Abstract
Guard cells represent a highly differentiated cell type within the epidermis of plant leaves and stems. They respond to many endogenous and environmental signals and thereby modify the size of the stomatal pore they surround. We identified a novel gene that is highly expressed in guard cells of potato (Solanum tuberosum). It encodes a repetitive proline (Pro)-rich protein of 54 kD (491 amino acids) and was named StGCPRP (S. tuberosum guard cell Pro-rich protein). StGCPRP has a bipartite structure. The C-terminal part of StGCPRP contains a high percentage (46%) of Pro residues organized in distinct repetitive sequence motifs, whereas its extended N terminus is essentially free of Pros. StGCPRP represents the first member of a novel class of hybrid Pro-rich proteins that we designated NHyPRPs. In young but not in mature leaves, StGCPRP transcripts were also present at high levels in mesophyll cells (in addition to guard cells), indicating developmental regulation of StGCPRP gene expression. In addition, StGCPRP expression is regulated by environmental factors, as shown by a decrease in StGCPRP transcript levels under drought stress. Two proteins similar to StGCPRP were found to be encoded by the Arabidopsis genome, indicating that NHyPRPs are more widely distributed in higher plants.
Stomatal guard cells are highly specialized and differentiated plant cells that play a critical role in metabolism by modulating gas exchange in photosynthetic tissues. They respond to various endogenous and environmental signals, including hormones (e.g. abscisic acid), light, atmospheric CO2 concentration, and humidity (Willmer and Fricker, 1996). Stomata have been studied extensively over the last few years using a variety of methods, including: (a) electrophysiology to characterize ion channels in the guard cell plasma membrane and vacuolar membrane (e.g. Schroeder, 1992; Assmann, 1993); (b) ion imaging on single guard cells to uncover the role of second messengers such as Ca2+ in stomatal responses (Webb et al., 1996); (c) biochemical studies to compare the protein composition in guard cells with that in other leaf cells (e.g. Ohya and Shimazaki, 1989; Cotelle, 1997); and (d) gas exchange measurements, which allow the analysis of stomatal opening and closing responses in whole leaves and plants (Zeiger et al., 1987; Mansfield et al., 1990). In contrast, rather limited emphasis has so far been given to a molecular biological analysis of guard cell function and differentiation and only relatively few genes have been identified that contribute to the highly specialized nature of guard cells (for review, see Müller-Röber et al., 1998).
The cell wall of guard cells is subjected to extreme forces during swelling and deswelling while stomata open and close. The stomatal cell wall has a highly organized morphology and its biochemical composition and physical properties differ from those of other leaf cells (Sack, 1987; Willmer and Fricker, 1996). The chemical composition of the guard cell wall determines its mechanical and storage properties. For example, deposition of lignin into the stomatal cell wall would increase the wall's rigidity. The wall of guard cells in some species has been reported to be rich in pectins (Sack, 1987), which, due to their negative charges, may significantly contribute to temporary storage of potassium ions during stomatal movements. In general, the walls of plant cells also contain various types of proteins, including those highly enriched in Pro residues. During development, environmental stress, and pathogen infection, the composition and structure of the cell wall is continuously rearranged due to enzyme action (Cassab and Varner, 1988). Pro-rich proteins are thought to participate in these processes and may therefore have important biological functions. Here we demonstrate that guard cells of potato (Solanum tuberosum) express a gene that encodes a novel type of repetitive Pro-rich protein (RPRP).
RPRPs are a subgroup of the Hyp-rich glycoproteins (HRGPs), a major class of structural proteins present in the primary cell wall of higher plants. HRGPs can be subdivided into five groups (Kieliszewski and Lamport, 1994; Cassab, 1998): (a) the extensins, characterized by multiple copies of the amino acid motif S(Hyp)4; (b) RPRPs, which lack this motif; (c) gum arabic glycoproteins; (d) arabinogalactan proteins (AGPs); and (e) chimeric proteins such as potato lectin. The repeat sequences of RPRPs vary considerably between plant species and between RPRPs present in an individual species. The common feature of these proteins is the massive presence of Pro residues that occur in repeating structures of at least two consecutive Pros. Expression of RPRP and extensin genes has been found to be regulated developmentally and cell type specifically on the level of transcription and in response to wounding and pathogen attack (Reiter, 1994). Immunolocalization of RPRPs has shown their preferential deposition in the cell wall of tissues that become lignified during development, such as vascular tissues, stems, and roots (Ye and Varner, 1991; Ye et al., 1991). It has been proposed that RPRPs are cross-linked to extensins, allowing them to lock the cellulose microfibrils within the three-dimensional network of the cell wall (Carpita and Gibeaut, 1993). Although these studies indicated that RPRPs are involved in strengthening the cell wall, their precise function has yet to be established.
We were interested in the molecular and genetic aspects of guard cell biology. To identify novel genes preferentially expressed in stomatal guard cells, we employed a differential screening protocol using radiolabeled cDNA derived from either epidermal fragment or whole leaf mRNA of potato. Epidermal fragments are highly enriched for guard cells (Kopka et al., 1997). We report the cloning of a cDNA, StGCPRP (S. tuberosum guard cell Pro-rich protein) encoding a novel type of RPRP. The StGCPRP protein contains unique sequence motifs in its Pro-rich part, as well as an extended N terminus, defining a novel type of hybrid RPRPs that we designate NHyPRPs. The StGCPRP gene is strongly expressed in guard cells, indicating that its encoded protein has an important function in the stomatal cell wall.
MATERIALS AND METHODS
Enzymes and Chemicals
Enzymes used for DNA restriction and modification were purchased from Boehringer Mannheim (Mannheim, Germany) and New England Biolabs (Danvers, MA). Sequencing primers were obtained from TibMolbiol (Berlin). Unless otherwise indicated, other chemicals were purchased from Boehringer Mannheim, Merck (Darmstadt, Germany), or Sigma-Aldrich (St. Louis).
Plant Material
Potato (Solanum tuberosum L. cv Desirée) plants were obtained from Saatzucht Fritz Lange (Bad Schwartau, Germany) and were grown in the green house with 16 h of light at 22°C and 8 h of darkness at 15°C, 60% humidity. Plants were usually harvested after 6 weeks of growth. For in situ hybridizations, leaves of 3-week-old plants were used. To induce drought stress, potato plants were treated as described by Kopka et al. (1997). For CO2 treatments, potato plants were grown from in vitro-propagated plantlets for 6 weeks in two separate growth chambers floated with air containing 350 μL L−1 CO2. The CO2 concentration was then increased in one of the chambers to 750 μL L−1 for 4 d.
