Abstract
Os-GRF1 (Oryza sativa-GROWTH-REGULATING FACTOR1) was identified in a search for genes that are differentially expressed in the intercalary meristem of deepwater rice (Oryza sativa L.) internodes in response to gibberellin (GA). Os-GRF1 displays general features of transcription factors, contains a functional nuclear localization signal, and has three regions with similarities to sequences in the database. One of these regions is similar to a protein interaction domain of SWI2/SNF2, which is a subunit of a chromatin-remodeling complex in yeast. The two other domains are novel and found only in plant proteins of unknown function. To study its role in plant growth, Os-GRF1 was expressed in Arabidopsis. Stem elongation of transformed plants was severely inhibited, and normal growth could not be recovered by the application of GA. Our results indicate that Os-GRF1 belongs to a novel class of plant proteins and may play a regulatory role in GA-induced stem elongation.
Plants adjust their final height to the prevailing environmental conditions by increasing or decreasing their growth rate in response to external and internal signals. To increase yield, the height of crop plants is often manipulated either genetically or by application of plant growth regulators. GA plays a major role in regulating stem or internodal elongation, as evident from physiological studies and from the phenotype of mutants impaired in GA biosynthesis or perception (Hooley, 1994; Swain and Olszewski, 1996; Ross et al., 1997; Harberd et al., 1998). Despite its complexity, the GA biosynthetic pathway has been well characterized (Hedden and Proebsting, 1999). In contrast, much remains to be learned about the GA signal transduction pathway that leads to stem elongation and other GA-regulated processes. Genetic analysis of GA-response mutants of Arabidopsis and cloning of the respective genes led to the identification of GA signal transduction components that appear to be negative regulators of GA action. Two putative transcription factors with high sequence similarity to each other, GAI (Peng et al., 1997) and RGA (Silverstone et al., 1998), were shown to mediate responses to GA, and their functions appear to be partially overlapping (Harberd et al., 1998; Silverstone et al., 1998). Overexpression of SHI, which also encodes a putative transcription factor, led to a semi-dominant dwarf phenotype (Fridborg et al., 1999). The product of the SPY gene shows significant similarity to O-linked GlcNAc transferases of animals and may regulate the activity of proteins of the GA signaling pathway by post-translational modifications (Jacobsen et al., 1996; Thornton et al., 1999).
Identification of genes whose expression is controlled by GA complements characterization of GA response mutants as an approach to elucidate the mode of action of GA. Genes whose transcript levels change following treatment with GA may encode proteins that act downstream of the GA signal transduction pathway and that may play a crucial role in regulating growth. One group of such genes encodes enzymes of GA biosynthesis and metabolism. As part of a negative feedback mechanism, the expression of these genes changes in response to GA (Hedden and Proebsting, 1999; Thomas et al., 1999). Whereas several GA-regulated genes of the cereal aleurone layer have been studied in detail (Bethke et al., 1997), relatively few GA-controlled genes have been identified in elongating stems. They include genes in tomato (Shi et al., 1992; Jacobsen and Olszewski, 1996), pea (Wu et al., 1993), and Arabidopsis (Phillips and Huttly, 1994). The function of these genes in GA-regulated stem elongation, however, is unknown.
Deepwater rice (Oryza sativa L.) is grown in Southeast Asia and has the capacity to elongate very rapidly when it is inundated by flood waters during the monsoon season (Kende et al., 1998). This growth response is based on internodal elongation and results at least in part from an ethylene-mediated increase in the ratio of an endogenous growth promoter (GA) and growth inhibitor (abscisic acid [ABA]) (Hoffmann-Benning and Kende, 1992). The ultimate growth-promoting hormone in internodes of deepwater rice is GA (Raskin and Kende, 1984), and the primary site of GA action is the intercalary meristem at the base of the growing internode (Sauter et al., 1993). Applied GA increases the rate of cell production in the intercalary meristem and promotes the growth of cells to three times their normal length in the internodal elongation zone (Raskin and Kende, 1984; Sauter and Kende, 1992; for the anatomy of the rice stem, see Fig. 3E). Unlike the shoot apical meristem of dicotyledoneous plants, from which growth of the stem and the formation of leaves and floral organs originate, the intercalary meristem of grasses contributes to stem elongation only. Therefore, GA-induced stem growth can be investigated in rice without interference by other developmental processes that occur in the shoot apical meristem.
Figure 3.
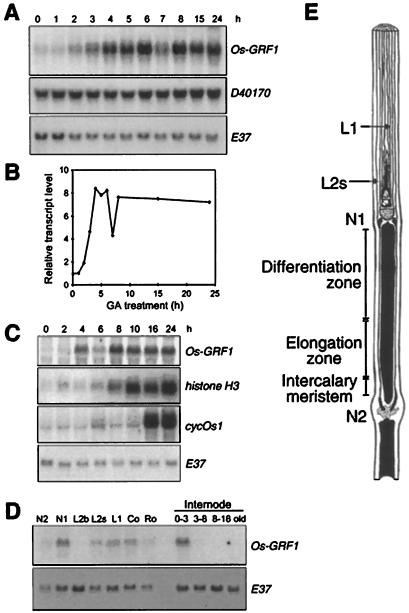
Expression of the Os-GRF1 gene in rice. A, Rice stem sections were incubated in 50 μm GA3 for the times indicated above the lanes. Expression of Os-GRF1 and D40170, a rice EST that contains both the QLQ and WRC motifs, was determined in the intercalary meristem by RNA gel-blot analysis (20 μg of total RNA per lane). E37 served as loading control. B, Quantitative analysis of Os-GRF1 expression shown in A using a PhosphorImager. The values were normalized to the E37 loading control. C, Expression of Os-GRF1 in submerged plants. Whole plants were submerged for the times indicated above the lanes. Expression of the Os-GRF1, histone H3, and cycOs1 genes in the intercalary meristem was determined by RNA gel-blot analysis (20 μg of total RNA per lane). E37 served as loading control. D, Tissue-specific expression of Os-GRF1. N2, Second highest node; N1, highest node containing the shoot apex; L2b, basal 2 cm of second youngest leaf blade; L2s, basal 2 cm of second youngest leaf sheath; L1, youngest leaf; Co, coleoptile 3 d after germination; Ro, root 3 d after germination; 0–3, internodal region 0 to 3 mm above N2 containing the intercalary meristem; 3–8, internodal region 3 to 8 mm above N2 containing the elongation zone; 8–18, internodal region 8 to18 mm above N2 containing the upper part of the elongation zone and the differentiation zone; old, oldest part of the internode. E37 served as loading control. E, Schematic representation of a rice stem section containing the uppermost internode.
