Abstract
Background
Streptococcus pyogenes or group A streptococcus (GAS) is an important human pathogen responsible for a broad range of infections, from uncomplicated to more severe and invasive diseases with high mortality and morbidity. Epidemiological surveillance has been crucial to detect changes in the geographical and temporal variation of the disease pattern; for this purpose the M protein gene (emm) gene typing is the most widely used genotyping method, with more than 200 emm types recognized. Molecular epidemiological data have been also used for the development of GAS M protein-based vaccines.
Methods
The aim of this paper was to provide an updated scenario of the most prevalent GAS emm types responsible for invasive infections in developed countries as Europe and North America (US and Canada), from 1st January 2000 to 31st May 2017. The search, performed in PubMed by the combined use of the terms (“emm”) and (“invasive”) retrieved 264 articles, of which 38 articles (31 from Europe and 7 from North America) met the inclusion criteria and were selected for this study. Additional five papers cited in the European articles but not retrieved by the search were included.
Results
emm1 represented the dominant type in both Europe and North America, replaced by other emm types in only few occasions. The seven major emm types identified (emm1, emm28, emm89, emm3, emm12, emm4, and emm6) accounted for approximately 50–70% of the total isolates; less common emm types accounted for the remaining 30–50% of the cases. Most of the common emm types are included in either one or both the 26-valent and 30-valent vaccines, though some well-represented emm types found in Europe are not.
Conclusion
This study provided a picture of the prevalent emm types among invasive GAS (iGAS) in Europe and North America since the year 2000 onward. Continuous surveillance on the emm-type distribution among iGAS infections is strongly encouraged also to determine the potential coverage of the developing multivalent vaccines.
Keywords: group A streptococcus invasive disease, Streptococcus pyogenes, emm types, Europe, North America, group A streptococci
Introduction
Streptococcus pyogenes or group A streptococcus (GAS) is a human Gram-positive bacterial species that either can exist as commensal or can be responsible for a broad range of infections, ranging from uncomplicated throat and skin infections to more severe and invasive diseases, such as bacteremia, soft tissue infections, necrotizing fasciitis, and septic shock; it represents, therefore, on a global scale, an important cause of morbidity and mortality (1). In addition, symptomatic infections can be involved in alterations of the immune system leading to post-streptococcal autoimmune sequelae, such as acute rheumatic fever (ARF) and chronic rheumatic heart disease (RHD), and post-streptococcal glomerulonephritis. ARF/RHD is an uncommon disease today in most high-income countries but it remains the major cause of acquired heart disease in adolescents and young adults in the developing world, responsible for at least 350,000 premature deaths per year (2).
Due to the size and clinical severity of the GAS disease burden, epidemiological surveillance has been crucial to detect changes in the disease pattern in various populations. Typing of collected bacterial isolates has been an important part of the epidemiological surveillance of GAS disease. Several different methods have been described and are available for GAS typing (3, 4). Classical serotyping methods based on the different forms of M, T, and OF surface antigens have been replaced by sequence typing of the N-terminal part of the M protein (emm) gene in the late 1990s (4–6), and it is now the most widely used genotyping method for GAS. So far, more than 200 emm types have been identified (http://www.cdc.gov/streplab/MProteinGene-typing.html), indicating that the M protein is the most polymorphic bacterial protein. Large epidemiological studies on pharyngitis and invasive disease have been performed worldwide using the emm sequence typing (3, 7, 8). DNA molecular typing techniques that consider multiple genome markers have been also used for GAS genotyping, such as pulsed-field gel electrophoresis (9) and multi locus sequence typing (10). These methodologies have proved to be of particular importance to define the clonal structure of particular GAS populations (11). Recently, emm-cluster typing system, which groups most emm types into 48 different functional emm clusters on the basis of their structural properties, and multiple-locus variable-number of tandem repeats analysis have been proposed as promising additional GAS typing tools (12, 13). The recent advances in whole-genome sequencing (WGS) technologies with reduced costs and turnaround times, along with the development of bioinformatics’ tools able to manage the large amount of generated data, made this technology accessible to reference microbiology (14). Recently, WGS coupled with the appropriate bioinformatic pipelines proved to be a reliable tool for the assignment of emm types and subtypes from genomic data (15–17).
Available molecular epidemiological data have been used for the design of a GAS vaccine. Despite the lack of licensed GAS vaccines, several vaccine candidates have been considered and they can be divided into M protein-based and non-M protein-based vaccines. Among the M-protein-based vaccines under clinical trial investigation there are the multivalent 26-valent and the 30-valent formulations as well as the conserved M protein vaccines (18–20); the non-M-protein-based candidate vaccines, at various stages of development, include either cell wall or several secreted virulence factors (21, 22).
The present review aimed to provide a picture of the emm-type distribution among invasive GAS (iGAS) strains in high-income countries of the Western world, such as Europe and North America, retrieved from the literature since the year 2000. This study had two prominent objectives: to update the picture on the most prevalent emm types causing invasive infections for epidemiological purposes and, second, to provide a scenario of the possible efficacies of the under-development M-based vaccine formulations.
Methodology
Search Strategy and Study Selection Criteria
We searched for studies that described the epidemiology of invasive GAS based on emm typing by the use of a systematic approach. Searches were done in PubMed Medline for papers published from 1st January 2000 to 31st May 2017 by the combined use of the search terms “emm” and “invasive.” The search was planned in order to include only GAS isolates responsible for invasive infections according to the case definition (23). Exclusion criteria included studies that involved other beta-hemolytic streptococci, not population-based surveys, outbreak, and case-report studies, surveillance studies focused to specific emm types or limited to the analysis of antibiotic-resistant strains. Reports considering small numbers of strains (approximately not more than 40 GAS strains) were also not considered, except for those papers that were the only representative of a given European Country. Moreover, all studies that were part of major cited studies and reviews were also excluded from the analysis. Those studies for which clear data were not available, such as studies with uncertainty on the time period of the collection of strains, on the origin of strains, on the emm-type distribution, on the geographical origin, or studies involving strains collected across 2000 but starting earlier in 1990s and for which emm types after 2000 were not precisely provided, were not considered in this review. Finally, the search was restricted to only papers in English language. Overall, 264 articles were retrieved. A total of 38 articles, of which 31 and 7 from European and North American studies, respectively, were selected by the criteria described above. Five additional articles, all European studies, which were cited in the references although not retrieved by using the search criteria, were also included.
Data Extraction and Analysis
A database was created to record the country, the period (years) of isolation, the geographic area involved (local, regional, or a nationwide survey), the group ages of patients affected by iGAS, the overall number of genotyped iGAS and the emm-type distribution. Only one isolate per patient was included and the relative frequency of each emm type was determined or extrapolated from each study. All the information obtained was incorporated into an Excel file and summarized in Table S1 in Supplementary Material for European studies and Table S2 in Supplementary Material for North American studies.
Results
Europe
All countries located in the European continent were considered. Therefore, besides the 28 European Union (EU) countries, Iceland, Liechtenstein, Norway, and Switzerland were included in the search.
