 | 4 | 26 | 1 | 0 | 0.00 | 1 |
| Cattle–chamois | 19 | 18 | 0 | 1 | 7 | 4 | 0 | 3 | 26 | 1 | 0 | 0.46 | 1 |
| Cattle–roe deer | 22 | 21 | 0 | 1 | 4 | 1 | 0 | 3 | 26 | 1 | 0 | 0.69 | 1 |
| Cattle–fallow deer | 13 | 13 | 0 | 0 | 13 | 9 | 0 | 4 | 26 | 1 | 0 | 0.00 | 1 |
| Cattle–red deer | 19 | 19 | 0 | 0 | 7 | 3 | 0 | 4 | 26 | 1 | 0 | 0.46 | 1 |
| Cattle–sika deer | 11 | 11 | 0 | 0 | 15 | 11 | 0 | 4 | 26 | 1 | 0 | −0.15 | NA |
| Sheep–cattle | 19 | 18 | 0 | 1 | 21 | 10 | 6 | 5 | 40 | 1 | 0 | −0.05 | NA |
| Sheep–mouflon | 27 | 22 | 5 | 0 | 13 | 6 | 1 | 6 | 40 | 0.85 | 0.15 | 0.35 | 1 |
| Sheep–ibex | 28 | 23 | 4 | 1 | 12 | 5 | 2 | 5 | 40 | 0.88 | 0.13 | 0.40 | 1 |
| Sheep–goat | 22 | 17 | 5 | 0 | 18 | 11 | 1 | 6 | 40 | 0.85 | 0.15 | 0.10 | 1 |
| Sheep–chamois | 31 | 26 | 4 | 1 | 9 | 2 | 2 | 5 | 40 | 0.88 | 0.13 | 0.55 | 1 |
| Sheep–roe deer | 26 | 26 | 0 | 0 | 14 | 2 | 6 | 6 | 40 | 1 | 0 | 0.30 | 0.647 |
| Sheep–fallow deer | 13 | 12 | 0 | 1 | 27 | 16 | 6 | 5 | 40 | 1 | 0 | −0.35 | NA |
| Sheep–red deer | 22 | 22 | 0 | 0 | 18 | 6 | 6 | 6 | 40 | 1 | 0 | 0.10 | 0.647 |
| Sheep–sika deer | 10 | 10 | 0 | 0 | 30 | 18 | 6 | 6 | 40 | 1 | 0 | −0.50 | NA |
| Mouflon–cattle | 16 | 16 | 0 | 0 | 16 | 10 | 5 | 2 | 32 | 1 | 0 | 0.00 | NA |
| Mouflon–sheep | 27 | 22 | 5 | 0 | 5 | 4 | 0 | 2 | 32 | 0.84 | 0.16 | 0.69 | 1 |
| Mouflon–ibex | 23 | 20 | 3 | 0 | 9 | 6 | 1 | 3 | 32 | 0.9 | 0.1 | 0.44 | 1 |
| Mouflon–goat | 21 | 17 | 4 | 0 | 11 | 9 | 1 | 2 | 32 | 0.87 | 0.13 | 0.31 | 1 |
| Mouflon–chamois | 27 | 24 | 3 | 0 | 5 | 2 | 2 | 2 | 32 | 0.9 | 0.1 | 0.69 | 1 |
| Mouflon–roe deer | 26 | 26 | 0 | 1 | 6 | 0 | 5 | 1 | 32 | 1 | 0 | 0.63 | 0.677 |
| Mouflon–fallow deer | 14 | 14 | 0 | 0 | 18 | 12 | 5 | 2 | 32 | 1 | 0 | −0.13 | NA |
| Mouflon–red deer | 22 | 21 | 0 | 1 | 10 | 5 | 5 | 1 | 32 | 1 | 0 | 0.38 | 0.677 |
| Mouflon–sika deer | 11 | 11 | 0 | 0 | 21 | 15 | 5 | 2 | 32 | 1 | 0 | −0.31 | NA |
| Ibex–cattle | 14 | 14 | 0 | 0 | 19 | 9 | 4 | 6 | 33 | 1 | 0 | −0.15 | NA |
| Ibex–sheep | 28 | 23 | 4 | 1 | 5 | 0 | 0 | 5 | 33 | 0.85 | 0.15 | 0.70 | 1 |
| Ibex–mouflon | 23 | 20 | 3 | 0 | 10 | 3 | 1 | 6 | 33 | 0.88 | 0.12 | 0.39 | 1 |
| Ibex–goat | 18 | 15 | 3 | 0 | 15 | 8 | 1 | 6 | 33 | 0.88 | 0.12 | 0.09 | 1 |
| Ibex–chamois | 32 | 23 | 3 | 6 | 1 | 0 | 1 | 0 | 33 | 0.88 | 0.12 | 0.94 | 1 |
| Ibex–roe deer | 23 | 23 | 0 | 0 | 10 | 0 | 4 | 6 | 33 | 1 | 0 | 0.39 | 0.704 |
| Ibex–fallow deer | 11 | 11 | 0 | 0 | 22 | 12 | 4 | 6 | 33 | 1 | 0 | −0.33 | NA |
| Ibex–red deer | 18 | 18 | 0 | 0 | 15 | 5 | 4 | 6 | 33 | 1 | 0 | 0.09 | 0.704 |
| Ibex–sika deer | 8 | 8 | 0 | 0 | 25 | 15 | 4 | 6 | 33 | 1 | 0 | −0.52 | NA |
| Goat–cattle | 13 | 13 | 0 | 0 | 9 | 4 | 5 | 0 | 22 | 1 | 0 | 0.18 | 0.545 |
| Goat–sheep | 22 | 17 | 5 | 0 | 0 | 0 | 0 | 0 | 22 | 0.77 | 0.23 | 1.00 | 1 |
| Goat–mouflon | 21 | 17 | 4 | 0 | 1 | 0 | 1 | 0 | 22 | 0.81 | 0.19 | 0.91 | 1 |
