Abstract
Storage of human retinal pigment epithelium (hRPE) can contribute to the advancement of cell-based RPE replacement therapies. The present study aimed to improve the quality of stored hRPE cultures by identifying storage medium additives that, alone or in combination, contribute to enhancing cell viability while preserving morphology and phenotype. hRPE cells were cultured in the presence of the silk protein sericin until pigmentation. Cells were then stored for 10 days in storage medium plus sericin and either one of 46 different additives. Individual effects of each additive on cell viability were assessed using epifluorescence microscopy. Factorial design identified promising additive combinations by extrapolating their individual effects. Supplementing the storage medium with sericin combined with adenosine, L-ascorbic acid and allopurinol resulted in the highest cell viability (98.6 ± 0.5%) after storage for three days, as measured by epifluorescence microscopy. Flow cytometry validated the findings. Proteomics identified 61 upregulated and 65 downregulated proteins in this storage group compared to the unstored control. Transmission electron microscopy demonstrated the presence of melanosomes after storage in the optimized medium. We conclude that the combination of adenosine, L-ascorbic acid, allopurinol and sericin in minimal essential medium preserves RPE pigmentation while maintaining cell viability during storage.
Introduction
Age-related macular degeneration (AMD) is a leading cause of blindness in the developed world and is characterized by impairment and loss of the retinal pigment epithelium (RPE)1. Due to the lack of treatment options for the dry type of AMD, which affects 85% of patients, replacement of the RPE has been proposed as a future therapy for this disease2–11. Expectations for the application of RPE transplants to treat retinal diseases are high, and several studies have shown that this approach can restore subretinal anatomy and improve visual function2. A recent review by Nommiste et al.12 covers several proof-of-principle studies investigating the efficacy of different cell sources and transplantation techniques. The RPE cells used in these studies are derived from primary human stem cells, induced pluripotent stem cells and several other sources, and have been transplanted to the subretinal space by means of suspensions, strips or patches on coated polymers13,14. While the strategies for cell replacement are improving, the production of cell sheets that fulfill the requirements for transplantation is complex15 and will likely lead to centralization of specialized culture laboratories. The ability to store RPE successfully is necessary in order to transport the tissue from the culture laboratories to the transplantation clinics and make widespread use of RPE replacement therapies possible12. An established storage method would not only allow for transportation, but also make quality control and microbiological testing of the tissue possible16. With continued improvement of RPE tissue engineering approaches, and more than 20 million patients suffering from AMD worldwide17, an upcoming need for improved storage and transportation methods for cultured RPE is anticipated. An above-freezing temperature storage system as suggested by our research group circumvents the need for cryoprotectants, which are known to inflict freezing injury to tissues at both high and low cooling rates18–20.
After testing nine different storage temperatures between 4 °C and 37 °C, we found that hRPE cultures stored at 4 °C in a storage medium containing 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)- and sodium bicarbonate-buffered Minimum Essential Medium (MEM) preserved the greatest number of viable cells (unpublished data). An earlier study showed that the addition of 1% sericin to the cell culture medium enhanced hRPE cell maturation, most notably by increasing cell pigmentation21. The MEM storage medium is a defined basal medium that mainly consists of inorganic salts, vitamins and glucose. We therefore investigated the effects of supplementing this medium with many different additives, including sericin to preserve the differentiated state of the cells. The effects of the 46 individual supplements on viability of hRPE cell cultures were analyzed after ten days of storage at 4 °C. Some additives were selected based on their known or proposed effects on viability or antioxidant function in cultures of RPE or other cell types21–32, while others were chosen based on effects demonstrated in pilot experiments. Most of the additives have, to our knowledge, never been tested in the current setting. The effects of a total of 32 different combinations of the five most promising additives were simulated using a factorial design experiment. The single best combination of additives was selected for further study by additional experiments to assess its effects on phenotype and morphology.
Results
Effect of Individual and Combined Storage Medium Additives on Viability of hRPE
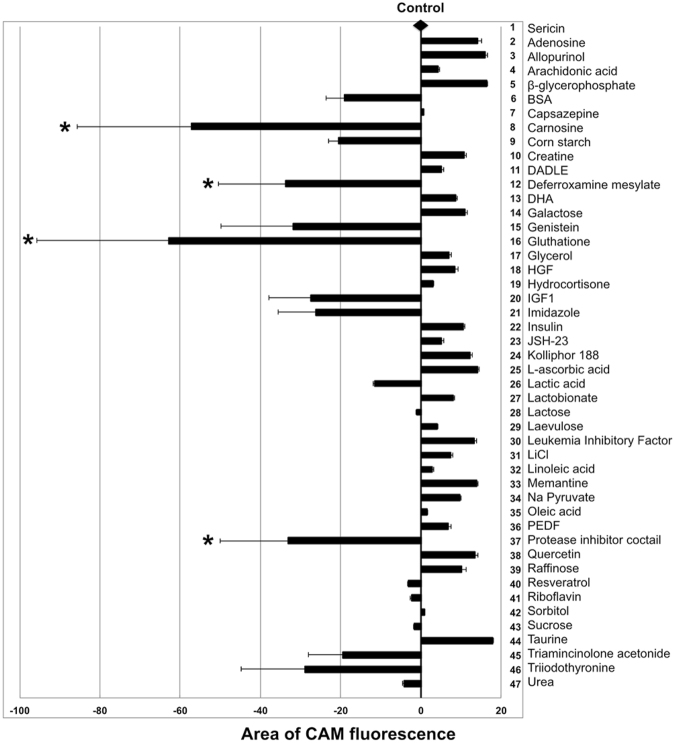
hRPE were seeded in complete EpiCM on Nunclon Δ surface plates and cultured for two days before replacing EpiCM with modified DMEM (hereafter named ≪differentiation medium≫) containing 1% sericin for 14 days. The cells had then developed pigmentation as demonstrated earlier21, and were stored for 10 days in storage medium plus sericin and either one of 46 different additives. The control group, containing sericin, was stored without additional additives. Cell survival following 10 days of storage in all 47 experimental groups (N = 3) was assessed by calcein-acetoxymethyl ester (CAM) fluorescence using ImageJ software to measure the culture well area covered by CAM-stained live hRPE cells. The results are presented in Fig. 1. Cells stored in MEM containing 1% sericin served as the control. Control cells covered 73.5 ± 22.3% of the culture well area. In comparison, cells that had not been stored covered 99.2 ± 0.1% of the culture well area. No single storage medium additive contributed to increasing the CAM-stained culture well area significantly compared to the control. One-way ANOVA revealed that two additives significantly reduced cell viability (carnosine and glutathione), while the Student’s t-test revealed that four additives significantly reduced cell viability (carnosine, glutathione, deferroxamine mesylate and protease inhibitor cocktail) compared to the control.
Figure 1.
Cell viability after ten days of storage as measured by area of CAM fluorescence. The control line drawn from the black diamond represents the area of calcein-acetoxymethyl ester (CAM) fluorescence obtained in the control group (N = 18), where 1% sericin was added to the MEM-based storage medium. Other bar points are representations of CAM area fluorescence for each additive (N = 3) supplemented to MEM in the presence of 1% sericin. Resulting effects are displayed as either increasing or decreasing CAM area fluorescence compared to the control line. The addition of carnosine, deferroxamine mesylate, glutathione or the protease inhibitor cocktail to the storage medium significantly reduces cell viability as measured by CAM area fluorescence (*, P < 0.05). Error bars represent the standard deviation of mean values. BSA: bovine serum albumin; DADLE: [D-Ala2, D-Leu5]-Enkephalin; DHA: docosahexaenoic acid; HGF: hepatocyte growth factor; IGF1: insulin-like growth factor 1; JSH-23: 4-methyl-1-N-(3-phenylpropyl)benzene-1,2-diamine; PEDF: pigment epithelium-derived factor.
To investigate whether combinations of additives could increase cell viability further, the five additives which provided the largest CAM fluorescence area (adenosine, allopurinol, β-glycerophosphate, L-ascorbic acid and taurine) were selected for factorial design experiments. Normality of the data was confirmed by Design-Expert, as was absence of significant outliers on residuals plots. The data on percentage of cell viability was then power transformed as recommended by the Design-Expert software before subsequent analysis. A significant model including all possible additive combinations was computed by Design-Expert software (Stat-Ease) using ANOVA (P = 0.047). No single additive supplemented individually to the storage medium had a significant impact on cell viability in the factorial design experiments. The combined effects of sericin, adenosine, allopurinol and L-ascorbic acid, however, provided the highest desirability regarding both CAM-stained culture well area and number of dead cells following storage (Figs 2 and 3).
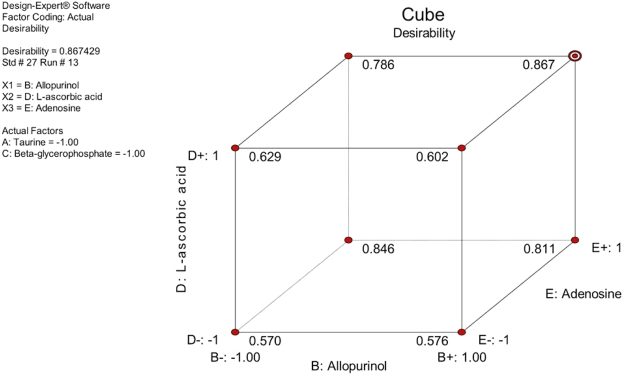
Figure 2.
