Abstract
Peroxisomal β-oxidation (pβo) is a highly conserved fat metabolism pathway involved in the biosynthesis of diverse signaling molecules in animals and plants. In Caenorhabditis elegans, pβo is required for the biosynthesis of the ascarosides, signaling molecules that control development, lifespan, and behavior in this model organism. Via comparative mass spectrometric analysis of pβo mutants and wildtype, we show that pβo in C. elegans and the satellite model P. pacificus contributes to life stage-specific biosynthesis of several hundred previously unknown metabolites. The pβo-dependent portion of the metabolome is unexpectedly diverse, e.g. intersecting with nucleoside and neurotransmitter metabolism. Cell type-specific restoration of pβo in pβo-defective mutants further revealed that pβo-dependent sub-metabolomes differ between tissues. These results suggest that interactions of fat, nucleoside, and other primary metabolism pathways can generate structural diversity reminiscent of that arising from combinatorial strategies in microbial natural product biosynthesis.
Graphical Abstract
INTRODUCTION
Peroxisomal β-oxidation (pβo) is one of the major fatty acid degradation pathways in eukaryotes, generating heat, NADH, and acetyl- or propionyl CoA. However, it has become apparent that lipid metabolism in the peroxisomes plays a central role in diverse signaling processes in animals 1–2 and plants.3–4 In nematodes, pβo is involved in the biosynthesis of the ascarosides, a nematode-specific family of small molecule signals that are involved in almost every aspect of worm behavior and development5 as well as in interactions of worms with other organisms (Figure 1a).6–8 Simple ascarosides consist of the dideoxysugar ascarylose and a fatty acid-like side chain of varying length. Additional moieties derived from other conserved primary metabolic pathways, e.g. amino acid, neurotransmitter, or nucleoside metabolism, may be attached to positions 2 and 4 of ascarylose and carboxy terminus of the fatty acid (Figure 1a). This strategy of modular assembly creates a combinatorial library of chemical messages, not unlike human language, where words are constructed from different combinations of letters or phonemes. To date, more than 30 different ascarosides have been identified in wildtype C. elegans.5 In addition, structurally and functionally diverse ascarosides have been identified from the satellite model organism Pristionchus pacificus 9–10 and other nematode species.11–12
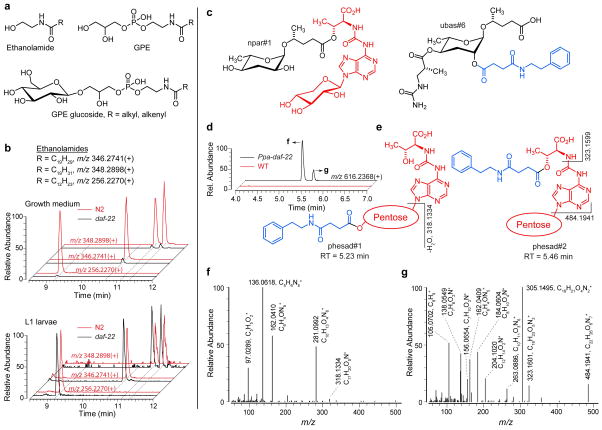
Figure 1.