| Goat–ibex | 18 | 15 | 3 | 0 | 4 | 2 | 2 | 0 | 22 | 0.85 | 0.15 | 0.64 | 1 |
| Goat–chamois | 20 | 17 | 1 | 2 | 2 | 0 | 2 | 0 | 22 | 0.94 | 0.06 | 0.82 | 1 |
| Goat–roe deer | 17 | 17 | 0 | 0 | 5 | 0 | 5 | 0 | 22 | 1 | 0 | 0.55 | 0.545 |
| Goat–fallow deer | 9 | 9 | 0 | 0 | 13 | 8 | 5 | 0 | 22 | 1 | 0 | −0.18 | NA |
| Goat–red deer | 14 | 14 | 0 | 0 | 8 | 3 | 5 | 0 | 22 | 1 | 0 | 0.27 | 0.545 |
| Goat–sika deer | 5 | 5 | 0 | 0 | 17 | 12 | 5 | 0 | 22 | 1 | 0 | −0.55 | NA |
| Chamois–cattle | 19 | 18 | 0 | 1 | 25 | 13 | 4 | 8 | 44 | 1 | 0 | −0.14 | NA |
| Chamois–sheep | 31 | 26 | 4 | 1 | 13 | 5 | 0 | 8 | 44 | 0.89 | 0.11 | 0.41 | 1 |
| Chamois–mouflon | 27 | 24 | 3 | 0 | 17 | 7 | 1 | 9 | 44 | 0.91 | 0.09 | 0.23 | 1 |
| Chamois–ibex | 32 | 23 | 3 | 6 | 12 | 8 | 1 | 3 | 44 | 0.91 | 0.09 | 0.45 | 1 |
| Chamois–goat | 20 | 17 | 3 | 0 | 24 | 14 | 1 | 9 | 44 | 0.91 | 0.09 | −0.09 | NA |
| Chamois–roe deer | 30 | 30 | 0 | 0 | 14 | 1 | 4 | 9 | 44 | 1 | 0 | 0.36 | 0.771 |
| Chamois–fallow deer | 14 | 14 | 0 | 0 | 30 | 17 | 4 | 9 | 44 | 1 | 0 | −0.36 | NA |
| Chamois–red deer | 24 | 24 | 0 | 0 | 20 | 7 | 4 | 9 | 44 | 1 | 0 | 0.09 | 0.771 |
| Chamois–sika deer | 11 | 11 | 0 | 0 | 33 | 20 | 4 | 9 | 44 | 1 | 0 | −0.50 | NA |
| Roe deer–cattle | 22 | 21 | 0 | 1 | 23 | 15 | 4 | 6 | 45 | 1 | 0 | −0.02 | NA |
| Roe deer–sheep | 26 | 26 | 0 | 0 | 19 | 10 | 4 | 7 | 45 | 1 | 0 | 0.16 | 0.8 |
| Roe deer–mouflon | 26 | 26 | 0 | 1 | 19 | 10 | 4 | 6 | 45 | 1 | 0 | 0.16 | 0.8 |
| Roe deer–ibex | 23 | 23 | 0 | 0 | 22 | 13 | 4 | 7 | 45 | 1 | 0 | 0.02 | NA |
| Roe deer–goat | 17 | 17 | 0 | 0 | 28 | 19 | 4 | 7 | 45 | 1 | 0 | −0.24 | NA |
| Roe deer–chamois | 30 | 30 | 0 | 0 | 15 | 6 | 4 | 7 | 45 | 1 | 0 | 0.33 | 0.8 |
| Roe deer–fallow deer | 21 | 18 | 3 | 0 | 24 | 18 | 0 | 8 | 45 | 0.92 | 0.08 | −0.07 | NA |
| Roe deer–red deer | 34 | 29 | 4 | 1 | 11 | 7 | 1 | 5 | 45 | 0.9 | 0.1 | 0.51 | 0.951 |
| Roe deer–sika deer | 20 | 16 | 4 | 0 | 25 | 20 | 0 | 7 | 45 | 0.9 | 0.1 | −0.11 | NA |
| Fallow deer–cattle | 13 | 13 | 0 | 0 | 12 | 5 | 5 | 4 | 25 | 1 | 0 | 0.04 | NA |
| Fallow deer–sheep | 13 | 12 | 0 | 1 | 12 | 6 | 5 | 3 | 25 | 1 | 0 | 0.04 | NA |
| Fallow deer–mouflon | 14 | 14 | 0 | 0 | 11 | 4 | 5 | 4 | 25 | 1 | 0 | 0.12 | 0.565 |
| Fallow deer–ibex | 11 | 11 | 0 | 0 | 14 | 7 | 5 | 4 | 25 | 1 | 0 | −0.12 | NA |
| Fallow deer–goat | 9 | 9 | 0 | 0 | 16 | 9 | 5 | 4 | 25 | 1 | 0 | −0.28 | NA |
| Fallow deer–chamois | 14 | 14 | 0 | 0 | 11 | 4 | 5 | 4 | 25 | 1 | 0 | 0.12 | 0.565 |
| Fallow deer–roe deer | 25 | 18 | 3 | 6 | 0 | 0 | 1 | −1 | 25 | 0.86 | 0.14 | 1.00 | 0.909 |
| Fallow deer–red deer | 22 | 18 | 4 | 0 | 3 | 0 | 1 | 4 | 25 | 0.82 | 0.18 | 0.76 | 1 |
| Fallow deer–sika deer | 16 | 12 | 4 | 0 | 9 | 6 | 0 | 5 | 25 | 0.82 | 0.18 | 0.28 | 1 |
| Red deer–cattle | 19 | 19 | 0 | 0 | 20 | 12 | 4 | 4 | 39 | 1 | 0 | −0.03 | NA |
| Red deer–sheep | 22 | 22 | 0 | 0 | 17 | 9 | 4 | 4 | 39 | 1 | 0 | 0.13 | 0.771 |