Several GA-regulated genes have been identified in deepwater rice internodes. They can be grouped into three categories: (a) genes whose products function in the cell cycle; (b) genes encoding proteins that act as cell wall-loosening factors; and (c) genes whose role in stem growth is unknown (Kende et al., 1998; Van der Knaap, 1998). We report here the identification of a novel gene, Os-GRF1 (Oryza sativa-GROWTH-REGULATING FACTOR1), whose expression increases rapidly in the intercalary meristem of deepwater rice internodes in response to GA and submergence. The protein encoded by Os-GRF1 shares features common to transcription factors and transcriptional activators. Significant amino acid sequence similarity has been found to hypothetical plant proteins in the Arabidopsis genome database and to proteins encoded by plant cDNAs. To investigate its role in GA-mediated growth, Os-GRF1 was expressed in Arabidopsis. Stem elongation of the transformed plants was greatly inhibited and was not reversed by the application of GA. Our results indicate that Os-GRF1 encodes a novel plant-specific protein that may be involved in transcriptional regulation and in mediating GA-induced stem elongation.
MATERIALS AND METHODS
Plant Material
Seeds of deepwater rice (Oryza sativa L. cv Pin Gaew 56) were obtained from the International Rice Research Institute (Los Baños, Philippines). Submergence experiments were conducted with 12-week-old plants according to the method of Métraux and Kende (1983). For treatment with GA, 20-cm-long stem sections containing the growing internode were excised and placed into a 50 μm GA3 solution (Raskin and Kende, 1984). Incubation was allowed to proceed for the periods indicated, and then the intercalary meristem was excised, frozen immediately, and stored at −80°C until use. Seeds of Arabidopsis ecotype Columbia were provided by Dr. Pamela Green and Linda Danhoff (Michigan State University).
Identification of Os-GRF1 and
A differentially displayed 222-bp cDNA fragment was identified using the primers T12MG and OPA04 (Van der Knaap and Kende, 1995). A full-length cDNA, Os-GRF1, was obtained by screening an intercalary-meristem-specific cDNA library with the differentially displayed product. Sequence analysis was performed at the W.M. Keck Facility, Yale University, New Haven, CT. The sequences were aligned using Sequencher, version 3.0 (Gene Codes Corporation, Ann Arbor, MI). The rice expressed sequence tag (EST) D40170, whose deduced amino acid sequence shows similarity to Os-GRF1, was obtained from the National Institute of Agrobiological Resources, Tsukuba, Japan. Sequence and RNA gel-blot analysis of D40170 indicated that it was a partial cDNA.
Nuclear Localization of Os-GRF1
To facilitate subcloning of Os-GRF1, restriction enzyme sites were introduced by PCR with Pwo polymerase (Boehringer) using the following primers: 5′-TCGGTCTAGAGGCGGTCGGTCGACGCTGAA-3′ and 5′-TCATTGTGGATCCGGGAGGTGGTGGTGATC-3′. The PCR product corresponded to the entire coding region of Os-GRF1, except for the bases encoding the last five amino acids. This PCR product was inserted into pMF6, a monocot-specific transient assay transformation vector, in frame with the coding region of the reporter protein β-glucuronidase (GUS), yielding the pOs-GRF1::GUS construct (Varagona et al., 1992). Onion epidermal cells were transformed and nuclear localization of GUS was assayed according to the method of Varagona et al. (1992).
RNA Gel-Blot Analyses
Total RNA was isolated according to the method of Puissant and Houdebine (1990). Twenty micrograms of RNA per sample was analyzed by standard RNA gel blotting (Ausubel et al., 1987). The following DNA fragments were used as probes: an insert containing the terminal 737 bp from the 3′ end of Os-GRF1 (starting at the codon for Gln-318 [see Fig. 1A] and excluding the poly-A tail), and the inserts of D40170, of E37, and of a partial histone H3 cDNA clone (Van der Knaap and Kende, 1995). Fifty nanograms of template DNA was labeled in the presence of α-[32P]dCTP (3,000 Ci/mmol, NEN Life Science Products, Boston) using a random primer labeling kit (Boehringer Mannheim/Roche, Basel). A RNA probe of cycOs1, which contained the 3′ untranslated region and most of the region encoding a mitotic cyclin (Sauter et al., 1995), was prepared in the presence of α-[32P]UTP (800 Ci/mmol, NEN). A RNA probe was also prepared from the 737-bp cDNA fragment of Os-GRF1 to detect the transcripts in Figure 3C. Hybridization and washing conditions for the RNA gel blots were as described by Van der Knaap et al. (1999). The radioactivity on blots was quantified by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, CA).
Figure 1.
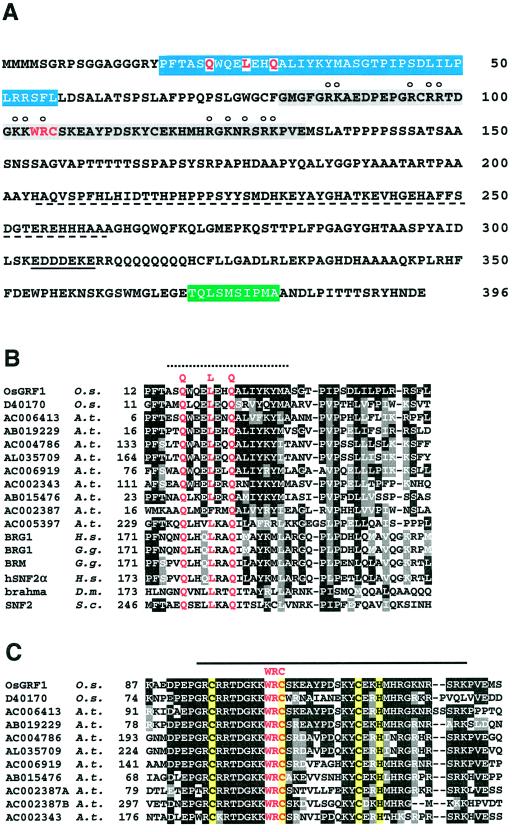
Sequence analysis of Os-GRF1. A, Amino acid sequence of Os-GRF1. The QLQ domain is boxed in blue, the WRC domain in gray, and the TQL domain in green. The His-rich region is underlined with dashes, and the acidic region with a solid line. In the WRC domain, amino acids that may be involved in nuclear localization are indicated by circles above the respective letters. B, Alignment of the QLQ domain with that of other organisms (multiple alignment by Clustal W; Thompson et al., 1994; http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html). O.s., Oryza sativa (accession no. for Os-GRF1 is AF201895); A.t., Arabidopsis; H.s., Homo sapiens (accession nos. for BRG1 and hSNF2α are P51532 and S45251, respectively); G.g., Gallus gallus (accession nos. for BRG1 and BRM are X91637 and X91638, respectively); D.m., Drosophila melanogaster (accession no. for brahma is P25439); S.c., Saccharomyces cerevisiae (accession no. for SNF2 is P22082). The dotted line denotes a region of a predicted α-helix. C, Alignment of the WRC domain by Clustal W. The letters on yellow background comprise the potential metal-binding motif CX9CX10CX2H. The solid line denotes the region rich in basic amino acids.