Differential cDNA Library Screening and DNA Sequence Analysis
An oriented cDNA library in λ-ZAP II (Stratagene, La Jolla, CA) was constructed using poly(A+) RNA from epidermal fragments of fully developed potato leaves. 2 × 105 plaque forming units of the library were differentially screened using 32P-labeled first-strand cDNA prepared from poly(A+) RNA of epidermal fragments and mature leaves. Phage clones giving intense signals with the epidermal fragment probe but not with the leaf probe were selected and subjected to two further rounds of differential screening until single phage plaques were obtained. Phages were in vivo-excised with helper phage (ExAssist, Stratagene) and plasmids were isolated using standard procedures (Sambrook et al., 1989). Escherichia coli strain XL-1 Blue (Stratagene) was used for most experimental procedures. Genomic Southern-blot analysis was essentially performed as described previously (Fieuw et al., 1995) using the full-length StGCPRP cDNA as 32P-labeled hybridization probe.
DNA sequencing was performed using the T7 sequencing kit from Pharmacia Biotech (Heidelberg). Analysis of similarity to entries in the GenBank database was performed using the BLAST server at the National Center for Biological Information (NCBI) and the programs FastA, PileUp, and BLAST of the GCG software package version 8 (Genetics Computer Group, Madison, WI). Signal peptide prediction was carried out as described previously (http://genome.cbs.dtu.dk/services/; Nielsen et al., 1997). For the analysis of hydrophobicity, amino acid composition, and charge, the programs PEPWINDOW (Kyte-Doolittle prediction) and PEPSTATS of the GCG software package were used. Secondary structure was predicted by computer programs as described previously (Levin et al., 1986; Deleage and Roux, 1987; Gibrat et al., 1987; Geourjon and Deleage, 1995) and transmembrane spanning regions were predicted by the program TMpred provided by the ExPASy internet server (http://www.expasy.ch; Wilkins et al., 1999).
RNA-Blot Experiments
Epidermal fragments were isolated as described by Kopka et al. (1997) and were homogenized by grinding them for 5 min in an electric mortar device (MM 2000, Retsch, Haan, Germany) that was precooled with liquid nitrogen. RNA from epidermal fragments was isolated as described previously (Kopka et al., 1997) using a CsCl cushion-based method. RNA from other tissues was isolated as described by Logemann et al. (1987). Leaf RNA was isolated by hand-grinding leaves under liquid nitrogen for approximately 30 s. Under these conditions guard cells of mature leaves remained mostly intact, resulting in RNA preparations essentially free of guard cell RNA. RNA-blot analysis was performed as described by Kopka et al. (1997) using either the complete StGCPRP cDNA or a 5′ end of it (omitting the region encoding the Pro-rich C-terminal domain) as 32P-labeled hybridization probe. Thirty to 40 μg of total RNA was loaded per lane. Signal intensities were quantified using a phosphor imager (Molecular Dynamics, Krefeld, Germany) and ImageQuant software, version 3.3 (Molecular Dynamics).
To exclude that the strong StGCPRP expression in guard cells was induced by wounding during the preparation of epidermal fragments, the following control experiment was performed. Leaves of different developmental stages were harvested from 6-week-old plants and, after the removal of major veins with a razor blade, were cooled for 20 min (i.e. the time usually needed to prepare epidermal fragments) in water supplemented with ice. Subsequently, leaves were homogenized in a blender (see Kopka et al., 1997). Epidermal fragments were removed by sieving through a 220-μm nylon sieve, and RNA was isolated from the epidermal fragment-free homogenate. Control leaves were directly frozen in liquid nitrogen after cutting from the plant, and RNA was prepared after grinding the frozen tissue in a mortar.
In Situ Hybridization
In situ hybridizations were performed as described by Van de Wiel et al. (1990), with the following modifications: leaves were fixed in 4% (v/v) paraformaldehyde and 0.25% (v/v) glutaraldehyde in 0.01 m sodium phosphate buffer, pH 7.2, at room temperature. One microgram/microliter tRNA from yeast (Merck) and 2 μg/μL poly(A+) RNA (Merck) were added to the hybridization solution. The RNase treatment after hybridization was performed with 50 μg/mL RNase A and the slides were washed subsequently six times for 15 min in buffer without RNase. Washes were performed down to 2× SSC.
To exclude cross-hybridization between the StGCPRP probe and unrelated mRNA sequences encoding Pro-rich proteins, a DNA fragment spanning nucleotides 1 through 505 of the StGCPRP cDNA was generated by PCR and cloned into vector pBluescript SK (Stratagene). This fragment encodes the N-terminal region of the StGCPRP protein, which is essentially free of Pro residues. Antisense and sense RNA probes were produced using 35S-labeled α-dUTP (Amersham, Braunschweig, Germany).
RESULTS
Cloning of a cDNA Encoding a Pro-Rich Protein from Potato Guard Cells
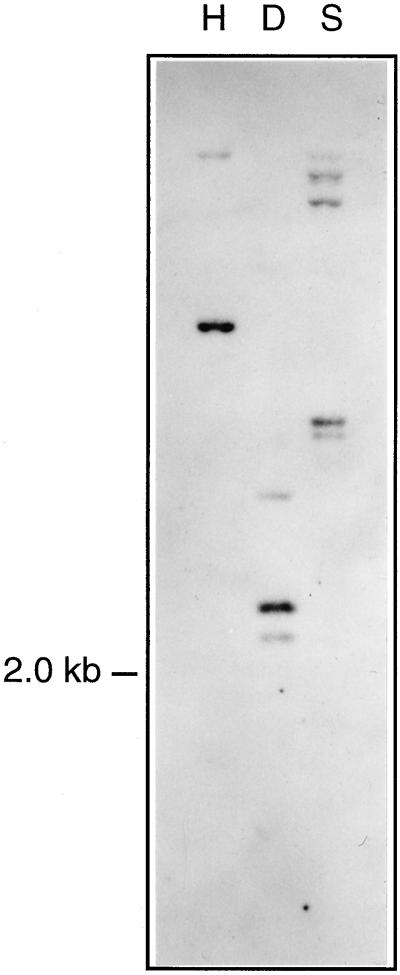
A differential screening approach was employed to identify genes preferentially expressed in stomatal guard cells of potato (for details, see “Materials and Methods”). An epidermal fragment cDNA library was hybridized against radiolabeled first-strand cDNA obtained from whole-leaf mRNA, followed by a second hybridization against cDNA established from epidermal fragment RNA (epidermal fragments are a rich source for guard cells; Kopka et al., 1997). Purified phage clones preferentially hybridizing to the epidermal fragment probe were processed for further analysis. One class of cDNAs was found to be highly abundant among the clones selected. The nucleotide sequence of the longest cDNA of this class was determined and deposited in the GenBank database under accession number AJ000997. The 1,892-bp long cDNA codes for a protein of 491 amino acids (Fig. 1) with a calculated molecular mass of 54 kD. The deduced protein shows similarity to previously identified plant RPRPs, and was therefore named StGCPRP. Southern-blot analysis performed under stringent hybridization conditions indicated that StGCPRP is encoded by a single or low-copy gene (Fig. 2).
