Abstract
A novel antisense transcript was identified in the human telomerase reverse transcriptase (hTERT) promoter region, suggesting that the hTERT promoter is bidirectional. This transcript, named hTERT antisense promoter-associated (hTAPAS) RNA, is a 1.6 kb long non-coding RNA. hTAPAS transcription is initiated 167 nucleotides upstream of the hTERT transcription start site and is present in both the nucleus and the cytoplasm. Surprisingly, we observed that a large fraction of the hTERT polyadenylated RNA is localized in the nucleus, suggesting this might be an additional means of regulating the cellular abundance of hTERT protein. Both hTAPAS and hTERT are expressed in immortalized B-cells and human embryonic stem cells but are not detected in normal somatic cells. hTAPAS expression inversely correlates with hTERT expression in different types of cancer samples. Moreover, hTAPAS expression is not promoted by an hTERT promoter mutation (-124 C>T). Antisense-oligonucleotide mediated knockdown of hTAPAS results in an increase in hTERT expression. Conversely, ectopic overexpression of hTAPAS down regulates hTERT expression, suggesting a negative role in hTERT gene regulation. These observations provide insights into hTAPAS as a novel player that negatively regulates hTERT expression and may be involved in telomere length homeostasis.
Keywords: lncRNA, TERT, cancer, bidirectional transcription
1. Introduction
Telomerase activity and telomere length have important implications in human disease and aging [1]; elevated telomerase activity has been detected in most human cancers [2,3,4]. Expression of human telomerase reverse transcriptase (hTERT), the enzymatic component of telomerase, is tightly regulated at the transcriptional level through epigenetic modifications in the promoter region [5], as well through alternative splicing [6,7,8]. Abundance of hTERT is a rate-limiting step in modulating telomerase activity [9]. While normally expressed in the germline and stem cells, up-regulated hTERT is essential for the continual proliferation and long-term viability of cells in many cancers [1,4]. Recurrent mutations in the hTERT promoter region, at -66 or -88 nucleotides (nts) relative to the hTERT transcriptional start site (TSS), are among the most common somatic mutations in many types of cancer, including melanomas, glioblastoma multiforme, hepatocellular carcinomas, and bladder cancers [7,10,11].
The hTERT promoter has many features characteristic of bidirectional promoters, such as the absence of a TATA box and high GC content [12], as well as binding sites for the ETS transcription factor GA binding protein (GABP) [13,14]. Recent data demonstrate that the mutant hTERT promoter can be bound and activated by GABP [15], suggesting it may induce bidirectional transcription of an antisense transcript.
We have previously reported retroviral activation of an antisense transcript upstream of TERT in chicken B-cell lymphomas, named TERT antisense promoter-associated (TAPAS) RNA [16]. Truncated TAPAS RNA is up regulated in chicken B-cell lymphomas. Here, we identify and characterize a human TAPAS (hTAPAS) RNA transcript in many types of human cancer and in several cell lines, using bioinformatics and experimental analyses. The hTAPAS transcript spans approximately 1.6 kb and has a single unspliced exon. Further, the absence of any conserved large open reading frames (ORFs) with protein domain homology suggests that this transcript is a previously unidentified long non-coding RNA (lncRNA).
We observe that hTAPAS expression is inversely correlated with hTERT expression in human cancers in The Cancer Genome Atlas (TCGA). Furthermore, we do not observe any activation of hTAPAS expression with the hTERT promoter mutation in a mini-gene construct expressed in human embryonic kidney (HEK-293) cells. Knocking down of hTAPAS via antisense-oligonucleotides increases hTERT expression, and ectopic overexpression of hTAPAS down regulates hTERT expression. This suggests that hTAPAS is involved in negatively regulating hTERT expression. Our work confirms the existence of an antisense lncRNA, hTAPAS, upstream of hTERT that exhibits negative regulation of hTERT expression. Moreover, we observe that nearly half of the hTERT transcript is localized in the nucleus, suggesting this might serve as an additional way of regulating the cellular abundance of hTERT protein.
2. Results
2.1. An Antisense Transcript Is Expressed Upstream of hTERT
Deep sequencing of transcriptomes from chickens revealed an antisense transcript upstream of TERT, which is an alternatively spliced, polyadenylated, lncRNA named TAPAS [16]; this suggests that the TERT promoter is bidirectional. To determine whether a similar transcript is expressed in humans, we first examined RNA sequencing data from the Encyclopedia of DNA Elements (ENCODE) Consortium [17]. An antisense RNA in the hTERT promoter region was readily observed in two different human B-cell tumor lines (GM12878 and OCI-LY7) (Figure 1 and Figure S1). Moderate expression levels of this transcript were also observed in a human embryonic stem cell line (H1-hESC), and to a lesser extent in a human hepatocellular carcinoma cell line (HepG2) (Figure S1). However, this transcript was not detected in HeLa (human cervical carcinoma) and K562 (human leukemic) cell lines (Figure S1). All of these cell lines expressed hTERT (Figure S1).
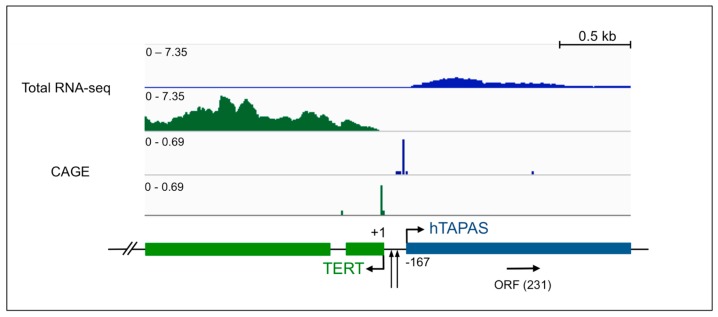
Figure 1.