Genomic DNA Analyses
Rice genomic DNA was isolated from a CsCl gradient according to the method of Ausubel et al. (1987). Arabidopsis genomic DNA was isolated in hot (65°C) hexadecyltrimethylammonium bromide (CTAB) buffer (2% [w/v] CTAB, 1.4 m NaCl, 20 mm EDTA, 100 mm Tris-HCl, pH 8.0, and 0.2% [v/v] β-mercaptoethanol), and incubated at 65°C for 30 min. After chloroform extraction, the DNA was precipitated with 0.6 volume of isopropanol. The pellet was dissolved in Tris-EDTA, and RNA was removed by RNase A treatment. After organic extraction, the DNA was precipitated with one-third volume of 7.5 m NH4-acetate and 2.5 volumes of ethanol, and subsequently dissolved in the appropriate amount of Tris-EDTA. Five micrograms of rice DNA and 500 ng of Arabidopsis DNA were analyzed by standard DNA gel blotting (Ausubel et al., 1987). Hybridization and washing conditions were as described for the hybridization of random-primer-labeled probes to RNA gel blots (Van der Knaap et al., 1999). The 737-bp cDNA fragment of Os-GRF1 was used as a probe for the rice DNA gel blot, and the full-length Os-GRF1 cDNA for the Arabidopsis DNA gel blot.
Transformation of Arabidopsis
Os-GRF1 was inserted in sense orientation into the XbaI-ClaI site of the binary vector pGA643 (An et al., 1988). For cloning purposes, the construct thus created, pGA::GRF1, lacked the 428 bp from the 3′ end of Os-GRF1, leaving only 74 bp of its 3′ untranslated region. Agrobacterium tumefaciens strain GV3101 (C58C1 RifR) pMP90 (GmR) was transformed with pGA::GRF1 or pGA643. A. tumefaciens containing pGA::GRF1 or pGA643 were vacuum infiltrated into Arabidopsis ecotype Columbia or ecotype Landsberg erecta, as described by Van Hoof and Green (1996). The seeds collected were surface-sterilized and plated on Murashige and Skoog (MS) medium (Life Technologies/Gibco-BRL, Cleveland) containing 1% (w/v) Suc, 10 mm 2-(N-morpholino)-ethanesulfonic acid (MES), 0.8% (w/v) phytagar (Life Technologies/Gibco-BRL), 500 μg/mL vancomycin, and 50 μg/mL kanamycin (Sigma). After 2 weeks, transgenic plants were transferred to soil and grown in a growth chamber at 20°C in 16-h light (100 μmol m−2 s−1)/8-h dark cycles. The severe female sterile lines were maintained by backcrossing to wild-type Arabidopsis using the transgenic plants as male parents.
Experiments with Transgenic Plants
Seeds were imbibed and kept for 4 d at 4°C in the dark. They were allowed to germinate on MS plates containing 50 μg/mL kanamycin and grown, unless otherwise indicated, under the conditions described above. The germination times of line 13 and of two control transformed lines were measured with 50 to 110 seeds each per MS plate (n = 6) in a growth chamber as described above. To determine growth of the hypocotyl, seeds were germinated on MS plates placed in a slanted position under continuous light (100 μmol m−2 s−1) at 25°C. After 3 d, the length of the hypocotyls was measured in increments of 0.25 mm using a dissecting microscope. Petiole and pedicel lengths were determined 33 d after germination. Petioles of the three longest rosette leaves and the first three pedicels per plant were measured.
RESULTS
Identification of Os-GRF1
Using differential display of mRNA, we identified a 222-bp cDNA fragment corresponding to a transcript whose level increased in the intercalary meristem of rice within 2.5 h of treatment with GA (Van der Knaap and Kende, 1995). A rice cDNA library prepared from the intercalary meristem was screened with this cDNA, and a full-length 1,891-bp clone, Os-GRF1, was isolated. The predicted 43-kD protein encoded by Os-GRF1 contains a Pro/Ser/Thr-rich region, a His-rich region, a region rich in acidic residues, a homopolymeric sequence of Gln, and a putative bipartite nuclear localization signal (NLS) (Hicks and Raikhel, 1995) (Fig. 1A). These features are reminiscent of transcription factors (Mitchell and Tjian, 1989; Gerber et al., 1994). Database searches indicated weak similarities to a range of transcription factors, transcriptional activators, and homeo-domain proteins. Using the BLASTP program and the NR database (Altschul et al., 1997), we found the highest similarity to several hypothetical proteins in the Arabidopsis genome database. The highest similarity to known proteins was to the chicken BRM and BRG1 and the human BRG1 and SNF2α proteins (Fig. 1B). These proteins are related to yeast SWI2/SNF2, a subunit of a large chromatin-remodeling complex (Peterson and Tamkun, 1995).
Three regions of Os-GRF1 showing similarities to sequences in the database have been identified (Fig. 1A). The first region close to the N terminus contains the motif QX3LX2Q and will be referred to as the QLQ domain. A region following the QLQ domain contains the sequence WRC and will be called the WRC domain. The third region close to the carboxyl end will be referred to as the TQL domain. The amino acid alignments of the QLQ and WRC domains with the corresponding regions of other proteins are shown in Figure 1, B and C. The QLQ domain has similarities to a domain involved in protein-to-protein interactions in SWI2/SNF2 and related proteins (Treich et al., 1995) (Fig. 1B). The WRC domain shows higher similarity to other proteins than does the QLQ domain (Fig. 1, B and C). It contains a putative NLS and three Cys and one His residues in the sequence CX9CX10CX2H, which may function in metal binding (Fig. 1, A and C). Both the QLQ and WRC motifs are present in the same plant proteins shown in the alignments of Figure 1, B and C. Regions of lower similarity containing the WRC motif were identified in searches of the Arabidopsis genomic database. However, these hypothetical proteins did not have the QLQ domain (not shown). The TQL domain (Fig. 1A) was also found in two entries of the Arabidopsis database (genomic sequences with accession nos. AC004786, which was also isolated in our cDNA library screen, and AL035709, which is identical to an EST with accession no. N95873; see Fig. 1, B and C). The 10 amino acids of the TQL domain distinguish Os-GRF1 and the proteins encoded by the two above-cited cDNAs from the other proteins containing the QLQ and WRC domains.