| Red deer–mouflon | 22 | 21 | 0 | 1 | 17 | 10 | 4 | 3 | 39 | 1 | 0 | 0.13 | 0.771 |
| Red deer–ibex | 18 | 18 | 0 | 0 | 21 | 13 | 4 | 4 | 39 | 1 | 0 | −0.08 | NA |
| Red deer–goat | 14 | 14 | 0 | 0 | 25 | 17 | 4 | 4 | 39 | 1 | 0 | −0.28 | NA |
| Red deer–chamois | 24 | 24 | 0 | 0 | 15 | 7 | 4 | 4 | 39 | 1 | 0 | 0.23 | 0.771 |
| Red deer–roe deer | 34 | 29 | 3 | 2 | 5 | 2 | 1 | 2 | 39 | 0.91 | 0.09 | 0.74 | 0.943 |
| Red deer–fallow deer | 22 | 17 | 5 | 0 | 17 | 13 | 0 | 4 | 39 | 0.86 | 0.14 | 0.13 | 1 |
| Red deer–sika deer | 20 | 15 | 4 | 1 | 19 | 16 | 0 | 3 | 39 | 0.89 | 0.11 | 0.03 | 1 |
| Sika deer–cattle | 11 | 11 | 0 | 0 | 10 | 5 | 5 | 0 | 21 | 1 | 0 | 0.05 | 0.524 |
| Sika deer–sheep | 10 | 10 | 0 | 0 | 11 | 6 | 5 | 0 | 21 | 1 | 0 | −0.05 | NA |
| Sika deer–mouflon | 11 | 11 | 0 | 0 | 10 | 5 | 5 | 0 | 21 | 1 | 0 | 0.05 | 0.524 |
| Sika deer–ibex | 8 | 8 | 0 | 0 | 13 | 8 | 5 | 0 | 21 | 1 | 0 | −0.24 | NA |
| Sika deer–goat | 5 | 5 | 0 | 0 | 16 | 11 | 5 | 0 | 21 | 1 | 0 | −0.52 | NA |
| Sika deer–chamois | 11 | 11 | 0 | 0 | 10 | 5 | 5 | 0 | 21 | 1 | 0 | 0.05 | 0.524 |
| Sika deer–roe deer | 20 | 16 | 4 | 0 | 1 | 0 | 1 | 0 | 21 | 0.8 | 0.2 | 0.90 | 1 |
| Sika deer–fallow deer | 16 | 12 | 3 | 1 | 5 | 4 | 1 | 0 | 21 | 0.84 | 0.16 | 0.52 | 1 |
| Sika deer–red deer | 20 | 15 | 5 | 0 | 1 | 1 | 0 | 0 | 21 | 0.76 | 0.24 | 0.90 | 1 |
G, generalist; S, specialist; L, liability index; L*, adapted liability index.
For 60 out of 90 compared ruminant species pairs the liability index was ≥0. The mean number of shared generalist helminth species was 17 (range 5–30, median: 17), the mean number of shared specialists was 1.17 (range 0–5, median: 0), the mean number of unique generalists was 8.4 (range 0–23, median: 8.0), and the mean number of USs was 1.8 (range 0–6, median: 1). The adapted liability index (L*) for the 59 ruminant combinations ranged from 0.55 (goat–cattle, goat–roe deer, and goat–red deer) to 1.0 (n = 23 pairs of ruminants) with a mean of 0.9 and a median of 0.94. No calculation of an adapted liability index was possible for 27 species pairs (Table 2).
The ratio of shared generalist helminth species between ruminant species pairs ranged from 0.77 (goat–sheep) to 1.0 (n = 58.0 species pairs) with a mean of 0.96 and median of 1. The ratio of shared specialist helminths between ruminant species pairs ranged from 0 (n = 58 pairs) to 0.23 (goat–sheep) with a mean of 0.04 and median of 0 (Table 2).
Discussion
The transmission of pathogens including helminths between host species of different taxa has implications in several directions, including ecology and epidemiology. In our work, we addressed the question whether possible transmission of helminths from one host species to another sharing the same environment can be estimated by calculating different indices, testing this assumption by comparison with the available literature.