An antisense RNA, named human TAPAS RNA, is expressed in the human telomerase reverse transcriptase (hTERT) promoter region. Normalized and stranded RNA sequencing (RNA-seq) (Bedgraph) transcription coverage for hTERT (green) and hTAPAS (blue) expression, as well as corresponding cap analysis gene expression (CAGE) start sites, are depicted for the human B-cell line GM12878. The schematic below denotes Reverse Transcription PCR (RT-PCR) validation of hTAPAS transcript from human embryonic kidney (HEK-293) cells by tiling arrays. The first two exons of hTERT and an approximately 1.6 kb long hTAPAS gene, located 167 nts upstream of the hTERT transcriptional start site are depicted. Arrowheads between hTERT and hTAPAS represent sites of point mutations in the hTERT promoter, located 124 or 146 nucleotides upstream of the hTERT translational start site.
To better determine the 5′ end of hTAPAS, we used 5′ cap analysis gene expression (CAGE) data from the ENCODE Consortium for the GM12878 cell line [17]. 5′ CAGE data specifically identifies the 5′ ends of capped transcripts, indicating TSSs. We observe a strong signal starting 167 nts upstream of the annotated hTERT TSS and in the opposite orientation (Figure 1). Taken together, the RNA sequencing (RNA-seq) and 5′ CAGE data from the ENCODE Consortium suggest that an antisense transcript, hereafter referred to as the hTERT antisense promoter-associated (hTAPAS) RNA, was detected in the hTERT promoter region in human embryonic stem cells and in some but not all cancer cell lines. Interestingly, the hTERT promoter mutations found in several tumor types, -124 C>T or -146 C>T [7,10,11], lie in between the TSS for hTERT and hTAPAS (Figure 1).
From TCGA we downloaded normal and tumor tissue RNA-seq read alignments of approximately 3800 samples from eight different cancer types, (Table S1), and reconstructed transcripts de novo to identify annotated and novel transcripts (Figures S2 and S3A). In addition to identifying hTERT transcript isoforms, we identified an approximately 1.6 kb long hTAPAS transcript at the locus predicted by the ENCODE data, in some tumors (Figure S3A). Low expression levels were observed for both hTAPAS and hTERT in corresponding normal tissues from cancer patients (Figure S3B).
Finally, to experimentally validate expression of this transcript, we performed a tiling array Reverse Transcription PCR (RT-PCR), using RNA from HEK-293 cells, and again identified an approximately 1.6 kb long hTAPAS RNA (Figure 1). We also detected the hTAPAS transcript in HeLa and hepatocellular carcinoma (SNU-449 and SNU-475) cells via RT-PCR analysis. Based on these results and the ENCODE Consortium RNA-seq and CAGE data, we predict that hTAPAS is about 1.6 kb long with a single unspliced exon and is transcribed starting at 167 nts upstream of the hTERT TSS.
2.2. hTAPAS Exhibits Features of a Long Non-Coding RNA
There is a small ORF in the middle of hTAPAS RNA (231 nts) (Figure 1). However, this ORF is not conserved and lacks any protein domain homology, implicating this transcript as a lncRNA. Unlike hTERT exons, the hTAPAS transcribed locus does not exhibit evolutionarily conserved elements as determined by phastCons scores (Figure S4A) [18]. Positions representing the hTERT exons had a mean phastCons score of 0.204, hTAPAS locus positions had a mean score of 0.048, and random positions had a mean score of 0.047. hTAPAS also shows evidence of faster evolution compared to neutral evolution rates as determined by phyloP scores (Figure S4A) [19]. phyloP scores were aggregated for hTAPAS, hTERT exons, and random positions. Positions representing hTERT exons had a mean phyloP score of 0.089 (positive scores indicate conservation), the hTAPAS locus had a mean score of −0.081, and random positions had a mean score of −0.1236 (negative scores indicate faster evolution compared to a neutral evolution rate). Thus, similar to the previously reported chicken TAPAS transcript, hTAPAS lacks ORFs with known protein domain homology and exhibits no protein-coding potential based on phyloCSF scores (Figure S4B) [20]. Based on these features, we propose the hTAPAS transcript to be a lncRNA.
2.3. Sub-Cellular Localization of hTAPAS and hTERT RNAs
To determine the sub-cellular localization of hTAPAS RNA, RNA-seq data was analyzed for polyadenylated (polyA+) and non-polyadenylated (polyA-) RNAs from cytoplasmic and nuclear fractions. We found evidence for both polyA+ and polyA- populations of hTAPAS in a human B-cell line (GM12878) (Figure 2A). The polyadenylated transcript was present in both nuclear and cytoplasmic fractions of the B-cell line. On the other hand, the polyA- transcript was absent from the cytoplasm. This suggests that a fraction of hTAPAS lncRNA is exported from the nucleus to the cytoplasm after being polyadenylated.
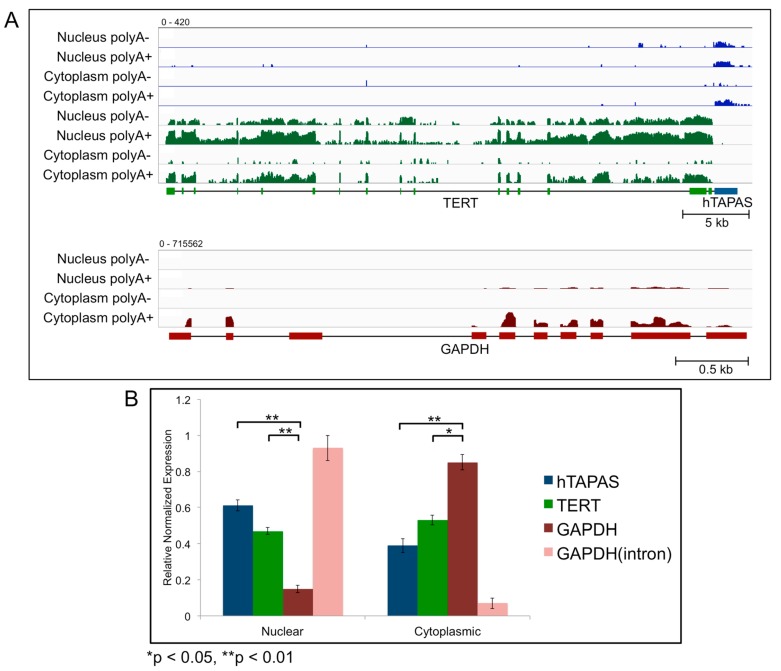
Figure 2.