Nuclear Localization of Os-GRF1
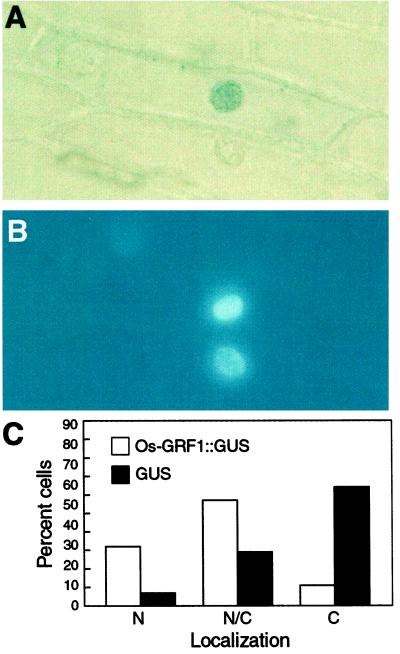
To address the cellular localization of Os-GRF1, we prepared a construct containing Os-GRF1 fused in-frame to the reporter gene encoding GUS. This construct was introduced into onion epidermal cells by biolistic bombardment, and the fusion protein was localized by histochemical staining. Blue staining was found to be associated with nuclei (Fig. 2A), a result confirmed by coincident staining with the DNA-specific dye 4′,6-diamino-phenylindole (DAPI) (Fig. 2B). Of the 79 onion cells counted, 32% showed exclusive nuclear localization of the fusion protein, whereas only 7% of the control nuclei contained GUS (Fig. 2C). These results indicate that Os-GRF1 contains a functional NLS.
Figure 2.
Nuclear localization of Os-GRF1. A, Localization of the Os-GRF1::GUS fusion protein in the nucleus of an onion epidermal cell. The staining reaction was performed with the GUS substrate 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-gluc). B, Staining of the nuclei shown in A with the DNA-specific dye DAPI. C, Localization of Os-GRF1::GUS and of GUS (control) in the nucleus (N), nucleus and cytoplasm (N/C), and cytoplasm (C). Seventy-nine and 240 cells were scored to determine the distribution of Os-GRF1::GUS and GUS, respectively.
Expression of Os-GRF1 and Gene Copy Number in Rice
The transcript level of Os-GRF1 was determined in growing internodes by RNA gel-blot analysis. The signals were quantified by PhosphorImager analysis and normalized for equal loading relative to the transcript level of E37, a gene whose expression did not change significantly over the course of the experiment (Van der Knaap and Kende, 1995) (Fig. 3, A and B). The level of Os-GRF1 mRNA in the intercalary meristem of stem sections increased more than 4-fold within 3 h of GA treatment and 8-fold within 4 h (Fig. 3, A and B). The transcript level of D40170, a rice EST that contains both the QLQ and WRC motifs, did not change in response to GA (Fig. 3A).
The expression pattern of Os-GRF1 was also investigated in the intercalary meristem of submerged plants (Fig. 3C). Os-GRF1 mRNA levels increased within 4 h of submergence and reached a maximum after 8 h. The increase in transcript levels of Os-GRF1 was compared with that of the histone H3 transcript and of the cyclin transcript cycOs1. Histone H3 is a marker for the S-phase of the cell cycle (Van der Knaap and Kende, 1995) and cycOs1 for the entry into the M-phase (Sauter et al., 1995; Sauter, 1997). The signals for the histone H3 and cycOs1 transcripts were quantified and normalized for equal loading with E37. A greater than 3-fold increase in histone H3 transcript level was found 8 h after the start of submergence, and an increase in cycOs1 mRNA was observed after 16 h. Because the signal for Os-GRF1 could not be detected by PhosphorImager analysis at the 0-h time point, the Os-GRF1 signals were not quantified. It is clear, however, that the increase in the expression of the Os-GRF1 gene preceded expression of the histone H3 and of cycOs1 genes. This indicates that changes in Os-GRF1 transcript levels occur during submergence before changes in the cell cycle activity are apparent. A transient decrease in Os-GRF1 transcript levels was observed reproducibly in both GA-treated stem sections and in submerged plants. The significance of this temporary decline is not known.
In the internode, the expression of Os-GRF1 was only detected in the intercalary meristem (Fig. 3D), which is the primary site of GA action (Sauter et al., 1993). No transcript was found in the elongation zone, in the differentiation zone, or in the oldest part of the internode. High transcript levels were also detected in the shoot apex. Lower expression of Os-GRF1 was found in the coleoptile and in the youngest leaf. Weak expression was seen in the second-highest node below the growing internode, in the basal part of the second-youngest leaf sheath, and in the root. The anatomy of the rice internode is shown in Figure 3E.
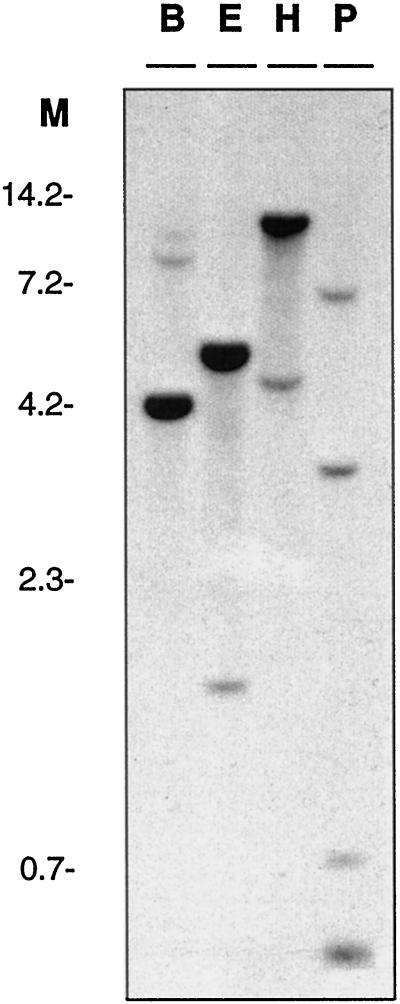
DNA gel-blot analysis was employed to investigate whether genes related to Os-GRF1 exist in rice. The hybridization probe was derived from the 3′ region of Os-GRF1 and did not include the regions encoding the QLQ and WRC domains. Three digests produced one predominant and at least one additional fainter band (Fig. 4). The probe used contained a PstI restriction site, which explains the presence of several bands in the lane containing PstI-digested DNA. The results indicate that the rice genome may contain at least one additional gene that hybridizes to the Os-GRF1 probe at high stringency. The same genomic DNA blot was hybridized to D40170, the rice EST that also contains the QLQ and WRC motifs. The banding pattern of the DNA gel blot was different from that obtained with Os-GRF1 as a probe (results not shown). None of the fainter bands on the blot shown in Figure 4 hybridized to D40170.