Liability index (20), BI, and standard variance of the BI (19) were calculated to determine which helminth species are likely to occur in the most abundant ruminant species in Austria.
Literature search focused only on reports of helminths in these species in Austria and provided 27 articles dating back to the year 1969. As actual comprehensive data about helminth parasites of ruminants in Austria is scarce, we first aimed to provide an overview on this. The results of our analysis suggest, however, that more helminth species may occur and that the host range of described helminth species is larger than described so far. Since the number of available publications for Austria was rather low the evaluation of the data must be seen as preliminary; nevertheless we could provide calculations which can be used to test hypotheses arising from them.
The liability index (20) was used to test which pairs of hosts are most likely to be susceptible for the same parasites. In general, the results of the liability index calculation were as expected: phylogenetically closely related ruminants with similar habitat requirements (e.g., sheep and mouflon) share a high liability index value (≥0.5 = a ruminant pair shares ≥ 75% of their parasites), whereas phylogenetically more distant ruminants with distinct habitats (e.g., chamois and sika deer) share a low liability index (≤0 = a ruminant pair shares less than 50% of all parasites). The outcome of this analysis was unexpectedly low for the pairs cattle–goat (0) and sheep–cattle (−0.05). As the liability index is influenced by the number of helminth species detected which is correlated with the number of hosts examined (e.g., fallow deer and mouflon are less well examined than other species), an adaptation of the liability index was developed. It was hypothesized that ruminant species pairs with a liability index ≥0 will additionally share generalist parasites which have the ability to infect hosts belonging to different host families. Furthermore, it was assumed that ruminant species pairs belonging to the same tribe/genus with a liability index ≥0 will additionally share specific helminths which have the ability to infect closely related host species.
As a measure of host specificity, the BI and its standard variance (18, 19) were calculated for the 73 described helminth species. It can essentially be assumed that host specificity of parasites increases with decreasing taxonomic distinctness between their host species (18). Out of 73 helminth species recorded, 42 were identified as generalists, and 8 were classified as specialists. Four of the generalist species (Chabertia ovina, Oesophagostomum venulosum, Haemonchus contortus, and Trichostrongylus axei) were described in all 10 ruminants occurring in Austria. Overall, 38 parasites may have the ability to infect more ruminant species than previously described. These should be in the focus for further investigations.
The adapted liability index was calculated for 59 pairs of ruminant species (liability index ≥0), by including the 42 generalist helminths and 8 specialist helminth species. Compared with the (conventional) liability index, the results of the adapted liability index provided higher values in all categories. An increase compared with the value of the conventional liability index was expected, as mainly generalist helminth species were identified in ruminants in Austria. From these findings we conclude that Austrian ruminants have a far more higher risk to be infected by generalist helminths than previously reported [see Table S1 in Supplementary Material for literature and Ref. (16)].A particularly interesting result of this analysis is that based on these calculations cattle were shown to be susceptible to all generalist parasites of all other ruminants in Austria.
The ratio of shared generalists and specialists also revealed tendencies of specialist helminths to occur in a pair of ruminant species. Generally, the ratio shows that mainly generalists (range: 0.77–1.00; mean: 0.96), and only a few specialists (range: 0.00–0.23; mean: 0.04) are shared by ruminant species pairs in Austria. The latter finding indicates the difficulties in unequivocal assigning “specialism” or “generalism” to a particular helminth species.
Overview about the helminth fauna of wild and domestic ruminants in Austria is provided in older literature (23, 35, 36). Additional work on analyzing the helminth fauna of ruminants in the country is necessary to complete the helminth inventory of Austrian ruminants and to further test the hypothesis arising from our analyses.
From an epidemiological point of view, (pre-)alpine mountain pastures can be considered as environment where transmission of helminth parasites between the same and different species are fostered by different factors. The practice of pasturing cattle, goats, and sheep originating from different farms and regions on shared mountain pasture over centuries favors the occurrence of a divergent local population of parasitic helminths. Wild ruminant species that occur in these habitats, such as red deer, chamois, and ibex, may further contribute to the cross-transmission of helminths in such areas by increasing the number of helminth species and sustaining infection cycles.
By contrast, lowland pasture areas of central and eastern Austria are mostly grazed by single species and are usually fenced, limiting the contact between domestic and wild ruminants. Differing spatial host distribution patterns limit the availability of hosts in specific locations, and the actual infection status of a host species may therefore be very different from what is proposed by a model only taking into account susceptibility and not spatial distribution of the host(s) (37–39). While some of the wild ungulate species included in the present study have a rather limited range (s. introduction) others are widely distributed in Austria; however, data on the exact range and density of wild ungulate species are not available and therefore could not be included in the index calculations.
From an epidemiological point of view, the specific risk for a herd or flock of domestic ruminants to become infected with helminths derived from other host species is therefore determined by the presence or absence of the latter in a particular area. This is also highlighted by the unexpectedly low L index for cattle-goat (Table 2), as these species are usually not pastured together. Thus, management of domestic animals (including range restrictions) can be a confounding factor for the estimation of the actual transmission risk between these hosts. By contrast, wild ruminants are free-ranging and may graze large geographic areas including domestic animals’ pastures where it is frequently observed (40).