Figure 1.
Amino acid sequence of the StGCPRP protein as deduced from its corresponding cDNA sequence. The N-terminal signal peptide is written in bold type. The six DxK motifs present in the N-terminal part of the protein as well as one similar ExK motif are underlined. Pro-rich repeats (partially overlapping) present within the Pro-rich domain are highlighted (see also Table I).
Figure 2.
Southern-blot analysis of the StGCPRP gene. Genomic DNA (50 μg/lane) was restricted with the enzymes HindIII (H), DraI (D), and SpeI (S) and hybridized against radiolabeled StGCPRP cDNA. Single restriction sites are present for HindIII and SpeI, and two sites are present for DraI in the corresponding StGCPRP cDNA.
StGCPRP Protein Has an Unusual Bipartite Structure
Figure 1 shows the amino acid sequence of the StGCPRP protein as deduced from its cDNA sequence. A distinct domain starting at amino acid position 160 is highly enriched for Pro residues (46%) and also contains many Lys (15%) and Val (12%) residues. This Pro-rich domain is N-terminally preceded by a domain essentially lacking Pro residues. The two parts of the protein are highly different in their charge values, with the N terminus being slightly negatively charged (charge value of −3, isoelectric point [pI] of 6, as calculated with the PEPSTATS program) and the Pro-rich domain being strongly positively charged (charge value of +45, pI of 11). This bipartite feature of StGCPRP is in accordance with the secondary structure of the protein, as predicted by four different computer programs, which indicated a mixture of helical and wheel structures at the N terminus and a coil structure in the Pro-rich domain (data not shown). The extreme N terminus of StGCPRP appeared to be highly hydrophobic, indicating the presence of a signal peptide (Fig. 1). No further transmembrane regions could be identified using the TMpred program.
As indicated in Figure 1 and summarized in Table I, four different types of repetitive motifs 12 to 18 amino acids long are present in the Pro-rich domain of StGCPRP. Apart from these repeats, several stretches typically containing the sequence motif PPx were detected (x mostly representing the amino acids Phe or Leu). According to rules developed by Kieliszewski and Lamport (1994), several of the Pro residues in StGCPRP may be subject to post-translational modifications, including hydroxylations or glycosylations. Hydroxylations may occur at the following positions: (a) at Pro residues within the dipeptide PV (present 24 times in StGCPRP); (b) at the first and last Pro within the motif PKPP (motif present at amino acid positions 265, 277, and 316); (c) at all Pros when present in blocks of at least three (11 times in StGCPRP). Furthermore, hydroxyPros occurring in pairs (11 times in StGCPRP) may be glycosylated with arabinosyl residues (Kieliszewski and Lamport, 1994).
Table I.
Pro-rich repeats present in StGCPRP
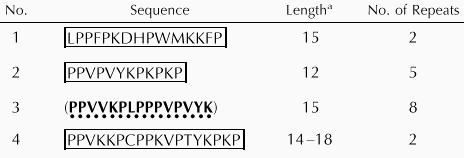 |
No. of amino acids.
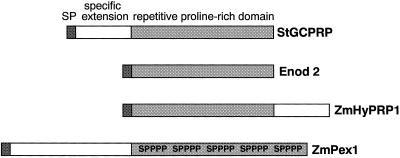
The N-terminal extension of StGCPRP contains seven copies of a D/ExK motif. It is difficult to ascertain whether such a small motif is a true functional motif. A similar but Pro-rich element, PEPK, was previously identified as a highly repetitive segment in WPRP1 from wheat (Raines et al., 1991) and in ZmPRP from maize (Vignols et al., 1999). Figure 3 shows a schematic presentation of the domain structure of StGCPRP and compares it with structures of other HRGPs. Within its Pro-rich domain, StGCPRP shows highest similarities to the maize (49%) and wheat (42%) proteins. StGCPRP is the first protein within the group of RPRPs containing a marked N-terminal extension. The only other class of HRGPs in which members with an extended N terminus exist are the extensins; however, extensins are characterized by the presence of multiple S(Hyp)4 motifs that are completely absent from StGCPRP.
Figure 3.
Structure of StGCPRP and representative examples of other repetitive Pro-rich proteins. Enod2 (Franssen et al., 1987) is a typical RPRP with Pro-rich repeats along the whole protein. Maize ZmHyPRP1 (Josè-Estanyol et al., 1992) is a member of the so-called hybrid RPRPs (see “Discussion”). Maize ZmPex1 (Rubinstein et al., 1995) is an extensin with multiple S(Hyp)4 motifs (shown as SPPPP) within the Pro-rich domain. Pex1 is one of two examples of extensins harboring an N-terminal extension. Note that the S(Hyp)4 motif is absent from RPRPs. SP, Signal peptide.
Using the StGCPRP sequence as a bait, two homologous Arabidopsis BAC clones (T9A14.50, accession no. AL035656, and F26H11.10, accession no. AC006264) were identified, both of which harbored sequences coding for RPRPs with a bipartite structure but were smaller in size (448 and 321 amino acids, respectively). Within the N-terminal extension, the two Arabidopsis proteins share 68% identical (81% similar) amino acids, and both exhibited around 44% identity (65% similarity) to the N-terminal part of StGCPRP (not shown). Notably, these extensions are also similar in size, being around 150 to 160 amino acids long. As in StGCPRP, the C-terminal parts of the Arabidopsis RPRPs are highly enriched for Pro residues, indicating a more general occurrence of these proteins in higher plants. Interestingly, the C-terminal regions exhibit considerable size variation (they are approximately 290 and 160 amino acids long, respectively, in the two Arabidopsis proteins, and 330 amino acids long in StGCPRP) and they are less conserved (in terms of defined sequence elements) than the N termini. Variation in sequence motifs is also known from other RPRPs.