Figure 4.
DNA gel-blot analysis of Os-GRF1. Rice genomic DNA was digested with BamHI (B), EcoRI (E), HindIII (H), or PstI (P). The blot was probed with a random-prime-labeled insert derived from the 3′ 737-bp region of Os-GRF1. Lane M, Molecular size markers (kb). The washing conditions were 0.1× SSC, 0.1% (w/v) SDS at 65°C.
Expression of Os-GRF1 in Arabidopsis: DNA and RNA Gel-Blot Analyses
To further investigate the function of Os-GRF1 in growth, Os-GRF1 was expressed in Arabidopsis under control of the cauliflower mosaic virus 35S promoter. The transgenic plants obtained were derived from independent transformation events, as determined by DNA gel-blot analysis (Fig. 5A). In Arabidopsis ecotype Columbia, a total of 17 independent kanamycin-resistant T1 plants were collected. The phenotype of the transgenic plants is described in the section below. The appearance of more than one band on the DNA gel blot signified the presence of more than one T-DNA insert in the genome. DNA gel-blot analysis of plants with the most severe phenotype (lines 11, 12, 13, and 23) showed a single band, indicating the presence of one copy of the transgene. The severe phenotype of these lines was transmitted to the T2 generation in a Mendelian fashion. A single band was also observed in the T1 plants 16 and 22.2, which also showed a severe phenotype. However, no offspring were obtained from line 22.2, and the progeny of line 16 showed variations in phenotype ranging from wild type to severe. The remaining transformed lines contained at least two inserts and showed less severe (lines 18 and 22.1) or wild-type (lines 5, 7, 8, and 15) phenotypes.
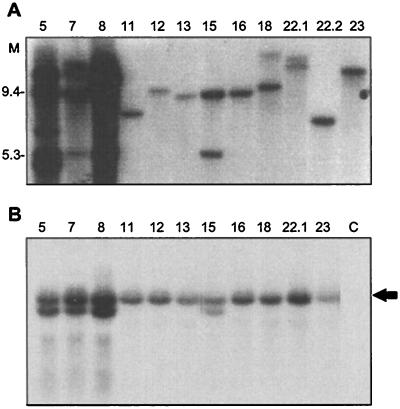
Figure 5.
Copy number and expression of Os-GRF1 in Arabidopsis. A, DNA gel-blot analysis of BamHI-digested DNA from the individual T1 lines denoted above the lanes. The blot was hybridized with a random-prime-labeled probe derived from the full-length Os-GRF1 cDNA. B, Os-GRF1 transcript levels in individual T2 lines. The RNA gel blot was hybridized with a random-prime-labeled insert derived from the 3′ 737-bp region of Os-GRF1. The arrow indicates the position of the full-length Os-GRF1 mRNA. C, Control plants transformed with the vector alone.
The expression of the transgene was determined by hybridizing Os-GRF1 to RNA isolated from 2-week-old T2 seedlings transformed with the Os-GRF1 construct or with the vector alone (Fig. 5B). Equal RNA loading was confirmed by ethidium bromide-staining of the ribosomal RNA (results not shown). The abundance of the full-length Os-GRF1 transcript did not correlate with the severity of the phenotype in either the T1 or the T2 generations. An additional band corresponding to mRNA of slightly shorter length was only found in plants with an apparent wild-type phenotype (lines 5, 7, 8, and 15). Similar results were obtained with RNA isolated from mature leaves and in experiments using other regions of Os-GRF1 as a probe (Van der Knaap, 1998). Thus, the copy number of the transgene was inversely correlated with the stability and severity of the phenotype, whereas the transcript level was not correlated to the severity of the phenotype. The phenotype of plants from line 13 is described below. The same phenotype was observed in T1 and T2 plants of lines 11, 12, and 23 and in T1 plants of lines 16 and 22.2.
The Phenotype of Arabidopsis Plants Expressing Os-GRF1
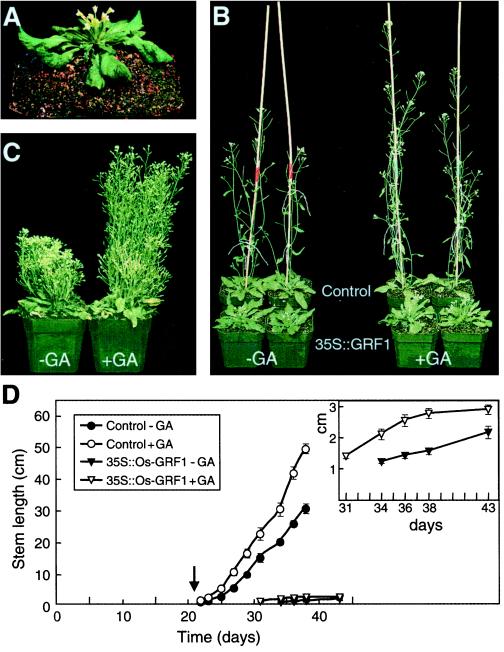
The most striking phenotype of plants expressing Os-GRF1 was the development of flowers inside the rosettes without concomitant stem elongation (Fig. 6, A and B). Control plants transformed with the vector alone had elongated inflorescence stems 7.5 weeks after germination, while stems of plants transformed with Os-GRF1 had barely started to elongate by that time (Fig. 6B). Whereas wild-type Arabidopsis plants stopped growing approximately 8 weeks after germination, the lateral shoots of plants expressing Os-GRF1 continued to grow for another 3 months to a final height that was one-half that of wild-type plants (Fig. 6C). The growth habit was atypical, however, showing severely reduced apical dominance (Fig. 6C) and, occasionally, fasciated and bifurcated stems (Van der Knaap, 1998). Plants expressing Os-GRF1 also had curly rosette leaves (Fig. 6A) and altered gynoecia. The carpels were only partially fused, the septum dividing the ovule was missing, and the stigma surface was sometimes replaced by a leaf-like outgrowth. The plants exhibited female sterility and reduced male fertility (Van der Knaap, 1998).