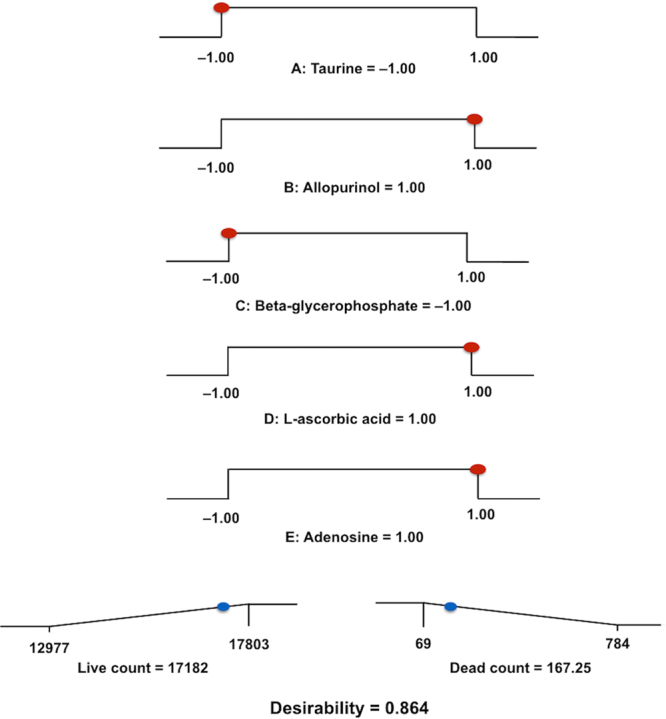
Factorial design analysis. Factorial design analysis of the five most promising additives providing a ramp display showing individual graphs for each additive in the most desirable storage medium combination. Presence of additive was set as “1”, while absence of additive was set as “−1”. The dot on each ramp represents the factor setting or response prediction for the resulting combination.
Figure 3.
Cube plot illustrating the predicted response as a function of the three additives that created the most desirable effect. The plot shows how three factors (B, D, E) combine to affect the response. All values are predicted. Maximum desirability is reached at settings B+ , D+ and E+ (allopurinol, L-ascorbic acid and adenosine).
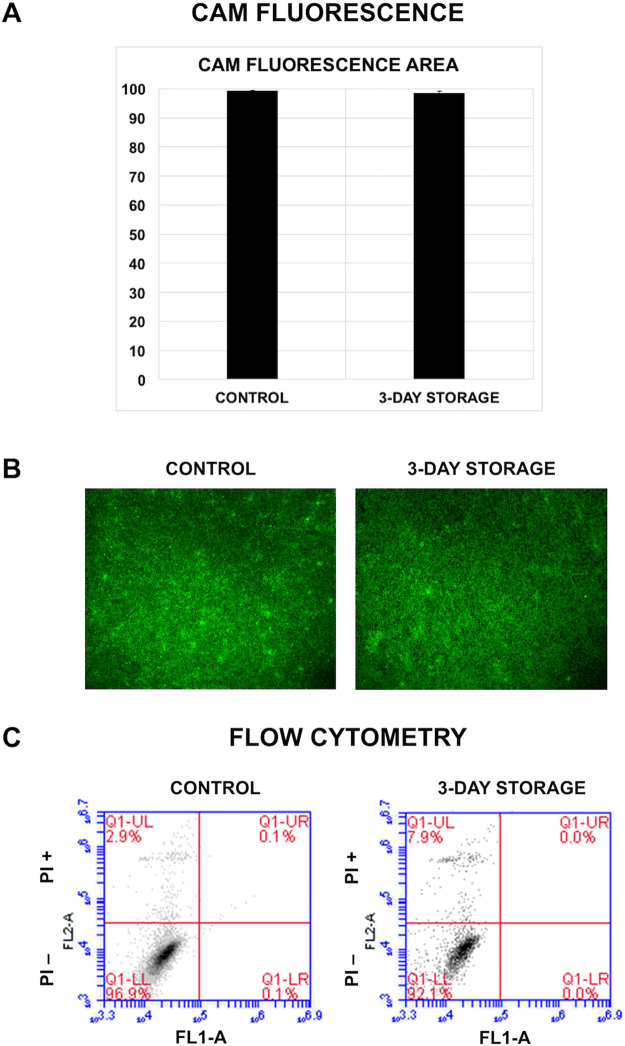
To confirm factorial design results, viability analysis using CAM area calculation was employed for the optimal additive combination (MEM supplemented with sericin, adenosine, allopurinol and L-ascorbic acid). hRPE cells (N = 3) were cultured at 37 °C and stored for three days at 4 °C in the optimal additive combination before being compared to control cells (N = 6) that had not been stored. Viability was similar between the groups, with a mean CAM fluorescence area of 99.2 ± 0.1% for control cells and 98.6 ± 0.5% for stored cells, respectively (Fig. 4A,B).
Figure 4.
Viability of hRPE stored in the optimal combination of storage medium additives for three days. hRPE were analyzed by (A,B) quantitative fluorescence and (C) flow cytometry. (A) Cell viability as measured by area of calcein-acetoxymethyl ester (CAM) fluorescence, demonstrating similar results between groups (N = 6). Error bars represent the standard deviation of mean values. (B) Representative photomicrographs demonstrating similar CAM labeling between groups. (C) Representative flow cytometry plots of dead cells by propidium iodide exclusion in control cells and cells stored for three days (N = 3). The plots demonstrate a relatively low cell death rate in the stored group.
Validation of Viability Data Using Flow Cytometry
The viability of hRPE stored in the optimal mix (MEM supplemented with sericin, adenosine, allopurinol and L-ascorbic acid) for three days was validated using flow cytometry with propidium iodide (PI) staining. PI passes through permeable cell membranes of dead cells and stains double-stranded DNA. PI bound to 3.1 ± 0.5% of control cells and 7.8 ± 2.5% of stored cells (P < 0.05), yielding a viability of 96.8 ± 0.5% and 92.1 ± 2.5%, respectively (Fig. 4C). While the difference was statistically significant (P = 0.03), these results support the CAM fluorescence area viability data showing only a small change in cell loss in cultures stored using the optimal combination of additives.
pH Measurement
pH of the storage medium was assessed using pH indicator paper and demonstrated pH in the physiological area (pH = 7.4).
Morphology of Optimal Combination hRPE
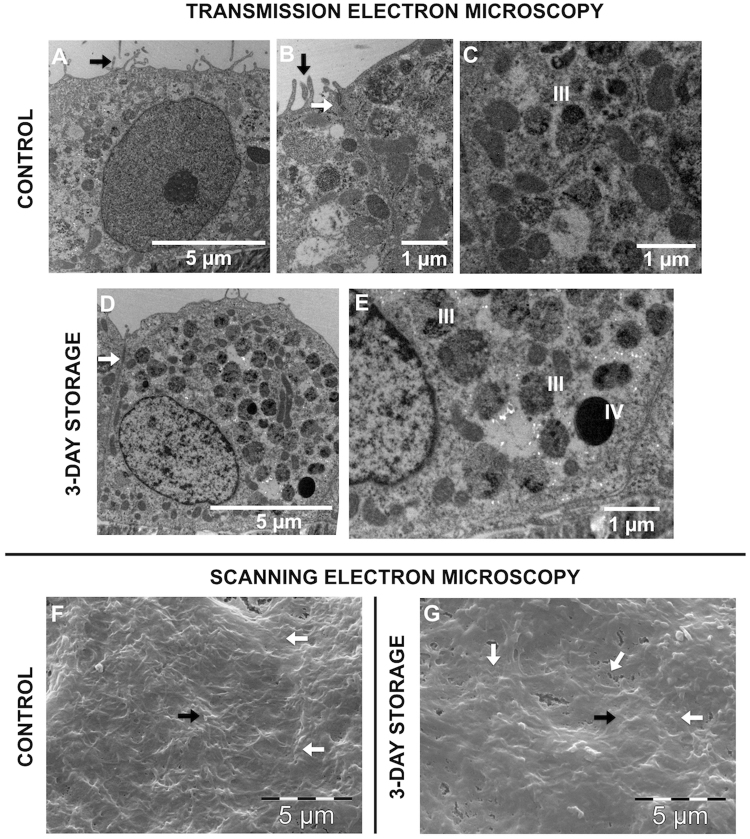
Both light microscopy, scanning electron microscopy and transmission electron microscopy were performed to investigate the effect of the optimal combination of storage medium additives on the morphology of hRPE. Control cells were cultured to confluence and obtained the characteristic morphology comprising a hexagonal cell shape and cytoplasmic pigmentation (Fig. 5). The same features were observed in hRPE cells that had been stored for three days using the optimal combination of storage medium additives, indicating that hRPE can be stored in this additive combination while retaining a classic RPE morphology.
Figure 5.
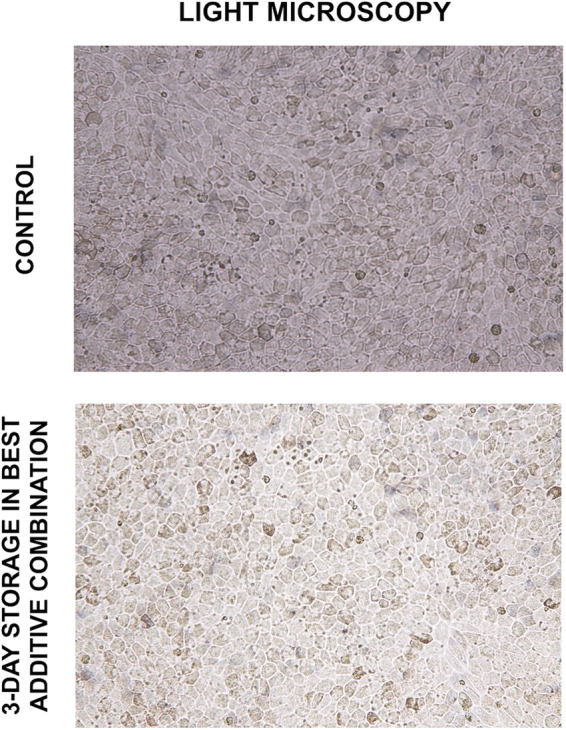
Representative sample from light microscopy observations before and after three days of storage using the best additive combination. The photomicrographs reflect the presence of melanized hRPE cells in both groups and show the classic hexagonal distribution of mature hRPE monolayers.