Sub-cellular localization of hTERT and hTAPAS RNAs (A) Normalized and stranded RNA-seq (Bedgraph) transcription coverage for hTERT (green) and hTAPAS (blue) expression are depicted for the polyadenylated (polyA+) and non-polyadenylated (polyA-) transcripts in the nuclear and cytoplasmic fractions, from the human B-cell cell line (GM12878). All plus strand (blue) and minus strand (green) track signals are depicted on a log-scale (0–420). Transcription coverage for corresponding transcripts of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (red) are depicted as a control, on a log scale (0–715562). Represented data are from the ENCODE Consortium. (B) Abundance of hTAPAS, hTERT and GAPDH (mRNA and pre-mRNA) transcripts in the nucleus and cytoplasm cellular fractions determined by quantitative Reverse Transcription PCR (qRT-PCR) after sub-cellular fractionation of HEK-293 cells.
Surprisingly, a substantial fraction of the hTERT transcript was also observed in the nucleus of GM12878 cells (Figure 2A). A similar sub-cellular distribution of hTERT RNA was also seen in H1-hESC, HepG2, HeLa and K562 cell lines (data not shown). This was an unexpected observation since hTERT is a protein-coding gene and would be expected to be localized mostly in the cytoplasm. As expected, the control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was predominantly polyadenylated and cytoplasmic (Figure 2A).
We observed similar sub-cellular localization of hTERT and hTAPAS RNAs in both the nucleus and cytoplasm, after sub-cellular fractionation of HEK-293 cells and RNA quantitation by quantitative Reverse Transcription PCR (qRT-PCR) (Figure 2B). Again, GAPDH mRNA and pre-mRNA transcripts were mainly observed in the cytoplasmic and nuclear fractionations, respectively (Figure 2B). Further, we tested the possibility that hTAPAS RNA might be sequestering hTERT RNA in the nucleus. However, knocking down of hTAPAS via antisense oligonucleotides did not alter the cellular localization of hTERT in HEK-293 cells (data not shown).
2.4. hTAPAS Expression Negatively Correlates with hTERT Expression in Cancer Patients
Antisense lncRNAs transcribed from bidirectional promoters have been known to be involved in regulation of the associated sense transcripts [21,22]. We thus investigated the relationship between the expression of hTAPAS and hTERT in TCGA database [23]. First, we analyzed hTAPAS and hTERT expression in primary tumor samples from eight different types of cancer by RNAseq (Table S1, Figure 3).
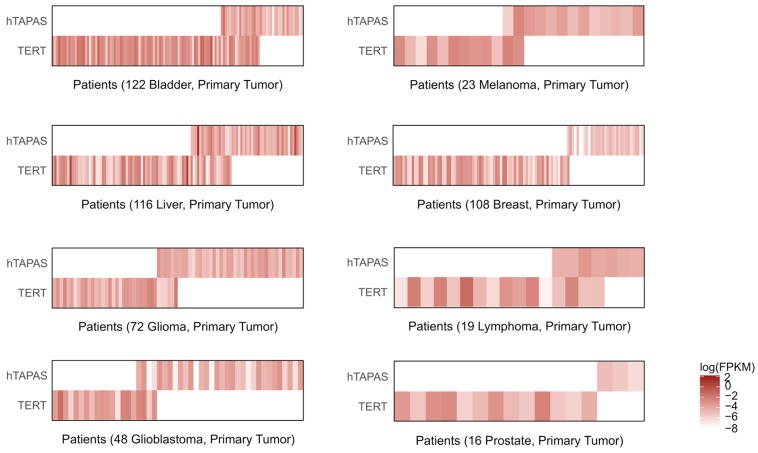
Figure 3.
hTERT and hTAPAS expression are inversely correlated in primary tumors. RNA-seq data samples from TCGA, which have detectable expression levels for either hTERT or hTAPAS were analyzed in the eight different cancers (melanomas, gliomas, glioblastomas, hepatocellular carcinomas, bladder, lymphoma, prostate and breast cancers). A majority of these tumor samples express either hTERT or hTAPAS, but not both. The heat-maps depict corresponding expression levels of hTAPAS and hTERT among individual patient samples from the eight different cancers. Expression is represented as log-transformed Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values (white indicates undetectable expression).
hTERT RNA detection ranged from 3 to 50% in the different types of primary tumors (Figure 3 and Figure S5A). Lymphomas had the highest percentage of patients expressing detectable levels of hTERT, followed by bladder and liver cancers, while prostate cancers had very few patients expressing detectable levels of hTERT. Tumor samples expressing detectable hTAPAS RNA ranged from 1% (prostate) to 20% (glioblastoma) (Figure S5B). In addition a few of the liver tumors expressed high levels of hTAPAS (Figure S3B).
In the primary tumor samples in which at least one of these transcripts was detected, fewer than 20% expressed both hTAPAS and hTERT RNAs (Figure 3). Co-expression of hTERT and hTAPAS RNAs was nearly undetectable in breast and prostate cancers (Figure 3 and Figure S5C). The highest levels of co-expression (4–8%) of hTERT and hTAPAS RNAs were observed in primary tumors of lymphomas, bladder and liver cancers (Figure S5C). This pattern is suggestive of an antagonistic relationship between hTAPAS and hTERT expression.
2.5. Knockdown of hTAPAS up Regulates hTERT Expression in HEK-293 Cells
To test whether loss of hTAPAS RNA can alter hTERT expression, targeted knockdown of hTAPAS was performed in a HEK-293 cell line, using oligonucleotides antisense to the hTAPAS transcript (570 to 594 nts from the hTAPAS TSS). Cells transfected with a scrambled oligonucleotide showed no significant change in either hTAPAS or hTERT expression; however, transfection with an hTAPAS antisense oligonucleotide (ASO) resulted in a 4-fold knockdown of hTAPAS and a significant 1.8-fold increase in hTERT expression (using primers in hTERT exons 13 and 14) (Figure 4). We also measured expression of the hTERT transcript with primers in the catalytic reverse transcriptase domain (exons 7 and 8). Similarly, we detected a 2.1-fold increase in expression of these hTERT transcripts. These results suggest that hTAPAS lncRNA functions to negatively regulate hTERT expression.