Figure 6.
Growth of Arabidopsis plants expressing Os-GRF1 (Line 13). A, Five-week-old plants that flowered without concomitant bolting and showed curly leaves. B, GA3 (1 μg) was applied in a 25-μL drop of a 0.01% (v/v) Tween 20 solution to the center of rosettes of 3-week-old plants transformed with Os-GRF1 or with the vector alone (control) before bolting of the control plants had started. This treatment was repeated three times. For the minus GA treatment, GA3 was omitted from the Tween 20 solution. The photograph was taken when the plants were 7.5 weeks old. C, Plants transformed with Os-GRF1 were sprayed with 0.02% (v/v) Tween 20 solution with or without 50 μm GA3 beginning 21 d after germination and then twice a week for 22 d. The plants were grown for an additional month without further hormone treatment. The primary stem had died off when the photograph was taken, but secondary shoots had continued to grow. D, Growth curve of the primary stem of plants (n = 7) treated with GA3 as described in C. The arrow denotes the start of hormone treatment, which was repeated twice a week for 17 and 22 d for the control plants and for plants expressing Os-GRF1, respectively. The growth of control plants was measured until d 38 after germination; that of the plants transformed with Os-GRF1 until d 43. The inset is a magnification of the growth curve for the primary stem of Os-GRF1-expressing plants treated with and without GA.
The timing of germination, flowering, and stem growth was compared between control plants and plants expressing Os-GRF1 (Table I). In Os-GRF1 transformants, germination was delayed by 19 h, average flowering time by 3 d, and the onset of stem growth by 13 d. Even though the long delay in stem growth was the most conspicuous result of Os-GRF1 expression, developmental processes in general appeared to lag. Os-GRF1 was also expressed in Arabidopsis ecotype Landsberg erecta. The phenotype was similar to that observed in the Columbia background, but was in some transformants so severe that stem elongation and flower development never occurred (results not shown).
Table I.
Germination, flowering, and bolting time of Arabidopsis transformed with 35S∷Os-GRF1 or with the vector alone (control)
| Developmental Process | Control | 35S∷Os-GRF1 |
|---|---|---|
| Germinationa | 64.1 (0.8)d | 82.7 (1.5) |
| Floweringb | 21.7 (1.2) | 24.3 (3.6) |
| Boltingc | 22.6 (1.2) | 35.9 (1.9) |
Germination time was measured in hours from the time the seeds were placed in the growth chamber until 90% of the seeds had green, open cotyledons.
Flowering time was measured in days from the time of germination until the first flower had opened.
Bolting time was measured in days from the time of germination until the inflorescence stem had elongated by 1 cm.
The nos. in parentheses denote the se.
The Effect of GA on Growth of Arabidopsis Plants Expressing Os-GRF1
Because Os-GRF1 was identified in a screen for GA-induced genes in rice internodes, we investigated the effect of applied GA on several growth processes in Arabidopsis plants that expressed Os-GRF1. GA promoted elongation of the inflorescence stem in plants expressing Os-GRF1 slightly, while enhancing stem growth in control plants considerably (Figs. 6, B and D). Thus, applied GA could not restore growth of the primary stem of plants expressing Os-GRF1 to that of control plants. However, in older Os-GRF1-expressing plants, which show greatly reduced apical dominance, GA promoted elongation of lateral shoots markedly (Fig. 6C).
We investigated the effect of Os-GRF1 expression and GA on growth processes other than stem elongation (Table II). In plants expressing Os-GRF1, the hypocotyl was slightly longer than in control plants but growth of the petiole and pedicel was not significantly affected. The promotion of hypocotyl elongation by GA was comparable in control plants and in plants expressing Os-GRF1, and GA-induced pedicel growth might have been even greater in plants expressing Os-GRF1 than in the control plants (Table II). Applied GA increased the length of the petiole to the same extent in plants expressing OsGRF1 and in control plants (Table II). Treatment with GA did not reduce fasciation of the stem and did not ameliorate defects in apical dominance and floral development (Van der Knaap, 1998).
Table II.
Elongation of the hypocotyl, petiole, and pedicel in Arabidopsis transformed with 35S∷Os-GRF1 or with the vector alone (control)
| Organ | Control
|
35S∷Os-GRF1
|
||
|---|---|---|---|---|
| −GA | +GA | −GA | +GA | |
| Hypocotyla | 1.13 (0.03)c | 1.66 (0.04) | 1.50 (0.04) | 1.90 (0.03) |
| Petioleb | 1.20 (0.05) | 1.47 (0.04) | 1.04 (0.03) | 1.47 (0.05) |
| Pedicelb | 0.88 (0.02) | 1.15 (0.03) | 0.82 (0.02) | 1.98 (0.04) |
Hypocotyl length (in mm) of seedlings grown on MS medium with or without 5 μm GA3.
The petiole and pedicel length (in cm) of plants sprayed twice a week with 50 μm GA3 in 0.01% (w/v) Tween-20 (+GA) or with 0.01% (w/v) Tween-20 (−GA).
The nos. in parentheses denote the se.
DISCUSSION
Os-GRF1 Is the Prototype Member of a Novel Class of Plant-Specific Genes
We identified a novel plant-specific gene, Os-GRF1, whose transcript level increases in internodes of deepwater rice in response to GA and submergence. The protein encoded by Os-GRF1 has three regions with similarities to sequences in the database. Of these, the N-terminal QLQ domain is the only one with similarity to known proteins, the highest similarity being to proteins related to SWI2/SNF2 of yeast. SWI2/SNF2 is a subunit of a 2-MD, ATP-dependent chromatin-remodeling complex (Peterson and Tamkun, 1995). The similarity between the N-terminal region of yeast SWI2/SNF2 and the corresponding Drosophila, human, and chicken proteins is restricted to two short sequences called domain 1 and 2 in the Drosophila ortholog brahma (Tamkun et al., 1992). The QLQ domain of Os-GRF1 corresponds to domain 1, which in yeast is responsible for interaction with SNF11, another component of the SWI/SNF complex (Treich et al., 1995). The similarity of the QLQ domain of Os-GRF1 to domain 1 of SWI2/SNF2 indicates that it too may be involved in protein-to-protein interactions.
