Abstract
Both common and rare polymorphisms within ABCA7 have been associated with Alzheimer’s disease (AD). In particular, the rare AD associated polymorphism rs200538373 was associated with altered ABCA7 exon 41 splicing and an AD risk odds ratio of ~1.9. To probe the role of this polymorphism in ABCA7 splicing, we used minigene studies and qPCR of human brain RNA. We report aberrant ABCA7 exon 41 splicing in the brain of a carrier of the rs200538373 minor C allele. Moreover, minigene studies show that rs200538373 acts as a robust functional variant in vitro. Lastly, although the ABCA7 isoform with an extended exon 41 is predicted to undergo nonsense mediated RNA decay, this was not supported by qPCR analyses, which showed relatively normal ABCA7 mRNA levels in the carrier of the rs200538373 minor C allele. In summary, rs200538373 is a functional polymorphism that alters ABCA7 exon 41 splicing without grossly altering the level of ABCA7 mRNA.
Keywords: Alzheimer’s, ABCA7, Genetics, SNP, Splicing
Introduction
AD currently affects as many as 5.1 million people in the United States [1]. As ‘baby boomers’ age, AD prevalence is projected to triple by the year 2050 unless a treatment is found [1]. Since pharmacologic agents based on genetic mechanisms are more likely to successfully transition to drugs approved by the Food and Drug Administration [2], we seek to elucidate the actions underlying AD-associated SNPs.
Single nucleotide polymorphisms (SNP)s in ABCA7 were identified as AD risk factors in several studies [3–6]. These SNPs included several common SNPs that were associated with modest AD risk, including rs3764650, rs4147914, rs3752246, and rs4147929 [3–5]. These SNPs were also found to associate with ABCA7 expression, cortical and hippocampal atrophy, as well as β-secretase activity in cells expressing the amyloid-β protein precursor (AβPP)-Swedish mutation [3, 7–9]. In addition to these common ABCA7 SNPs, several rare ABCA7 SNPs, including rs200538373, were associated with AD at odds ratios as high as 1.9 [6].
The role of ABCA7 function itself in AD is unclear. Recent findings using human brain show ABCA7 is expressed at low levels in several cell types, including neurons and microglia [10]. ABCA7 has been implicated in lipid transport, phagocytosis and Aβ homeostasis [11–17].
Here, we sought to better understand the role of the rare SNP, rs200538373, which was associated with ABCA7 exon 41 splicing in blood [6]. We extend these prior findings by reporting an aberrant 14 bp extension of exon 41 in the brain of an individual that was heterozygous for this SNP. The hypothesis that this SNP is functional was supported by in silico modeling and by an ABCA7 minigene study, which demonstrated that rs200538373 acts to alter exon 41 splicing. Lastly, ABCA7 expression in a carrier of the minor allele of rs200538373 was similar to that of other brain samples; this finding is inconsistent with hypothesized nonsense mediated RNA decay for this isoform, suggesting that the likely action for this SNP is altered splicing leading to a truncated ABCA7 protein.
Materials and Methods
Ethics statement
This work was conducted under the approval of the University of Kentucky Institutional Review Board.
Human brain tissue
RNA was purified from human anterior cingulate brain samples (supplied by the University of Kentucky AD Center Neuropathology Core) and converted to cDNA as previously described [18–20]. The AD status of the brain donors was determined by the AD Center Neuropathology and Clinical Cores by using guidelines that included evaluation of neurofibrillary tangles and neuritic senile plaques as well as cognitive status [21–23]. Age at death for the cognitively intact, i.e. non-AD donors, was 82.6 ± 1.6 (mean ± SE, n = 28) while AD donors were 81.7 ± 1.2 (mean ± SE, n = 28). The post-mortem interval (PMI) for non-AD and AD donors was 2.7 ± 0.2 (n = 28) hours and 3.4 ± 0.1 (n = 28), respectively.
Genotyping
DNA samples were genotyped for the indicated polymorphisms by using FAM and VIC dye-labeled probes (Invitrogen, Carlsbad, CA) and TaqMan polymerase chain reaction (PCR) (Bio-Rad, Hercules, CA).