Figure 4.
hTAPAS knockdown in HEK-293 cells results in increased hTERT expression. hTERT and hTAPAS expression was determined by qRT-PCR in HEK-293 cells transfected with mock, scrambled oligos, and antisense oligonucleotides (ASO) targeted to hTAPAS. Use of ASO resulted in a 4-fold knock down of hTAPAS expression, relative to scrambled. hTERT expression was analyzed using primers spanning its exons 13–14 or exons 7–8 (in the Reverse Transcriptase domain).
2.6. hTAPAS Expression Is Not Affected by the hTERT Promoter Mutation
The mutant hTERT promoter can be bound and activated by GABP [15], suggesting it may induce bidirectional transcription of an antisense transcript [13,14]. Therefore, we asked whether this hTERT promoter mutation affects hTAPAS expression. We first analyzed RNA-seq data from eight hepatocellular carcinoma cell lines, four with and four without the hTERT promoter mutation -124 C>T (Figure S6) [6]. Consistent with previous reports [15], hTERT exhibited an average of approximately 2-fold higher expression in the cells with the promoter mutation, relative to the cells with the wild-type promoter (Figure S6). In contrast, no obvious difference in expression of hTAPAS was apparent between cell lines with the absence or presence of the point mutation (Figure S6). Furthermore, in cells with a wild-type hTERT promoter, hTERT and hTAPAS had similar expression levels; however, in cells with the presence of the point mutation, hTERT expression was approximately 2 to 4 fold higher than hTAPAS expression (Figure S6).
Next, we generated constructs to ectopically express the hTERT promoter region, with a luciferase reporter in lieu of hTERT (Figure 5A). We introduced the hTERT promoter point mutation (-124 C>T) into this construct to determine its influence on luciferase activity (as an indicator of hTERT expression) and hTAPAS expression in HEK-293 cells. We observed that luciferase activity was up regulated approximately 2-fold by the presence of the point mutation (Figure 5B), consistent with previously reported effects of the point mutation on hTERT expression. However, hTAPAS expression was not altered by the presence of the point mutation (Figure 5C). Moreover, in the ENCODE cell line HepG2, which has an hTERT promoter mutation, we observed hTERT expression but poor hTAPAS expression levels (Figure S1). Taken together, our data suggest that the hTERT promoter mutation significantly increases expression of hTERT but not of hTAPAS.
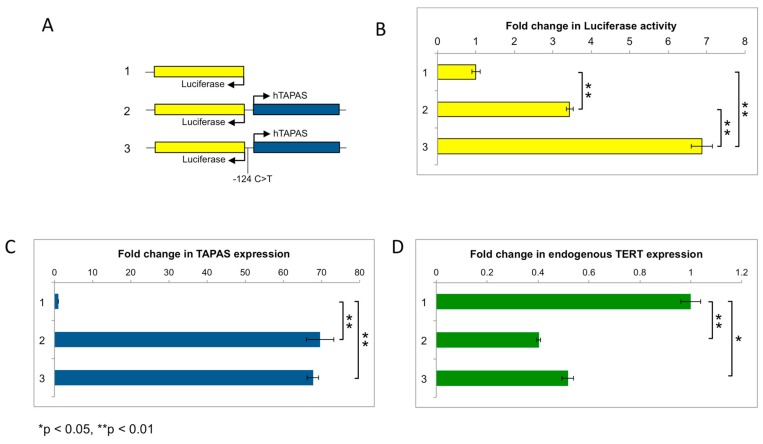
Figure 5.
Presence of the hTERT promoter mutation does not alter hTAPAS expression. (A) Schematic representation of an empty control vector (1), and hTAPAS insert with a wild-type (2) or mutant (3) hTERT promoter region, in a luciferase reporter vector. (B) Fold-change in luciferase activity observed for the different constructs relative to empty vector. (C) Fold-change in hTAPAS or (D) endogenous hTERT expression levels observed for the different constructs relative to empty vector.
As a consequence of transfecting these luciferase constructs into HEK-293 cells, hTAPAS was ectopically over-expressed approximately 70-fold relative to endogenous levels (Figure 5C). hTAPAS over-expression resulted in down regulation of endogenous hTERT expression approximately 2-fold in HEK-293 cells. (Figure 5D). This expression pattern is consistent with our observations of hTAPAS knockdown resulting in increased hTERT expression levels (Figure 4). Therefore, these observations support our proposal that hTAPAS is involved in negatively regulating hTERT expression.
3. Discussion
Telomerase activity in somatic cells is limited by the availability of hTERT protein and thus expression of hTERT is tightly regulated [24]. At the transcriptional level, hTERT is regulated by many transcription factors, such as c-MYC, ETS, and SP1/SP3, as well as through hTERT promoter mutations which can cause epigenetic modifications in the promoter region [7,25]. Extensive alternative splicing of the hTERT transcript has also been shown to generate inactive and dominant negative hTERT variants that decrease telomerase activity [6,7,8]. In this work, we identify a novel antisense lncRNA in the hTERT promoter region, hTAPAS, which functions as an additional negative regulator of hTERT expression. We also observed that a substantial portion of the hTERT transcript was localized in the nucleus, suggesting this might be an additional means of regulating the cellular abundance of catalytic hTERT by limiting its translation.
Many lncRNAs play a role in transcriptional regulation [26]. For example, XIST acts in cis to recruit the polycomb repressive complex 2 (PRC2) to chromosome X, causing gene silencing [27], while HOTAIR acts in trans to repress the expression of genes in the HoxD gene cluster [28]. It has been proposed that antisense lncRNAs transcribed from bidirectional promoters may be involved in regulation of their associated sense transcripts [21,28,29]. Such an arrangement may allow for tighter transcriptional regulation. We observed that hTAPAS RNA over-expression down-regulated hTERT expression in trans. We also observed that knocking down hTAPAS RNA up regulated hTERT RNA expression. This suggests a role of the hTAPAS RNA in the transcriptional regulation of hTERT. hTAPAS might recruit epigenetic machinery to regulate hTERT expression, similar to other well-studied lncRNAs [30]. In primary tumors in TCGA, we typically saw expression of either hTERT or hTAPAS, but not both. This suggests the possibility of promoter occlusion in the hTERT bidirectional promoter region. Furthermore, a substantial fraction of hTAPAS transcripts are exported to the cytoplasm. We cannot rule out its possible translation, as has been previously observed for other lncRNAs [31]. This suggests, that in addition to negative regulation of hTERT transcription, hTAPAS might be involved in other cellular functions in the nucleus and/or cytoplasm.