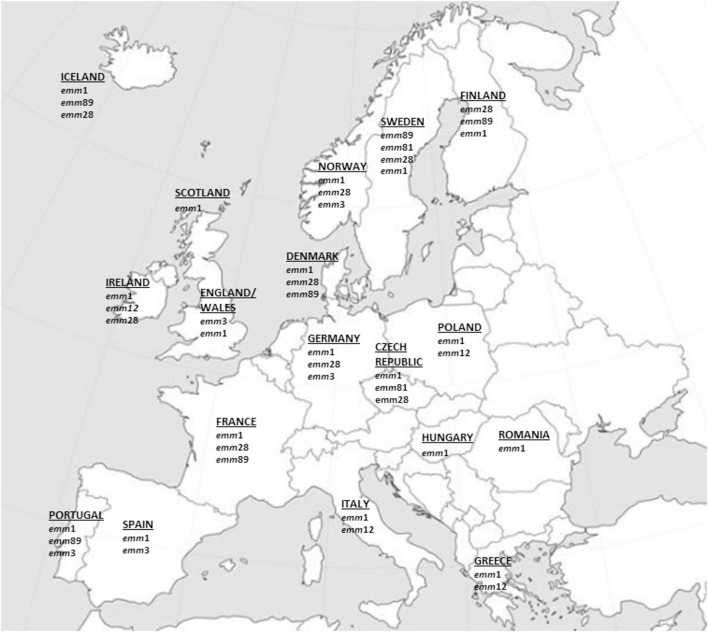
No data on iGAS emm types were retrieved from 15 countries (Austria, Belgium, Bulgaria, Croatia, Cyprus, Estonia, Latvia, Lithuania, Luxembourg, Malta, Netherlands, Slovakia, Slovenia, Liechtenstein, and Switzerland) while for the other 17 countries at least one study on iGAS emm-type distribution could be retrieved and taken into consideration. The findings of the most prevalent emm types encountered in each European Country are summarized in the Figure 1 and in Table S1 in Supplementary Material.
Figure 1.
Most prevalent invasive GAS emm types in Europe since 2000 onward. The major emm types (accounting for approximately ≥10% of all emm types) from European epidemiologic surveys that met the criteria used in this review are represented.
Northern Countries
Denmark
Three studies were available for Denmark covering the period 2001–2011. Overall, the major emm types were emm1, 28, 89, and 3, with the latter commonly found only since the year 2003 onward. The most recent study described a nationwide laboratory-based surveillance between years 2005 and 2011 and it analyzed 910 iGAS isolates (24). A total of 58 different emm types were identified, with emm types 1 (26%), 28 (17%), 3 (13%), 89 (11%), and 12 (9%) representing 76% of all isolates. emm1 predominated in most years, except in the year 2010 when emm28 was the most prevalent, and 2011 when emm1 predominated along with emm3 (24).
Immediately before, in the period 2003–2004, another national active surveillance study on 278 iGAS infections, as a part of the Strep-EURO project, demonstrated that emm28 (26%) and emm1 (24%) accounted for approximately 50% of strains, followed by emm3 (11%), emm89 (7%), and emm12 (5.5%) (25). Fluctuation of the emm-type distribution was observed over time but emm28 that was constant over time, whereas emm1 decreased and emm3 increased (25).
Another report was a nationwide prospective surveillance study performed between January 2001 and August 2002 on GAS collected from patients with invasive infections, noninvasive infections, and asymptomatic carriers. This study revealed that among the 200 invasive isolates, a total of 27 different emm types were found, of which emm1, emm28, emm12, emm6, and emm4 accounted for 69% of the total emm types (26). emm1 and emm28 alone constituted 32 and 20% of the total emm types, respectively, with emm1 that significantly increased over 2002 (26).
Finland
Four studies met the criteria used in this review. These encompassed the period 2000–2013 and the major emm types were emm28, 89, and 1. The most recent paper was a nationwide study conducted during the period 2008–2013 on 1,112 iGAS (27). A total of 72 different emm types were identified, of which emm28 (26%), emm89 (17%), and emm1 (12%) were the most common types (27). An increase of erythromycin and clindamycin resistance was observed at that time that paralleled with the emergence of the novel clone emm33, detected for the first time in 2012 (27). This study revealed that emm89 increased from 2008 to 2013, emm1 decreased while emm84, that was common in 2008, was not detected in 2013 (27).
Another nationwide survey was performed to assess the population-based incidence and outcomes of pediatric iGAS infections (28). During the period of 1996–2010, a total of 151 children with iGAS infection were identified (28). Overall, 60 isolates were genotyped and emm1 strains were the most prevalent in the years 2000–2010, showing a typical fluctuation by herd immunity, followed by emm28, emm12, and emm89 (28).
In 2006, an increase of iGAS disease was observed (29). Therefore, a nationwide surveillance study was conducted in the period 2004–2007. A total of 1,318 cases of iGAS were identified displaying 46 different emm types. The most common emm types were 28 (21%), 1 (16%), 84 (10%), 75 (7%), and 89 (6%), accounting for 60% of isolates (29). Fluctuations in emm-type distribution were observed over the study period.
One regional study on the emm-type distribution among invasive beta-hemolytic streptococci, including GAS, conducted in 1-year period (2008–2009) in Pirkanmaa Health District (western Finland) revealed that among 50 iGAS blood or deep tissue isolates the most common emm types were emm77 (32%), emm28 (22%), emm89 (14%), and emm1 (8%) (30).
Iceland
Only one study was retrieved by using the chosen criteria; it comprised 288 iGAS cases from 1975 to 2012 (31). Overall, 226 GAS isolates were emm typed, of which 132 isolates were recovered since 2000 onward; they displayed a total of 11 different emm types with emm types 1 (26%), 89 (17%), 28 (14%), 12 (11%), 4 (8%), 3 (6%), and 22 (6%), as the more prevalent (31). emm1 strains most likely caused severe infections, emm4 was significantly more common among children, whereas emm28 was identified solely among adults (31).
Norway
Two nationwide and three regional surveillance studies on iGAS in Norway have been reported since 2000. The studies comprised iGAS collected over the period 2000–2014 and the major emm types identified were emm1, 28, 3, and 89.
The most recent national study regarded an epidemiological surveillance on iGAS infections from 2010 to 2014 (32). Overall, 756 iGAS isolates were analyzed and 52 different emm types were identified (32). emm1 (27%), followed by emm89 (12%), emm12 (9%), emm3 (8%), and emm28 (8%) were the most prevalent types and accounted for 64% of all isolates (32).
The second nationwide study was conducted on 262 invasive isolates collected during the years 2006 and 2007. Overall, 29 different emm types, with 37 emm subtypes, were identified (33). The most prevalent emm types were emm28 (19.5%), emm1 (14.1%), emm82 (14.1%), emm12 (11.8%), emm4 (8.0%), emm3 (6.1%), and emm87 (5.7%). A significant increase in the prevalence of emm6, overrepresented in males, was noted (33). Comparing the emm-type distribution between 2007 and 2014, emm28 dominated in 2007 then decreased, while emm1 increased over time (32, 33). Five out of the six most frequent emm types in 2007 remained among the top six emm types from 2010 to 2014, but emm82, the third most frequent emm type in 2007 was completely absent in the years 2010–2014, replaced by emm89 which increased (32, 33).
The three regional epidemiological studies on iGAS were conducted in Western Norway. The largest regional study was focused on all beta-hemolytic groups (A, C, and G streptococci) isolated over a 15-years period (1999–2013) (34). Among the 209 iGAS, 26 different emm types and 34 emm subtypes were identified (34). The most frequent emm types were consistent along the entire period of study and they were emm1 (23.9%), emm28 (14.3%), emm3 (13.9%), emm89 (6.2%), emm11 (5.7%, responsible for an outbreak), and emm4 (5.2%) (34). The other two studies considered a lower number of strains and involved beta-hemolytic group A, C, and G streptococci. One study was performed on necrotizing soft tissue infections caused by beta-hemolytic group A, C, and G streptococci during the period 2000–2009 (35). Overall, 42 iGAS isolates were analyzed and the most prevalent emm types were, in decreasing order, emm1 (31%), emm3 (11.9%), and emm4 (9.5%) (35). The other study was on invasive A, C, and G streptococcal infections observed in the years 2006–2009 and involved a total of 59 iGAS (36). The six most common emm types accounted for 81% of the isolates and they were emm28 (22%), emm3 (17%), emm1 (15%), emm12 (10%), emm82 (10%), and emm89 (7%), with the first three emm types responsible for more than 50% of the infections, associated with severe outcome (36).