Transmission electron microscopy demonstrated that the degree of melanization in cells stored in the optimal additive combination maintained or even exceeded that of control cells (Fig. 6A–E), thereby supporting the findings made by light microscopy. Intercellular tight junctions were present both between control cells and between cells stored in the optimal additive combination (Fig. 6B,D). Microvilli were demonstrated in both groups, both by transmission and scanning electron microscopy (Fig. 6A,B,F,G).
Figure 6.
Ultrastructure of stored hRPE cells compared to control. Transmission and scanning electron microscope photomicrographs of control cells that have not been stored (A–C,F) and hRPE cells that have been stored at 4 °C in the optimal additive combination for three days (D,E,G). Intercellular tight junctions are present both between control cells and between cells stored in the optimal additive combination (white arrows in B,D). Microvilli are present in both groups, demonstrated both by transmission and scanning electron microscopy (black arrows in A,B,F,G). Photomicrographs C and E demonstrate the presence of melanosomes at stage III (some melanin pigment deposited onto internal striations) in the control group and stage III and IV (fully melanized) in the storage group. Cell borders are indicated by white arrows (F,G).
Proteomic Analysis of hRPE Using the Optimal Additive Combination
Proteomic analysis was performed to investigate the effect of the optimal combination of storage medium additives on the hRPE proteome. hRPE cells stored in the optimal storage medium combination were compared to control cells that had not been stored. Of 3902 identified proteins, 126 were differentially expressed applying t-test with P < 0.05 (Tables 1 and 2). A total of 65 proteins (1.7%) were downregulated during storage for three days in the optimal additive mix, while 61 proteins (1.6%) were upregulated during storage in the optimal additive mix (Fig. 7). The distribution of differentially expressed proteins was similar between the groups (Fig. 8).
Table 1.
Significantly upregulated proteins during storage (low in control cells).
| Gene Symbol | Gene Description | Biological function |
|---|---|---|
| ACOX1 | Peroxisomal acyl-coenzyme A oxidase 1 | Desaturation of acyl-CoAs to 2-trans-enoyl-CoAs |
| ALDH18A1 | Delta-1-pyrroline-5-carboxylate synthase | Synthesis of proline, ornithine and arginine |
| AP2A1 | AP-2 complex subunit alpha-1 | Component of the adaptor protein complex 2; clathrin-dependent endocytosis |
| ARHGAP1 | Rho GTPase-activating protein 1 | GTPase activator for Rho, Rac and Cdc42 |
| ARMT1 | Protein-glutamate O-methyltransferase | Formation of gamma-glutamyl methyl ester residues |
| BCLAF1 | Bcl-2-associated transcription factor 1 | Death-promoting transcriptional repressor |
| DAZAP1 | DAZ-associated protein 1 | RNA-binding protein; possibly required in spermatogenesis |
| DBI | Acyl-CoA-binding protein | Possible intracellular carrier of acyl-CoA esters |
| DBT | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial | Conversion of alpha-keto acids to acyl-CoA and CO2 |
| DDX23 | Probable ATP-dependent RNA helicase DDX23 | Pre-mRNA splicing |
| DNAJC3 | DnaJ homolog subfamily C member 3 | Unfolded protein response during endoplasmic reticulum stress |
| DSP | Desmoplakin | Anchoring of intermediate filaments to desmosomes |
| EIF6 | Eukaryotic translation initiation factor 6 | Prevents the association of the 60S ribosomal subunit with the 40S subunit |
| EPM2AIP1 | EPM2A-interacting protein 1 | Unknown |
| EZR | Ezrin | Connection of cytoskeletal structures to the plasma membrane; formation of microvilli |
| FARSB | Phenylalanine–tRNA ligase beta subunit | Regulatory tRNA ligase beta subunit |
| FBN2 | Fibrillin-2 | Component of extracellular calcium-binding microfibrils; regulation of elastic fibers |
| FLOT1 | Flotillin-1 | Possible scaffolding protein within caveolar membranes; formation of caveolae |
| FUCA2 | Plasma alpha-L-fucosidase | Hydrolyzation of glycoproteins |
| GATM | Glycine amidinotransferase, mitochondrial | Synthesis of creatine precursor guanidinoacetate |
| GDAP2 | Ganglioside-induced differentiation-associated protein 2 | Unknown |
| HIST1H4A | Histone H4 | Core nucleosome component |
| HNRNPD | Heterogeneous nuclear ribonucleoprotein D0 | RNA-binding protein |
| HNRNPH1 | Heterogeneous nuclear ribonucleoprotein H | Pre-mRNA processing |
| HNRNPM | Heterogeneous nuclear ribonucleoprotein M | Pre-mRNA binding protein |
| HSD17B2 | Estradiol 17-beta-dehydrogenase 2 | Interconversion of testosterone and androstenedione; estradiol and estrone |
| KIAA1468 | LisH domain and HEAT repeat-containing protein KIAA1468 | Unknown |
| KTN1 | Kinectin | Kinesin-driven vesicle motility |
| LRPPRC | Leucine-rich PPR motif-containing protein, mitochondrial | Nuclear and mitochondrial RNA metabolism |
| LRRC8A | Volume-regulated anion channel subunit LRRC8A | Essential component of the volume-regulated anion channel |
| LRSAM1 | E3 ubiquitin-protein ligase LRSAM1 | Regulation of signaling pathways, cell adhesion, self-ubiquitylation, and cargo sorting during receptor endocytosis |
| MRPL28 | 39S ribosomal protein L28, mitochondrial | Component of the 39S mitochondrial ribosome subunit |
| MRRF | Ribosome-recycling factor, mitochondrial | Release of ribosomes from mRNA |
| MYH9 | Myosin-9 | Cytokinesis, cell shape, secretion |
| MYO7A | Unconventional myosin-VIIa | Intracellular movements |
| NUDT19 | Nucleoside diphosphate-linked moiety X motif 19 | Hydrolysis various CoA esters |
| NUP155 | Nuclear pore complex protein Nup155 | Component of the nuclear pore complex |
| PDHB | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | Conversion of pyruvate to acetyl-CoA and CO2 |
| POLR2E | DNA-directed RNA polymerases I, II, and III subunit RPABC1 | Subunit of RNA polymerase II |
| PRCP | Lysosomal Pro-X carboxypeptidase | Cleavage of C-terminal amino acids |
| PRDX2 | Peroxiredoxin-2 | Involved in redox regulation of the cell |
| PRDX3 | Thioredoxin-dependent peroxide reductase, mitochondrial | Involved in redox regulation of the cell |
| PRKCSH | Glucosidase 2 subunit beta | Beta-subunit of glucosidase II, an ER glycan-processing enzyme |
| PRKDC | DNA-dependent protein kinase catalytic subunit | DNA double-strand break repair |
| PRPF8 | Pre-mRNA-processing-splicing factor 8 | Assembly of spliceosomal proteins |
| PRPS2 | Ribose-phosphate pyrophosphokinase 2 | Synthesis of phosphoribosylpyrophosphate (PRPP), essential for nucleotide synthesis |
| PTCD3 | Pentatricopeptide repeat domain-containing protein 3, mitochondrial | Mitochondrial RNA-binding protein |
| PTPRA | Receptor-type tyrosine-protein phosphatase alpha | Regulation of integrin signaling, cell adhesion and proliferation |
| RAB7A | Ras-related protein Rab-7a | Key regulator in endo-lysosomal trafficking |
| RPL18 | 60S ribosomal protein L18 | Component of the ribosomal 60S subunit |
| RPL37A | 60S ribosomal protein L37a | Component of the ribosomal 60S subunit |
| SLC25A3 | Phosphate carrier protein, mitochondrial | Transport of phosphate groups from the cytosol to the mitochondrial matrix |
| SOSTDC1 | Sclerostin domain-containing protein 1 | Bone morphogenetic protein antagonist |
| SRP54 | Signal recognition particle 54 kDa protein | Transfer of presecretory protein from ribosomes to TRAM (translocating chain-associating membrane protein) |
| STAG2 | Cohesin subunit SA-2 | Component of the cohesin complex |
| SUCLG2 | Succinyl-CoA ligase [GDP-forming] subunit beta, mitochondrial | Citric acid cycle |
| SULT1A1 | Sulfotransferase 1A1 | Sulfate conjugation of catecholamines, phenolic drugs and neurotransmitters |
| TOR1A | Torsin-1A | Protein folding, processing, stability and localization |
| TRA2B | Transformer-2 protein homolog beta | Pre-mRNA splicing |
| UBA1 | Ubiquitin-like modifier-activating enzyme 1 | Ubiquitin conjugation |
| VPS18 | Vacuolar protein sorting-associated protein 18 homolog | Vesicle-mediated protein trafficking to lysosomal compartments |
Table 2.