Splicing assay
ABCA7 exon 41 splicing was assessed by PCR coupled to polyacrylamide gel electrophoresis (PAGE). Reactions contained a sense primer corresponding to sequence within exon 40, i.e., 5′-CCGTGGGCAGAGGATG-3′ and an antisense primer corresponding to exon 42 sequence, i.e., 5′-TCGGATTGAGGGCAGTATC-3′. Each 20 μL reaction mixture contained ~20 ng of cDNA, 25 pmole of each primer, and Platinum Taq (ThermoFisher) and was subjected to a PCR profile of 30 cycles at 95°C for 15 s, 59°C for 30 s, and 72°C for 20 s. PCR products were separated on 10% polyacrylamide gels and detected by SYBR gold fluorescence (ThermoFisher) as per manufacturer’s protocol. Each sample was analyzed twice and reactions lacking cDNA template were analyzed in parallel to check for PCR product contamination.
Maximum entropy scores
The sequences of the ABCA7 exon 41 isoforms were scored for 5′ splice site favorability by using an online prediction tool ((http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html [24]. Briefly, this algorithm was trained on large datasets of human splice sites to calculate a log odds ratio for a splicing score for input sequence [24]. A higher score correlates with a more favorable splice site [24, 25].
RNA splicing assay
ABCA7 minigenes for each rs200538373 allele were generated by PCR and contained exon 41, intron 41, and exon 42 in their entirety, cloned into pcDNA3.1 (ThermoFisher). Minigene construction used a sense primer corresponding to sequence at the start of exon 41, i.e., 5′-TGTTTTGGGCTGCTGG-3′ and an antisense primer corresponding to sequence at the end of exon 42, i.e., 5′-CTGGGCAACCTGGGC-3′. Sequencing confirmed inserts were accurate and complete for each allele and differed only for rs200538373 alleles. Human Be(2)-M17 neuroblastoma cells (ATCC, Manassas, VA) were maintained in Opti-MEM I reduced-serum medium supplemented to 10% fetal bovine serum, 50 U/ml penicillin and 50 μg/ml streptomycin with humidified 5% carbon dioxide at 37°C. For transfections, cells (1×106) were plated in 350 μL media in a 24-well plate, allowed to grow 24 hrs and then transfected in triplicate by using 2 μg of allele-specific ABCA7 minigene vector and Lipofectamine 3000, as per manufacturer’s protocol (ThermoFisher). Forty-eight hours after transfection, RNA was prepared and reverse transcribed using random hexamers and Superscript III as described previously [26]. ABCA7 expression from each minigene was detected by PCR using a pcDNA3.1 vector-derived forward primer (5′-ACTAGTCCAGTGTGGTGGAATTGCC-3′) and exon 42-derived reverse primer (5′-TCGGATTGAGGGCAGTATC-3′).
Quantitative PCR assay
The quantification of ABCA7, synaptophysin, ITGAM and AIF1 expression in these samples has been described previously [7, 26, 27]. Briefly, 20 μL reactions contained 1μM of each primer, 1x PerfeCTa SYBR Green Super Mix (Quanta Biosciences), and approximately 20 ng cDNA generated from anterior cingulate RNA. PCR was conducted using an initial 2-minute incubation at 95°, followed by cycles of 10 seconds at 95°, 20 seconds at 60°, and 20 seconds at 72°. Experimental samples were amplified in parallel with serially diluted standards that were generated by PCR of cDNA followed by purification and quantitation by UV absorbance. Results from samples were compared relative to the standard curve to calculate copy number in each sample. Real time assays were performed twice and the average copy number was used for further analyses.
Statistical Analysis
ABCA7 expression was analyzed by linear regression (SPSS, version 23). Each model included AD status and rs3764650 genotype. In addition, since ABCA7 is expressed in both microglia and neurons [16, 17], we included expression of either (i) a neuronal gene (synaptophysin), (ii) microglial genes (geometric mean of ITGAM and AIF1), or (iii) both the neuronal and microglial genes (SPSS, v. 23). The geometric mean of ITGAM and AIF1 was used, as opposed to the arithmetic mean, because AIF1 levels were markedly higher than ITGAM levels. To generate a normal distribution of the data, the square root of the copy number data was used for the linear regression analyses; the square root approach was validated by the Shapiro-Wilk test for normality. We also analyzed the expression results for variation in sample mRNA content by dividing mRNA copy number by the geometric mean of two constitutively expressed genes, i.e., RPL32 and EIF4H [26]. To generate a normal distribution as assessed by Shapiro-Wilk, regression analyses used the square root of these values. Since our cohort of samples had only one rs3764650 minor allele homozygous individual, we used a dominant model for this analysis, i.e., rs3764650 minor allele carriers were considered as a single group.