Sweden
Only one study met the selected criteria; it was an enhanced surveillance study on iGAS (746 isolates) performed in the years 2002–2004, of which the majority (94%) were isolated from bacteremia (37). The most abundant emm types were emm89 (15.7%) and emm81 (14.5%), followed by emm28 (13.9%), emm1 (11.9%), emm12 (6.3%), emm77 (5.9%), and emm4 (5.9%).
England and Wales
Two studies from United Kingdom were included in this survey, of which one on a large collection of iGAS isolated in 2014, the other on a small collection of strains recovered in the early 2000s. The bigger study was conducted after an increase of scarlet fever episodes observed in 2014 in England and involved 252 iGAS. emm3 was the most prevalent emm type representing 28.2% of all iGAS, followed by emm1 (22.6%), emm89 (9.1%), emm12, and emm28 (7.1% each) (38).
The other report was a small surveillance study performed on blood GAS isolates obtained from injecting drug users compared to isolates from non-drug users at the Royal Sussex Hospital in Brighton over the period 2000–2003 (39). Overall, 44 iGAS were available for the study and the most frequent emm types were emm83 (25%), emm82 (18.2%), emm1 (15.9%), emm89 (13.6%), and emm87 (11.4%) (39). Differences in emm-types distribution were noted between injecting drug users and non-drug users, with emm82 and emm83 almost exclusively found in the injecting drug users population (39).
Scotland
Only one national study from Scotland was retrieved; it reported the distribution of GAS emm types from invasive and noninvasive sites from all age groups patients over a 4-years period (2011–2015). This study revealed that among 329 iGAS strains recovered from sterile sites emm1 (66% of all iGAS) was the most prevalent and strongly associated with iGAS infections (40). The other most common emm types identified in invasive infections were, in decreasing order, emm76 (7.4%), emm89 (6.7%), and emm3 (5.8%).
Ireland
The only study from Ireland that met the chosen criteria documented an increased incidence of iGAS over in the years 2012–2013; emm1 dominated over the entire study period (41). In 2012, 176 iGAS were genotyped and the most predominant types were emm1 (48.6%), emm12 (9.2%), and emm28 (7.3%). In 2013, a further increment in iGAS infections was noted and it was associated with a notable increase in emm3 isolates in the first half of 2013, reaching the rate of 22% from that of 4% observed in 2012 (41). The relative percentages of emm types found over the entire study period were not clearly indicated.
Eastern Countries
Czech Republic
The only study on iGAS was an epidemiological survey on 215 iGAS obtained from 34 hospitals throughout the Country, isolated during the years 2001–2005 (42). The emergence of the uncommon GAS emm53 type was reported, with the highest proportion observed during 2003, possessing resistance to macrolides (42). The most prevalent emm type was emm1, followed by emm81, emm28, emm53, emm3, and emm66, although the relative frequency of each emm type was not reported (42).
Hungary, Poland, and Romania
One national surveillance study from each Country has been retrieved according to the used criteria, all three including only a limited number of iGAS isolates.
The nationwide laboratory-based surveillance study from Hungary regarded the molecular characterization of 26 iGAS isolates obtained in the years 2004 and 2005. The most prevalent emm types encountered in this study were emm1 (50%) and emm80 (19.2%) (43).
The study from Poland was a national laboratory-based survey involving 17 different hospitals distributed in different part of the Country (44). The major limitation of this study for our analysis was the lack of data on the emm-type distribution obtained since 2000 onward, but considering that it was the only molecular study on iGAS from this country, it was included. Overall, 41 iGAS clinical isolates were obtained between the years 1997 and 2005, with a total of 23 different emm types identified, of which emm1 and emm12 (19.5% each) were the most frequent, followed by emm 81 (7.3%) (44).
A total of 33 iGAS obtained from 8 different Romanian districts during the years 2003–2004 were genotyped as a part of the Strep-Euro project; the predominant emm types were emm1 (15.1%), emm76, and emm81 (12.1% each) (45).
Western Countries
France
Several studies on the epidemiology and emm-type distribution of iGAS isolates recovered from 2000 to 2013 met the criteria used in this review. Overall, the most prevalent emm types were emm1, 28, and 89.
The first report was a molecular epidemiology study on iGAS isolated from French children between the years 1999 and 2006 (46). Among the 74 strains available for the study, 31 different emm types were identified, associated with specific virulence genes. emm types 1 (25.7%), 89 (9.5%), 3 and 4 (8.1% each), and 6, 12, and 28 (6.7% each) predominated (46).
Another report investigated the characteristics of GAS responsible for meningitis in adults from 2003 to 2013 (47). Overall, 63 GAS isolates were identified and four predominant emm types were found: emm1 (44%), emm28 (12.7%), and emm3 and emm6 (11.1% each) (47).
Four other studies on French iGAS strains isolated in the years 2006–2011 found emm1 as the predominant type. Two of these studies were nationwide surveillances on a large number of strains recovered from all age group patients in France (48, 49); in particular, one report assessed the epidemiology of 623 iGAS strains isolated in 2007, the other study involved more than 2,600 iGAS collected in the period 2007–2011. The most prevalent emm types were quite the same between studies, with slight differences: emm1 predominated, followed by emm28 or emm89, emm4, and emm12 (48, 49). The other two French studies were a report on 1,542 iGAS recovered from adults in the years 2006 and 2010 (50) and 125 iGAS strains isolated from children in the period 2009–2011 (51). The first study identified emm1 as the most common type accounting for 24% of the isolates, followed by emm28 (17%), emm89 (15%), emm4, emm3, and emm12 (5% each); the other study revealed that emm1 predominated (24.8%), followed by emm12 and emm28 (15.2% each), emm6 (12%), and emm3 (9.6%).
Germany
Two German studies have been included in this review, covering the years 2000–2009. emm types 1, 28, and 3 predominated. One major study was a nationwide laboratory-based surveillance to analyze the epidemiology of varicella-associated iGAS infections over the years 1996–2009 (52). Overall, 1,342 iGAS were analyzed and the most abundant emm type was emm1 (32.6%), followed by emm28 (13.8%), emm3 (8.3%), emm12 (6.1%), and emm89 (5.5%). emm1, emm12, and emm4 types, possessing a high rate of the virulence gene ssa, were significantly related to varicella-positive isolates (52).
The second nationwide laboratory-based surveillance study of iGAS was performed on a total of 586 strains, mostly recovered from blood and skin infections, during the years 2003–2007 (53). Also in this study, emm1 strongly dominated (30.5%) followed by emm28 (18.3%), emm3 (9.6%), and emm12 and emm89 (7% each) (53).
Southern Countries
Greece
Two national surveillance reports have been selected, covering the period 2003–2007, and they both indicated emm1 and emm12 as the most prevalent types. A multicenter laboratory-based surveillance study was conducted between the years 2003 and 2007; among the 138 iGAS available for genotyping the two most prevalent emm types were emm1 (28.2%), mainly isolated in adults, and emm12 (8.5%) (54). The other study was a multicenter surveillance involving a total of 101 isolates obtained between the years 2003 and 2005 and the most common emm types were emm1 (26.5%), emm12 (8.9%), emm4, emm6, and emm95 (5% each) (55).
Italy
Only one study from Italy was retrieved on iGAS isolates from 2000 onward, a 3-year (between 2003 and 2005) nationwide enhanced surveillance study as part of the StrepEuro surveillance; it comprised 89 iGAS, where emm1 largely predominated (19%), followed by emm12 (12%), emm3 (10%), emm4 (9%), emm18 (8%), and emm6 (5%) (56).