Significantly downregulated proteins during storage (high in control cells).
| Gene Symbol | Gene Description | Biological function |
|---|---|---|
| ABCA1 | ATP-binding cassette sub-family A member 1 | Transmembrane transport |
| ATP1A1 | Sodium/potassium-transporting ATPase subunit alpha-1 | Hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane |
| ATP1A3 | Sodium/potassium-transporting ATPase subunit alpha-3 | Hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane |
| ATP6V1C2 | V-type proton ATPase subunit C 2 | Subunit of the vacuolar ATPase |
| CD2AP | CD2-associated protein | Adapter protein between membrane proteins and the actin cytoskeleton |
| COASY | Bifunctional coenzyme A synthase | CoA biosynthetic pathway |
| COPZ1 | Coatomer subunit zeta-1 | Binds dilysine motifs, reversibly associates with Golgi non-clathrin-coated vesicles |
| CTTN | Src substrate cortactin | Organization of the actin cytoskeleton |
| EIF2S2 | Eukaryotic translation initiation factor 2 subunit 2 | Early protein synthesis |
| EIF4A3 | Eukaryotic initiation factor 4A-III | ATP-dependent RNA helicase |
| FAH | Fumarylacetoacetase | Tyrosine catabolism |
| FAM234A | Protein ITFG3/Protein FAM234A | Unknown |
| FERMT2 | Fermitin family homolog 2 | Scaffolding protein, activates integrin |
| FNDC3A | Fibronectin type-III domain-containing protein 3A | Spermatid-Sertoli adhesion in spermatogenesis |
| G3BP2 | Ras GTPase-activating protein-binding protein 2 | Probable scaffold protein, may be involved in mRNA transport |
| GDI2 | Rab GDP dissociation inhibitor beta | Regulates the GDP/GTP exchange reaction of Rab proteins |
| GLYR1 | Putative oxidoreductase GLYR1 | Promotes KDM1B demethylase activity |
| GNPDA1 | Glucosamine-6-phosphate isomerase 1 | Conversion of D-glucosamine-6-phosphate into D-fructose-6-phosphate and ammonium |
| GOLM1 | Golgi membrane protein 1 | Unknown |
| GPX8 | Probable glutathione peroxidase 8 | Unknown |
| HDDC2 | HD domain-containing protein 2 | Unknown |
| HSD17B10 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Mitochondrial tRNA maturation |
| KIF5B | Kinesin-1 heavy chain | Distribution of mitochondria and lysosomes |
| KPNA2 | Importin subunit alpha-1 | Nuclear protein import |
| KPNB1 | Importin subunit beta-1 | Nuclear protein import |
| LDHB | L-lactate dehydrogenase B chain | Synthesizes lactate from pyruvate |
| LIMCH1 | LIM and calponin homology domains-containing protein 1 | Unknown |
| LPL | Lipoprotein lipase | Hydrolysis of triglycerides of chylomicrons and very low density lipoproteins |
| LRRN1 | Leucine-rich repeat neuronal protein 1 | Unknown |
| MAP4 | Microtubule-associated protein 4 | Promotes microtubule assembly |
| MARS | Methionine–tRNA ligase, cytoplasmic | Ligation of methionine to tRNA molecules |
| MAT2B | Methionine adenosyltransferase 2 subunit beta | Regulatory subunit of S-adenosylmethionine synthetase 2 |
| MPI | Mannose-6-phosphate isomerase | Mannosyl transfer reactions |
| MRPL2 | 39 S ribosomal protein L2, mitochondrial | Component of the 39 S mitochondrial ribosome subunit |
| MYO1D | Unconventional myosin-Id | Intracellular movement |
| MYRIP | Rab effector MyRIP | Melanosome transport |
| NDUFB3 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 | Mitochondrial respiratory chain NADH dehydrogenase |
| NHP2 | H/ACA ribonucleoprotein complex subunit 2 | Ribosome biogenesis |
| NOV | Protein NOV homolog | Cell proliferation, adhesion, differentiation |
| PARK7 | Protein DJ-1 | Oxidative stress and cell death protection |
| PDAP1 | 28 kDa heat- and acid-stable phosphoprotein | PDGFA-stimulated fibroblast growth |
| PLS3 | Plastin-3 | Actin-bundling protein of intestinal microvilli, stereocilia, fibroblast filopodia |
| RABGAP1 | Rab GTPase-activating protein 1 | Unknown |
| RHOT2 | Mitochondrial Rho GTPase 2 | Mitochondrial trafficking |
| RPS13 | 40S ribosomal protein S13 | Component of the ribosomal 40S subunit |
| RTCB | tRNA-splicing ligase RtcB homolog | Subunit of tRNA-splicing ligase |
| SCPEP1 | Retinoid-inducible serine carboxypeptidase | Unknown |
| SEC23B | Protein transport protein Sec. 23B | ER-derived vesicle transport |
| SLC1A5 | Neutral amino acid transporter B(0) | Sodium-dependent amino acid transport |
| SLC7A5 | Large neutral amino acids transporter small subunit 1 | L-leucine transport across the blood-retinal barrier |
| SMARCD1 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 | Chromatin remodeling |
| SPR | Sepiapterin reductase | Reduction of pteridine derivatives |
| TKFC | Triokinase/FMN cyclase | Dihydroxyacetone phosphorylation |
| TNKS1BP1 | 182 kDa tankyrase-1-binding protein | Colocalizes with chromosomes in mitosis |
| TPRN | Taperin | Sensory epithelial protein associated with autosomal recessive deafness. |
| TSPAN4 | Tetraspanin-4 | Cell surface glycoprotein binding to integrin |
| TWF2 | Twinfilin-2 | Actin-binding protein involved in motile and morphological processes |
| TXNL1 | Thioredoxin-like protein 1 | Active thioredoxin |
| TYR | Tyrosinase | Formation of pigments, melanin production from tyrosine |
| UBE2O | E2 ubiquitin-conjugating enzyme | Monoubiquitination of target proteins |
| UNC13D | Protein unc-13 homolog D | Vesicle maturation during exocytosis |
| USP5 | Ubiquitin carboxyl-terminal hydrolase 5 | Cleaves multiubiquitin polymers |
| VCL | Vinculin | Actin binding protein involved in cell-matrix adhesion and cell-cell adhesion |
| VPS25 | Vacuolar protein-sorting-associated protein 25 | Sorting of ubiquitinated membrane proteins during endocytosis |
| YAP1 | Transcriptional coactivator YAP1 | Critical regulatory target of the Hippo signaling pathway |
Figure 7.
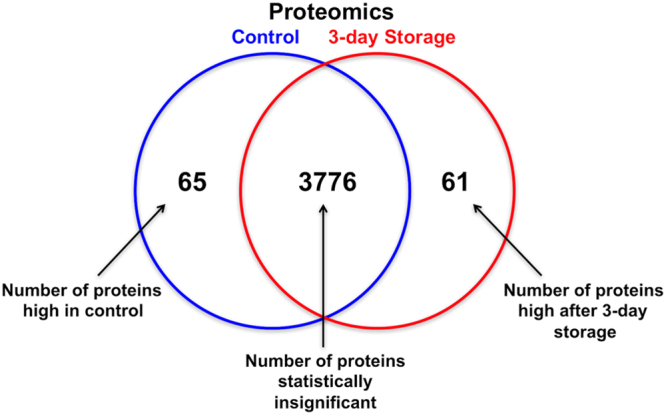
Protein expression in experimental groups. Venn diagram showing the distribution of proteins that are highly expressed in control cells, stored cells, and those which are statistically insignificant.
Figure 8.
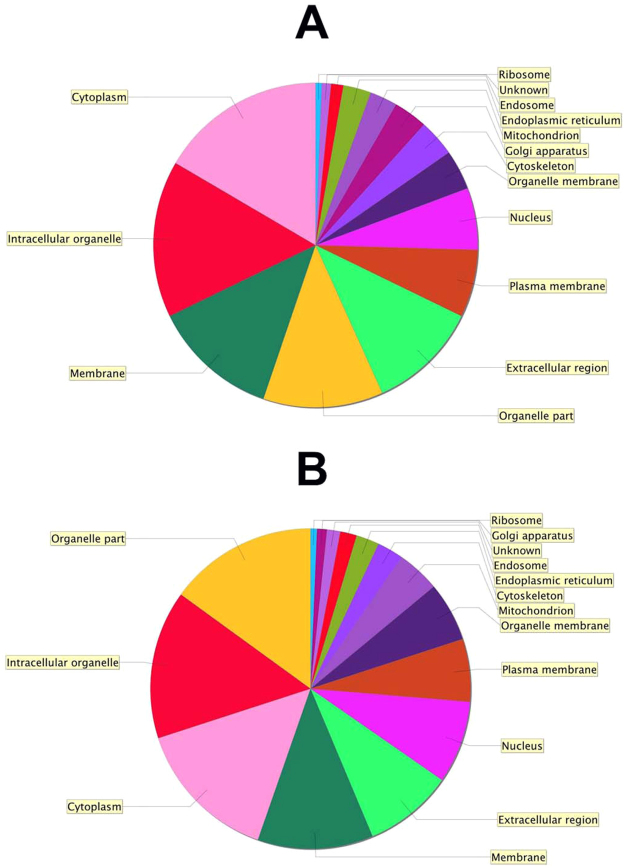
Distribution of protein functions. Gene ontology pie chart showing the distribution of protein functions in hRPE before (A) and after (B) storage according to their molecular functions as determined using Scaffold software with NCBI annotations.
The cytoskeleton-related proteins ezrin and desmoplakin, and perioxiredoxins 2 and 3, important antioxidant enzymes of the cytosol and mitochondria, respectively33, were upregulated during storage. Expression of vinculin and microtubule-associated protein 4 was reduced during storage. Vinculin is a membrane-associated protein that functions as a multiprotein linker to the actin cytoskeleton34, while microtubule-associated protein 4 is involved in crosslinking of microtubules to actin filaments35. The expression levels of several proteins associated with important RPE functions were specifically analyzed. The list of proteins was selected based on their known roles in visual pigment generation, phagocytosis and adhesion of RPE36–40. Only one of the selected proteins important for specific RPE functions had significantly changed regulation in the stored cells compared to the control (Table 3). Tyrosinase was slightly, but significantly downregulated in stored cells compared to control cells (fold change 0.8; P < 0.01).
Table 3.