Results
Rs200538373 is a rare intronic ABCA7 variant that was associated with AD risk and with exon 41 splicing in blood [6]. To confirm and extend this finding, we determined rs200538373 genotypes in a set of 57 cDNA samples generated from AD and non-AD brains; this effort identified a single sample that was heterozygous for rs200538373 (G/C) while the remainder were major allele (G/G) homozygous. The frequency of the rs200538373 minor C allele in our cohort was 0.8%, similar to that reported previously in European datasets [6, 28].
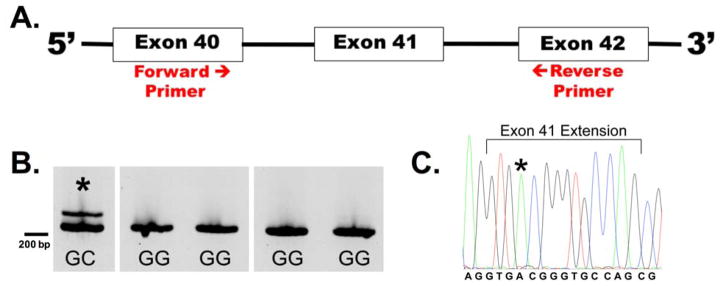
To explore the effects of the C allele of rs200538373, we sought to confirm whether aberrant splicing occurs in the brain of an individual with this allele. To assess splicing, cDNA from the rs200538373 heterozygous sample as well as several rs200538373 major allele homozygous samples was subjected to RT-PCR with amplicons spanning exon 40 to exon 42 (Figure 1A). We observed the expected product along with an additional longer amplicon for the sample that was heterozygous for rs200538373 (Figure 1B). Sanger sequencing of this longer amplicon found that exon 41 was extended 14 bp into the typical intron 41. Interestingly, the sequence electropherogram shows the rs200538373 minor C allele but not the major G allele (Figure 1C). Although not quantitative, this suggests that this abnormal splice form is largely produced from the minor allele of the ABCA7 pre-mRNA. The 14 bp inclusion would encode an in-frame UGA termination codon beginning at position 2 in the extension and is therefore predicted to produce a truncated ABCA7 protein.
Figure 1.
Aberrant ABCA7 exon 41 splicing in an rs200538373 heterozygous sample. A). RNA purified from the brains of a rs200538373G/C individual and several rs200538373G/G individuals was reverse transcribed and subjected to PCR with primers corresponding to sequences within exons 40 and 42. The amplicon size for normal splicing was 222 bp. B). The expected splice product as well as a longer ABCA7 isoform was consistently detected only in the rs200538373G/C individual (denoted by *). C). Sanger sequencing of the longer ABCA7 isoform found that exon 41 was extended by 14 bp relative to the expected isoform. This sequence from the rs200538373 heterozygous individual includes the last two bp of exon 41, the 14 bp extension of exon 41, and the first two bp of exon 42. Only the rs200538373 minor C allele is observed (marked by asterisk) (blue=C, black=G, green=A, red=T).
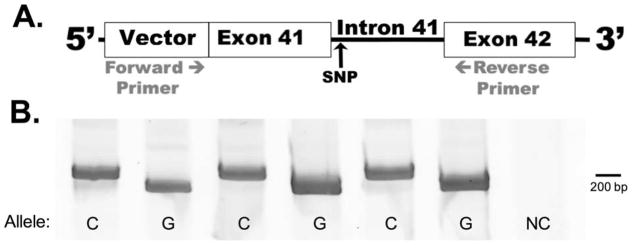
To test directly whether rs200538373 modulates exon 41 splicing, we generated minigenes for each ABCA7 allele. The constructs included all of exon 41, intron 41, and exon 42 (Figure 2A). Since ABCA7 is expressed in neurons, as well as other brain cell types [10, 29, 30], we tested the minigene splicing in the human neural cell line Be(2)-M17. We found that the transcript from the minigene with the rs200538373 minor C allele was spliced to generate exon 41 with the 14 bp extension (Figure 2B). Conversely, the minigene with the rs200538373 major G allele was spliced to generate exon 41 at the usual 5′ consensus splice site. The effect of the SNP in this assay was very robust, with minimal “normal” isoform generated by the minigene carrying the minor C allele and minimal extended isoform generated by the minigene carrying the major G allele.
Figure 2.
Rs200538373 is a functional SNP that modulates ABCA7 exon 41 splicing. A). Cells were transfected in triplicate with constructs that expressed ABCA7 exon 41, intron 41, and exon 42 and that carried with the rs200538373 major G or minor C allele. These transcripts also included vector-derived 5′ and 3′ sequence. B). RT-PCR analyses with a forward primer targeting vector sequence and a reverse primer targeting exon 42 detect normal splicing (192 bp amplicon) from the major G allele minigene and aberrant splicing (206 bp amplicon) in the minor C allele minigene. Note that ‘C’ and ‘G’ labels indicate rs200538373 alleles while ‘NC’ labels a negative control sample wherein cells were transfected in parallel with an irrelevant (GFP) gene. This result was replicated in three independent experiments.