The WRC domain has features that indicate a function in transcriptional control and DNA binding. The Os-GRF1::GUS fusion protein is targeted to the nucleus of onion epidermal cells, most likely by a NLS that is located in the WRC domain. The WRC domain also contains Cys and His residues in the sequence CX9CX10CX2H, a motif that may be involved in metal binding. A large class of metal-binding proteins chelates zinc (Berg and Shi, 1996; Borden and Freemont, 1996). These zinc-binding domains or zinc fingers are often present in transcription factors, where they may be involved in DNA binding. The amino acid sequence of the WRC domain and the spacing of the Cys and His residues do not fall into any of the known classes of zinc fingers (Berg and Shi, 1996; Borden and Freemont, 1996). However, HRT, a barley transcriptional repressor protein, has recently been shown to contain three novel zinc-binding motifs of the CX9CX10CX2H type (Raventós et al., 1998).
Although the amino acids between the three Cys residues and the His residue in the CX9CX10CX2H motif of Os-GRF1 and HRT are not conserved, the spacing of the Cys and His residues is identical in both. In several classes of transcription factors, an NLS was found to overlap with or to be adjacent to a DNA-binding domain (Varagona and Raikhel, 1994; Hicks and Raikhel, 1995). Since the WRC domain may include both a NLS and a zinc finger, it is likely to function in DNA binding. The region that follows the WRC domain possesses sequences rich in Pro/Ser/Thr, His, and acidic amino acids, as well as a homopolymeric stretch of Gln. These too are features found in transcription factors and transcriptional activators (Mitchell and Tjian, 1989; Gerber et al., 1994). The third conserved motif of Os-GRF1, TQL, is the distinguishing characteristic of a subset of plant proteins with the QLQ and WRC domains. The TQL domain has not been recognized before in plant proteins, and its function is not yet known.
Os-GRF1 may therefore belong to a family of transcriptional regulatory proteins that are characterized by the QLQ and WRC motifs. The QLQ motif appears to be present in all eukaryotes. In contrast, the WRC motif appears to be unique to plants. The QLQ and WRC domains occur often in the same plant protein and may constitute a functional unit, the QLQ domain, which mediates homo- and/or heterodimerization between family members of this class of proteins and the WRC domain acting in DNA binding. Alternatively, the QLQ domain may be essential in the interaction with other proteins that are part of the transcriptional machinery.
Role of Os-GRF1 in Plant Growth and Development
RNA gel-blot analysis showed that Os-GRF1 is specifically expressed in regions of the rice plant that contain meristematic tissues. In the rice internode, Os-GRF1 transcripts were only detected in the intercalary meristem, which is the primary site of GA action (Sauter et al., 1993). The basal level of Os-GRF1 mRNA was very low in the intercalary meristem but increased sharply after 2 h of GA treatment and 4 h of submergence. GA induces internodal elongation with a lag time of 40 min, and the initial growth response is due to increased cell elongation. Enhanced cell division activity commenced approximately 4 h after the start of treatment with GA (Sauter and Kende, 1992). The increase in the transcript level of Os-GRF1 was apparent after cell elongation had started but before the increase in transcript levels of the cell-cycle-related genes encoding histone H3 and the mitotic cyclin cycOs1 was evident. The time course of GA-induced cell elongation and cell division activity in the intercalary meristem suggests that Os-GRF1 may be involved in mediating an early event in GA-promoted entry into the cell cycle.
To further investigate the role of Os-GRF1 in growth, we transformed Arabidopsis with Os-GRF1. Rather than promoting growth, expression of Os-GRF1 in Arabidopsis led to severely reduced elongation of the primary inflorescence stem. This phenotype could have resulted from a gain-of-function with respect to one component of growth and, thereby, to an imbalance between growth-related processes. For example, expression of Os-GRF1 in Arabidopsis may lead to excessive cell proliferation in the shoot apical meristem and, as a consequence, to the observed growth inhibition and fasciation (Van der Knaap, 1998). Alternatively, transformation of Arabidopsis with the rice gene may yield a dominant-negative phenotype. Os-GRF1 may displace the Arabidopsis GRF1 from a protein complex, thereby inactivating the latter.
The dwarf phenotype of Arabidopsis plants expressing Os-GRF1 was not reversed by application of GA. Other growth processes, such as hypocotyl, petiole, and pedicel elongation, were promoted by GA to a similar extent as in control plants. Because stem growth was severely inhibited but other growth processes were not affected, it appears that expression of Os-GRF1 in Arabidopsis disrupts functioning of the shoot apical meristem. GA not only plays a role in promoting stem, hypocotyl, and petiole elongation, but also in regulating germination, flowering, male fertility, and phase changes. Whereas stem elongation, germination, and male fertility were affected in Arabidopsis plants expressing Os-GRF1, other GA-controlled functions appeared to be normal. These observations indicate that GRF1 is not involved in regulating a basic reaction shared by all GA responses. Processes in which GA is not known to play a role, such as carpel and leaf development, were also affected by expression of Os-GRF1 in Arabidopsis (van der Knaap, 1998). It is not known whether these pleiotropic effects reflect the actual role of GRF1 in plant development, or whether they are caused by ectopic expression of the heterologous gene in Arabidopsis and interference with signal transduction pathways that are regulated by GRF1-like proteins.
In conclusion, we have identified a new family of putative regulatory proteins in plants that are typified by the QLQ and WRC motifs. The prototype member of this family, Os-GRF1, is targeted to the nucleus, and several of its structural features indicate that it may play a role in controlling transcriptional activity. Both the expression pattern of Os-GRF1 in rice and the phenotype of Arabidopsis plants transformed with Os-GRF1 indicate that Os-GRF1 may be involved in mediating GA-dependent stem growth. Elucidating the function of Os-GRF1 will contribute to our understanding of GA action and the regulation of stem elongation.
ACKNOWLEDGMENTS
We thank Dr. Marguerite Varagona and Emily Avila-Teeguarden (then at New Mexico State University, Las Cruces) for performing the nuclear localization experiments; Cassandra Zylstra for technical help in making measurements of Arabidopsis plants; Drs. Anthony Bleecker (University of Wisconsin, Madison), Michael Thomashow, and Steven Triezenberg (both at Michigan State University, East Lansing) for critical reading of the manuscript; and the National Institute of Agrobiological Resources, Tsukuba, Japan, for providing the rice EST D40170.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN9722915) and by the U.S. Department of Energy (grant no. DE–FG02–91ER20021).