The StGCPRP Gene Is Strongly Expressed in Guard Cells
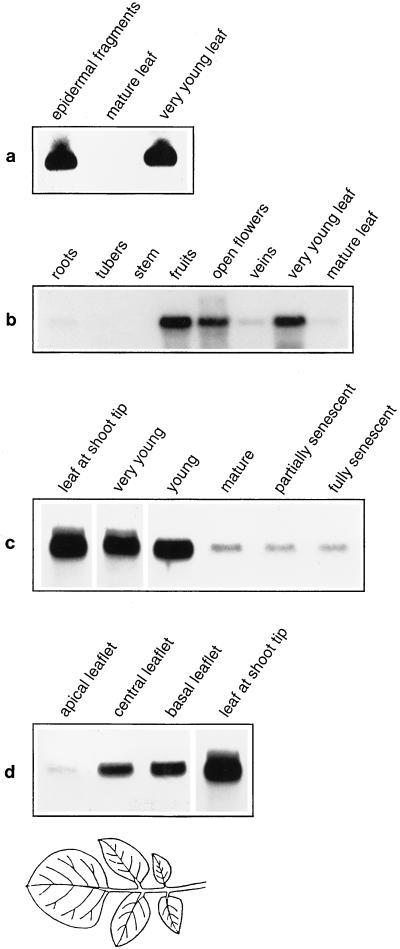
To confirm that StGCPRP is expressed in stomatal guard cells, we initially performed northern-blot experiments. RNA was extracted from epidermal fragments, as well as from fully developed leaves of greenhouse-grown potato plants. The result of a typical experiment is shown in Figure 4a. Very high StGCPRP mRNA levels were detected in epidermal fragments, indicating strong StGCPRP gene transcription in guard cells. By contrast, much lower StGCPRP transcript levels were present in mature (i.e. fully developed) leaves, confirming a differential expression in guard cells and leaves. However, StGCPRP also appeared to be strongly expressed during the early stages of leaf development (Fig. 4a), as well as in flowers and potato fruits, whereas expression was significantly weaker in stems, roots, and tubers (Fig. 4b). Expression was also low in vascular tissues (veins), which distinguishes StGCPRP from many other RPRP genes.
Figure 4.
Northern-blot analysis of StGCPRP transcript levels in potato tissues. a, RNA extracted from epidermal fragments, from mature leaves, and from very young leaves (harvested from the shoot apex). Note that epidermal fragments originated from mature leaves. b, RNA isolated from various potato tissues. The northern blot indicates strong expression of the StGCPRP gene in flowers and developing fruits. c, RNA extracted from leaves of different developmental stages. d, RNA from different leaflets of the most developed leaves from a 6-week-old plant. The position of the leaflets is schematically indicated below the autoradiogram. For comparison, signal intensitiy is shown for a leaf harvested from the shoot tip.
Regulation of StGCPRP Gene Expression during Leaf Development
As shown above, StGCPRP transcript levels were very low in fully developed leaves, but were very high in young leaves. To further analyze StGCPRP mRNA levels during leaf development, total RNA was isolated from whole leaves of different developmental stages, ranging from very young leaves (located close to the shoot tip) to senescent leaves and was probed in northern-blot experiments using the StGCPRP cDNA as a hybridization probe. As can be seen in Figure 4c, expression of StGCPRP was highest in the youngest leaves. The amount of StGCPRP mRNA decreased progressively when leaves matured and was lowest in fully developed leaves. No further change in StGCPRP transcript level was apparent at the early or late stages of leaf senescence.
A mature potato leaf is composed of several (usually five to seven) leaflets of different developmental stages. The tip leaflet constitutes the oldest part of the compound leaf, whereas the youngest leaflets (which occur pairwise) are found at the leaf base. To determine whether StGCPRP expression varies between these different parts of a leaf, RNA was isolated from basal, central, and apical leaflets of the most developed (but still not senescent) leaves from 6-week-old potato plants, and probed in a northern-blot experiment against the StGCPRP cDNA. As seen in Figure 4d, almost no StGCPRP transcript was present in the tip leaflet. In contrast, StGCPRP mRNA was easily detected in central and basal leaflets, i.e. in the younger parts of the leaf. Transcript levels in these tissues were lower than those in the youngest (whole) leaves harvested from the shoot tip (Fig. 4d, right lane). These data demonstrate that StGCPRP is not only expressed in stomatal guard cells but that its expression is also developmentally regulated throughout leaf development.
mRNA in Situ Hybridization Confirms Expression of StGCPRP in Guard Cells
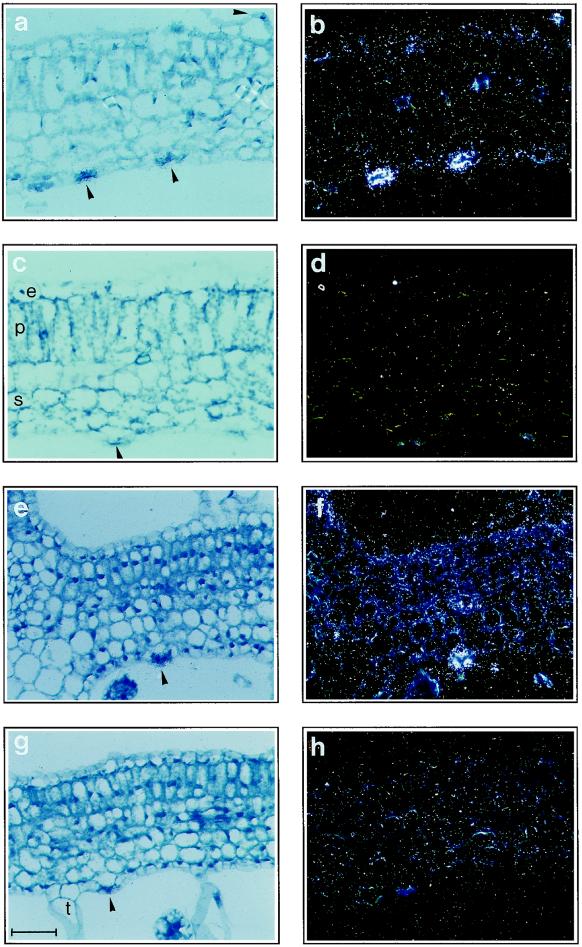
To confirm expression of StGCPRP in guard cells, mRNA in situ hybridization was performed on cross-sections of potato leaves. When hybridized to the StGCPRP antisense probe, very intense signals were observed in guard cells of mature leaves, whereas only background signals (or signals slightly above background) were detected in mesophyll cells (Fig. 5, a and b). StGCPRP mRNA was detected in abaxial and in adaxial stomatal guard cells. Significant StGCPRP RNA levels were also found within the epidermal cell layer of mature leaves (Fig. 5b), although expression was much weaker than in guard cells. Only background signals were detected when the sense RNA probe was used for hybridizations (Fig. 5, c and d), indicating specificity of the reaction. We further analyzed StGCPRP mRNA levels in cross-sections of young potato leaves (Fig. 5, e–h), and found that StGCPRP is also strongly expressed in guard cells of immature leaves (Fig. 5, e and f). In addition, StGCPRP-specific hybridization signals were detected in various other cell types of the young leaf, including pallisade and spongy mesophyll cells (Fig. 5f). In leaves of intermediate developmental stages, the levels of detectable mRNA in epidermal and mesophyll cells were lower than in very young leaves but still stronger than in mature leaves (data not shown). In situ hybridization results confirmed the expression pattern observed in northern-blot experiments (see above).