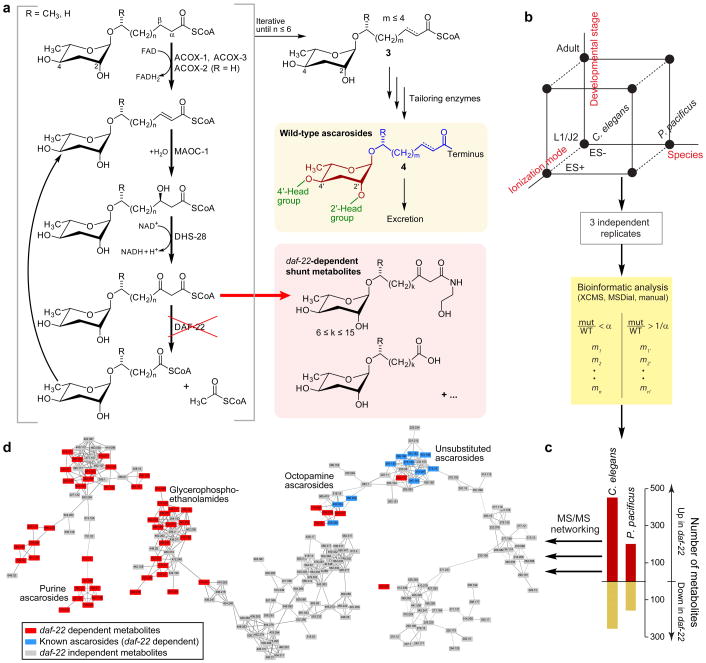
Characterization of the daf-22-dependent portion of the C. elegans and P. pacificus metabolomes. a) Model for the biosynthesis of ascarosides. Iterative 4-step β-oxidation of long-chain ascarosides produces short-chain precursors of modular ascarosides that further functionalized at the carboxy terminus or at positions 2′ and/or 4′. daf-22 loss-of-function results in build-up of shunt metabolites, including long chain ascarosides and their ethanolamide derivatives. b) Schematic of experimental approach and analysis pipeline to detect and identify differential metabolites. Two species at two different developmental stages were analyzed in ESI+ and ESI− mode, resulting in 8 data sets (represented by black dots) c) Total number of daf-22-dependent metabolites in C. elegans and P. pacificus. d) Partial representation of MS/MS network (ES−) showing known ascarosides and newly discovered daf-22-dependent metabolites as part of diverse networks that represent a wide range of different metabolite families. See Supporting Information for full networks (Figure S2 and S3).
In ascaroside biosynthesis, iterative shortening of the fatty acid side chain via pβo generates ascarosides with short 3–11 carbon side chains from long-chain (side chains of up to ~30 carbons13) lipid-like precursors (Figure 1a).14–16 The thiolase DAF-22 catalyzes the final step in the four-step β-oxidation cycle and is thus required for the biosynthesis of all short-chain ascarosides.15, 17 Therefore, daf-22 worms or metabolite extracts are often used as negative controls in assays that investigate the biological functions of ascarosides.18–20 As a consequence of impaired pβo, long-chain ascarosides and their ethanolamide derivatives accumulate in daf-22 mutant worms as shunt metabolites.13, 15, 21 Prior work further reported downregulation of endocannabinoid-like N-acylethanolamines in pβo–defective mutants.21
Several approaches have been used for the identification of ascaroside-based signaling molecules. The first members of the ascaroside family were isolated via activity-guided fractionation.22–24 Subsequently, 2D NMR-based comparative metabolomics (DANS) of wildtype and daf-22 mutant worms was used as an untargeted approach to discover additional ascarosides;21, 25–26 however, because of NMR-specific sensitivity limitations only a few additional compounds could be identified in this manner. More recently, it was noticed that ascarosides yield characteristic MS/MS fragmentation patterns, which enabled discovery of additional signaling molecules in C. elegans13–15 and other nematodes10, 27 via targeted approaches.
Whereas the requirement of pβo for ascaroside biosynthesis is well established, previous studies could not estimate the scope of pβo in C. elegans or clarify whether this evolutionarily conserved pathway is involved in the biosynthesis of compounds other than ascarosides in nematodes. High resolution LC-MS instrumentation has enabled detection of 10,000s of mass features in the metabolomes of animals, including C. elegans, most of which have not yet been characterized biochemically or functionally. We set out to map the effect of abrogating pβo on the entire worm metabolome. Specifically, we asked (1) are there undiscovered families of ascarosides; (2) are ascarosides the only products of pβo or are there other pβo-dependent metabolite families; and (3) is pβo function tissue-specific? To address these questions we employed untargeted comparisons of wildtype and daf-22 exo-metabolomes using LC-MS followed by bioinformatic analysis to identify differential peaks and their high resolution MS/MS-based characterization.28 Coupled with validation of selected representative structures via total synthesis or isolation, this approach provided a robust framework for elucidating direct and indirect effects of pβo abrogation and annotating a large number of previously uncharacterized C. elegans and P. pacificus metabolites.