Shortening of telomeres limits the replicative potential of most primary human cells and serves a tumor-suppressive function. Conversely, telomerase expression in the germline, in somatic cells during early embryogenesis, and in cancer cells promotes telomere length homeostasis [2,32,33,34,35,36]. The preferential elongation of short telomeres and maintenance of telomere length homeostasis requires a low cellular concentration of telomerase [37]. This is achieved via expression of hTERT over a limited range, approximately 500 hTERT protein molecules per cell [37,38]. Telomere length regulation and telomerase expression in multicellular organisms is thought to have evolved by opposing selective pressures to suppress tumor formation on one side, while not promoting premature cellular senescence in highly proliferative tissues on the other side. Similar to hTERT expression, hTAPAS expression is observed in the germline, cancer cell lines and tumor samples, and is absent in normal tissues. Since hTAPAS is not expressed in normal tissues of cancer patients, it does not appear to be involved in negatively regulating hTERT expression in somatic cells. Therefore, it is possible that as a regulator of hTERT expression, hTAPAS might facilitate the maintenance of hTERT expression within the narrow range required for telomere length homeostasis in cancer cells and stem cells.
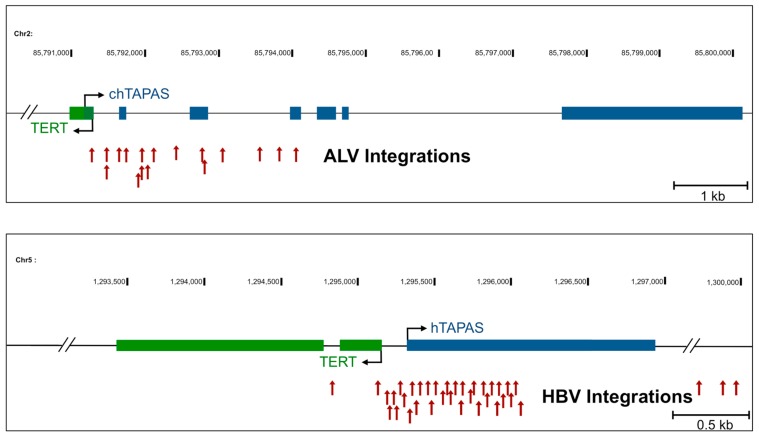
We have previously reported the presence of numerous clonally expanded integrations of the avian leukosis virus (ALV) in the TERT promoter region in chicken B-cell lymphomas, associated with slightly elevated TERT expression [39,40]. We have also shown that proviral integrations in these tumors are in the antisense orientation relative to TERT and are driving the over-expression of chicken TAPAS lncRNA [16]. In this work we find evidence for an upstream antisense transcript in the hTERT promoter region. Similar to elevated expression levels of chicken TAPAS in B-cell lymphomas, we observe that hTAPAS also is expressed in immortalized human B-cell lines. Interestingly, the hTERT promoter region is also reported to be the most common integration site for hepatitis B virus integrations (HBV) in hepatocellular carcinomas [41,42,43,44]. Approximately 20% of these integrations are in the hTERT promoter region [41]. These integrations near hTERT are thought to confer an early clonal advantage during chronic HBV infection. Moreover, HBV integrations lead to up-regulated hTERT expression and are associated with poorer survival rates in infected patients with these integrations [42]. Of note, the frequency of hTERT promoter point mutations is significantly lower in hepatocellular carcinomas bearing HBV integrations in the hTERT promoter region [41].
Interestingly, nearly all of the reported ALV and HBV integrations in the hTERT promoter region are present within the 5′ end of chicken TAPAS and hTAPAS, respectively (Figure 6). The majority of the ALV integrations in the chicken TERT promoter region are in the same transcriptional orientation as TAPAS [16]. In contrast, HBV integrations are in a mixed transcriptional orientation with respect to hTAPAS [41,42,43,44]. The prevalence of viral integrations within TAPAS in multiple types of cancers in different organisms suggests that these integrations are selected for during oncogenesis. These viral integrations might confer a proliferative advantage and make cells predisposed to oncogenic transformation. Since hTAPAS negatively regulates hTERT expression and HBV integrations would likely disrupt hTAPAS expression, this could promote hTERT expression. These observations, therefore, suggest functional implications of hTAPAS in tumorigenesis.
Figure 6.
TAPAS is a viral integration hotspot in chicken and human tumors. The schematic represents the hTERT promoter region, including the transcription start sites (black arrows) for hTERT and TAPAS, in the chicken (top) and human (bottom) genomes. This region is depicted as a viral integration hotspot for the avian leukosis virus (ALV) and hepatitis B virus (HBV), respectively in tumors. Red arrowheads represent the integration sites for ALV or HBV in this region.
Like human telomerase, chicken telomerase is down regulated in most somatic tissues [45]. Furthermore, chicken telomeres shorten with age, and telomerase activity is important for oncogenesis [46]. In contrast, the mouse TERT enzyme is active in normal somatic cells [47]. This discrepancy between humans and mice is important because telomerase activation is a critical step in the human oncogenic process, with aberrant telomerase activation seen in most human cancers [3,4]. Further, no lncRNA expression has been observed in the TERT promoter region of mouse embryos (data not shown). This absence of a TAPAS transcript might facilitate increased TERT expression in mice. Therefore, chicken serves as an advantageous model to study oncogenic events of TERT signaling and transcriptional regulation.