As the occurrence of the same parasites in different host species does not necessarily indicate transmission from one species to another (22), the calculated scenarios were compared with the results of experimental infection studies. In summary, results of 14 transmission experiments [cf. Table 1 and Ref. (41), for literature] support the calculated model. Especially, trichostrongylids (most of all H. contortus) could frequently be transmitted between different species of domestic and wild ungulates, but also O. venulosum, Nematodirus helvetianus, and Trichuris ovis and lungworms of the genus Dictyocaulus were cross-transmitted. Red deer and roe deer were most frequently implemented in the cross-transmission between wild and domestic ruminants (Table 1). Apart from the host species mentioned in this study, M. capillaris was transmissible from goats to bighorn sheep (30) and from white-tailed deer (Odocoileus virginianus) to cattle and lambs in an inoculation experiment (41).
In total, 22 helminths (Table 1) were successfully transmitted between several ruminant species. Seventeen out of these 22 helminth species were previously defined as generalists, one species (M. capillaris) was transmitted between sheep and goat and previously identified as tribe specialist. Cooperia curticei and Cooperia punctata, which were unassigned in our prediction due to the lack of data, were attributed further to the generalists, as successful transmission between cattle, sheep, and red deer was shown (25, 26, 28, 34, 42). No attribution could be done for two helminths, Nematodirus roscidus and N. helvetianus. Overall the results of the transmission experiments are in accordance with the predictions made in the present analysis, but further analyses will be necessary to evaluate and confirm all cases properly.
Since many helminth taxa have notoriously poor morphological characteristics (even at the adult stage) misidentification should be considered as a potential confounding effect of studies determining the helminth fauna of wildlife. DNA barcoding could provide additional data that can be used to exactly determine the helminth fauna of different hosts but data are scarce (43). Cryptic parasite species have been suggested previously [for review, see Ref. (44)], including species of trichostrongylids from ungulates (45–47). Especially specimens of the Trichostrongylus and members of the Ostertagiinae subfamily are notoriously difficult to determine to species level, and a number of different names (valid or invalid) can be found in the literature which further complicate species assignment (48). T. axei seems to be a real generalist as has been shown by transmission experiments (see above), and this is corroborated by its genetic population structure with high diversity and high gene flow between sympatric hosts (49). In lungworms of the genus Dictyocaulus from wild ungulates from Hungary different gene flow rates suggested different host range capacities in the three species Dictyocaulus eckerti, Dictyocaulus capreolus, and Dictyocaulus viviparus with D. eckerti being the most generalist species, while D. capreolus seems to have a cryptic genetic structure (50). As red deer was shown to harbor lungworms, which were genetically distinct from the other Dictyocaulus species, a new name, Dictyocaulus cervi, was proposed recently (51).
Although considerable genetic differences were found in H. contortus of sheep, goat, chamois, roe deer, and ibex in alpine areas, the lack of correlation between different mitochondrial clusters with host species indicates that H. contortus is a generalist parasite species (52).
Another genus that is known for its diversity despite morphological similarities is Trichuris, and it has previously been suggested that T. discolor may be a species complex (53).
In the trematode species found in our search, different levels of genetic diversity were found; Fasciola hepatica is highly divergent (54) whereas Fascioloides magna is an imported fluke in Austria (55) with limited genetic diversity (56).
These examples of differing genetic population structures of helminths, implying the presence of cryptic species with different host preferences, are not exhaustive but highlight the difficulties in determining the exact associations of species/genotypes to host species and the underlying transmission scenarios. Genotyping seems to be the appropriate prerequisite for correct assignment of parasites to their hosts. The “Austrian Barcode of Life” initiative is dedicated to barcode all major taxa of animals, including helminths, in Austria (ABOL6). It has contributed so far with 112 sequences from 23 different helminths for DNA barcodes at the cytochrome oxidase I mitochondrial locus (compared with 5 sequences that were available for Austria in the Barcode of Life Data Systems7).
With the present study and the proposed model we intend to guide future research interest toward the generalist helminths species of ruminants in Austria to unravel their true host associations and population structures. This is not only of ecological interest but also of relevance for veterinary medicine as the effective control of helminths in livestock also depends on the range of parasites in wildlife. In principle, wild ungulates are not legally accessible for anthelmintic treatment [although exceptions have been made for, e.g., control of F. magna in Austria, see Ref. (55)]. Therefore pasture contamination by helminth eggs or larvae from feces of, e.g., free-ranging deer may be considerable and must be taken into account when judging infection risks for domestic livestock. This is particularly important in areas where unfenced pastures are utilized for transhumance in alpine regions, but also applies to lowland pasture with high densities of roe deer populations crossing fields regularly. Such interphases between wild and domestic animals are of particular importance for the exchange of parasite communities and host utilization by parasites and should also be in the focus of studies testing the proposed model for ruminant helminths in Austria.
Author Contributions
JW, SR, and AJ planned the project; JW drafted the manuscript, AJ and SR revised it. All the authors agreed on the final version of the submitted manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was funded by the Institute of Parasitology, Vetmeduni Vienna.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/articles/10.3389/fvets.2018.00030/full#supplementary-material.