Figure 5.
In situ hybridization to demonstrate StGCPRP mRNA in cross-sections of leaves. a, b, e, and f, Hybridization with the StGCPRP antisense probe; c, d, g, and h, hybridization with the StGCPRP sense control probe. a through d, Mature leaves; e through h, very young leaves. Note strong expression in guard cells (arrowhead) as well as expression in mesophyll cells (f). e, Epidermal cell layer; p, pallisade parenchyma cells; s, spongy parenchyma cells; t, trichome. Bar = 50 μm.
Moderate Drought Stress Reduces StGCPRP Transcript Levels
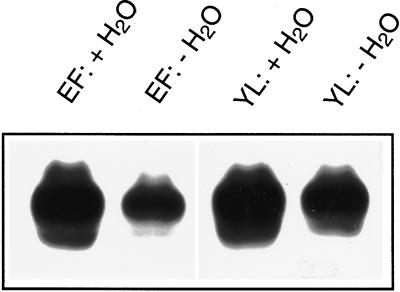
Guard cells are known to respond to moderate drought stress by stomatal closing (Willmer and Fricker, 1996), and this has also been demonstrated for guard cells from potato (Kopka et al., 1997). In addition, it has recently been recognized that drought leads to complex changes in guard cell gene expression (e.g. Kopka et al., 1997), which may go along with alterations of stomatal properties (Raschke, 1987; Hirasawa et al., 1995). When drought stress was applied to plants by withholding water for 4 d, StGCPRP transcript levels were markedly decreased (by almost 70%) in epidermal fragments isolated from mature leaves (Fig. 6), indicating that in guard cells drought stress targets the StGCPRP gene. Similarly, StGCPRP mRNA levels were reduced (by approximately 50%) in young leaves harvested from the shoot apex of moderately droughted potato plants (Fig. 6).
Figure 6.
Effect of moderate drought stress on StGCPRP transcript levels in epidermal fragments (EF) and very young leaves (YL). Tissues were harvested from well-watered plants (+H2O) or from plants moderately drought-stressed for 4 d (−H2O). Conditions of drought treatments are described in detail in Kopka et al. (1997).
It is well known that an elevation of atmospheric CO2 concentration induces stomatal closure in many plant species (Morison, 1987; Mansfield et al., 1990). StGCPRP gene expression was unaffected by increasing the ambient CO2 concentration from 350 to 750 ppm for 4 d (data not shown).
DISCUSSION
StGCPRP Defines a Novel Type of Pro-Rich Proteins
We described the isolation and characterization of a cDNA encoding a novel RPRP from potato, which we designated StGCPRP. The StGCPRP cDNA codes for a protein of 491 amino acids, which harbors two parts, an N-terminal part (amino acids 1–159) almost free of Pro-residues and a C-terminal Pro-rich domain (amino acids 160–491).
The group of HRGPs can be divided into five subgroups (see introduction; Kieliszewski and Lamport, 1994; Cassab, 1998). The StGCPRP protein shows similarities to members of two subgroups, i.e. extensins and RPRPs. Extensins contain at least one copy (but usually many) of the sequence motif S(Hyp)4. StGCPRP completely lacks this motif and containes only a single Ser residue in its Pro-rich domain (Fig. 1). Although StGCPRP contains a high percentage of Pro residues, several features clearly distinguish it from currently known members of the RPRP family.
First, StGCPRP does not exhibit comprehensive sequence homology to known RPRPs. Similarity to RPRPs is mainly due to the massive amount of Pro residues (46%) within the Pro-rich domain of StGCPRP. The most similar repetitive Pro-rich proteins, i.e. WPRP1 from wheat (Raines et al., 1991) and ZmPRP from maize (Vignols et al., 1999), have a similarly high percentage of Pro residues and also a high Lys content. The similarity between StGCPRP and these other proteins is solely based on the presence of these amino acids but not on defined sequence motifs, indicating that they are divergent members of the RPRP subgroup.
Second, the StGCPRP Pro-rich domain harbors four different repeat units 12 to 18 amino acids long. In strong contrast, wheat WPRP1 and many other RPRPs, such as Enod2 (Franssen et al., 1987) and SbPRP1–3 from soybean (Hong et al., 1987, 1989; Datta and Marcus, 1990) consist of multiple repeat units of a single short amino acid motif, which are distributed throughout the whole protein.
Third, StGCPRP contains a unique N terminus that essentially lacks Pro residues. Josè and Puigdomènech (1993) further subdivided RPRPs into three classes: (a) proteins with Pro-rich repeats along the whole protein but lacking cysteines (e.g. Enod2 and WPRP1); (b) RPRPs that are non-repetitive and contain only short stretches enriched for Pros (e.g. in sunflower SF18; Evrard et al., 1991); (c) HyPRP consisting of a repetitive Pro-rich domain at the N terminus and a hydrophobic Cys-rich domain at the C terminus (e.g. in maize ZmHyPRP1; Josè-Estanyol et al., 1992). Although the StGCPRP identified here also contains a bipartite structure, it is clearly different from other proteins belonging to the HyPRP subgroup. The Pro-poor part of StGCPRP is located at the N terminus and is not rich in Cys residues nor does it exhibit hydrophobic features (with the exception of the signal peptide). We conclude that StGCPRP represents the first member of a previously unknown class of Pro-rich proteins. We named this class the NHyPRPs to demonstrate that its members are hybrid Pro-rich proteins containing an extended Pro-poor domain at their N terminus. An extended N terminus poor in Pro residues has so far only been observed in the two extensins, Pex1 from maize (Rubinstein et al., 1995; see Fig. 3) and ISG from Volvox (Ertl et al., 1992). However, their N-termini do not show any similarity with the N terminus of StGCPRP, and, as discussed above, StGCPRP is undoubtedly not a member of the extensin family. Two proteins with a structure similar to potato StGCPRP could be deduced from Arabidopsis BAC sequences (see “Results”), indicating that NHyPRPs are not unique to potato but are more widely distributed in higher plants.