Portugal
Two major consecutive surveillance studies involving iGAS have been performed in the period 2000–2009. emm1, emm3, emm6, and emm89 represented the four most common types, with differences in their order of frequency.
The most recent study was conducted on iGAS isolated between 2006 and 2009 (57). A total of 191 iGAS were characterized with emm1 (29.3%), emm89 (12.6%), emm3 (10.5%), emm6 (7.8%), accounting for 60% of the isolates (57). In this study, it was observed an intra-clonal diversity of the superantigen genes profiles, with evidence of specific superantigen gene loss and acquisition (57).
The second study was a multicenter laboratory-based surveillance of iGAS conducted over a period immediately before the aforementioned survey, between the years 2000 and 2005 (11). Among 160 non-duplicate iGAS isolates, the six most common emm types were emm1 (20%), emm3 (9.3%), emm89 (8.1%), emm6, emm28, and emm64 (6.9% each) (11).
Spain
Only one study met the criteria and it was a regional survey on invasive strains recovered from two regions in Spain between the years 1998 and 2009. Among the 215 isolates, emm1, associated with speA and ssa genes, largely predominated (27.9%) and was responsible for the majority of fatal outcomes, followed by emm3 (9.8%), emm4 (6.5%), and emm28, emm12, and emm89 (6% each) (58).
North America
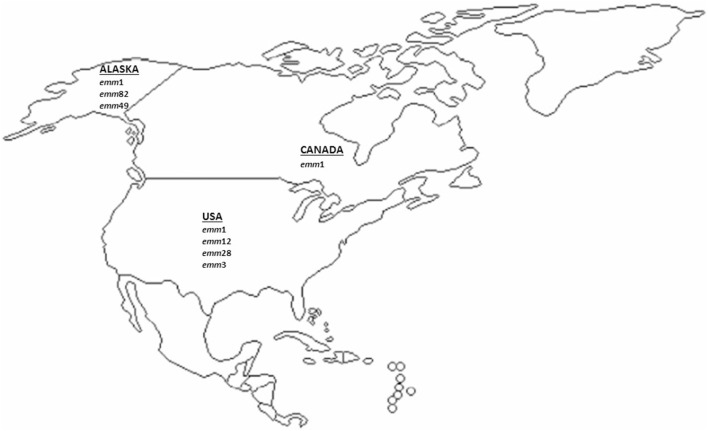
Four and three studies on emm-type distribution of iGAS from US including Alaska and from Canada, respectively, were considered in this review. The results of the emm-type distribution and their relative frequencies reported in North America are depicted in Figure 2 and in Table S2 in Supplementary Material.
Figure 2.
Most prevalent invasive GAS emm-types described in North America (US and Canada) since 2000 onward. The major emm types (accounting for approximately ≥10% of all emm types) from the US and Canadian studies included in this analysis are represented.
United States
Three major studies on iGAS collected by the Centers for Disease Control and Prevention’s Active Bacterial Core surveillance, a population-based network, including geographically different states from US, were retrieved. Altogether, these three studies encompassed a time period of 13 years, from 2000 to 2012. emm1 was the predominant type in all studies and four other emm types, namely emm3, emm12, emm28, and emm89, were among the most common types, with some fluctuations over time.
The most recent survey was conducted on 8,200 iGAS obtained over the period 2005–2012 in which emm1 was the most prevalent type (22%), followed by emm12 (9%), emm28 (8%), emm89, and emm3 (7% each) (59). emm types included in the currently proposed 30-valent vaccine accounted for 91% of the cases (59).
Another study was performed on 4,350 iGAS recovered in 10 different US sites during the period 2000–2004 (8). emm1 largely dominated at a rate of 18.2%, followed by emm3, emm28, emm12 (9% each), and emm89 (6%), all together accounting for 55% of the isolates (8). In this study, the emm types included in the proposed 26-valent vaccine accounted for 79% of all cases (8).
The first population-based study conducted on iGAS recovered since 2000 onward involved nine different States and 1,061 invasive strains recovered during the years 2000 and 2001 (60). The six most prevalent emm types were emm1 (18.2%), emm3 (10.2%), emm12 (8.5%), emm28 (7.9%), emm82 (5.9%), and emm89 (5.5%), which accounted for approximately 55% of the isolates (60).
In Alaska, the Arctic Investigations Program set up a population-based laboratory surveillance program on iGAS infections during the period 2001–2013 (61). Overall, 516 cases of iGAS infections were reported, and 422 isolates were available for microbiological analysis and typing. A total of 51 different emm types were identified, of which emm1 was the most common (11.1%) followed by emm82 (8.8%), emm49 (7.8%), emm12, and emm3 (6.6% each), emm89 (6.2%), and emm108 (5.5%) (61). The emm types included in the 30-valent M protein vaccine accounted for 78% of isolates (61).
Canada
Overall, three studies on molecular epidemiology of iGAS infections were found, encompassing a 9-years period (between 2006 and 2014), with large geographical differences in the emm-type distribution.
A recent epidemiological study on iGAS strains from remote indigenous communities in Northwestern Ontario, over the period 2009–2014, indicated that among 46 invasive isolates, 14 different emm types were identified, with emm-type variability observed over time. The most common emm types were emm114 (17.4%), emm11 (15.2%), emm118 (13.0%), emm68, and emm82 (10.9% each) (62).
Another regional surveillance study of iGAS in the province of Ontario started because of an outbreak of iGAS infections due to emm59 strains occurring in Thunder Bay District, North-Western Ontario in 2008. All iGAS obtained in Thunder Bay District from 2011 to 2013 were studied and they were compared to iGAS strains recovered during the same period from the metropolitan area of Toronto/Peel and the province of Ontario (63). Most iGAS cases isolated in the Thunder Bay District were caused by strains belonging to skin or generalist emm types, while those from the province of Ontario and the Toronto metropolitan area were caused by emm types frequently associated with invasive GAS infections (63). In particular, among the 66 iGAS obtained from Thunder Bay District the six most prevalent emm types were emm87 (12.3%), emm82 (10.8), emm1, emm101, and emm83 (9.2% each), and emm114 (7.7%). By contrast, from the metropolitan area of Toronto/Peel and the rest of province of Ontario emm1 predominated (23.5%), followed by emm89 (12.7%), emm3 (11.3%), emm12 (8.2%), and emm28 (5.7%) (63).
The third study was a nationwide surveillance conducted from January 2006 to December 2009 and included 4143 iGAS obtained from 10 provinces and 3 territories in Canada (64). Among all iGAS cases, 539 (13%) were attributed to emm59, mainly circulating in the province of Ontario. emm1, emm28, emm3, emm89, and emm12 were other well-represented types, although the relative percentages of each emm type were not provided (64).
Discussion and Conclusion
In this review, we provided a picture of the most prevalent emm types among iGAS reported in Europe and North America from the year 2000 to May 2017 (last updated on June 1st 2017). It is interesting to note that the major emm types were almost the same in all European countries and in US. emm1 largely represented the dominant type in all countries, and in only few cases it was replaced by other common emm types, such as emm28, reported in Denmark, Finland, and Norway, emm89 and emm77 reported in Sweden and Finland, respectively, and emm3 in the United Kingdom. Other common emm types were emm28, emm89, emm3, emm12, emm4, and emm6, with differences in their prevalence among countries. Some “uncommon” emm types were sporadically encountered as prevalent types, as for emm84 and emm119 in Finland; emm11 in Norway; emm75 in Finland, Norway, and Spain; emm81 in Finland, Sweden, Czech Republic, Hungary, Poland, and Romania; emm82 in Norway and Alaska; emm66 in Norway and Czech Republic; emm53 in Finland and Czech Republic; emm18 in Italy; emm64 in Portugal; emm49 and emm108 in Alaska; and emm59 in Canada. Interestingly, emm81 was commonly found in all Eastern European countries, suggesting the likely spread of this type between neighboring countries.