LncRNAs, as well as bidirectional transcription, have arisen as novel players in tumorigenesis, warranting a need for further research of their role in regulating cancer development. The mutant hTERT promoter can be bound and activated by GABP, an ETS transcription factor [15], which has been shown to induce bidirectional transcription in other loci [13,14]. However, we observe no association of hTAPAS expression with the presence of a point mutation, in transfected HEK-293 cells or liver cancer cell lines. We observed that the most common point mutation in the hTERT promoter region (-124 C>T) does not affect hTAPAS expression. However, we did not test whether the -146 C>T point mutation affected hTAPAS expression. Additionally, we observe hTAPAS expression in the absence of detectable hTERT expression in some of the TCGA samples, but not in cell lines. This variation in the hTAPAS expression profile between primary tumor samples and cell lines could be due to the differences in the genomic features and microenvironments between cell lines and tumor samples [48,49].
Reactivated hTERT expression is reported in 90% of all human cancers [3,4]. However, through our analysis from the TCGA we detected hTERT expression in only 3–50% of analyzed samples. While previous studies have used qRT-PCR to observe hTERT transcript levels, our analysis involves the use of total RNA-seq data from primary tumors. The detection and quantification sensitivity of RNA-seq is dependent on the read depth. Even at an above average coverage of 100 million reads, RNA-seq suffers from greatly reduced quantification and detection sensitivity compared to qRT-PCR [50]. Therefore, the differences in our observations compared to previous work can be explained by the method used for detection of hTERT expression.
Activation of hTERT expression is a crucial step in the progression of many cancers, and understanding the molecular mechanisms of such activation is important for understanding oncogenesis. Recent genome-wide analyses have highlighted that somatic point mutations in the hTERT promoter are among the most common mutations in human cancer and are known to drive hTERT expression [51]. The hTERT promoter region is also the site of viral integrations in both chicken (ALV) and human (HBV) tumors (Figure 6). We observe that in many tumors there is activation of a previously uncharacterized antisense transcript in the hTERT promoter region, which we call hTAPAS. This lncRNA negatively regulates hTERT expression and, thus, is implicated in oncogenesis and telomere homeostasis.
4. Materials and Methods
4.1. Analysis of RNA Sequencing and CAGE Data
Total and cellular fractionated RNA-seq data for analysis of expression of hTAPAS and hTERT in human cells were downloaded from the ENCODE Consortium and TCGA [17,23]. The ENCODE accession number for the total RNA-seq data analyzed for GM12878 is ENCSR889TRN, for OCI-LY7 is ENCSR001HHK, for H1-hESC is ENCSR537BCG, for HepG2 is ENCSR181ZGR, for HeLa-S3 is ENCSR552EGO, and for K562 is ENCSR000AEL. The RNA-seq data for sub-cellular fractions of GM12878 are available through the ENCODE RNA Dashboard (hg19) [17]. Data available through the ENCODE Data Coordination Center, and TCGA and represent normalized, stranded RNA-seq signal. RNA-Seq libraries for liver cancer cell lines were prepared in duplicate using the TruSeq stranded mRNA library kit according to manufacturers directions and sequenced on the Illumina HiSeq platform (San Diego, CA, USA). Two biological replicates were analyzed for each data set. The abundances of transcripts (fragments per kilobase per million, FPKM) were estimated and compared using Cufflinks [52]. CAGE data for analysis of hTAPAS and hTERT TSSs in GM12878 cells were downloaded from the ENCODE Consortium (ENCODE accession number ENCSR000CKA).
4.2. Evolutionary Conservation and Coding Potential Analysis
phastCons, phyloCSF and phyloP scores, based on multi-way alignment of 20 mammalian genomes, were aggregated over the hTAPAS locus (chr5:1295329-1296947, 1619 positions), hTERT exons (NM_198253 exons annotated in hg38, 4,013 positions), and an intergenic sample of 10,000 positions from the hTAPAS/hTERT locus (chr5:1247464-1308783).
4.3. Fractionation Experiments
Sub-cellular fractionation of cultured HEK-293 cells was performed as follows. Cells were incubated in five volumes of isotonic lysis buffer and incubated on ice for 10 min. 1% NP40 was added with gentle inversion, followed by centrifugation at 5500 rcf for 30 s. The supernatant (cytoplasmic fraction) was removed into a separate tube. The pellet was re-suspended in five volumes of isotonic lysis buffer, followed by centrifugation at 1500 rcf for five min. The supernatant was added to the cytoplasmic fraction and the pellet was used as the nuclear fraction.
4.4. RNA Isolation and Quantitative Reverse Transcription PCR
RNA from total cell and sub-cellular fractions was extracted using Trizol (Thermo Fisher, Scientific, Carlsbad, CA, USA). DNase I (Ambion) treated RNA was reverse transcribed with Maxima H minus reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) and oligo(dT)18 primer and/or random hexamers. hTAPAS was validated by a primer tiling array. All the PCR primers were commercially synthesized (Integrated DNA Technologies, Inc., San Diego, CA, USA). qRT-PCR was performed using iQ SYBR green Supermix (Thermo Fisher Scientific, Austin, TX, USA) on a Bio-Rad C1000 thermal cycler/CFX96 Real-Time System. The expression of hTAPAS RNA was measured using primers TGTAGCTGAGGTCGGCAAAC and GGTGCGAGGCCTGTTCAAAT. hTERT expression was measured using primers GTGCTGCAGCTCCCATTTCAT and GCTTTCAGGATGGAGTAGCAGA. Expression was normalized to the expression of GAPDH using exon junction primers AATCCCATCACCATCTTCCA and TGGACTCCACGACGTACTCA. qRT-PCR was performed in triplicate, with each sample present in technical duplicate during each run. The results were normalized using the comparative threshold cycle (CT) method.