Effect of storage on the expression of some proteins associated with RPE-specific functions. Fold change represents changes in cells stored for three days compared to control cells.
| Gene Symbol | Gene Description | Role in RPE | Fold change | P value (T test) |
|---|---|---|---|---|
| RPE65 | Retinoid isomerohydrolase | Visual pigment regeneration | 1.1 | 0.21 |
| RLBP1 (CRALBP) | Retinaldehyde-binding protein 1 | Visual cycle | 1 | 0.93 |
| TYR | Tyrosinase | Pigmentation | 0.8 | 0.0038 |
| PMEL | Melanocyte protein PMEL | Pigmentation | 1 | 0.65 |
| TYRP1 | 5,6-dihydroxyindole-2-carboxylic acid oxidase | Pigmentation | 0.9 | 0.32 |
| TYRP2 | L-dopachrome tautomerase | Pigmentation | 0.8 | 0.23 |
| MFGE8 | Lactadherin | Phagocytosis | 1 | 0.81 |
| ZO-1 | Tight junction protein ZO-1 | Tight junctions | 1 | 0.90 |
| OCLN | Occludin | Tight junctions | 0.9 | 0.75 |
| KRT18 | Keratin, type I cytoskeletal 18 | Cytoskeleton | 0.9 | 0.08 |
Discussion
The present study indicates that the storage viability of hRPE cells can be increased by supplementing the serum-free MEM-based storage medium containing sericin with a combination of three additional additives, while maintaining a differentiated morphology and with only slight phenotypic changes. A total of 47 individual additives were studied, including 32 combinations of the five most promising additives using a full-factorial design experiment. Herein, the five most promising storage media additives (adenosine, allopurinol, β-glycerophosphate, L-ascorbic acid and taurine) were investigated simultaneously. Compared to one-factor-at-a-time (OFAT) studies, factorial experiments have several advantages41. First, they require less time, material, and number of experiments, making them more cost-effective. Second, they yield better estimates of the effects of each factor because all observations are used to calculate the effect of each individual variable. Third, they reveal interactions between factors and thus permit the exploration of optimal combinations over the entire repertoire of substances. Hence, compared to OFAT studies, which vary only one factor at a time, factorial experiments simultaneously inspecting several factors are far more efficient when analyzing the effect of two or more variables.
The full-factorial experiment revealed that adenosine, allopurinol and L-ascorbic acid together provided the most desirable additive combination with regard to cell viability. This finding was controlled using CAM fluorescence measurements and validated by flow cytometry. The combined effects of these additives on hRPE storage have not been described earlier, but their individual effects on many cellular processes have been widely studied. Adenosine is a purine nucleoside which has been shown to participate in the regulation of inflammatory responses by limiting inflammatory tissue destruction42. Adenosine binds G protein-coupled adenosine receptors43, and A3 receptor activation has been demonstrated to protect retinal cultures against neurodegeneration44. Activation of the ATP receptor P2X7 is known to induce death of retinal ganglion cells, but simultaneous intravitreal injection of an A3 receptor agonist can prevent the P2X7-associated cell death45. P2X7 overactivation results in dysregulated calcium signaling and is involved in the age-related dysfunction and degeneration of RPE cells46. This suggests that overactive purinergic signaling may contribute to the geographic atrophy seen in dry AMD47. The activation of adenosine receptors and inhibition of P2X7 is considered clinically relevant for the prevention of cell death in several eye diseases, including AMD47. Whether the beneficial effect of adenosine on preventing P2X7-associated cell death is responsible for providing increased hRPE cell viability, or other mechanisms are at play, warrants further study.
Allopurinol is a xanthine oxidase inhibitor that reduces the production of uric acid and is being investigated for management of reperfusion injury. It has been shown to prevent postasphyxial changes in newborn pig retinas48 and has been successfully used in the treatment of autoimmune uveitis in an experimental setting49. Allopurinol administered to RPE cell cultures in high doses has been demonstrated to prevent free-radical-induced cell damage50. Its proposed effect on quenching free radicals might have contributed to enhancing cell viability of cultured hRPE cells during storage in the present study.
It has been established that high levels of antioxidant vitamins can significantly reduce the risk of advanced AMD and its associated vision loss in patients with intermediate or advanced AMD51. The addition of ascorbic acid to primary RPE cell cultures in vitro has been demonstrated to provide a dose-related downregulation of early-response proteins that are triggered by oxidative stress52. In a study using the RPE cell line ARPE-19, however, ascorbic acid was not shown to protect the cells from hydroxyl radical induced cell death53. Yet other studies have shown that ascorbic acid supplementation can protect RPE cells from hypoxic damage54 and reduce vision cell loss from damaging light55. However, the latter effect might be attributable to ascorbic acid preventing excessive shedding of rod outer segments upon light exposure56. The effect of ascorbic acid in the present study might be similar to that of allopurinol in that it reduces the oxidative stress burden.
Our research group recently demonstrated that sericin induces melanogenesis of hRPE cells through activation of the NF-κB pathway21. Sericin has been shown to inhibit tyrosinase57, and proteomic analysis in the present study confirmed that tyrosinase expression is slightly reduced in cells stored in the optimal additive combination in the presence of sericin. The expression of other pigment-related proteins (premelanosome protein 17, tyrosinase related protein 1 and tyrosinase related protein 2) was maintained during storage using the optimal additive combination. Tyrosinase is the main rate-limiting melanogenesis enzyme, catalyzing the formation of dihydroxyphenylalanine (L-DOPA) from L-tyrosine58. However, light microscopy and TEM demonstrated the presence of melanized cells and melanosomes in stored cell cultures. While phase contrast and transmission electron microscopy can determine the presence of melanosomes, these are not satisfactory methods by which to objectively determine the level of pigmentation. Future studies warrant the use of other methods, i.e. spectrophotometry or modified scanning devices as demonstrated by Lane et al.59.
In a study by Vugler et al.60 investigating RPE cells differentiated from human embryonal stem cells (HESC-RPE), a larger number of stage 4 melanosomes were displayed; however, these cells were of a different origin and were cultured under very different conditions than used in the present study. For instance, the HESC-RPE were cultured on Matrigel for five weeks. Polarization was evident with basally oriented nuclei like in our cells, but apical microvilli were more developed in this study than is shown in our cultures. Both the cell source and culture length might be of essence in order to further enhance differentiation61–63. Ultrastructure is presented in great detail in a study by Carr et al.64, who demonstrated that co-culture of HESC-RPE with human retina leads to maturation-associated morphological alterations. Herein, the presence of melanosomes, tight junctions and microvilli is demonstrated. Similar findings are made in control cells and cells stored in the optimal additive combination in this study (Fig. 6).
Pyruvate has been shown to induce pigmentation of ARPE-19 cells cultured in DMEM with high glucose61. In our study, the basic storage medium was supplemented with pyruvate, which might have contributed further to the increased pigmentation demonstrated in both the current and earlier studies by our research group. Although several culture protocols using hESCs or iPSCs have successfully produced differentiated and pigmented RPE cells, they are usually more time-consuming62,63. The use of sericin might contribute in shortening the culture period. The focus on the differentiation process is critical, as its efficiency is considered crucial to the economic feasibility of regenerative therapy using RPE cells59.
The expression of the tight junction protein ZO-1 was maintained during storage, as demonstrated by proteomics analysis and transmission electron microscopy. Cultured cells established the classic hexagonal distribution of mature hRPE monolayers. The RPE, being a polarized monolayer, is dependent on functional intercellular tight junctions to maintain high transepithelial resistance, secure cellular barrier function and regulate paracellular permeability65–68. Hence, the present study confirms earlier findings, but still indicates that hRPE cells can retain features of a mature phenotype when stored in the optimal additive combination.
The cytoskeleton-related proteins ezrin and desmoplakin were upregulated during storage. Ezrin is a cortical cytoskeleton protein which localizes to epithelial microvilli69. Loss of ezrin function as demonstrated in ezrin knockout mice leads to substantial reduction in RPE apical microvilli and retarded photoreceptor development69. Desmoplakin is necessary for the anchoring of keratin at cell-cell contacts70, and thus important for the regulation of desmosomal adhesion strength71. It functions as a tumor suppressor72, and a decrease in desmosomal protein expression is associated with poor prognosis in several cancers73–75. Loss-of-function mutations in desmosomal proteins have been associated with clinical syndromes involving the skin, heart, hair and immune system76–79. Upregulation of these proteins during storage might indicate that stored cells maintain robust cytoskeletal functions.
Third passage hRPE cells were employed in this study. The increased tendency of epithelial to mesenchymal transition with increasing passages of RPE cells has been demonstrated by Grisanti et al. They showed a large disparity between passage 2 RPE and passage 10 RPE, where cells of the higher passages transdifferentiate and lose differentiated RPE properties80. While there is a wide consensus regarding the advantages of using early-passage RPE cells to avoid this phenomenon in culture, an exact passage number has not been defined. In a study by Ganti et al. investigating vitreous modulation of gene expression in low-passage hRPE, cells from passages 3–6 were termed “early-passage”81. Based on the observed benefits of early passage cell lines, we selected third passage hRPE cells for this study.
In conclusion, the current study demonstrates that the storage medium additive combination of sericin, adenosine, allopurinol and L-ascorbic acid successfully maintains hRPE cell viability during storage while preserving the characteristic hRPE morphology and proteome. The effects of the individual additives are not thoroughly understood, but previous research points to free radical scavenging mechanisms as possible explanations for these findings.
Future studies should investigate the effect of increased storage duration on hRPE cells in the optimal combination medium, and ideally expand the scope to RPE derived from different sources, including primary human stem cells and induced pluripotent stem cells. This could provide valuable knowledge when establishing a storage protocol for clinical use.
Methods
Supplies
Primary hRPE and complete epithelial cell medium (EpiCM) were purchased from ScienCell Research Laboratories (San Diego, CA). Dulbecco’s Modified Eagle’s Medium (high glucose, with pyruvate; hereafter named DMEM), Minimal Essential Medium, heat-inactivated fetal bovine serum (FBS), N1 growth supplement, taurine, triiodothyronine, non-essential amino acids, glutamine-penicillin-streptomycin, hydrocortisone, propidium iodide (PI), phosphate-buffered saline (PBS) and 4′,6-diamidino-2-phenylindole (DAPI) were obtained from Sigma Aldrich (St Louis, MO). Nunclon Δ surface plates, pipettes and other routine plastics were purchased from VWR International (West Chester, PA). The calcein-acetoxymethyl ester (CAM)/ethidium homodimer 1 (EH-1) viability kit was purchased from Invitrogen. The 47 additives used in the study are listed in Supplementary Information, Table S1.