Overall, the major emm types (emm1, emm28, emm89, emm3, emm12, and emm4, and emm6) accounted to only up 70% of the total isolates. Therefore, it is of some concern that several other different emm types that are less represented nevertheless accounted for the remaining 30–50% of the cases. Some of these minor emm types could emerge and become among the most prevalent ones in the future.
In North America, emm1 was the most prevalent type in most national studies from US, followed by emm12, emm28, and emm3 but in regional remote communities in Canada other uncommon emm types predominated, such as emm11, emm87, emm101, emm114, and emm118.
It is important to be aware of the increase in iGAS infections with associated mortality observed in the last years in several European countries (40, 65–67), reinforcing the need of an European multinational surveillance network that could compensate the scattered available information on the iGAS disease burden (68).
Data on the emm-type distribution of population-based GAS surveillance have been also used for the development of M-multivalent GAS vaccine candidates. The 26-M-valent and 30-M-valent vaccines have been developed in order to maximize the “coverage” of circulating emm types (19). Epidemiologic surveys suggest that the 26-valent vaccine would provide good coverage of circulating GAS strains in industrialized countries (over 72%) but poor coverage in many developing countries due to differences in emm distribution (3). Similarly, the 30-valent vaccine has a limited coverage in many developing countries where GAS infections are endemic but this inconvenience is likely mitigated by the demonstration that the 30-valent vaccine induces protection not only against the emm types contained in the vaccine but it also cross-reacts to some non-vaccine emm types (69).
It is of some concern that some emm types found to be well-represented in some European studies are not included into the vaccine formulations, as is the case of the emm types 53, 64, 66, 84, 108, and 119; on the other end, sporadic emm types as emm 49, 81, and 82 that were identified in the European studies or emm types 87, 101, and 118 isolated in regional Canadian surveys are included in one or the other of the two vaccines (19, 70).
Fluctuations in emm-type distribution have been attributed to multiple factors, such as different frequencies of the most prevalent circulating clones, influence of the herd immunity, different clinical manifestations of iGAS infections associated with specific emm types, age, racial, and seasonal differences (25, 28, 32). Recently, seasonal, geographic, and temporal trends of specific emm clusters associated with iGAS infections have been found, due to a probable different capacity for transmission or infection (71).
The factors contributing to the fluctuations and/or success of specific epidemic clones in invasive diseases have always gained a priority interest in term of prevention control and the study of the dynamics of GAS population. The predominance of particular emm types in invasive disease could likely be a consequence of the high prevalence in the entire population, as demonstrated for emm1, suggestive of the better success of specific clones as well-adapted human colonizers (72). In the last few years, the use of WGS has proved to be a useful way to investigate the evolutionary history of these highly successful GAS epidemic or pandemic clones belonging to specific emm types. Specific virulence factors, such as streptococcal pyrogenic exotoxin (spe) A and its alleles, streptokinase, streptolysin O (slo), NAD glycohydrolase (nga), have been associated with shaping and spreading of successful strains. For example, the emergence of the emm1 epidemic clone has been associated with three consecutive gene transfer events, which is the acquisition of Dnase sdaD2, expression of speA2 and upregulation of the virulence factors slo and nga (73). Similarly, for emm89, the emergence of a third clonage lineage (clade 3 clone) by modification in the nga/slo locus and loss of the capacity to synthesize the capsule is the cause of an ongoing epidemic of invasive infections in Europe and North America (74). Another recent study from England reported an increase in iGAS infections associated with emm3 isolates. By analysis of the whole-genome single-nucleotide polymorphism-based phylogeny the main factor responsible for this upsurge seemed to be associated with the expansion of a genetic lineage characterized by the presence of a prophage carrying speC exotoxin and spd1 DNAase genes and the loss of two other prophages considered typical of the emm3 strains (75).
Notwithstanding the importance of loss or acquisition of genes by horizontal transfer (mostly mediated by bacteriophage or pathogenicity islands) as a fundamental evolutionary forces shaping the structure of GAS genomes, it is also evident that there is not a strict correlation between defined patterns of exogenous elements-associated virulence genes and invasiveness or disease severity. The main contribution to the generation and expansion of emm-type specific invasive clones seems to be more related to mutations in some important genetic regulatory circuits governing the global expression of GAS virulence. Among them, there are the two components system CovR/S (76), the regulator of protease B ropB (77), the regulator of Cov RocA (78), the Fas system (79), and the RofA-like protein IV regulator RivR (80).
In this context, it is clear how emm type (i.e., M serotype) continues to be the GAS epidemiological reference marker for tracking the clonal radiation of epidemic lineage from a common genetic background. The recent advances of WGS technologies with reduction in terms of costs and reduced turnaround times have been proposed as the method of choice to type strains, GAS included. The design of bioinformatics components to emulate current methods, however, is still laboratory-restricted and standardization of different steps still needs improvements to ensure backward compatibility with the classical “gold standard” typing methods and schemes (15, 17). The potential of WGS data analysis is huge as already demonstrated also for GAS where temporal and geographic relatedness between GAS isolates could be deduced (16). These new “omics” approaches may also provide rapid assessment of outbreaks by discriminating between closely related isolates, and produce extensive data on the virulome, including the associated regulatory mechanisms, and the antibiotic resistome (81, 82). Hopefully, this review can be a reference for all those who work in the field of molecular epidemiology of GAS and may represent a milestone also to those who are approaching the problem using the recent next generation sequencing methodologies.
In any case, continuous surveillance of the emm-types distribution of iGAS is strongly encouraged to monitor the prevalent emm types responsible for the disease burden and the potential coverage of the under-development multivalent vaccines.
Author Contributions
GG and RC conceived the project. GG and RC searched the database for potentially eligible articles, extracted the data, and performed the analyses. GG, LV, and RC interpreted the results and wrote the manuscript. All the authors reviewed the final version of the manuscript prior to submission for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/articles/10.3389/fpubh.2018.00059/full#supplementary-material.