References
- 1.Bützler W. Cervus elaphus Linnaeus, 1758 – Rothirsch. In: Bohlken H, Niethammer J, Krapp F, editors. Handbuch der Säugetiere Europas: Band II: Paarhufer-Artiodactyla (Suidae, Cervidae, Bovidae). Wiesbaden: AULA-Verlag; (1986). 115, 125 p. [Google Scholar]
- 2.von Lehmann E, Sägesser H. Capreolus capreolus Linnaeus, 1758-Reh. In: Bohlken H, Niethammer J, Krapp F, editors. Handbuch der Säugetiere Europas: Band II: Paarhufer-Artiodactyla (Suidae, Cervidae, Bovidae). Wiesbaden: AULA-Verlag; (1986). 240, 254 p. [Google Scholar]
- 3.Sägesser H, Krapp F. Rupicapra rupicapra (Linnaeus, 1758)-Gemse, Gams. In: Bohlken H, Niethammer J, Krapp F, editors. Handbuch der Säugetiere Europas: Band II: Paarhufer-Artiodactyla (Suidae, Cervidae, Bovidae). Wiesbaden: AULA-Verlag; (1986). p. 324, 37–8. [Google Scholar]
- 4.Röhrs M. Ovis ammon musimon (Pallas, 1811)-Mufflon. In: Bohlken H, Niethammer J, Krapp F, editors. Handbuch der Säugetiere Europas: Band II: Paarhufer-Artiodactyla (Suidae, Cervidae, Bovidae). Wiesbaden: AULA-Verlag; (1986). p. 439–42, 444. [Google Scholar]
- 5.Nievergelt B, Zingg R. Capra ibex Linnaeus, 1758-Steinbock. In: Bohlken H, Niethammer J, Krapp F, editors. Handbuch der Säugetiere Europas: Band II: Paarhufer-Artiodactyla (Suidae, Cervidae, Bovidae). Wiesbaden: AULA-Verlag; (1986). p. 392–3, 397–8. [Google Scholar]
- 6.Heidemann G. Cervus dama (Linnaeus, 1758)-Damhirsch. In: Bohlken H, Niethammer J, Krapp F, editors. Handbuch der Säugetiere Europas: Band II: Paarhufer-Artiodactyla (Suidae, Cervidae, Bovidae). Wiesbaden: AULA-Verlag; (1986). p. 144–7, 151. [Google Scholar]
- 7.Krapp F, Niethammer J. Cervus nippon Temminck, 1836-Sikahirsch. In: Bohlken H, Niethammer J, Krapp F, editors. Handbuch der Säugetiere Europas: Band II: Paarhufer-Artiodactyla (Suidae, Cervidae, Bovidae). Wiesbaden: AULA-Verlag; (1986). p. 163–7, 169. [Google Scholar]
- 8.Wilson DE, Reeder DM. Mammal Species of the World. A Taxonomic and Geographic Reference. 3rd ed Baltimore: Johns Hopkins Univ. Press; (2005). [Google Scholar]
- 9.di Stefano G, Petronio C. Systematics and evolution of the Eurasian Plio-Pleistocene tribe Cervini (Artiodactyla, Mammalia). Geol Romana (2002) 36:311–34. [Google Scholar]
- 10.Emerson BC, Tate ML. Genetic analysis of evolutionary relationships among deer (subfamily Cervinae). J Hered (1990) 84(4):266–73. 10.1093/oxfordjournals.jhered.a111338 [DOI] [PubMed] [Google Scholar]
- 11.Senn HV, Pemberton JM. Variable extent of hybridization between invasive sika (Cervus nippon) and native red deer (C. elaphus) in a small geographical area. Mol Ecol (2009) 18(5):862–76. 10.1111/j.1365-294X.2008.04051.x [DOI] [PubMed] [Google Scholar]
- 12.Reimoser F, Reimoser S. Ungulates and their management in Austria. In: Apollonio M, Andersen R, Putman R, editors. European Ungulates and their Management in the 21st Century. Cambridge: Cambridge University Press; (2010). p. 338–56. [Google Scholar]
- 13.Geiter O, Homma S, Kinzelbach R. Bestandsaufnahme und Bewertung von Neozoen in Deutschland. Umweltforschungsplan des Bundesministeriums Für Umwelt, Naturschutz und Reaktorsicherheit. Forschungsbericht (2002) 296(89):901. [Google Scholar]
- 14.Ueckermann E, Hansen P. Das Damwild: Biologie, Hege und Jagd. Frankfurt: Kosmos-Verlag; (2002). [Google Scholar]
- 15.Bibi F. A multi-calibrated mitochondrial phylogeny of extant Bovidae (Artiodactyla, Ruminantia) and the importance of the fossil record to systematics. BMC Evol Biol (2013) 13(1):166. 10.1186/1471-2148-13-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kutzer E, Knaus E. Untersuchungen über die Endoparasitenfauna eines Rehbestandes in der freien Wildbahn. Z Jagdwiss (1969) 15(2):62–72. 10.1007/BF01917819 [DOI] [Google Scholar]
- 17.Rehbein S, Visser M, Jekel I, Silaghi C. Endoparasites of the fallow deer (Dama dama) of the Antheringer Au in Salzburg, Austria. Wien Klin Wochenschr (2014) 126(Suppl 1):37–41. 10.1007/s00508-014-0506-8 [DOI] [PubMed] [Google Scholar]
- 18.Poulin R, Mouillot D. Parasite specialization from a phylogenetic perspective: a new index of host specificity. Parasitology (2003) 126(5):473–80. 10.1017/S0031182003002993 [DOI] [PubMed] [Google Scholar]