Culture and Preparation of Cells
Third passage hRPE were seeded (20,000 cells/cm2) in complete EpiCM on 96-well Nunclon Δ surface plates and cultured under routine conditions of 95% air and 5% CO2 at 37 °C. After two days, EpiCM was replaced with modified DMEM (hereafter named «differentiation medium») containing 4.5 g/L glucose, pyruvate, 1% sericin, and 1% penicillin-streptomycin. Cells were then cultured for 14 days in differentiation medium until pigmentation, as demonstrated in an earlier study21. The culture medium was changed every two or three days.
Storage of hRPE cells
Cells were cultured in the differentiation medium for 14 days, until cells were confluent and >20% of cells were pigmented as visually determined by phase contrast microscopy. The differentiation medium was then removed and the cultures were rinsed with PBS before addition of storage medium. The storage medium consisted of 0.3 mL MEM, 25 mM 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES), 22.3 mM sodium bicarbonate, 50 µg/mL gentamycin, and 1% sericin. A total of 46 different additives were individually supplemented to the storage medium and sterile-filtered (pore size 0.2 μm) before being added to the culture wells (N = 3) using a Biomek® 4000 Laboratory Automation Workstation (Beckman Coulter, Inc., Brea, CA). All cultures were stored at 4 °C for ten days, without change or addition of storage medium. The storage containers were custom-built as reported elsewhere82. pH measurements of the storage medium were performed using pH indicator paper.
Viability Analysis using Quantitative Immunofluorescence
Cell viability was analyzed after 10 days of storage by incubating the stored cells with PBS containing 1.0 μM CAM and 1.0 μM EH-1 for 30 min. CAM is enzymatically cleaved into the green fluorescent calcein inside living cells. EH-1 is a membrane-impermeable dye that binds to DNA of dead cells. Area of fluorescence was calculated for all additive groups using epifluorescence microscopy and custom-made macros with ImageJ software (National Institutes of Health, Bethesda, MD). In detail, photomicrographs were captured at 200x magnification at five predetermined locations in each culture well using a Nikon Eclipse Ti fluorescence microscope (Nikon Instruments, Tokyo, Japan) with a DS-Qi1 black-and-white camera (Nikon Instruments) and a motorized microscope stage. Identical exposure length and gain were used for all compared groups, while keeping the image brightness within the camera’s dynamic range.
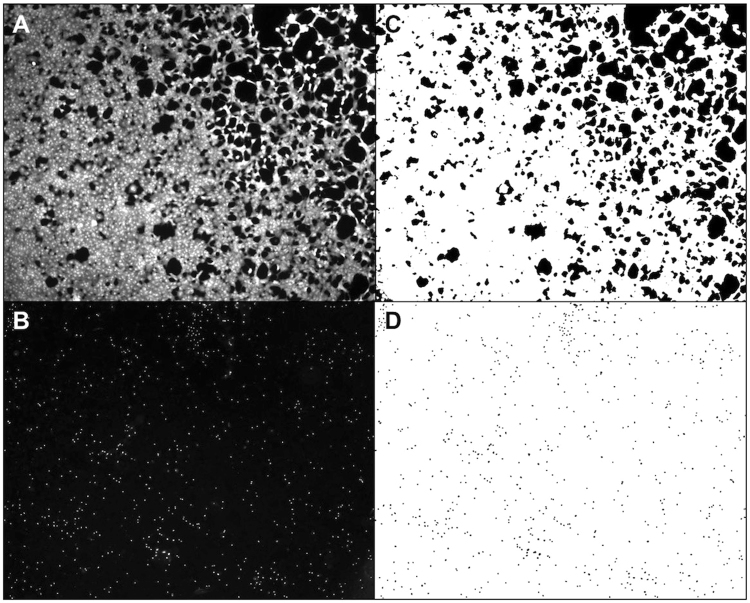
ImageJ software was used to subtract unevenly transmitted light from all 16-bit photomicrographs using the “Subtract Background” -command. All photomicrographs were converted to binary photos before the “Area Fraction” -command was used to measure the culture well area covered by CAM-stained cells. The number of EH-1 stained nuclei was automatically counted using the “Analyze Particles” -command (Fig. 9).
Figure 9.
Quantification of viable cells. Following storage, hRPE cells were stained with calcein-acetoxymethyl ester (CAM) (A) to visualize viable cells and ethidium homodimer-1 (EH-1) (B) to identify dead cells. Images of CAM-stained (C) or EH-1-stained (D) cells were segmented by ImageJ based on the fluorescence intensity. To compare the amount of live and dead cells between groups, ImageJ quantified the area (white) of the viable cells (C) and counted the number of particles that represented dead cell nuclei (D).
Factorial Design
A factorial design experiment is a complex statistical design offering the possibility to study more than one factor at a time by creating a simulation of combined factor effects. Factorial design using Design-Expert (Stat-Ease, Inc., Minneapolis, MN) was employed to identify the most promising combination of storage medium additives. The five best additives from the viability analysis were included as independent variables (adenosine, allopurinol, β-glycerophosphate, L-ascorbic acid and taurine), with area of CAM fluorescence and the number of dead cells as the two dependent variables. The combined results of two end points were studied. However, the «Importance» tool was employed to set relative priorities for the two variables. The importance of viability (CAM fluorescence area) was emphasized over cell death (number of dead cells). The two-level full-factorial design included replicates of all 32 possible combinations of the five additives. Data were fitted to a full quadratic model. ANOVA was used to calculate the adjusted significance of both models (viability and death) in Design-Expert (P = 0.0047 and P = 0.036, respectively).
Flow Cytometry
Flow cytometry was used to validate the viability results. Cells were cultured in T25 cell culture flasks following the aforementioned protocol. Control cells (N = 3) and cells stored in the optimal additive combination (1% sericin, 5 mmol/L adenosine, 50 μg/mL L-ascorbic acid and 1 mM allopurinol) (N = 3) for three days were compared. Propidium iodide (PI), which binds to double-stranded DNA of dead cells, was added to the culture medium of both culture groups at a concentration of 2.5 μg/300 μL sample and cells were returned to the incubator for 15 minutes. Cells were then rinsed with PBS, trypsinized for 2–3 minutes, then washed and re-suspended in ice-cold HBSS +4% FBS. Samples were kept on ice and analyzed using the BD Accuri C6 bench top flow cytometer. PI is excited by the 588 nm laser and is detected in filter 616//23 (FL3).
Transmission Electron Microscopy
Both unstored cultures and samples of hRPE stored for three days in MEM storage medium with the optimal additive combination (1% sericin, 5 mmol/L adenosine, 50 μg/mL L-ascorbic acid and 1 mM allopurinol) were processed for transmission electron microscopy (TEM) analysis as described earlier83. In essence, a Leica Ultracut Ultramicrotome (Leixa, Wetzlar, Germany) was used to cut ultrathin sections, which were examined using a CM120 transmission electron microscope (Philips, Amsterdam, the Netherlands).
Statistical Analysis
Statistical analysis was performed using IBM SPSS Statistics for Macintosh version 22.0 (IBM Corp, Armonk, NY). A one-way analysis of variance with Tukey’s post-hoc comparisons was used for statistical evaluation of the viability results. The Student’s t-test was used to compare two groups. P values below 0.05 were considered significant.
Proteomics
The proteome of hRPE cells stored in the optimal storage medium combination was analyzed and compared to control cells that had not been stored. The proteome analyses were performed as previously described84. Briefly, the proteins of cell lysates were digested in-solution with trypsin. The generated peptides were analyzed by LC-MS using a nano-UHPLC connected to a Q Exactive mass spectrometer. Proteins were identified using the Mascot search engine and Scaffold software (version Scaffold_4.7.3, Proteome Software Inc., Portland, OR) was used for further data analysis and label-free quantification. Scaffold was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Peptide Prophet algorithm85 with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm86. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Distribution of protein functions in hRPE before and after storage was determined using Scaffold software with annotations downloaded from the NCBI web database.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on request.