References
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis (2005) 5:685–94. 10.1016/S1473-3099(05)70267-X [DOI] [PubMed] [Google Scholar]
- 2.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. Lancet (2012) 380:2095–128. 10.1016/S0140-6736(12)61728-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis (2009) 9:611–6. 10.1016/S1473-3099(09)70178-1 [DOI] [PubMed] [Google Scholar]
- 4.Creti R, Gherardi G, Berardi A, Baldassarri L. Pathogenesis of Streptococcus in humans. In: Singh SK, editor. Human Emerging and Re-Emerging Infections: Viral and Parasitic Infections. (Vol. I), Hoboken, NJ: John Wiley & Sons, Inc; (2015). 795 p. [Google Scholar]
- 5.Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR product for routine and accurate typing of group A streptococci. J Clin Microbiol (1996) 34:953–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Facklam R, Beall B, Efstratiou A, Fischetti V, Johnson D, Kaplan E, et al. emm typing and validation of provisional M types for group A streptococci. Emerg Infect Dis (1999) 5:247–53. 10.3201/eid0502.990209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamagni TL, Efstratiou A, Vuopio-Varkila J, Jasir A, Schalén C, Strep-EURO The epidemiology of severe Streptococcus pyogenes associated disease in Europe. Euro Surveill (2005) 10:179–84. 10.2807/esm.10.09.00563-en [DOI] [PubMed] [Google Scholar]
- 8.O’Loughlin RE, Roberson A, Cieslak PR, Lynfield R, Gershman K, Craig A, et al. The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000–2004. Clin Infect Dis (2007) 45:853–62. 10.1086/521264 [DOI] [PubMed] [Google Scholar]
- 9.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol (1995) 33:2233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGregor KF, Spratt BG, Kalia A, Bennett A, Bilek N, Beall B, et al. Multilocus sequence typing of Streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. J Bacteriol (2004) 186:4285–94. 10.1128/JB.186.13.4285-4294.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friães A, Ramirez M, Melo-Cristino J, Portuguese Group for the Study of Streptococcal Infections Non-outbreak surveillance of group A streptococci causing invasive disease in Portugal identified internationally disseminated clones among members of a genetically heterogeneous population. J Clin Microbiol (2007) 45:2044–7. 10.1128/JCM.00496-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanderson-Smith M, De Oliveira DM, Guglielmini J, McMillan DJ, Vu T, Holien JK, et al. A systematic and functional classification of Streptococcus pyogenes that serves as a newtool for molecular typing and vaccine development. J Infect Dis (2014) 210:1325–38. 10.1093/infdis/jiu260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imperi M, Pittiglio V, D’Avenio G, Gherardi G, Ciammaruconi A, Lista F, et al. A new genotyping scheme based on MLVA for inter-laboratory surveillance of Streptococcus pyogenes. J Microbiol Methods (2016) 127:176–81. 10.1016/j.mimet.2016.06.010 [DOI] [PubMed] [Google Scholar]
- 14.Loman NJ, Constantinidou C, Chan JZM, Halachev M, Sergeant M, Penn CW, et al. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol (2012) 10:599–606. 10.1038/nrmicro2850 [DOI] [PubMed] [Google Scholar]
- 15.Athey TBT, Teatero S, Li A, Marchand-Austin A, Beall BW, Fittipaldi N. Deriving group A Streptococcus typing information from short-read whole-genome sequencing data. J Clin Microbiol (2014) 52:1871–6. 10.1128/JCM.00029-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chochua S, Metcalf BJ, Li Z, Rivers J, Mathis S, Jackson D, et al. Population and whole genome sequence based characterization of invasive group A streptococci recovered in the United States during 2015. MBio (2017) 8:e01422–17. 10.1128/mBio.01422-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapatai G, Coelho J, Platt S, Chalker VJ. Whole genome sequencing of group A Streptococcus: development and evaluation of an automated pipeline for emm gene typing. PeerJ (2017) 5:e3226. 10.7717/peerj.3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNeil SA, Halperin SA, Langley JM, Smith B, Warren A, Sharratt GP, et al. Safety and immunogenicity of 26-valent group a Streptococcus vaccine in healthy adult volunteers. Clin Infect Dis (2005) 41:1114–22. 10.1086/444458 [DOI] [PubMed] [Google Scholar]
- 19.Dale JB, Penfound TA, Chiang EY, Walton WJ. New 30-valent M protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group A streptococci. Vaccine (2011) 29:8175–8. 10.1016/j.vaccine.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steer AC, Carapetis JR, Dale JB, Fraser JD, Good MF, Guilherme L, et al. Status of research and development of vaccines for Streptococcus pyogenes. Vaccine (2016) 34:2953–8. 10.1016/j.vaccine.2016.03.073 [DOI] [PubMed] [Google Scholar]
- 21.Steer AC, Batzloff MR, Mulholland K, Carapetis JR. Group A streptococcal vaccines: facts versus fantasy. Curr Opin Infect Dis (2009) 22:544–52. 10.1097/QCO.0b013e328332bbfe [DOI] [PubMed] [Google Scholar]
- 22.Bensi G, Mora M, Tuscano G, Biagini M, Chiarot E, Bombaci M, et al. Multi high-throughput approach for highly selective identification of vaccine candidates: the group A Streptococcus case. Mol Cell Proteomics (2012) 11:M111.015693. 10.1074/mcp.M111.015693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies HD, McGeer A, Schwartz B, Green K, Cann D, Simor AE, et al. Invasive group A streptococcal infections in Ontario, Canada. Ontario group A streptococcal study group. N Engl J Med (1996) 335:547–54. 10.1056/NEJM199608223350803 [DOI] [PubMed] [Google Scholar]
- 24.Lambertsen LM, Ingels H, Schønheyder HC, Hoffmann S, Danish Streptococcal Surveillance Collaboration Group 2011 . Nationwide laboratory-based surveillance of invasive beta-haemolytic streptococci in Denmark from 2005 to 2011. Clin Microbiol Infect (2014) 20:O216–23. 10.1111/1469-0691.12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luca-Harari B, Ekelund K, van der Linden M, Staum-Kaltoft M, Hammerum AM, Jasir A. Clinical and epidemiological aspects of invasive Streptococcus pyogenes infections in Denmark during 2003 and 2004. J Clin Microbiol (2008) 46:79–86. 10.1128/JCM.01626-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekelund K, Darenberg J, Norrby-Teglund A, Hoffmann S, Bang D, Skinhøj P, et al. Variations in emm type among group A streptococcal isolates causing invasive or noninvasive infections in a nationwide study. J Clin Microbiol (2005) 43:3101–9. 10.1128/JCM.43.7.3101-3109.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smit PW, Lindholm L, Lyytikäinen O, Jalava J, Pätäri-Sampo A, Vuopio J. Epidemiology and emm types of invasive group A streptococcal infections in Finland, 2008–2013. Eur J Clin Microbiol Infect Dis (2015) 34:2131–6. 10.1007/s10096-015-2462-2 [DOI] [PubMed] [Google Scholar]
- 28.Tapiainen T, Launonen S, Renko M, Saxen H, Salo E, Korppi M, et al. Invasive group A streptococcal infections in children: a nationwide survey in Finland. Pediatr Infect Dis J (2016) 35:123–8. 10.1097/INF.0000000000000945 [DOI] [PubMed] [Google Scholar]
- 29.Siljander T, Lyytikäinen O, Vähäkuopus S, Snellman M, Jalava J, Vuopio J. Epidemiology, outcome and emm types of invasive group A streptococcal infections in Finland. Eur J Clin Microbiol Infect Dis (2010) 29:1229–35. 10.1007/s10096-010-0989-9 [DOI] [PubMed] [Google Scholar]