To gain further insight into this finding, we used MaxEntScan::score5ss, a 5′ splice site prediction tool, to compare the normal 5′ consensus splice site in the presence of each rs200538373 allele and the aberrant splice site. We observe that the favorable score of the major G allele for rs200538373 at the normal splice site is reduced when the minor C allele of rs200538373 is present (Table 1). The aberrant splice site 14 bp further into the intron is unaffected by the SNP and has a splicing score that is competitive with the normal splice site when the G allele of rs200538373 is present. These results are consistent with rs200538373 being a functional SNP resulting in the normal 5′ consensus splice site being used when the G allele of rs200538373 is present and the aberrant splice site 14 bp further into the intron being used when minor C allele of rs200538373 is present.
Table 1.
Impact of rs200538373 on ABCA7 exon 41 splicing. This SNP is at position 5, in parenthesis, within intron 41 and is predicted to weaken the splice site, i.e., the prototypic splice donor sequence is CAG/gtragt (exon in upper case, intron in lower case, r = purine).
| Exon 41 Splicing | Splice Site Sequence (splice site at ^) | Splicing Score |
|---|---|---|
| Normal Splice Site Major G allele |
CAG^gtga(g)g | 10.07 |
| Normal Splice Site Minor C allele |
CAG^gtga(c)gggtgccaggtaggg | 7.66 |
| Aberrant Splice Site Extended Exon 41 |
CAGgtga(g/c)gggtgccag^gtaggg | 9.46 |
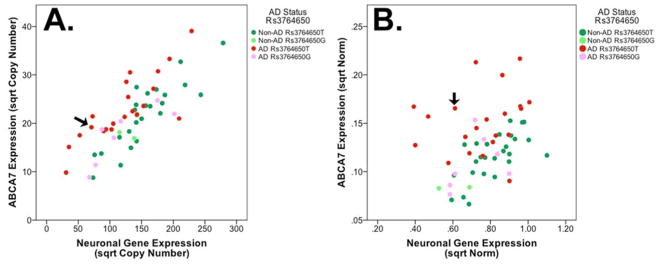
Lastly, we assessed indirectly whether the aberrant splicing and predicted premature termination codon observed with the minor C allele of rs200538373 may be associated with nonsense mediated RNA decay by comparing ABCA7 expression in rs200538373 major G allele homozygous individuals with that of the single rs200538373 heterozygous individual in our cohort of samples. For this effort, we established a quantitative model of ABCA7 expression.. This model included ABCA7 expression as well as AD status and rs3764650, a common AD-associated ABCA7 SNP [3–5]. ABCA7 expression was assessed previously by qPCR using primers in the constitutively spliced exons 44 and 45 [7] and those data are re-analyzed here. Since ABCA7 is expressed in neurons and microglia, we compared models that evaluated ABCA7 expression relative to expression of microglial and neuronal genes separately and together (Table 2). The ABCA7 expression model that used only the neuronal mRNA synaptophysin had slightly more explanatory power (Figure 3A, adjusted R2=0.700) than a model that included synapthophysin as well as microglial (ITGAM and AIF1) mRNAs (adjusted R2=0.697). A model that used only the microglial mRNAs had a lower goodness-of-fit (adjusted R2=0.323). Within each model, ABCA7 expression was increased significantly in AD individuals and decreased significantly in carriers of the minor rs3764650 allele (Table 2, Figure 3A). We also analyzed these data by normalizing mRNA copy numbers to two “housekeeping” mRNAs (Figure 3B). Although this model is less visually striking (adjusted R2 of 0.412), ABCA7 expression was significantly increased in AD (standardized β coefficient=0.559, p=3.0×10−6) and the minor allele of rs3764650 was associated with a significant decrease in ABCA7 expression (standardized β coefficient=−0.383, p=0.001). ABCA7 expression in the individual that was heterozygous for rs200538373 was similar to that of other samples (Figure 3A–B, arrow). Hence, the individual with the minor C allele of rs200538373 did not have a large decrease in ABCA7 expression, a finding we interpret as suggesting that nonsense mediated RNA decay does not grossly affect ABCA7 expression. In summary, this quantitative analysis of ABCA7 expression suggests that (i) the minor AD-protective allele of rs3764650 is associated with decreased ABCA7 expression, (ii) AD itself is associated with increased ABCA7 expression, and (iii) the rare AD-associated SNP rs200538373 acts through altered splicing without grossly affecting ABCA7 expression levels, a finding supported by another quite recent study [31].