- 19.Clarke KR, Warwick RM. A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar Ecol Prog Ser (2001) 216:265–78. 10.3354/meps216265 [DOI] [Google Scholar]
- 20.Walker JG, Morgan ER. Generalists at the interface: nematode transmission between wild and domestic ungulates. Int J Parasitol Parasites Wildl (2014) 3(3):242–50. 10.1016/j.ijppaw.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.RStudio Team. RStudio: Integrated Development for R. Boston, MA: RStudio, Inc. (2016). Available from: http://www.rstudio.com/ [Google Scholar]
- 22.Barth D, Dollinger P. Zur Wirtsspezifität der Magen-Darm-Nematoden von Reh, Schaf und Rind. Z Jagdwiss (1975) 21(3):164–82. [Google Scholar]
- 23.Kutzer E. Bedeutung Parasitärer Wechselinfektionen bei Haus- und Wildwiederkäuern. Monatsh Vetmed (1988) 43:577–80. [Google Scholar]
- 24.Bienioschek S, Rehbein S, Ribbeck R. Cross-infections between fallow deer and domestic ruminants with large lungworms (Dictyocaulus spp.). Appl Parasitol (1996) 37(4):229–38. [PubMed] [Google Scholar]
- 25.Porter DA. Cross transmission of parasitic worms between cattle and sheep. Am J Vet Res (1953) 14(53):550–4. [PubMed] [Google Scholar]
- 26.Borgsteede FHM. Experimental cross-infections with gastrointestinal nematodes of sheep and cattle. Z Parasitenkd (1981) 65(1):1–10. 10.1007/BF00926548 [DOI] [PubMed] [Google Scholar]
- 27.Smith HJ, Archibald RM. Cross transmission of bovine parasites to sheep. Can Vet J (1965) 6(4):91. [PMC free article] [PubMed] [Google Scholar]
- 28.Ten Doesschate SJ, Pomroy WE, Tapia-Escárate D, Scott I, Wilson PR. Establishment rate of cattle gastrointestinal nematodes in farmed red deer (Cervus elaphus). Vet Parasitol (2017) 243:105–8. 10.1016/j.vetpar.2017.06.016 [DOI] [PubMed] [Google Scholar]
- 29.Stoll NR. Observations on cattle nematode infections, with a demonstration of their secondary transmission to grazing sheep. J Parasitol (1936) 22(4):386–407. 10.2307/3271863 [DOI] [Google Scholar]
- 30.Foreyt WJ, Jenkins EJ, Appleyard GD. Transmission of lungworms (Muellerius capillaris) from domestic goats to bighorn sheep on common pasture. J Wildl Dis (2009) 45(2):272–8. 10.7589/0090-3558-45.2.272 [DOI] [PubMed] [Google Scholar]
- 31.Tapia-Escárate D, Pomroy WE, Scott I, Wilson PR, Lopez-Villalobos N. Establishment rate of sheep gastrointestinal nematodes in farmed red deer (Cervus elaphus). Vet Parasitol (2015) 209(1–2):138–41. 10.1016/j.vetpar.2014.12.035 [DOI] [PubMed] [Google Scholar]
- 32.Johnson M, Mackintosh CG, Labes RE, Taylor MJ, Wharton DA. Dictyocaulus species: cross infection between cattle and red deer. N Z Vet J (2003) 51(2):93–8. 10.1080/00480169.2003.36346 [DOI] [PubMed] [Google Scholar]
- 33.Wetzel R, Fortmeyer HP. Experimentelle Untersuchungen über die Wirtsspezifität parasitischer Rundwürmer des jagdbaren Wildes. Z Parasitenkd (1964) 25(2):108–34. [DOI] [PubMed] [Google Scholar]
- 34.Koutz FR, Rebrassier RE. The infection of cattle with gastrointestinal parasites of sheep. J Am Vet Med Assoc (1946) 109(836):359–61. [PubMed] [Google Scholar]
- 35.Kutzer E, Hinaidy HK. Die Parasiten der wildlebenden Wiederkäuer Österreichs. Z Parasitenkd (1969) 32(4):354–68. [DOI] [PubMed] [Google Scholar]
- 36.Kutzer E. Beziehungen und Übertragungsmöglichkeiten zwischen den Parasiten der Haus- und Wildwiederkäuer. Anblick (1969) 24:7–12. [Google Scholar]
- 37.Krasnov BR, Mouillot D, Shenbrot GI, Khokhlova IS, Poulin R. Beta-specificity: the turnover of host species in space and another way to measure host specificity. Int J Parasitol (2011) 41(1):33–41. 10.1016/j.ijpara.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 38.Poulin P, Krasnov BR, Mouillot D. Host specificity in phylogenetic and geographic space. Trends Parasitol (2011) 27(8):355–61. 10.1016/j.pt.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 39.Pojmańska T, Niewiadomska K. New trends in research on parasite host specificity: a survey of current parasitological literature. Ann Parasitol (2012) 58(2):57–61. [PubMed] [Google Scholar]
- 40.Köstenberger K, Tichy A, Bauer K, Pless P, Wittek T. Associations between fasciolosis and milk production, and the impact of anthelmintic treatment in dairy herds. Parasitol Res (2017) 116(7):1981–7. 10.1007/s00436-017-5481-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGhee MB, Nettles VF, Rollor EA, III, Prestwood AK, Davidson WR. Studies on cross-transmission and pathogenicity of Haemonchus contortus in white-tailed deer, domestic cattle and sheep. J Wildl Dis (1981) 17(3):353–64. 10.7589/0090-3558-17.3.353 [DOI] [PubMed] [Google Scholar]