Several HRGPs have been shown to be located in the cell wall (Stafstrom and Staehelin, 1988). Computer predictions indicate that StGCPRP contains an N-terminal peptide of 26 amino acids, which may serve as a targeting signal for transport into the cell wall. Several components of the cell wall are known to be binding partners of HRGPs. The Tyr residues of HRGPs are essential for cross-linking to each other by isodityrosine bonds (Cooper and Varner, 1984; Fry, 1986). Analysis of StGCPRP suggests that this protein may also be cross-linked to cell wall components:
The amino acid motif VYK occurs several times in StGCPRP. The VYK motif was found in extensins known to be selectively cross-linked by an isolated 4.6 peroxidase isoform in vitro (Schnabelrauch et al., 1996). However, the VYK motif alone was not sufficient for cross-linking. High levels of hydroxylation and glycosylation were additional prerequisites for the cross-linking activity by this peroxidase isoform. Bradley et al. (1992) demonstrated an elicitor-induced and peroxidase-mediated insolubilization of VYK-containing RPRPs within the plant cell wall, indicating that cross-linking behavior is not restricted to extensins. Although direct biochemical evidence is missing for StGCPRP up to now, rules given by Kieliszewski and Lamport (1994) support the view that more than one-third of the Pro residues of StGCPRP may be hydroxylated and several of these may also be glycosylated with arabinosyl residues (see “Results”).
The Pro-rich domain of StGCPRP has a strong positive charge (+45), potentially allowing ionic interactions (potentially mediated by the many Lys residues) with acidic cell wall pectins (Showalter and Rumeau, 1990).
Keller (1993) proposed cross-linking between RPRPs and GRPs via isodityrosine bonds or ionic bridges between Lys residues of RPRPs and Gln residues of GRPs. We have recently identified cDNAs encoding two different GRPs of potato guard cells (U. Menke and B. Müller-Röber, unpublished data), indicating that interaction between RPRPs (StGCPRP) and GRPs may also occur in this cell type.
StGCPRP Transcripts Are Most Prominent in Stomatal Guard Cells and in Young Leaves
StGCPRP was initially identified as a gene strongly expressed in epidermal fragments but not in mature leaves of potato. Upon closer inspection, however, we found that StGCPRP transcript levels were also high in very young leaves, demonstrating that StGCPRP expression is not only cell specifically, but also developmentally regulated in whole leaves. This view is supported by the observation that within expanded leaves, high StGCPRP transcript levels are present in young leaflets, whereas low mRNA levels are found in older leaflets present at the leaf tip (Fig. 4d). In situ hybridizations proved that StGCPRP mRNA is most abundant in guard cells (Fig. 5). In mature leaves, StGCPRP transcript was also detected in other epidermal cells, although expression was much weaker. In young leaves, StGCPRP transcripts could also be detected in a variety of other leaf cells, including palisade and spongy parenchyma cells.
From the RNA expression data (northern-blot and in situ hybridization experiments) we conclude that StGCPRP gene expression changes from a more general expression at early stages of leaf development to a rather restricted expression in guard cells of mature leaves. To our knowledge, high expression in guard cells has never been reported previously for any other HRGP. In fact, many HRGP genes do not appear to be expressed in leaves or guard cells at all. In contrast, HRGP gene expression is often strong in hypocotyls and roots or styles (Josè and Puigdomènech, 1993; Keller, 1993). Wyatt et al. (1992) demonstrated by in situ hybridization that within mature soybean leaves, the SbPRP3 gene is exclusively expressed in the epidermal cell layer, but no information was provided with respect to guard cells. Developmental regulation of RPRP gene expression has been observed in many cases (for review, see Keller, 1993), e.g. for maize ZmHyPRP (Josè-Estanyol et al., 1992; Josè-Estanyol and Puigdomènech, 1998), soybean SbPRP1 and SbPRP2 (Wyatt et al., 1992), and wheat WPRP1 (Raines et al., 1991). A common feature of many RPRPs is their accumulation in actively growing tissues (Showalter and Rumeau, 1990). Similarly, in potato we detected StGCPRP mRNA in developing fruits, indicating that StGCPRP may be functionally linked to growth as well. Many extensin and RPRP genes respond to wounding by an elevation of corresponding transcript levels. We found that StGCPRP transcript levels did not change in response to wounding in young or mature leaves of potato (data not shown).
Recently, genes almost exclusively expressed in guard cells have been identified, including kst1 from potato and kat1 from Arabidopsis, which both encode inwardly rectifying potassium channels (Müller-Röber et al., 1995; Nakamura et al., 1995). In these cases expression is restricted to guard cells, even in very young leaves and cotyledons, i.e. these genes do not appear to be expressed in leaf cells other than guard cells at any stage of leaf development, indicating that the underlying transcription machinery (or at least part of it) is also restricted to guard cells. We have recently identified the cis-regulatory elements of the kst1 promoter required for guard-cell-specific gene expression (G. Plesch, T. Ehrhardt, and B. Müller-Röber, unpublished data). The fact that StGCPRP is strongly expressed in mesophyll cells of immature leaves, but only weakly (if at all) in mesophyll cells of mature leaf tissues, with a prominent expression in guard cells throughout all stages of leaf development, may indicate a further pathway activating the expression of genes in stomata. The StGCPRP gene and its promoter may therefore serve as a further molecular marker for the analysis of guard cell (and leaf) differentiation and its underlying transcriptional regulation.
The high level of StGCPRP expression in guard cells during all stages of leaf development indicates an important function of the StGCPRP protein in this cell type. Because StGCPRP is likely to be a cell wall protein, it may help to cope with the mechanical stress imposed on guard cells during stomatal movements. StGCPRP mRNA levels decreased in response to moderate drought stress in guard cells as well as in young leaves. Complex changes in cell wall proteins in response to drought stress have also been reported. For example, in soybean, a 28-kD protein specifically located in the cell wall of young tissues was found to be up-regulated in response to drought, whereas reduction of a 70-kD protein was observed in mature tissues (Bozarth et al., 1987). In growing zones of soybean seedlings drought reduces transcript levels of an extensin gene and of a GRP gene, whereas SbPRP1 transcripts accumulated under these conditions (Creelman and Mullet, 1991). Although the response of StGCPRP gene expression to drought was not specific to guard cells, our data demonstrate that guard cells contain the machinery to regulate gene expression in response to environmental factors. We and others have recently demonstrated that a variety of other genes encoding transporters and enzymes of metabolic pathways are subject to environmental regulation in the highly specialized guard cells (e.g. Taylor et al., 1995; Kopka et al., 1997).