Table 2.
Evaluation of cell-type specific reference gene expression in ABCA7 expression model. The neuronal reference gene was synaptophysin while the microglial reference was the geometric mean of ITGAM and AIF1 expression. The model with both cell types used the geometric mean of synaptophysin and the microglial geometric mean.
| Reference Gene Type | Adjusted R2 | AD Beta Coefficient (p value) | Rs3764650 Beta Coefficient (p value) | Reference Gene Beta Coefficient (p value) |
|---|---|---|---|---|
| Neuronal | 0.700 | 0.322 (1.7×10−4) | −0.236 (0.003) | 0.840 (7.1×10−15) |
| Microglial | 0.323 | 0.240 (0.050) | −0.257 (0.029) | 0.556 (1.6×10−5) |
| Both Cell Type | 0.697 | 0.337 (3.9×10−4) | −0.234 (0.008) | 0.805 (1.6×10−12) |
Figure 3.
ABCA7 expression is associated with AD status and rs3764650 but appears unaffected by rs200538373. A). When ABCA7 expression was analyzed by comparing ABAC7 mRNA copy numbers relative to a neuronal mRNA, expression appeared to correlate with rs3764650 and AD status. For both A and B, the arrow points to the single sample that was heterozygous for rs200538373. Additionally, rs3764650T refers to rs3764650T/T homozygous samples while rs3764650G refers to rs3764650G/T heterozygous samples as well as a single rs3764650G/G homozygous sample. B). ABCA7 expression relative to AD status and rs3764650 when mRNA copy numbers were normalized to RPL32 and EIF4H housekeeping mRNAs. For both A and B, the square root of values was used to generate a normal distribution of the data.
Discussion
The primary findings of this report are that abnormal ABCA7 exon 41 splicing is found in the brain of an individual carrying the minor C allele of rs200538373 and that rs200538373 is a functional SNP. Additional findings include that rs200538373 does not appear associated with a gross decline in ABCA7 expression, suggesting that NMD does not grossly impact ABCA7 expression for this individual, and a confirmation that ABCA7 expression is decreased with the minor allele of rs3764650 and increased in AD. As such, this report confirms and extends a report that rs200538373 was associated with ABCA7 exon 41 splicing in blood [6] and our prior report that AD and rs3764650 was associated with expression per se [7].
The mechanisms underlying AD genetic risk factors are currently under intense investigation. Common ABCA7 SNPs such as rs3764650 have been associated with a modest increase in AD risk (odds ratio: 1.23) while rare SNPs that involve premature stop termination codons, such as rs200538373, are associated with increased AD risk (odds ratio: 1.91) [4, 6, 32, 33]. In our original report examining rs3764650, we found that the minor allele of rs3764650 was associated with a decrease in ABCA7 expression with a standardized beta-coefficient of −0.375 [7]. Here, we re-analyzed these ABCA7 data by using a model that incorporated levels of mRNAs specific to microglia and neurons. This analysis produced a similar result, with the rs3764650 minor allele being associated with reduced ABCA7 expression. Hence, modestly reduced ABCA7 expression correlates with a modest increase in the AD odds ratio. In contrast, the rare rs200538373 was associated with aberrant exon 41 splicing that was predicted to produce a premature translation termination codon [6]. In pursuing the effect of this SNP, we observed a very robust SNP effect on exon 41 splicing in vitro and that the extended exon 41 isoform was clearly present in the brain of the rs200538373 minor allele carrier. That noted, the in vivo finding was limited because we identified only a single sample with the SNP minor allele. An additional limitation of this study was that we were unable to identify isoform specific qPCR primers and hence were not able to quantify the extended exon 41 splice variant. Our semi-quantitative gel-based analysis suggested that this extended exon 41 isoform was present at a level approaching that of the normal ABCA7 isoform. Hence, the effect of the SNP may approach 50% of ABCA7 mRNA containing a premature stop codon. This truncated ABCA7 protein would likely represent a loss of function because the encoded ABCA7 protein would lack its second ATPase domain. This effect could be greater if the truncated ABCA7 acts as a dominant negative; this possibility is supported by reports that similarly truncated ABCA1 proteins act as a dominant negative in dimer formation with full-length ABCA1 [34–36]. In summary, we interpret these results overall as showing that rs3764650, a common SNP associated with a modest reduction in ABCA7 expression, leads to a small increase in AD risk. The increase in ABCA7 observed in AD may represent a compensatory attempt to increase ABCA7 and thereby reduce AD risk or may reflect other inflammation related processes. In contrast, rs200538373, a rare SNP associated with a more robust reduction in ABCA7, leads to a larger increase in AD risk. Overall, these results appear to essentially demonstrate a genetic dose response between functional impact and AD risk.