Electronic supplementary material
Author Contributions
L.P., T.P.U., C.J. and J.R.E. supervised the project. L.P., S.R., A.Z.K., B.T. and J.R.E. performed the experiments. L.P., S.R., A.Z.K., B.T., E.M. and J.R.E. analyzed the data. L.P., T.P.U., J.P.B. and J.R.E. wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
There is a competing financial interest. A patent application based on results obtained in this study has been filed. There are none non-financial competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24121-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 2.da Cruz L, Chen FK, Ahmado A, Greenwood J, Coffey P. RPE transplantation and its role in retinal disease. Prog Retin Eye Res. 2007;26:598–635. doi: 10.1016/j.preteyeres.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Lund RD, et al. Subretinal transplantation of genetically modified human cell lines attenuates loss of visual function in dystrophic rats. Proc Natl Acad Sci USA. 2001;98:9942–9947. doi: 10.1073/pnas.171266298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGill TJ, et al. Preservation of vision following cell-based therapies in a model of retinal degenerative disease. Vision Res. 2004;44:2559–2566. doi: 10.1016/j.visres.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 5.Sheridan CM, et al. Replacement of the RPE monolayer. Eye. 2009;23:1910–1915. doi: 10.1038/eye.2008.420. [DOI] [PubMed] [Google Scholar]
- 6.Yaji N, Yamato M, Yang J, Okano T, Hori S. Transplantation of tissue-engineered retinal pigment epithelial cell sheets in a rabbit model. Biomaterials. 2009;30:797–803. doi: 10.1016/j.biomaterials.2008.10.045. [DOI] [PubMed] [Google Scholar]
- 7.Falkner-Radler CI, et al. Human retinal pigment epithelium (RPE) transplantation: outcome after autologous RPE-choroid sheet and RPE cell-suspension in a randomised clinical study. Br J Ophthalmol. 2011;95:370–375. doi: 10.1136/bjo.2009.176305. [DOI] [PubMed] [Google Scholar]
- 8.Coffey PJ, et al. Long-term preservation of cortically dependent visual function in RCS rats by transplantation. Nat Neurosci. 2002;5:53–56. doi: 10.1038/nn782. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Lu B, Wood P, Lund RD. Grafting of ARPE-19 and Schwann cells to the subretinal space in RCS rats. Invest Ophthalmol Vis Sci. 2005;46:2552–2560. doi: 10.1167/iovs.05-0279. [DOI] [PubMed] [Google Scholar]
- 10.Zhang T, et al. Photoreceptors repair by autologous transplantation of retinal pigment epithelium and partial-thickness choroid graft in rabbits. Invest Ophthalmol Vis Sci. 2009;50:2982–2988. doi: 10.1167/iovs.08-2131. [DOI] [PubMed] [Google Scholar]
- 11.Sauve Y, Pinilla I, Lund RD. Partial preservation of rod and cone ERG function following subretinal injection of ARPE-19 cells in RCS rats. Vision Res. 2006;46:1459–1472. doi: 10.1016/j.visres.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Nommiste B, et al. Stem cell-derived retinal pigment epithelium transplantation for treatment of retinal disease. Prog Brain Res. 2017;231:225–244. doi: 10.1016/bs.pbr.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Ramsden CM, et al. Stem cells in retinal regeneration: past, present and future. Development. 2013;140:2576–2585. doi: 10.1242/dev.092270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsden C, da Cruz L, Coffey PJ. Stemming the Tide of Age-Related Macular Degeneration: New Therapies for Old Retinas. Invest Ophthalmol Vis Sci. 2016;57:ORSFb1–ORSFb3. doi: 10.1167/iovs.15-18643. [DOI] [PubMed] [Google Scholar]
- 15.Ahmad S, Osei-Bempong C, Dana R, Jurkunas U. The culture and transplantation of human limbal stem cells. J Cell Physiol. 2010;225:15–19. doi: 10.1002/jcp.22251. [DOI] [PubMed] [Google Scholar]
- 16.Utheim TP, et al. Sterility control and long-term eye-bank storage of cultured human limbal epithelial cells for transplantation. Br J Ophthalmol. 2009;93:980–983. doi: 10.1136/bjo.2008.149591. [DOI] [PubMed] [Google Scholar]
- 17.Chopdar A, Chakravarthy U, Verma D. Age related macular degeneration. BMJ. 2003;326:485–488. doi: 10.1136/bmj.326.7387.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pegg DE. The History and Principles of Cryopreservation. Semin Reprod Med. 2002;20:5–13. doi: 10.1055/s-2002-23515. [DOI] [PubMed] [Google Scholar]
- 19.Wang AW, Zhang H, Ikemoto I, Anderson DJ, Loughlin KR. Reactive oxygen species generation by seminal cells during cryopreservation. Urology. 1997;49:921–925. doi: 10.1016/S0090-4295(97)00070-8. [DOI] [PubMed] [Google Scholar]
- 20.Honda S, Weigel A, Hjelmeland LM, Handa JT. Induction of telomere shortening and replicative senescence by cryopreservation. Biochem Biophys Res Commun. 2001;282:493–498. doi: 10.1006/bbrc.2001.4585. [DOI] [PubMed] [Google Scholar]
- 21.Eidet JR, et al. The Silk-protein Sericin Induces Rapid Melanization of Cultured Primary Human Retinal Pigment Epithelial Cells by Activating the NF-kappaB Pathway. Sci Rep. 2016;6:22671. doi: 10.1038/srep22671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsao L-I, Su T-P. Hibernation-induction peptide and cell death: [D-Ala2, D-Leu5]enkephalin blocks Bax-related apoptotic processes. European Journal of Pharmacology. 2001;428:3. doi: 10.1016/S0014-2999(01)01346-2. [DOI] [PubMed] [Google Scholar]
- 23.Vecchio L, et al. DADLE induces a reversible hibernation-like state in HeLa cells. Histochem Cell Biol. 2006;125:193–201. doi: 10.1007/s00418-005-0085-x. [DOI] [PubMed] [Google Scholar]
- 24.Radtke C, et al. TRPV channel expression in human skin and possible role in thermally induced cell death. J Burn Care Res. 2011;32:150–159. doi: 10.1097/BCR.0b013e318203350c. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee PK, Marcheselli VL, Serhan CN, Bazan NG. Neuroprotectin D1: a docosahexaenoic acid-derived docosatriene protects human retinal pigment epithelial cells from oxidative stress. Proc Natl Acad Sci USA. 2004;101:8491–8496. doi: 10.1073/pnas.0402531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antony R, Lukiw WJ, Bazan NG. Neuroprotectin D1 Induces Dephosphorylation of Bcl-xL in a PP2A-dependent Manner during Oxidative Stress and Promotes Retinal Pigment Epithelial Cell Survival. The Journal of Biological Chemistry. 2010;285:8. doi: 10.1074/jbc.M109.095232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansoor S, et al. Inhibition of apoptosis in human retinal pigment epithelial cells treated with benzo(e)pyrene, a toxic component of cigarette smoke. Invest Ophthalmol Vis Sci. 2010;51:2601–2607. doi: 10.1167/iovs.09-4121. [DOI] [PubMed] [Google Scholar]
- 28.King RE, Kent KD, Bomser JA. Resveratrol reduces oxidation and proliferation of human retinal pigment epithelial cells via extracellular signal-regulated kinase inhibition. Chem Biol Interact. 2005;151:143–149. doi: 10.1016/j.cbi.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Pintea A, et al. Antioxidant effect of trans-resveratrol in cultured human retinal pigment epithelial cells. J Ocul Pharmacol Ther. 2011;27:315–321. doi: 10.1089/jop.2010.0144. [DOI] [PubMed] [Google Scholar]
- 30.Sheu SJ, Liu NC, Chen JL. Resveratrol protects human retinal pigment epithelial cells from acrolein-induced damage. J Ocul Pharmacol Ther. 2010;26:231–236. doi: 10.1089/jop.2009.0137. [DOI] [PubMed] [Google Scholar]
- 31.Dajas F. Life or death: neuroprotective and anticancer effects of quercetin. J Ethnopharmacol. 2012;143:383–396. doi: 10.1016/j.jep.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Kook D, et al. The protective effect of quercetin against oxidative stress in the human RPE in vitro. Investigative ophthalmology & visual science. 2008;49:1712–1720. doi: 10.1167/iovs.07-0477. [DOI] [PubMed] [Google Scholar]
- 33.Chidlow G, Wood JP, Knoops B, Casson RJ. Expression and distribution of peroxiredoxins in the retina and optic nerve. Brain Struct Funct. 2016;221:3903–3925. doi: 10.1007/s00429-015-1135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeMali KA, Barlow CA, Burridge K. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J Cell Biol. 2002;159:881–891. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohan R, John A. Microtubule-associated proteins as direct crosslinkers of actin filaments and microtubules. IUBMB Life. 2015;67:395–403. doi: 10.1002/iub.1384. [DOI] [PubMed] [Google Scholar]
- 36.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 37.Smith-Thomas L, et al. Human ocular melanocytes and retinal pigment epithelial cells differ in their melanogenic properties in vivo and in vitro. Curr Eye Res. 1996;15:1079–1091. doi: 10.3109/02713689608995139. [DOI] [PubMed] [Google Scholar]
- 38.Rizzolo LJ. Development and Role of Tight Junctions in the Retinal Pigment Epithelium. Int Rev Cytol. 2007;258:195–234. doi: 10.1016/S0074-7696(07)58004-6. [DOI] [PubMed] [Google Scholar]
- 39.Lu F, Yan D, Zhou X, Hu D-N, Qu J. Expression of melanin-related genes in cultured adult human retinal pigment epithelium and uveal melanoma cells. Mol Vis. 2007;13:2066–2072. [PubMed] [Google Scholar]
- 40.Tamiya S, Liu L, Kaplan HJ. Epithelial-mesenchymal transition and proliferation of retinal pigment epithelial cells initiated upon loss of cell-cell contact. Invest Ophthalmol Vis Sci. 2010;51:2755–2763. doi: 10.1167/iovs.09-4725. [DOI] [PubMed] [Google Scholar]
- 41.Czitrom, V. One-Factor-at-a-Time Versus Designed Experiments. Amer Statist. 53, 126-131 (1999).