The StGCPRP gene appears to encode the first protein of the stomatal cell wall. As stomatal development requires a highly coordinated sequence of reactions involving alterations in cell wall structure (e.g. Sack, 1987; Zhao and Sack, 1999), StGCPRP and its proposed interacting partners (such as other cell wall proteins; see above) may provide molecular and cytological markers for further studies of guard cell differentiation.
ACKNOWLEDGMENTS
We thank Gunnar Plesch for helpful comments on the manuscript and Joseph Bergstein for photography. We also thank Katharina Pawlowski for supporting our in situ hybridizations, which were performed within the framework of the European Molecular Biology Organization Practical Course on in Situ Hybridization and Cytochemistry in Plants carried out in Wageningen, The Netherlands, 1995. We acknowledge Uwe Sonnewald and his colleagues (IPK Gatersleben, Germany) for growing plants under elevated CO2.
Footnotes
This work was supported by the Max-Planck Society and the Deutsche Forschungsgemeinschaft (grant nos. Mu 1199/1–1 and Mu 1199/1–2).
LITERATURE CITED
- Assmann SM. Signal transduction in guard cells. Annu Rev Cell Biol. 1993;9:345–375. doi: 10.1146/annurev.cb.09.110193.002021. [DOI] [PubMed] [Google Scholar]
- Bozarth CS, Mullet JE, Boyer JS. Cell wall proteins at low water potentials. Plant Physiol. 1987;85:261–267. doi: 10.1104/pp.85.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, Kjellbom P, Lamb C. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell. 1992;70:21–30. doi: 10.1016/0092-8674(92)90530-p. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Cassab GI. Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:281–309. doi: 10.1146/annurev.arplant.49.1.281. [DOI] [PubMed] [Google Scholar]
- Cassab GI, Varner JE. Cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:321–353. doi: 10.1146/annurev.arplant.49.1.281. [DOI] [PubMed] [Google Scholar]
- Cooper JB, Varner JE. Cross-linking of soluble extensin in isolated cell walls. Plant Physiol. 1984;76:414–417. doi: 10.1104/pp.76.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotelle V. Étude de precessus de phosphorylation de protéines impliqués dans la régulation des mouvements stomatiques. PhD thesis. France: University of Toulouse; 1997. [Google Scholar]
- Creelman RA, Mullet JE. Water deficit modulates gene expression in growing zones of soybean seedlings: analysis of differentially expressed cDNAs, a new beta-tubulin gene, and expression of genes encoding cell wall proteins. Plant Mol Biol. 1991;17:591–608. doi: 10.1007/BF00037046. [DOI] [PubMed] [Google Scholar]
- Datta K, Marcus A. Nucleotide sequence of a gene encoding soybean repetitive proline-rich protein 3. Plant Mol Biol. 1990;14:285–286. doi: 10.1007/BF00018570. [DOI] [PubMed] [Google Scholar]
- Deleage G, Roux B. An algorithm for protein structure prediction based on class prediction. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- Ertl H, Hallmann A, Wenzel S, Sumper M. A novel extensin that may organize extracellular matrix biogenesis in Volvox carteri. EMBO J. 1992;11:2055–2062. doi: 10.1002/j.1460-2075.1992.tb05263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard JL, Jako C, Saint-Guily A, Weil JH, Kuntz M. Anther-specific, developmentally regulated expression of genes encoding a new class of proline-rich proteins in sunflower. Plant Mol Biol. 1991;16:271–281. doi: 10.1007/BF00020558. [DOI] [PubMed] [Google Scholar]
- Fieuw S, Müller-Röber B, Galvez S, Willmitzer L. Cloning and expression analysis of the cytosolic NADP+-dependent isocitrate dehydrogenase from potato: implications for nitrogen metabolism. Plant Physiol. 1995;107:905–913. doi: 10.1104/pp.107.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen HJ, Nap JP, Gloudemans T, Stiekema W, van Dam H, Govers F, Louwerses J, van Kammen A, Bisseling T. Characterization of cDNA for nodulin-75 of soybean: a gene product involved in early stages of root nodule development. Proc Natl Acad Sci USA. 1987;84:4495–4499. doi: 10.1073/pnas.84.13.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. Crosslinking of matrix polymers in the growing cell walls of angiosperms. Annu Rev Plant Physiol. 1986;37:165–186. [Google Scholar]
- Geourjon C, Deleage G. Significant improvements in protein secondary structure prediction by prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- Gibrat JF, Garnier J, Robson B. Further developments of protein structure prediction using information theory: new parameters and consideration of residue pairs. J Mol Biol. 1987;198:425–443. doi: 10.1016/0022-2836(87)90292-0. [DOI] [PubMed] [Google Scholar]
- Hirasawa T, Wakabayashi K, Touya S, Ishihara K. Stomatal responses to water deficit and abscisic acid in leaves of sunflower (Helianthus annuusL.) grown under different conditions. Plant Cell Physiol. 1995;36:955–964. [Google Scholar]
- Hong JC, Nagao RT, Key JL. Characterization and sequence analysis of a developmentally regulated putative cell wall protein gene isolated from soybean. J Biol Chem. 1987;262:8367–8376. [PubMed] [Google Scholar]
- Hong JC, Nagao RT, Key JL. Developmentally regulated expression of soybean proline-rich cell wall protein genes. Plant Cell. 1989;1:937–943. doi: 10.1105/tpc.1.9.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josè M, Puigdomènech P. Transley review no. 55: structure and expression of genes coding for structural proteins of the plant cell wall. New Phytol. 1993;125:259–282. doi: 10.1111/j.1469-8137.1993.tb03881.x. [DOI] [PubMed] [Google Scholar]
- Josè-Estanyol M, Puigdomènech P. Rapid changes induced in developmental programmes of the maize embryo detected by analysis of expression of genes encoding proline-rich proteins. FEBS Lett. 1998;422:400–402. doi: 10.1016/s0014-5793(98)00030-1. [DOI] [PubMed] [Google Scholar]