Considering the translational impact of these findings, agents that increase ABCA7 expression would be expected to reduce AD risk, especially for those individuals with the minor allele of rs3764650. Since ABCA7 expression is increased in AD, we speculate that a pharmacologic intervention to reduce AD risk would likely have to begin well before disease onset. With regards to rs200538373 and ABCA7 splicing, we note that in this age of personalized medicine, several drugs that target splicing are in human trials or have been approved for human use. Some of these agents involve peripheral tissue, such as eteplirsen which targets splicing in a form of Duchene muscular dystrophy [37]. However, other agents target splicing in the central nervous system, with the drug nusinersen, which was recently FDA approved [38] for spinal muscular atrophy, being the most robust representative [39]. As science moves forward, agents that target aberrant splicing for AD may emerge.
In summary, our primary finding is an apparent dose response between the functional impact of AD-associated SNPs and AD risk. This finding is based upon (i) the observation that rs200538373, which is associated with robust AD risk, appears to have a robust effect on splicing in vitro with aberrant splicing found in the brain of a minor allele carrier and (ii) the observation that rs3764650, which is associated with modest AD risk, is associated with a modest reduction in ABCA7 expression.
Acknowledgments
The authors acknowledge the NIH (R01-AG045775) and the BrightFocus Foundation (A2014210S) for funding this work, as well as the University of Kentucky AD Center Neuropathology Core for tissue (P30-AG028383). The authors also acknowledge Elsevier Press for allowing data re-use to generate Figure 3. The authors appreciate David Fardo, Ph.D., for statistical consulting.
Footnotes
Conflict of Interest Statement
The authors have no conflict of interest to report.
References
- 1.National Institute on Aging NIoH, U.S. Department of Health and Human Services. Alzheimer’s Disease Progress Report: Advancing Research Toward a Cure. 2014–2015. [Google Scholar]
- 2.Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, Cardon LR, Whittaker JC, Sanseau P. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 3.Allen M, Zou F, Chai HS, Younkin CS, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, Maharjan S, Nguyen T, Ma L, Malphrus KG, Palusak R, Lincoln S, Bisceglio G, Georgescu C, Schultz D, Rakhshan F, Kolbert CP, Jen J, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Petersen RC, Graff-Radford NR, Dickson DW, Younkin SG, Ertekin-Taner N. Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology. 2012;79:221–228. doi: 10.1212/WNL.0b013e3182605801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinberg S, Stefansson H, Jonsson T, Johannsdottir H, Ingason A, Helgason H, Sulem P, Magnusson OT, Gudjonsson SA, Unnsteinsdottir U, Kong A, Helisalmi S, Soininen H, Lah JJ, DemGene, Aarsland D, Fladby T, Ulstein ID, Djurovic S, Sando SB, White LR, Knudsen GP, Westlye LT, Selbaek G, Giegling I, Hampel H, Hiltunen M, Levey AI, Andreassen OA, Rujescu D, Jonsson PV, Bjornsson S, Snaedal J, Stefansson K. Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease. Nat Genet. 2015;47:445–447. doi: 10.1038/ng.3246. [DOI] [PubMed] [Google Scholar]
- 7.Vasquez JB, Fardo DW, Estus S. ABCA7 expression is associated with Alzheimer’s disease polymorphism and disease status. Neurosci Lett. 2013;556:58–62. doi: 10.1016/j.neulet.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bamji-Mirza M, Li Y, Najem D, Liu QY, Walker D, Lue LF, Stupak J, Chan K, Li J, Ghani M, Yang Z, Rogaeva E, Zhang W. Genetic Variations in ABCA7 Can Increase Secreted Levels of Amyloid-beta40 and Amyloid-beta42 Peptides and ABCA7 Transcription in Cell Culture Models. J Alzheimers Dis. 2016 doi: 10.3233/JAD-150965. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez LM, Goukasian N, Porat S, Hwang KS, Eastman JA, Hurtz S, Wang B, Vang N, Sears R, Klein E, Coppola G, Apostolova LG. Common variants in ABCA7 and MS4A6A are associated with cortical and hippocampal atrophy. Neurobiol Aging. 2016;39:82–89. doi: 10.1016/j.neurobiolaging.2015.10.037. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, Duncan JA, 3rd, Cheshier SH, Shuer LM, Chang EF, Grant GA, Gephart MG, Barres BA. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim WS, Weickert CS, Garner B. Role of ATP-binding cassette transporters in brain lipid transport and neurological disease. J Neurochem. 2008;104:1145–1166. doi: 10.1111/j.1471-4159.2007.05099.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim WS, Guillemin GJ, Glaros EN, Lim CK, Garner B. Quantitation of ATP-binding cassette subfamily-A transporter gene expression in primary human brain cells. Neuroreport. 2006;17:891–896. doi: 10.1097/01.wnr.0000221833.41340.cd. [DOI] [PubMed] [Google Scholar]