4.5. hTAPAS Knockdown Experiments Using Antisense Oligonucleotides
ASO with phosphorothioate bonds were generated by Integrated DNA Technologies, Inc., San Diego, CA, USA. The ASO targeting hTAPAS binds 570 nts downstream from the transcription start site (GTGATTAACAGATTTGGGGTGGTTG). A scrambled ASO control was generated with matched base composition (GGTACGATTATTTCGGTTCGATTAGT). HEK-293 cells were transfected with 100 nM ASO using FuGene 6 (Promega, Madison, WI, USA) according to manufacturers protocol. Cells were then cultured for 48 h before being collected for measurement of RNA expression. hTAPAS primers flanked the ASO binding site (TGAGCAACCACCCCAAATCT, TTTCCCACCCTTTCTCGACG), hTERT primers spanned either the exon 13–14 junction (GTGCTGCAGCTCCCATTTCAT, GCTTTCAGGATGGAGTAGCAGA) or the exon 7–8 junction (GCGTAGGAAGACGTCGAAGA, ACAGTTCGTGGCTCACCTG). All expression was normalized to a housekeeping gene, GAPDH using primers AATCCCATCACCATCTTCCA and TGGACTCCACGACGTACTCA.
4.6. hTAPAS Constructs and Luciferase Assays
The hTERT promoter region, including hTAPAS, was cloned into the firefly luciferase construct pGL3-basic (Promega, Madison, WI, USA) using XhoI and Acc65I cloning sites. The hTERT promoter mutation (-124 C>T) was introduced by a quick-change PCR as described previously [53]. The pRL-CMV construct (Promega, Madison, WI, USA) was co-transfected into HEK-293 cells as a transfection control. 48 h after transfection, HEK-293 cells were harvested and assayed for firefly and Renilla luciferase activities by using the Dual-Luciferase Reporter Assay System (Promega).
4.7. Nucleotide Sequence Accession Number
The sequence of the hTAPAS cDNA, as validated by RT-PCR analysis, was submitted to GenBank under the accession number MG677549.
Acknowledgments
This work was supported by research grants from National Institutes of Health (NIH) (T32GM007231) and (R21AI28353). We would like to thank Nick Papadopoulos for his generosity in providing the RNA-seq data for the liver cancer cell lines SNU-182, Hep3B2, SNU-449, PLC/PRF/5, SNU-423, HepG2, SNU-475 and SNU-398. The results published here are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/) and accessed using the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). We thank the ENCODE Consortium and the ENCODE production laboratories who generated the RNA-seq (CSHL, Gingeras) and 5′ CAGE (Riken) datasets.
Supplementary Materials
The following are available online at www.mdpi.com/2311-553X/4/1/1/s1, Table S1: Number of RNA-seq samples from TCGA for each cancer type investigated for hTERT and hTAPAS expression; Figure S1: Normalized and stranded RNA-seq (Bedgraph) transcription coverage for hTERT and hTAPAS RNAs for different ENCODE cell lines; Figure S2: Pipeline of TCGA RNA-seq data analysis; Figure S3: StringTie gene models, and expression of hTAPAS and hTERT in tumor and normal tissues of eight cancer types; Figure S4: hTAPAS does not exhibit conservation or protein-coding potential; Figure S5: Proportion of primary tumor samples with detectable expression of hTERT, hTAPAS or both; Figure S6: hTAPAS and hTERT expression in eight different human liver cancer cell lines, with the absence (−) or presence (+) of hTERT promoter mutation (-124 C>T).
Author Contributions
S.M., M.A.F. and K.L.B. conceived and designed the experiments; S.M. and S.W. performed the experiments; S.M., M.A.F. and S.W. analyzed the data; K.L.B and J.T. contributed reagents/materials/analysis tools; S.M. and K.L.B. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Ethical Statement
The human cancer lines SNU-449 and SNU-475 were obtained from Nick Papadopoulos, Johns Hopkins School of Medicine. The human cancer cell lines HeLa and HEK-293 were obtained from American Type Culture Collection (ATCC), Manassas, VA, USA.
References
- 1.Blasco M.A. Telomeres and human disease: Ageing, cancer and beyond. Nat. Rev. Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 2.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 3.Garcia C.K., Wright W.E., Shay J.W. Human diseases of telomerase dysfunction: Insights into tissue aging. Nucleic Acids Res. 2007;35:7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shay J.W., Wright W.E. Role of telomeres and telomerase in cancer. Semin. Cancer Biol. 2011;21:349–353. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J., Zhao Y., Wang S. Chromatin and epigenetic regulation of the telomerase reverse transcriptase gene. Protein Cell. 2010;1:22–32. doi: 10.1007/s13238-010-0014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cevik D., Yildiz G., Ozturk M. Common telomerase reverse transcriptase promoter mutations in hepatocellular carcinomas from different geographical locations. World J. Gastroenterol. 2015;21:311–317. doi: 10.3748/wjg.v21.i1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Withers J.B., Ashvetiya T., Beemon K.L. Exclusion of Exon 2 Is a Common mRNA Splice Variant of Primate Telomerase Reverse Transcriptases. PLoS ONE. 2012;7:e48016. doi: 10.1371/journal.pone.0048016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong Y.-S., Wright W.E., Shay J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn S., Figl A., Rachakonda P.S., Fischer C., Sucker A., Gast A., Kadel S., Moll I., Nagore E., Hemminki K., et al. TERT Promoter Mutations in Familial and Sporadic Melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 11.Killela P.J., Reitman Z.J., Jiao Y., Bettegowda C., Agrawal N., Diaz L.A., Jr., Friedman A.H., Friedman H., Gallia G.L., Giovanella B.C., et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renaud S., Bosman F.T., Benhattar J. Implication of the exon region in the regulation of the human telomerase reverse transcriptase gene promoter. Biochem. Biophys. Res. Commun. 2003;300:47–54. doi: 10.1016/S0006-291X(02)02775-4. [DOI] [PubMed] [Google Scholar]
- 13.Collins P.J., Kobayashi Y., Nguyen L., Trinklein N.D., Myers R.M. The ets-related transcription factor GABP directs bidirectional transcription. PLoS Genet. 2007;3:2247–2255. doi: 10.1371/journal.pgen.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orekhova A.S., Rubtsov P.M. Bidirectional promoters in the transcription of mammalian genomes. Biochemistry. 2013;78:335–341. doi: 10.1134/S0006297913040020. [DOI] [PubMed] [Google Scholar]