- 30.Vähäkuopus S, Vuento R, Siljander T, Syrjänen J, Vuopio J. Distribution of emm types in invasive and non-invasive group A and G streptococci. Eur J Clin Microbiol Infect Dis (2012) 31:1251–6. 10.1007/s10096-011-1436-2 [DOI] [PubMed] [Google Scholar]
- 31.Olafsdottir LB, Erlendsdóttir H, Melo-Cristino J, Weinberger DM, Ramirez M, Kristinsson KG, et al. Invasive infections due to Streptococcus pyogenes: seasonal variation of severity and clinical characteristics, Iceland, 1975 to 2012. Euro Surveill (2014) 19:5–14. 10.2807/1560-7917.ES2014.19.17.20784 [DOI] [PubMed] [Google Scholar]
- 32.Naseer U, Steinbakk M, Blystad H, Caugant DA. Epidemiology of invasive group A streptococcal infections in Norway 2010–2014: a retrospective cohort study. Eur J Clin Microbiol Infect Dis (2016) 35:1639–48. 10.1007/s10096-016-2704-y [DOI] [PubMed] [Google Scholar]
- 33.Meisal R, Andreasson IK, Høiby EA, Aaberge IS, Michaelsen TE, Caugant DA. Streptococcus pyogenes isolates causing severe infections in Norway in 2006 to 2007: emm types, multilocus sequence types, and superantigen profiles. J Clin Microbiol (2010) 48:842–51. 10.1128/JCM.01312-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oppegaard O, Mylvaganam H, Kittang BR. Beta-haemolytic group A, C and G streptococcal infections in Western Norway: a 15-year retrospective survey. Clin Microbiol Infect (2015) 21:171–8. 10.1016/j.cmi.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 35.Bruun T, Kittang BR, de Hoog BJ, Aardal S, Flaatten HK, Langeland N, et al. Necrotizing soft tissue infections caused by Streptococcus pyogenes and Streptococcus dysgalactiae subsp. equisimilis of groups C and G in western Norway. Clin Microbiol Infect (2013) 19:E545–50. 10.1111/1469-0691.12276 [DOI] [PubMed] [Google Scholar]
- 36.Kittang BR, Bruun T, Langeland N, Mylvaganam H, Glambek M, Skrede S. Invasive group A, C and G streptococcal disease in western Norway: virulence gene profiles, clinical features and outcomes. Clin Microbiol Infect (2011) 17:358–64. 10.1111/j.1469-0691.2010.03253.x [DOI] [PubMed] [Google Scholar]
- 37.Darenberg J, Luca-Harari B, Jasir A, Sandgren A, Pettersson H, Schalén C, et al. Molecular and clinical characteristics of invasive group A streptococcal infection in Sweden. Clin Infect Dis (2007) 45:450–8. 10.1086/519936 [DOI] [PubMed] [Google Scholar]
- 38.Chalker V, Jironkin A, Coelho J, Al-Shahib A, Platt S, Kapatai G, et al. Genome analysis following a national increase in scarlet fever in England 2014. BMC Genomics (2017) 18:224. 10.1186/s12864-017-3603-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curtis SJ, Tanna A, Russell HH, Efstratiou A, Paul J, Cubbon M, et al. Invasive group A streptococcal infection in injecting drug users and non-drug users in a single UK city. J Infect (2007) 54:422–6. 10.1016/j.jinf.2006.10.004 [DOI] [PubMed] [Google Scholar]
- 40.Lindsay DS, Brown AW, Scott KJ, Denham B, Thom L, Rundell G, et al. Circulating emm types of Streptococcus pyogenes in Scotland: 2011–2015. J Med Microbiol (2016) 65:1229–31. 10.1099/jmm.0.000335 [DOI] [PubMed] [Google Scholar]
- 41.Meehan M, Murchan S, Bergin S, O’Flanagan D, Cunney R. Increased incidence of invasive group A streptococcal disease in Ireland, 2012 to 2013. Euro Surveill (2013) 18:20556. 10.2807/1560-7917.ES2013.18.33.20556 [DOI] [PubMed] [Google Scholar]
- 42.Strakova L, Motlova J, Jakubu V, Urbaskova P, Kriz P. Emergence of a novel macrolide-resistant Streptococcus pyogenes emm53 strain. Clin Microbiol Infect (2007) 13:443–5. 10.1111/j.1469-0691.2006.01658.x [DOI] [PubMed] [Google Scholar]
- 43.Krucsó B, Gacs M, Libisch B, Hunyadi ZV, Molnár K, Füzi M, et al. Molecular characterisation of invasive Streptococcus pyogenes isolates from Hungary obtained in 2004 and 2005. Eur J Clin Microbiol Infect Dis (2007) 26:807–11. 10.1007/s10096-007-0359-4 [DOI] [PubMed] [Google Scholar]
- 44.Szczypa K, Sadowy E, Izdebski R, Strakova L, Hryniewicz W. Group A streptococci from invasive-disease episodes in Poland are remarkably divergent at the molecular level. J Clin Microbiol (2006) 44:3975–9. 10.1128/JCM.01163-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luca-Harari B, Straut M, Cretoiu S, Surdeanu M, Ungureanu V, van der Linden M, et al. Molecular characterization of invasive and non-invasive Streptococcus pyogenes isolates from Romania. J Med Microbiol (2008) 57:1354–63. 10.1099/jmm.0.2008/001875-0 [DOI] [PubMed] [Google Scholar]
- 46.Bidet P, Courroux C, Salgueiro C, Carol A, Mariani-Kurkdjian P, Bonacorsi S, et al. Molecular epidemiology of the sil streptococcal invasive locus in group A streptococci causing invasive infections in French children. J Clin Microbiol (2007) 45:2002–4. 10.1128/JCM.00104-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plainvert C, Doloy A, Joubrel C, Maataoui N, Dmytruk N, Touak G, et al. Characterization of Streptococcus pyogenes isolates responsible for adult meningitis in France from 2003 to 2013. Diagn Microbiol Infect Dis (2016) 84:350–2. 10.1016/j.diagmicrobio.2015.12.006 [DOI] [PubMed] [Google Scholar]
- 48.Lepoutre A, Doloy A, Bidet P, Leblond A, Perrocheau A, Bingen E, et al. Epidemiology of invasive Streptococcus pyogenes infections in France in 2007. J Clin Microbiol (2011) 49:4094–100. 10.1128/JCM.00070-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plainvert C, Loubinoux J, Bidet P, Doloy A, Touak G, Dmytruk N, et al. [Epidemiology of Streptococcus pyogenes invasive diseases in France (2007–2011)]. Arch Pediatr (2014) 21:S62–8. 10.1016/S0929-693X(14)72262-6 [DOI] [PubMed] [Google Scholar]
- 50.Plainvert C, Doloy A, Loubinoux J, Lepoutre A, Collobert G, Touak G, et al. Invasive group A streptococcal infections in adults, France (2006–2010). Clin Microbiol Infect (2012) 18:702–10. 10.1111/j.1469-0691.2011.03624.x [DOI] [PubMed] [Google Scholar]
- 51.d’Humières C, Bidet P, Levy C, Béchet S, Bonacorsi S, Bingen E, et al. Comparative epidemiology of Streptococcus pyogenes emm-types causing invasive and noninvasive infections in French children by use of high-resolution melting-polymerase chain reaction. Pediatr Infect Dis J (2015) 34:557–61. 10.1097/INF.0000000000000677 [DOI] [PubMed] [Google Scholar]
- 52.Imöhl M, van der Linden M, Reinert RR, Ritter K. Invasive group A streptococcal disease and association with varicella in Germany, 1996–2009. FEMS Immunol Med Microbiol (2011) 62:101–9. 10.1111/j.1574-695X.2011.00788.x [DOI] [PubMed] [Google Scholar]
- 53.Imöhl M, Reinert RR, Ocklenburg C, van der Linden M. Epidemiology of invasive Streptococcus pyogenes disease in Germany during 2003–2007. FEMS Immunol Med Microbiol (2010) 58:389–96. 10.1111/j.1574-695X.2010.00652.x [DOI] [PubMed] [Google Scholar]
- 54.Zachariadou L, Stathi A, Tassios PT, Pangalis A, Legakis NJ, Papaparaskevas J, et al. Differences in the epidemiology between paediatric and adult invasive Streptococcus pyogenes infections. Epidemiol Infect (2014) 142:512–9. 10.1017/S0950268813001386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stathi A, Papaparaskevas J, Zachariadou L, Pangalis A, Legakis NJ, Tseleni-Kotsovili A, et al. Prevalence of emm types 1 and 12 from invasive Streptococcus pyogenes disease in Greece – results of enhanced surveillance. Clin Microbiol Infect (2008) 14:808–12. 10.1111/j.1469-0691.2008.02032.x [DOI] [PubMed] [Google Scholar]