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB. Plant Molecular Biology Manual A3. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. Binary vectors; pp. 1–19. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- Berg JM, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- Bethke PC, Schuurink R, Jones RL. Hormonal signalling in cereal aleurone. J Exp Bot. 1997;48:1337–1356. [Google Scholar]
- Borden KLB, Freemont PS. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- Fridborg I, Kuusk S, Moritz T, Sundberg E. The Arabidopsis dwarf mutant shiexhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell. 1999;11:1019–1031. doi: 10.1105/tpc.11.6.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber HP, Seipel K, Georgiev O, Höfferer M, Hug M, Rusconi S, Schaffner W. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- Harberd NP, King KE, Carol P, Cowling RJ, Peng J, Richards DE. Gibberellin: inhibitor of an inhibitor of … ? BioEssays. 1998;20:1001–1008. doi: 10.1002/(SICI)1521-1878(199812)20:12<1001::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Hedden P, Proebsting WM. Genetic analysis of gibberellin biosynthesis. Plant Physiol. 1999;119:365–370. doi: 10.1104/pp.119.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks GR, Raikhel NV. Protein import into the nucleus: an integrated view. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- Hoffmann-Benning S, Kende H. On the role of abscisic acid and gibberellin in the regulation of growth in rice. Plant Physiol. 1992;99:1156–1161. doi: 10.1104/pp.99.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley R. Gibberellins: perception, transduction and responses. Plant Mol Biol. 1994;26:1529–1555. doi: 10.1007/BF00016489. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Gibberellins regulate the abundance of RNAs with sequence similarity to proteinase inhibitors, dioxygenases and dehydrogenases. Planta. 1996;198:78–86. doi: 10.1007/BF00197589. [DOI] [PubMed] [Google Scholar]
- Kende H, van der Knaap E, Cho H-T. Deepwater rice: a model plant to study stem elongation. Plant Physiol. 1998;118:1105–1110. doi: 10.1104/pp.118.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métraux J-P, Kende H. The role of ethylene in the growth response of submerged deepwater rice. Plant Physiol. 1983;72:441–446. doi: 10.1104/pp.72.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAIgene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Tamkun JW. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- Phillips AL, Huttly AK. Cloning of two gibberellin-regulated cDNAs from Arabidopsis thaliana by subtractive hybridization: expression of the tonoplast water channel, γ-TIP, is increased by GA3. Plant Mol Biol. 1994;24:603–615. doi: 10.1007/BF00023557. [DOI] [PubMed] [Google Scholar]
- Puissant C, Houdebine LM. An improvement of the single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Biotechniques. 1990;8:148–149. [PubMed] [Google Scholar]
- Raskin I, Kende H. The role of gibberellin in the growth response of submerged deepwater rice. Plant Physiol. 1984;76:947–950. doi: 10.1104/pp.76.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raventós D, Skriver K, Schlein M, Karnahl K, Rogers SW, Rogers JC, Mundy J. HRT, a novel zinc finger, transcriptional repressor from barley. J Biol Chem. 1998;273:23313–23320. doi: 10.1074/jbc.273.36.23313. [DOI] [PubMed] [Google Scholar]
- Ross JJ, Murfet IC, Reid JB. Gibberellin mutants. Physiol Plant. 1997;100:550–560. [Google Scholar]
- Sauter M. Differential expression of a CAK (cdc2-activating kinase)-like protein kinase, cyclins and cdc2genes from rice during the cell cycle and in response to gibberellin. Plant J. 1997;11:181–190. doi: 10.1046/j.1365-313x.1997.11020181.x. [DOI] [PubMed] [Google Scholar]
- Sauter M, Kende H. Gibberellin-induced growth and regulation of the cell division cycle in deepwater rice. Planta. 1992;188:362–368. doi: 10.1007/BF00192803. [DOI] [PubMed] [Google Scholar]
- Sauter M, Mekhedov SL, Kende H. Gibberellin promotes histone H1 kinase activity and the expression of cdc2and cyclin genes during the induction of rapid growth in deepwater rice internodes. Plant J. 1995;7:623–632. doi: 10.1046/j.1365-313x.1995.7040623.x. [DOI] [PubMed] [Google Scholar]
- Sauter M, Seagull RW, Kende H. Internodal elongation and orientation of cellulose microfibrils and microtubules in deepwater rice. Planta. 1993;190:354–362. [Google Scholar]
- Shi L, Gast RT, Gopalraj M, Olszewski NE. Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J. 1992;2:153–159. [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T-P. The Arabidopsis RGAgene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Olszewski NE. Genetic analysis of gibberellin signal transduction. Plant Physiol. 1996;112:11–17. doi: 10.1104/pp.112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: a regulator of Drosophilahomeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton TM, Swain SM, Olszewski NE. Gibberellin signal transduction presents … the SPY who O-GlcNAc'd me. Trends Plant Sci. 1999;4:424–428. doi: 10.1016/s1360-1385(99)01485-5. [DOI] [PubMed] [Google Scholar]
- Treich I, Cairns BR, de los Santos T, Brewster E, Carlson M. SNF11, a new component of the yeast SNF-SWI complex that interacts with a conserved region of SNF2. Mol Cell Biol. 1995;15:4240–4248. doi: 10.1128/mcb.15.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Knaap E, Kende H. Identification of a gibberellin-induced gene in deepwater rice using differential display of mRNA. Plant Mol Biol. 1995;28:589–592. doi: 10.1007/BF00020405. [DOI] [PubMed] [Google Scholar]
- Van der Knaap E, Song W-Y, Ruan D-L, Sauter M, Ronald PC, Kende H. Expression of a gibberellin-induced leucine-rich repeat receptor-like protein kinase in deepwater rice and its interaction with kinase-associated protein phosphatase. Plant Physiol. 1999;120:559–570. doi: 10.1104/pp.120.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Knaap EKM. Identification and characterization of GA-induced genes in deepwater rice. PhD thesis. East Lansing: Michigan State University; 1998. [Google Scholar]
- Van Hoof A, Green PJ. Premature nonsense codons decrease the stability of phytohemagglutinin mRNA in a position-dependent manner. Plant J. 1996;10:415–424. doi: 10.1046/j.1365-313x.1996.10030415.x. [DOI] [PubMed] [Google Scholar]
- Varagona MJ, Raikhel NV. The basic domain in the bZIP regulatory protein Opaque2 serves two independent functions: DNA binding and nuclear localization. Plant J. 1994;5:207–214. doi: 10.1046/j.1365-313x.1994.05020207.x. [DOI] [PubMed] [Google Scholar]
- Varagona MJ, Schmidt RJ, Raikhel NV. Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell. 1992;4:1213–1227. doi: 10.1105/tpc.4.10.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L-L, Mitchell JP, Cohn NS, Kaufman PB. Gibberellin (GA3) enhances cell wall invertase activity and mRNA levels in elongating dwarf pea (Pisum sativum) shoots. Int J Plant Sci. 1993;154:280–289. doi: 10.1086/297108. [DOI] [PubMed] [Google Scholar]