RESULTS
daf-22 Deletion Abolishes Hundreds of Metabolites
DAF-22 catalyzes the last step of the four-step β-oxidation cycle and is therefore strictly required for pβo. To obtain a comprehensive overview of the contribution of pβo in worm metabolism, we analyzed two nematode species in parallel, C. elegans and the satellite model Pristionchus pacificus. The latter has two homologs of the C. elegans daf-22 gene, Ppa-daf-22.1 and Ppa-daf-22.2, as a result of a recent gene duplication event.29 For simplicity, we used a Ppa-daf-22.1; Ppa-daf-22.2 double mutant throughout unless otherwise noted. For both species, we compared wildtype and daf-22 loss-of-function mutant exo-metabolomes of arrested larval stages (L1 larvae in C elegans and J2 larvae in P pacificus,) and of cultures where metabolites accumulated during worm growth from synchronized L1 or J2 larvae to gravid adults (“growth medium”, Figure 1b). Characterization of L1/J2 metabolomes enabled sensitive detection of metabolites specifically produced during early larval arrest.30 Additionally, arrested larval samples do not contain added bacteria and thus have reduced sample complexity and minimal matrix effects. Overall, we noticed less variability among replicates of larval samples compared to growth medium samples (Figure S1).
We analyzed each of the four sample sets (arrested larval and growth medium samples for two species) by high resolution UPLC-MS in both negative and positive ionization modes, followed by automated feature detection. Several algorithms and software tools exist for extracting features from LC-MS data sets, but all of them have limitations.31–32 We compared XCMS33, using profile and centroided MS data, as well as MS-DIAL34 software (Table S1). In all cases, we found that manual curation of the software-generated list of differential features was necessary to remove false positives that upon visual inspection of raw data were not differential or did not represent peaks above background noise. In addition, features representing fragments, isotope peaks, and adducts were removed manually. To further test performance of our pipeline in detecting differential features, we examined how well software picked known ascarosides in our sample sets. From 18 ascarosides that we manually verified to be present in C elegans L1 samples, 15 were detected as differential using XCMS to analyze profile MS data. In C. elegans growth medium samples, manual analysis identified peaks representing 19 known ascarosides, whereas different feature detection algorithms picked between 9 and 16 of these (Table S1). Hence, we estimate that our approach allowed us to detect a majority of daf-22 dependent metabolites at the level of abundance of known ascarosides; however, it is likely that some metabolites of lower abundance were missed. For further analysis, we only considered MS peaks that were at least five-fold different between daf-22 and wildtype samples in all 3 replicates. These stringent criteria were employed to ensure a low false discovery rate and exclude metabolites whose abundances are only modestly (likely indirectly) affected by daf-22 loss of function. After quality control and dereplication, remaining MS peaks were put on inclusion lists for additional characterization via MS/MS.
Overall, combining L1 larval and growth medium metabolites, we detected 245 new unique molecules that were absent or downregulated in Cel-daf-22 mutants; 450 that were upregulated in Cel-daf-22; 150 that were absent or downregulated in Ppa-daf-22.1; Ppa-daf-22.2 mutants and 250 that were upregulated in Ppa-daf-22.1; Ppa-daf-22.2 (Figure 1c, Tables S2–S17). These numbers do not include previously identified C. elegans and P. pacificus ascarosides. We used these known ascaroside peaks (Table S18) for quality control during data acquisition and analysis but otherwise excluded them from the presentation, except for daf-22 tissue specific rescue experiments (vide infra). To obtain an overview of the level of structural diversity represented by the detected daf-22-dependent peaks we used molecular networking based on the analysis of MS/MS fragmentation patterns35–37 (Figure 1d, S2, S3). In the resulting network, the known simple ascarosides form a well-defined cluster. Notably, many other clusters, each representing families of metabolites related on the basis of their MS/MS spectra, include daf-22-dependent members, indicating that pβo intersects with several other metabolic pathways.
In the subsequent sections, we discuss several main classes of daf-22-dependent metabolites and their proposed structures. From 245 daf-22-dependent molecules in C. elegans, we identified over 80 new ascarosides, which range from small modifications of known structures (Figure 2a) to compound families with new or unexpected structural features. In addition, we discovered a large number of daf-22-dependent non-ascaroside metabolites. Notably, the daf-22 dependent metabolomes of L1 and growth medium differed greatly; for example, of 116 and 141 daf-22 dependent metabolites detected in C. elegans starved L1 cultures and growth medium, respectively, only 39 were shared (Figure S4). A complete list of daf-22 dependent compounds and shunt metabolites detected in this study is available in the Supporting Information (Tables S2–S17).
Figure 2.