- 56.Creti R, Imperi M, Baldassarri L, Pataracchia M, Recchia S, Alfarone G, et al. emm types, virulence factors, and antibiotic resistance of invasive Streptococcus pyogenes isolates from Italy: what has changed in 11 years? J Clin Microbiol (2007) 45:2249–56. 10.1128/JCM.00513-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friães A, Lopes JP, Melo-Cristino J, Ramirez M, Portuguese Group for the Study of Streptococcal Infections Changes in Streptococcus pyogenes causing invasive disease in Portugal: evidence for superantigen gene loss and acquisition. Int J Med Microbiol (2013) 303:505–13. 10.1016/j.ijmm.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 58.Montes M, Ardanuy C, Tamayo E, Domènech A, Liñares J, Pérez-Trallero E. Epidemiological and molecular analysis of Streptococcus pyogenes isolates causing invasive disease in Spain (1998–2009): comparison with non-invasive isolates. Eur J Clin Microbiol Infect Dis (2011) 30:1295–302. 10.1007/s10096-011-1226-x [DOI] [PubMed] [Google Scholar]
- 59.Nelson GE, Pondo T, Toews KA, Farley MM, Lindegren ML, Lynfield R, et al. Epidemiology of invasive group A streptococcal infections in the United States, 2005–2012. Clin Infect Dis (2016) 63:478–86. 10.1093/cid/ciw248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z, Sakota V, Jackson D, Franklin AR, Beall B, Active Bacterial Core Surveillance/Emerging Infections Program Network . Array of M protein gene subtypes in 1064 recent invasive group A Streptococcus isolates recovered from the active bacterial core surveillance. J Infect Dis (2003) 188:1587–92. 10.1086/379050 [DOI] [PubMed] [Google Scholar]
- 61.Rudolph K, Bruce MG, Bruden D, Zulz T, Reasonover A, Hurlburt D, et al. Epidemiology of invasive group A streptococcal disease in Alaska, 2001 to 2013. J Clin Microbiol (2016) 54:134–41. 10.1128/JCM.02122-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bocking N, Matsumoto CL, Loewen K, Teatero S, Marchand-Austin A, Gordon J, et al. High incidence of invasive group A streptococcal infections in remote indigenous communities in Northwestern Ontario, Canada. Open Forum Infect Dis (2016) 4:ofw243. 10.1093/ofid/ofw243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Athey TB, Teatero S, Sieswerda LE, Gubbay JB, Marchand-Austin A, Li A, et al. High incidence of invasive group A Streptococcus disease caused by strains of uncommon emm types in Thunder Bay, Ontario, Canada. J Clin Microbiol (2016) 54:83–92. 10.1128/JCM.02201-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tyrrell GJ, Lovgren M, St Jean T, Hoang L, Patrick DM, Horsman G, et al. Epidemic of group A Streptococcus M/emm59 causing invasive disease in Canada. Clin Infect Dis (2010) 51:1290–7. 10.1086/657068 [DOI] [PubMed] [Google Scholar]
- 65.Zakikhany K, Degail MA, Lamagni T, Waight P, Guy R, Zhao H, et al. Increase in invasive Streptococcus pyogenes and Streptococcus pneumoniae infections in England, December 2010 to January 2011. Euro Surveill (2011) 16:19785. 10.2807/ese.16.05.19785-en [DOI] [PubMed] [Google Scholar]
- 66.Koutouzi F, Tsakris A, Chatzichristou P, Koutouzis E, Daikos GL, Kirikou E, et al. Streptococcus pyogenes emm types and clusters during a 7-year period (2007 to 2013) in pharyngeal and nonpharyngeal pediatric isolates. J Clin Microbiol (2015) 53:2015–21. 10.1128/JCM.00301-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Efstratiou A, Lamagni T. Epidemiology of Streptococcus pyogenes. In: Ferretti JJ, Stevens DL, Fischetti VA, editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City, OK: University of Oklahoma Health Sciences Center; (2016) p. 437–57. [PubMed] [Google Scholar]
- 68.Creti R. Have group A and B streptococcal infections become neglected diseases in Europe? Eur J Clin Microbiol Infect Dis (2017) 36:1063–4. 10.1007/s10096-017-2984-x [DOI] [PubMed] [Google Scholar]
- 69.Dale JB, Penfound TA, Tamboura B, Sow SO, Nataro JP, Tapia M, et al. Potential coverage of a multivalent M protein-based group A streptococcal vaccine. Vaccine (2013) 31:1576–81. 10.1016/j.vaccine.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu MC, Walls MA, Stroop SD, Reddish MA, Beall B, Dale JB. Immunogenicity of a 26-valent group A streptococcal vaccine. Infect Immun (2002) 70:2171–7. 10.1128/IAI.70.4.2171-2177.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smeesters PR, Laho D, Beall B, Steer AC, Van Beneden CA. Seasonal, geographic, and temporal trends of emm clusters associated with invasive group A streptococcal infections in US multistate surveillance. Clin Infect Dis (2017) 64:694–5. 10.1093/cid/ciw807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilkening RV, Federle MJ. Evolutionary constraints shaping Streptococcus pyogenes-host interactions. Trends Microbiol (2017) 25:562–72. 10.1016/j.tim.2017.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog (2006) 2:e5. 10.1371/journal.ppat.0020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beres SB, Kachroo P, Nasser W, Olsen RJ, Zhu L, Flores AR, et al. Transcriptome remodeling contributes to epidemic disease caused by the human pathogen Streptococcus pyogenes. MBio (2016) 3:e403–16. 10.1128/mBio.00403-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Al-Shahib A, Underwood A, Afshar B, Turner CE, Lamagni T, Sriskandan S, et al. Emergence of a novel lineage containing a prophage in emm/M3 group A Streptococcus associated with upsurge in invasive disease in the UK. Microb Genom (2016) 2:e000059. 10.1099/mgen.0.000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ikebe T, Ato M, Matsumura T, Hasegawa H, Sata T, Kobayashi K, et al. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog (2010) 6:e1000832. 10.1371/journal.ppat.1000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Friães A, Pato C, Melo-Cristino J, Ramirez M. Consequences of the variability of the CovRS and RopB regulators among Streptococcus pyogenes causing human infections. Sci Rep (2015) 5:12057. 10.1038/srep12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Danger JL, Cao TN, Cao TH, Sarkar P, Trevino J, Pflughoeft KJ, et al. The small regulatory RNA FasX enhances group A Streptococcus virulence and inhibits pilus expression via serotype-specific targets. Mol Microbiol (2015) 96:249–62. 10.1111/mmi.12935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kreikemeyer B, Boyle MD, Buttaro BA, Heinemann M, Podbielski A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol Microbiol (2001) 39:392–406. 10.1046/j.1365-2958.2001.02226 [DOI] [PubMed] [Google Scholar]
- 80.Cao TN, Liu Z, Cao TH, Pflughoeft KJ, Trevino J, Danger JL, et al. Natural disruption of two regulatory networks in serotype M3 group A Streptococcus isolates contributes to the virulence factor profile of this hypervirulent serotype. Infect Immun (2014) 82:1744–54. 10.1128/IAI.01639-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chalker VJ, Smith A, Al-Shahib A, Botchway S, Macdonald E, Daniel R, et al. Integration of genomic and other epidemiologic data to investigate and control a cross-institutional outbreak of Streptococcus pyogenes. Emerg Infect Dis (2016) 22:973–80. 10.3201/eid2206.142050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tagini F, Aubert B, Troillet N, Pillonel T, Praz G, Crisinel PA, et al. Importance of whole genome sequencing for the assessment of outbreaks in diagnostic laboratories: analysis of a case series of invasive Streptococcus pyogenes infections. Eur J Clin Microbiol Infect Dis (2017) 36:1173–80. 10.1007/s10096-017-2905-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.