- 42.Haskó G. Adenosine: an endogenous regulator of innate immunity. Trends in Immunology. 2004;25:33–39. doi: 10.1016/j.it.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 43.Fredholm BB, AP IJ, Jacobson KA, Linden J, Muller CE. International Union of Basic and Clinical Pharmacology. LXXXI. Nomenclature and classification of adenosine receptors–an update. Pharmacol Rev. 2011;63:1–34. doi: 10.1124/pr.110.003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galvao J, et al. Adenosine A3 receptor activation is neuroprotective against retinal neurodegeneration. Exp Eye Res. 2015;140:65–74. doi: 10.1016/j.exer.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 45.Hu H, et al. Stimulation of the P2X7 receptor kills rat retinal ganglion cells in vivo. Exp Eye Res. 2010;91:425–432. doi: 10.1016/j.exer.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, D. et al. Activation of P2X Receptors Induces Apoptosis in Human Retinal Pigment Epithelium. Invest Ophthalmol Vis Sci52, 1522-1530, https://doi.org/10.1167/ (2011). [DOI] [PMC free article] [PubMed]
- 47.Reichenbach A, Bringmann A. Purinergic signaling in retinal degeneration and regeneration. Neuropharmacology. 2016;104:194–211. doi: 10.1016/j.neuropharm.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Chemtob S, Roy M-S, Abran D, Fernandez H, Varma DR. Prevention of Postasphyxial Increase in Lipid Peroxides and Retinal Function Deterioration in the Newborn Pig by Inhibition of Cyclooxygenase Activity and Free Radical Generation. Pediatr Res. 1993;33:336–340. doi: 10.1203/00006450-199304000-00006. [DOI] [PubMed] [Google Scholar]
- 49.Augustin AJ, Loeffler KU, Sekundo W, Grus FH, Lutz J. Effects of systemically applied allopurinol and prednisolone on experimental autoimmune uveitis. Graefe’s Arch Clin Exp Ophthalmol. 1999;237:508–512. doi: 10.1007/s004170050270. [DOI] [PubMed] [Google Scholar]
- 50.Augustin AJ, Grus FH, Hunt S. Effects of allopurinol on free-radical-induced reduction of the proliferation of retinal pigment epithelial cells. Doc Ophthalmol. 1997;93:231–236. doi: 10.1007/BF02569063. [DOI] [PubMed] [Google Scholar]
- 51.AREDS. A. Randomized, Placebo-Controlled, Clinical Trial of High-Dose Supplementation With Vitamins C and E, Beta Carotene, and Zinc for Age-Related Macular Degeneration and Vision Loss: AREDS Report No. 8. Arch Ophthal. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin J, Thomas F, Lang JC, Chaum E. Modulation of oxidative stress responses in the human retinal pigment epithelium following treatment with vitamin C. J Cell Physiol. 2011;226:2025–2032. doi: 10.1002/jcp.22532. [DOI] [PubMed] [Google Scholar]
- 53.Zeitz O, Schlichting L, Richard G, Strauss O. Lack of antioxidative properties of vitamin C and pyruvate in cultured retinal pigment epithelial cells. Graefes Arch Clin Exp Ophthalmol. 2007;245:276–281. doi: 10.1007/s00417-006-0384-5. [DOI] [PubMed] [Google Scholar]
- 54.Castillo M, et al. Effects of Hypoxia on Retinal Pigmented Epithelium Cells: Protection by Antioxidants. Ophthalmic Res. 2002;34:338–342. doi: 10.1159/000067050. [DOI] [PubMed] [Google Scholar]
- 55.Organisciak DT, Jiang Y-L, Wang H-M, Bicknell I. The Protective Effect of Ascorbic Acid in Retinal Light Damage of Rats Exposed to Intermittent Light. Invest Ophthalmol Vis Sci. 1990;31:1195–1202. [PubMed] [Google Scholar]
- 56.Blanks JC, Pickford MS, Organisciak DT. Ascorbate Treatment Prevents Accumulation of Phagosomes in RPE in Light Damage. Invest Ophthalmol Vis Sci. 1992;33:2814–2821. [PubMed] [Google Scholar]
- 57.Kato N, et al. Silk protein, sericin, inhibits lipid peroxidation and tyrosinase activity. Biosci. Biotechnol. Biochem. 1998;62:145–147. doi: 10.1271/bbb.62.145. [DOI] [PubMed] [Google Scholar]
- 58.Reinisalo M, Putula J, Mannermaa E, Urtti A, Honkakoski P. Regulation of the human tyrosinase gene in retinal pigment epithelium cells: the significance of transcription factor orthodenticle homeobox 2 and its polymorphic binding site. Mol Vis. 2012;18:38–54. [PMC free article] [PubMed] [Google Scholar]
- 59.Lane A, et al. Engineering efficient retinal pigment epithelium differentiation from human pluripotent stem cells. Stem Cells Transl Med. 2014;3:1295–1304. doi: 10.5966/sctm.2014-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vugler A, et al. Elucidating the phenomenon of HESC-derived RPE: anatomy of cell genesis, expansion and retinal transplantation. Exp Neurol. 2008;214:347–361. doi: 10.1016/j.expneurol.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 61.Ahmado A, et al. Induction of differentiation by pyruvate and DMEM in the human retinal pigment epithelium cell line ARPE-19. Invest Ophthalmol Vis Sci. 2011;52:7148–7159. doi: 10.1167/iovs.10-6374. [DOI] [PubMed] [Google Scholar]
- 62.Klimanskaya I, et al. Derivation and Comparative Assessment of Retinal Pigment Epithelium from Human Embryonic Stem Cells Using Transcriptomics. Cloning and Stem Cells. 2004;6:217–245. doi: 10.1089/clo.2004.6.217. [DOI] [PubMed] [Google Scholar]
- 63.Hirami Y, et al. Generation of retinal cells from mouse and human induced pluripotent stem cells. Neurosci Lett. 2009;458:126–131. doi: 10.1016/j.neulet.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 64.Carr AJ, et al. Molecular characterization and functional analysis of phagocytosis by human embryonic stem cell-derived RPE cells using a novel human retinal assay. Mol Vis. 2009;15:283–295. [PMC free article] [PubMed] [Google Scholar]
- 65.Sonoda S, et al. A protocol for the culture and differentiation of highly polarized human retinal pigment epithelial cells. Nat Protoc. 2009;4:662–673. doi: 10.1038/nprot.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marmor MF. Mechanisms of fluid accumulation in retinal edema. Doc. Ophthalmol. 1999;97:239–249. doi: 10.1023/A:1002192829817. [DOI] [PubMed] [Google Scholar]
- 67.Ablonczy Z, et al. Human retinal pigment epithelium cells as functional models for the RPE in vivo. Invest Ophthalmol Vis Sci. 2011;52:8614–8620. doi: 10.1167/iovs.11-8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ablonczy Z, Crosson CE. VEGF modulation of retinal pigment epithelium resistance. Exp. Eye Res. 2007;85:762–771. doi: 10.1016/j.exer.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bonilha VL, Rayborn ME, Saotome I, McClatchey AI, Hollyfield JG. Microvilli defects in retinas of ezrin knockout mice. Exp Eye Res. 2006;82:720–729. doi: 10.1016/j.exer.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 70.Hobbs RP, Green KJ. Desmoplakin regulates desmosome hyperadhesion. J Invest Dermatol. 2012;132:482–485. doi: 10.1038/jid.2011.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huen AC, et al. Intermediate filament-membrane attachments function synergistically with actin-dependent contacts to regulate intercellular adhesive strength. J Cell Biol. 2002;159:1005–1017. doi: 10.1083/jcb.200206098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang L, et al. Desmoplakin acts as a tumor suppressor by inhibition of the Wnt/beta-catenin signaling pathway in human lung cancer. Carcinogenesis. 2012;33:1863–1870. doi: 10.1093/carcin/bgs226. [DOI] [PubMed] [Google Scholar]
- 73.Papagerakis S, et al. Altered desmoplakin expression at transcriptional and protein levels provides prognostic information in human oropharyngeal cancer. Hum Pathol. 2009;40:1320–1329. doi: 10.1016/j.humpath.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 74.Alazawi WO, Morris LS, Stanley MA, Garrod DR, Coleman N. Altered expression of desmosomal components in high-grade squamous intraepithelial lesions of the cervix. Virchows Arch. 2003;443:51–56. doi: 10.1007/s00428-003-0771-9. [DOI] [PubMed] [Google Scholar]
- 75.Hamidov Z, et al. Reduced expression of desmocollin 2 is an independent prognostic biomarker for shorter patients survival in pancreatic ductal adenocarcinoma. J Clin Pathol. 2011;64:990–994. doi: 10.1136/jclinpath-2011-200099. [DOI] [PubMed] [Google Scholar]
- 76.Carvajal-Huerta L. Epidermolytic palmoplantar keratoderma with woolly hair and dilated cardiomyopathy. J Am Acad Dermatol. 1998;39:481–421. doi: 10.1016/S0190-9622(98)70317-2. [DOI] [PubMed] [Google Scholar]
- 77.Norgett, E. E. et al. Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum Mol Genet. 9 (2000). [DOI] [PubMed]
- 78.Antonov NK, et al. Early-onset heart failure, alopecia, and cutaneous abnormalities associated with a novel compound heterozygous mutation in desmoplakin. Pediatr Dermatol. 2015;32:102–108. doi: 10.1111/pde.12484. [DOI] [PubMed] [Google Scholar]
- 79.McAleer MA, et al. Severe dermatitis, multiple allergies, and metabolic wasting syndrome caused by a novel mutation in the N-terminal plakin domain of desmoplakin. The Journal of allergy and clinical immunology. 2015;136:1268–1276. doi: 10.1016/j.jaci.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grisanti S, Guidry C. Transdifferentiation of Retinal Pigment Epithelial Cells From Epithelial to Mesenchymal Phenotype. Invest Ophthalmol Vis Sci. 1995;36:391–405. [PubMed] [Google Scholar]
- 81.Ganti R, Hunt RC, Parapuram SK, Hunt DM. Vitreous modulation of gene expression in low-passage human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2007;48:1853–1863. doi: 10.1167/iovs.06-0198. [DOI] [PubMed] [Google Scholar]
- 82.Pasovic L, et al. Optimization of Storage Temperature for Cultured ARPE-19 Cells. J Ophthalmol. 2013;2013:1–11. doi: 10.1155/2013/216359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raeder S, et al. Effect of limbal explant orientation on the histology, phenotype, ultrastructure and barrier function of cultured limbal epithelial cells. Acta Ophthalmol Scand. 2007;85:377–386. doi: 10.1111/j.1600-0420.2006.00873.x. [DOI] [PubMed] [Google Scholar]
- 84.Tran TT, Bollineni RC, Strozynski M, Koehler CJ, Thiede B. Identification of Alternative Splice Variants Using Unique Tryptic Peptide Sequences for Database Searches. J Proteome Res. 2017 doi: 10.1021/acs.jproteome.7b00126. [DOI] [PubMed] [Google Scholar]
- 85.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 86.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on request.