- 13.Jehle AW, Gardai SJ, Li S, Linsel-Nitschke P, Morimoto K, Janssen WJ, Vandivier RW, Wang N, Greenberg S, Dale BM, Qin C, Henson PM, Tall AR. ATP-binding cassette transporter A7 enhances phagocytosis of apoptotic cells and associated ERK signaling in macrophages. J Cell Biol. 2006;174:547–556. doi: 10.1083/jcb.200601030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Karl T, Garner B. Understanding the function of ABCA7 in Alzheimer’s disease. Biochem Soc Trans. 2015;43:920–923. doi: 10.1042/BST20150105. [DOI] [PubMed] [Google Scholar]
- 15.Sakae N, Liu CC, Shinohara M, Frisch-Daiello J, Ma L, Yamazaki Y, Tachibana M, Younkin L, Kurti A, Carrasquillo MM, Zou F, Sevlever D, Bisceglio G, Gan M, Fol R, Knight P, Wang M, Han X, Fryer JD, Fitzgerald ML, Ohyagi Y, Younkin SG, Bu G, Kanekiyo T. ABCA7 Deficiency Accelerates Amyloid-beta Generation and Alzheimer’s Neuronal Pathology. J Neurosci. 2016;36:3848–3859. doi: 10.1523/JNEUROSCI.3757-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim WS, Li H, Ruberu K, Chan S, Elliott DA, Low JK, Cheng D, Karl T, Garner B. Deletion of Abca7 increases cerebral amyloid-beta accumulation in the J20 mouse model of Alzheimer’s disease. J Neurosci. 2013;33:4387–4394. doi: 10.1523/JNEUROSCI.4165-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satoh K, Abe-Dohmae S, Yokoyama S, St George-Hyslop P, Fraser PE. ATP-binding cassette transporter A7 (ABCA7) loss of function alters Alzheimer amyloid processing. J Biol Chem. 2015;290:24152–24165. doi: 10.1074/jbc.M115.655076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling IF, Estus S. Role of SFRS13A in low-density lipoprotein receptor splicing. Hum Mutat. 2010;31:702–709. doi: 10.1002/humu.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou F, Gopalraj RK, Lok J, Zhu H, Ling IF, Simpson JF, Tucker HM, Kelly JF, Younkin SG, Dickson DW, Petersen RC, Graff-Radford NR, Bennett DA, Crook JE, Younkin SG, Estus S. Sex-dependent association of a common low-density lipoprotein receptor polymorphism with RNA splicing efficiency in the brain and Alzheimer’s disease. Hum Mol Genet. 2008;17:929–935. doi: 10.1093/hmg/ddm365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu H, Tucker HM, Grear KE, Simpson JF, Manning AK, Cupples LA, Estus S. A common polymorphism decreases low-density lipoprotein receptor exon 12 splicing efficiency and associates with increased cholesterol. Hum Mol Genet. 2007;16:1765–1772. doi: 10.1093/hmg/ddm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson PT, Kukull WA, Frosch MP. Thinking outside the box: Alzheimer-type neuropathology that does not map directly onto current consensus recommendations. J Neuropathol Exp Neurol. 2010;69:449–454. doi: 10.1097/NEN.0b013e3181d8db07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson PT, Braak H, Markesbery WR. Neuropathology and cognitive impairment in Alzheimer disease: a complex but coherent relationship. J Neuropathol Exp Neurol. 2009;68:1–14. doi: 10.1097/NEN.0b013e3181919a48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson PT, Abner EL, Scheff SW, Schmitt FA, Kryscio RJ, Jicha GA, Smith CD, Patel E, Markesbery WR. Alzheimer’s-type neuropathology in the precuneus is not increased relative to other areas of neocortex across a range of cognitive impairment. Neurosci Lett. 2009;450:336–339. doi: 10.1016/j.neulet.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 25.Eng L, Coutinho G, Nahas S, Yeo G, Tanouye R, Babaei M, Dork T, Burge C, Gatti RA. Nonclassical splicing mutations in the coding and noncoding regions of the ATM Gene: maximum entropy estimates of splice junction strengths. Hum Mutat. 