- 42.Samuel W, Zewde GG. Prevalence, risk factors, and distribution of Cysticercus tenuicollis in visceral organs of slaughtered sheep and goats in central Ethiopia. Trop Anim Health Prod (2010) 42(6):1049–51. 10.1007/s11250-010-9537-y [DOI] [PubMed] [Google Scholar]
- 43.Ondrejicka DA, Locke SA, Morey K, Borisenko AV, Hanner RH. Status and prospects of DNA barcoding in medically important parasites and vectors. Trends Parasitol (2014) 30(12):582–91. 10.1016/j.pt.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 44.de León GP-P, Nadler SA. What we don’t recognize can hurt us: a plea for awareness about cryptic species. J Parasitol (2010) 96(2):453–64. 10.1645/GE-2260.1 [DOI] [PubMed] [Google Scholar]
- 45.Blouin MS. Molecular prospecting for cryptic species of nematodes: mitochondrial DNA versus internal transcribed spacer. Int J Parasitol (2002) 32:527–31. 10.1016/S0020-7519(01)00357-5 [DOI] [PubMed] [Google Scholar]
- 46.Grillo V, Jackson F, Cabaret J, Gilleard JS. Population genetic analysis of the ovine parasitic nematode Teladorsagia circumcincta and evidence for a cryptic species. Int J Parasitol (2007) 37(3–4):435–47. 10.1016/j.ijpara.2006.11.014 [DOI] [PubMed] [Google Scholar]
- 47.Hoberg EP, Monsen KJ, Kutz S, Blouin MS. Structure, biodiversity, and historical biogeography of nematode faunas in holarctic ruminants: morphological and molecular diagnoses for Teladorsagia boreoarcticus n. sp. (Nematoda: Ostertagiinae), a dimorphic cryptic species in muskoxen (Ovibos moschatus). J Parasitol (1999) 85:910–34. [PubMed] [Google Scholar]
- 48.Wyrobisz A, Kowal J, Nosal P. Insight into species diversity of the Trichostrongylidae Leiper, 1912 (Nematoda: Strongylida) in ruminants. J Helminthol (2016) 90(6):639–46. 10.1017/S0022149X15001017 [DOI] [PubMed] [Google Scholar]
- 49.Archie EA, Ezenwa VO. Population genetic structure and history of a generalist parasite infecting multiple sympatric host species. Int J Parasitol (2011) 41(1):89–98. 10.1016/j.ijpara.2010.07.014 [DOI] [PubMed] [Google Scholar]
- 50.Ács Z, Hayward A, Sugár L. Genetic diversity and population genetics of large lungworms (Dictyocaulus, Nematoda) in wild deer in Hungary. Parasitol Res (2016) 115(9):3295–312. 10.1007/s00436-016-5088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pyziel AM, Laskowski Z, Demiaszkiewicz AW, Höglund J. Interrelationships of Dictyocaulus spp. in wild ruminants with morphological description of Dictyocaulus cervi n. sp. (Nematoda: Trichostrongyloidea) from red deer, Cervus elaphus. J Parasitol (2017) 103(5):506–18. 10.1645/16-75 [DOI] [PubMed] [Google Scholar]
- 52.Cerutti MC, Citterio CV, Bazzocchi C, Epis S, D’Amelio S, Ferrari N, et al. Genetic variability of Haemonchus contortus (Nematoda: Trichostrongyloidea) in alpine ruminant host species. J Helminthol (2010) 84(3):276–83. 10.1017/S0022149X09990587 [DOI] [PubMed] [Google Scholar]
- 53.Callejón R, Halajian A, de Rojas M, Marrugal A, Guevara D, Cutillas C. 16S partial gene mitochondrial DNA and internal transcribed spacers ribosomal DNA as differential markers of Trichuris discolor populations. Vet Parasitol (2012) 186(3–4):350–63. 10.1016/j.vetpar.2011.11.033 [DOI] [PubMed] [Google Scholar]
- 54.Walker SM, Prodöhl PA, Hoey EM, Fairweather I, Hanna REB, Brennan G, et al. Substantial genetic divergence between morphologically indistinguishable populations of Fasciola suggests the possibility of cryptic speciation. Int J Parasitol (2012) 42(13–14):1193–9. 10.1016/j.ijpara.2012.10.007 [DOI] [PubMed] [Google Scholar]
- 55.Sattmann H, Hörweg C, Gaub L, Feix AS, Haider M, Walochnik J, et al. Wherefrom and whereabouts of an alien: the American liver fluke Fascioloides magna in Austria: an overview. Wien Klin Wochenschr (2014) 126(1):23–31. 10.1007/s00508-014-0499-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Husch C, Sattmann H, Hörweg C, Ursprung J, Walochnik J. Genetic homogeneity of Fascioloides magna in Austria. Vet Parasitol (2017) 243:75–8. 10.1016/j.vetpar.2017.06.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.