- Josè-Estanyol M, Ruiz-Avila L, Puigdomènech P. A maize embryo-specific gene encodes a proline-rich and hydrophobic protein. Plant Cell. 1992;4:413–423. doi: 10.1105/tpc.4.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller B. Structural cell wall proteins. Plant Physiol. 1993;101:1127–1130. doi: 10.1104/pp.101.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA. Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 1994;5:157–172. doi: 10.1046/j.1365-313x.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- Kopka J, Provart N, Müller-Röber B. Potato guard cells respond to drying soil by a complex change in the expression of genes related to carbon metabolism and turgor regulation. Plant J. 1997;11:871–882. doi: 10.1046/j.1365-313x.1997.11040871.x. [DOI] [PubMed] [Google Scholar]
- Levin JM, Robson B, Garnier J. An algorithm for secondary structure determination in proteins based on sequence similartity. FEBS Lett. 1986;205:303–308. doi: 10.1016/0014-5793(86)80917-6. [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:21–26. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Mansfield TA, Hetherington AM, Atkinson CJ. Some current aspects of stomatal physiology. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:55–75. [Google Scholar]
- Morison JIL. Intercellular CO2 concentration and stomatal responses to CO2. In: Zeiger E, Farquhar GD, Cowan IR, editors. Stomatal Function. Stanford, CA: Stanford University Press; 1987. pp. 229–251. [Google Scholar]
- Müller-Röber B, Ehrhardt T, Plesch G. Molecular features of stomatal guard cells. J Exp Bot. 1998;49:293–304. [Google Scholar]
- Müller-Röber B, Ellenberg J, Provart N, Willmitzer L, Busch H, Becker D, Dietrich P, Hoth S, Hedrich R. Cloning and electrophysiological analysis of KST1, an inward-rectifying K+channel expressed in potato guard cells. EMBO J. 1995;14:2409–2416. doi: 10.1002/j.1460-2075.1995.tb07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura RL, McKendree WL, Jr, Hirsch RE, Seedbrook JC, Gaber RF, Sussman MR. Expression of an Arabidopsispotassium channel gene in guard cells. Plant Physiol. 1995;109:371–374. doi: 10.1104/pp.109.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, Heijne G. Identification of procaryotic and eucaryotic signal peptides and predicton of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Ohya T, Shimazaki K-I. Profiles of proteins in guard cell and mesophyll cell protoplasts from Vicia fabaL.: fractionation by sodium dodecylsulfate-polyacrylamide gel electrophoresis. Plant Cell Physiol. 1989;30:783–787. [Google Scholar]
- Raines CA, Lloyd JC, Shiaoman C, John UP, Murphy GJP. A novel proline-rich protein from wheat. Plant Mol Biol. 1991;16:663–670. doi: 10.1007/BF00023430. [DOI] [PubMed] [Google Scholar]
- Raschke K. Action of abscisic acid on guard cells. In: Zeiger E, Farquhar GD, Cowan IR, editors. Stomatal Function. Stanford, CA: Stanford University Press; 1987. pp. 253–279. [Google Scholar]
- Reiter WD. Structure, synthesis, and function of the plant cell wall. In: Meyerowitz EM, Sommerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 955–988. [Google Scholar]
- Rubinstein AL, Broadwater AH, Lowrey KB, Bedinger PA. Pex1, a pollen-specific gene with an extensin-like domain. Proc Natl Acad Sci USA. 1995;92:3086–3090. doi: 10.1073/pnas.92.8.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack FD. The development and structure of stomata. In: Zeiger E, Farquhar GD, Cowan IR, editors. Stomatal Function. Stanford, CA: Stanford University Press; 1987. pp. 59–89. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schnabelrauch LS, Kieliszewski M, Upham BL, Alizedeh H, Lamport DTA. Isolation of pl 4.6 extensin peroxidase from tomato cell suspension cultures and identification of Val-Tyr-Lys as putative intermolecular cross-link site. Plant J. 1996;9:477–489. doi: 10.1046/j.1365-313x.1996.09040477.x. [DOI] [PubMed] [Google Scholar]
- Schroeder JI. Plasma membrane ion channel regulation during abscisic acid-induced closing of stomata. Philos Trans R Soc Lond B. 1992;338:83–89. [Google Scholar]
- Showalter AM, Rumeau D. Molecular biology of plant cell wall hydroxyproline-rich glycoproteins. In: Adair WS, Mecham RP, editors. Organization and Assembly of Plant and Animal Extracellular Matrix. New York: Academic Press; 1990. pp. 247–281. [Google Scholar]
- Stafstrom JP, Staehelin LA. Antibody localisation of extensin in cell walls of storage roots. Planta. 1988;174:321–332. doi: 10.1007/BF00959517. [DOI] [PubMed] [Google Scholar]
- Taylor JE, Renwick KF, Webb AAR, McAinsh MR, Furini A, Bartels D, Quatrano RS, Marcotte WR, Jr, Hetherington AM. ABA-regulated promoter activity in stomatal guard cells. Plant J. 1995;7:129–134. doi: 10.1046/j.1365-313x.1995.07010129.x. [DOI] [PubMed] [Google Scholar]
- Van de Wiel C, Scheres B, Franssen H, van Lierop MJ, van Lammeren A, van Kammen A, Bisseling T. The early nodulin transcript ENOD2 is located in the nodule parenchyma (inner cortex) of pea and soybean root nodules. EMBO J. 1990;9:1–7. doi: 10.1002/j.1460-2075.1990.tb08073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignols F, José-Estanyol M, Caparrós-Ruiz D, Rigau J, Puigdomènech P. Involvement of a maize proline-rich protein in secondary cell wall formation as deduced from its specific mRNA localization. Plant Mol Biol. 1999;39:945–952. doi: 10.1023/a:1006129703262. [DOI] [PubMed] [Google Scholar]
- Webb AAR, McAinsh MR, Taylor JE, Hetherington AM. Calcium ions as intracellular second messengers in higher plants. In: Callow JA, editor. Advances in Botanical Research. Vol. 22. London: Academic Press; 1996. pp. 45–96. [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- Willmer C, Fricker M. Stomata. London: Chapman & Hall; 1996. [Google Scholar]
- Wyatt RE, Nagao RT, Key JL. Patterns of soybean proline-rich protein expression. Plant Cell. 1992;4:99–110. doi: 10.1105/tpc.4.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH, Song YR, Marcus A, Varner JE. Comparative localization of three classes of cell wall proteins. Plant J. 1991;1:175–183. doi: 10.1111/j.1365-313x.1991.00175.x. [DOI] [PubMed] [Google Scholar]
- Ye ZH, Varner JE. Tissue-specific expression of cell wall proteins in developing soybean tissues. Plant Cell. 1991;3:23–27. doi: 10.1105/tpc.3.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger E, Farquhar GD, Cowan IR. Stomatal Function. Stanford, CA: Stanford University Press; 1987. [Google Scholar]
- Zhao L, Sack FD. Ultrastructure of stomatal development in Arabidopsis (Brassicaceae) leaves. Am J Bot. 1999;86:929–939. [PubMed] [Google Scholar]