- 15.Bell R.J.A., Rube H.T., Kreig A., Mancini A., Fouse S.F., Nagarajan R.P., Choi S., Hong C., He D., Pekmezci M., et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348:1036–1039. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nehyba J., Malhotra S., Winans S., O’Hare T.H., Justice J., Beemon K. Avian Leukosis Virus Activation of an Antisense RNA Upstream of TERT in B-Cell Lymphomas. J. Virol. 2016;90:9509–9517. doi: 10.1128/JVI.01127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin M.F., Jungreis I., Kellis M. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakano C., Byun J.S., Di L.J., Gardner K. The dual lives of bidirectional promoters. Biochim. Biophys. Acta Gene Regul. Mech. 2012;1819:688–693. doi: 10.1016/j.bbagrm.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei W., Pelechano V., Järvelin A.I., Steinmetz L.M. Functional consequences of bidirectional promoters. Trends Genet. 2011;27:267–276. doi: 10.1016/j.tig.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang K., Creighton C.J., Davis C., Donehower L., Drummond J., Wheeler D., Ally A., Balasundaram M., Birol I., Butterfield Y.S.N., et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masutomi K., Yu E.Y., Khurts S., Ben-Porath I., Currier J.L., Metz G.B., Brooks M.W., Kaneko S., Murakami S., DeCaprio J.A., et al. Telomerase maintains telomere structure in normal human cells. Cell. 2003;114:241–253. doi: 10.1016/S0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 25.Heidenreich B., Kumar R. TERT promoter mutations in telomere biology. Mutat. Res. Mutat. Res. 2017;771:15–31. doi: 10.1016/j.mrrev.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Tsai M.-C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao J., Sun B.K., Erwin J.A., Song J.-J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.-C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotake Y., Nakagawa T., Kitagawa K., Suzuki S., Liu N., Kitagawa M., Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 31.Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J.S., Jackson S.E., Wills M.R., Weissman J.S. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B., Harley C.B., Shay J.W., Lichtsteiner S., Wright W.E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 33.Harley C.B., Futcher A.B., Greider C.W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 34.Ulaner G.A., Giudice L.C. Developmental regulation of telomerase activity in human fetal tissues during gestation. Mol. Hum. Reprod. 1997;3:769–773. doi: 10.1093/molehr/3.9.769. [DOI] [PubMed] [Google Scholar]
- 35.Vaziri H., Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol. 1998;8:279–282. doi: 10.1016/S0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 36.Wright W.E., Piatyszek M.A., Rainey W.E., Byrd W., Shay J.W. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 37.Cristofari G., Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xi L., Cech T.R. Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res. 2014;42:8565–8577. doi: 10.1093/nar/gku560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang F., Xian R.R., Li Y., Polony T.S., Beemon K.L. Telomerase reverse transcriptase expression elevated by avian leukosis virus integration in B cell lymphomas. Proc. Natl. Acad. Sci. USA. 2007;104:18952–18957. doi: 10.1073/pnas.0709173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Justice J.F., Morgan R.W., Beemon K.L. Common viral integration sites identified in avian leukosis virus- induced B-cell lymphomas. MBio. 2015;6:e01863-15. doi: 10.1128/mBio.01863-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buendia M.-A., Neuveut C. Hepatocellular Carcinoma. Cold Spring Harb. Perspect. Med. 2015;5:a021444. doi: 10.1101/cshperspect.a021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao L.-H., Liu X., Yan H.-X., Li W.-Y., Zeng X., Yang Y., Zhao J., Liu S.-P., Zhuang X.-H., Lin C., et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat. Commun. 2016;7:12992. doi: 10.1038/ncomms12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toh S.T., Jin Y., Liu L., Wang J., Babrzadeh F., Gharizadeh B., Ronaghi M., Toh H.C., Chow P.K.-H., Chung A.Y.-F., et al. Deep sequencing of the hepatitis B virus in hepatocellular carcinoma patients reveals enriched integration events, structural alterations and sequence variations. Carcinogenesis. 2013;34:787–798. doi: 10.1093/carcin/bgs406. [DOI] [PubMed] [Google Scholar]
- 44.Ding D., Lou X., Hua D., Yu W., Li L., Wang J., Gao F., Zhao N., Ren G., Li L., et al. Recurrent Targeted Genes of Hepatitis B Virus in the Liver Cancer Genomes Identified by a Next-Generation Sequencing-Based Approach. PLoS Genet. 2012;8:e1003065. doi: 10.1371/journal.pgen.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delany M.E., Daniels L.M. The chicken telomerase reverse transcriptase (chTERT): Molecular and cytogenetic characterization with a comparative analysis. Gene. 2004;339:61–69. doi: 10.1016/j.gene.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 46.Delany M.E., Krupkin A.B., Miller M.M. Organization of telomere sequences in birds: Evidence for arrays of extreme length and for in vivo shortening. Cytogenet. Cell Genet. 2000;90:139–145. doi: 10.1159/000015649. [DOI] [PubMed] [Google Scholar]
- 47.Hackett J.A., Greider C.W. Balancing instability: Dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene. 2002;21:619–626. doi: 10.1038/sj.onc.1205061. [DOI] [PubMed] [Google Scholar]
- 48.Goodspeed A., Heiser L.M., Gray J.W., Costello J.C. Tumor-Derived Cell Lines as Molecular Models of Cancer Pharmacogenomics. Mol. Cancer Res. 2016;14:3–13. doi: 10.1158/1541-7786.MCR-15-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Domcke S., Sinha R., Levine D.A., Sander C., Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Everaert C., Luypaert M., Maag J.L.V., Cheng Q.X., Dinger M.E., Hellemans J., Mestdagh P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 2017;7:1559. doi: 10.1038/s41598-017-01617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiba K., Johnson J.Z., Vogan J.M., Wagner T., Boyle J.M., Hockemeyer D. Cancer-associated TERT promoter mutations abrogate telomerase silencing. Elife. 2015;4:1–20. doi: 10.7554/eLife.07918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cossio M.L.T., Giesen L.F., Araya G., Pérez-Cotapos M.L.S., Vergara R.L., Manca M., Tohme R.A., Holmberg S.D., Bressmann T., Lirio D.R., et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 2012;7:562. doi: 10.1007/s13398-014-0173-7.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H., Naismith J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.