2004;23:67–76. doi: 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- 26.Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT, Estus S. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. 2013;33:13320–13325. doi: 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Rossi P, Buggia-Prevot V, Clayton BL, Vasquez JB, van Sanford C, Andrew RJ, Lesnick R, Botte A, Deyts C, Salem S, Rao E, Rice RC, Parent A, Kar S, Popko B, Pytel P, Estus S, Thinakaran G. Predominant expression of Alzheimer’s disease-associated BIN1 in mature oligodendrocytes and localization to white matter tracts. Mol Neurodegener. 2016;11:59. doi: 10.1186/s13024-016-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Del-Aguila JL, Fernandez MV, Jimenez J, Black K, Ma S, Deming Y, Carrell D, Saef B, Howells B, Budde J, Cruchaga C Alzheimer’s Disease Neuroimaging I. Role of ABCA7 loss-of-function variant in Alzheimer’s disease: a replication study in European-Americans. Alzheimers Res Ther. 2015;7:73. doi: 10.1186/s13195-015-0154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe-Dohmae S, Ikeda Y, Matsuo M, Hayashi M, Okuhira K, Ueda K, Yokoyama S. Human ABCA7 supports apolipoprotein-mediated release of cellular cholesterol and phospholipid to generate high density lipoprotein. J Biol Chem. 2004;279:604–611. doi: 10.1074/jbc.M309888200. [DOI] [PubMed] [Google Scholar]
- 30.Wang N, Lan D, Gerbod-Giannone M, Linsel-Nitschke P, Jehle AW, Chen W, Martinez LO, Tall AR. ATP-binding cassette transporter A7 (ABCA7) binds apolipoprotein A-I and mediates cellular phospholipid but not cholesterol efflux. J Biol Chem. 2003;278:42906–42912. doi: 10.1074/jbc.M307831200. [DOI] [PubMed] [Google Scholar]
- 31.Allen M, Lincoln SJ, Corda M, Watzlawik JO, Carrasquillo MM, Reddy JS, Burgess JD, Nguyen T, Malphrus K, Petersen RC, Graff-Radford NR, Dickson DW, Ertekin-Taner N. ABCA7 loss-of-function variants, expression, and neurologic disease risk. Neurol Genet. 2017;3:e126. doi: 10.1212/NXG.0000000000000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, Destefano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thornton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Hollingworth P, Ramirez A, Hanon O, Fitzpatrick AL, Buxbaum JD, Campion D, Crane PK, Baldwin C, Becker T, Gudnason V, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MJ, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuinness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannfelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltunen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P European Alzheimer’s Disease I, Genetic Environmental Risk in Alzheimer’s D, Alzheimer’s Disease Genetic C, Cohorts for H, Aging Research in Genomic E. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nature genetics. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagata KO, Nakada C, Kasai RS, Kusumi A, Ueda K. ABCA1 dimer-monomer interconversion during HDL generation revealed by single-molecule imaging. Proc Natl Acad Sci U S A. 2013;110:5034–5039. doi: 10.1073/pnas.1220703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorrenson B, Suetani RJ, Bickley VM, George PM, Williams MJ, Scott RS, McCormick SP. An ABCA1 truncation shows no dominant negative effect in a familial hypoalphalipoproteinemia pedigree with three ABCA1 mutations. Biochem Biophys Res Commun. 2011;409:400–405. doi: 10.1016/j.bbrc.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Stukas S, Robert J, Wellington CL. High-density lipoproteins and cerebrovascular integrity in Alzheimer’s disease. Cell Metab. 2014;19:574–591. doi: 10.1016/j.cmet.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Mendell JR, Goemans N, Lowes LP, Alfano LN, Berry K, Shao J, Kaye EM, Mercuri E Eteplirsen Study G, Telethon Foundation DMDIN. Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Ann Neurol. 2016;79:257–271. doi: 10.1002/ana.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corey DR. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat Neurosci. 2017 doi: 10.1038/nn.4508. [DOI] [PubMed] [Google Scholar]
- 39.Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, Yamashita M, Rigo F, Hung G, Schneider E, Norris DA, Xia S, Bennett CF, Bishop KM. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]