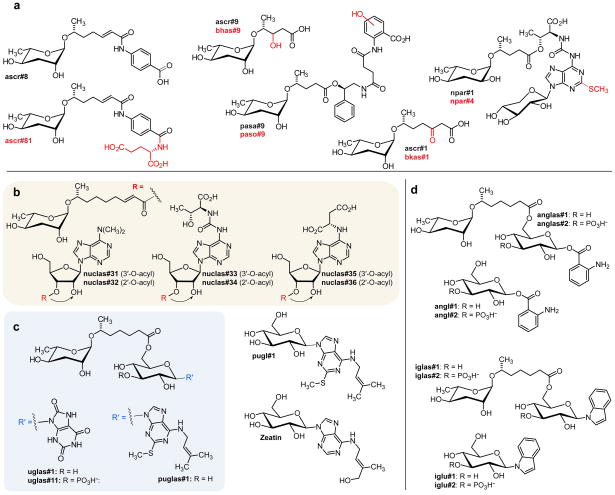
a) Examples of new ascaroside derivatives closely related to previously described compounds from C. elegans (ascr#9, ascr#8, ascr#1) and P. pacificus (npar#1, pasa#9). Known structures are in black, predicted new modifications are shown in red. The structure of ascr#81 was confirmed by total synthesis. b) Structures of ascarosylated ribonucleosides. Nuclas#31–36 occur as interconverting mixtures of the 2-O- and 3-O-ascarosylated isomers. c) Ascarosylated gluconucleosides uglas#1 and puglas#1, the highly abundant pugl#1, and the plant cytokinin zeatin. c) Anthranilic acid and indole derivative anglas#1 and iglas#1. Also shown are the previously described angl#1 and iglu#1,49 which are not daf-22-dependent.
It is important to note that most structural assignments were based on molecular formulae and mass spectrometric fragmentation patterns inferred from high-resolution MS/MS data. Because of the very large number of new compounds, full NMR spectroscopic characterization or total synthesis of all compounds was not feasible. Therefore, we implemented a tiered approach for structural assignments.38 For most detected daf-22 dependent compounds, high-resolution MS/MS data enabled assignments of molecular formulae. For the majority of compounds, detailed analysis of MS/MS patterns furthermore allowed either assignment to a specific compound class or proposal of a complete structure. Among the proposed structures, we selected specific examples of particularly intriguing or unusual compound families for total synthesis or chromatographic isolation and subsequent NMR spectroscopic characterization.
Ascaroside Glucosides and Nucleoside Combinations
Among daf-22 dependent metabolites in C. elegans growth medium, we detected several families of ascarosylated nucleoside derivatives (Figure 2b). This includes O-ascarosyl derivatives of N6,N6-dimethyladenosine (nuclas#31), N6-threonylcarbamoyladenosine (t6A, in nuclas#33), and succinyladenosine (nuclas#35), whose structures were confirmed via independent synthesis or isolation and subsequent NMR spectroscopic characterization (see Supporting Information). These nucleoside derivatives occur as pairs of isomers derived from ascaroside attachment at the 2′ and 3′ positions of ribose. We found that these isomers interconvert via trans-esterification, as previously described for similar acylated ribose derivatives.39
The nucleoside building blocks in these ascaroside derivatives appear to originate from several different branches of nucleoside metabolism. Dimethyladenosine is part of a conserved hairpin loop in the small-subunit rRNA,40 whereas t6A is a highly conserved nucleoside found directly adjacent to the anticodon triplet of a subset of tRNAs,41 and succinyladenosine represents an intermediate in the conversion of inosine 5′-phosphate into adenosine 5′-phosphate.42 Previously, it had been shown that t6A metabolism may feed into the biosynthesis of the dauer diapause-inducing paratoside npar#1 (Figure 2a) in P. pacificus, which, however, features an unusual xylose in place of the canonical ribose as in nuclas#33.10
In addition, we detected a family of ascaroside-nucleoside combinations derived from ascaroside esters of hexose-based nucleosides, e.g. puglas#1, which based on MS/MS analysis appeared to represent an O-ascarosylated derivative of N-glucosyl-2-methylthio-N6-isopentenyladenine (pugl#1, Figure 2c). Targeted analysis of C. elegans growth medium revealed that the parent nucleoside, pugl#1, which we fully characterized by NMR, is abundantly secreted, by both wildtype and daf-22 mutant worms. 2-Methylthio-N6-isopentenyladenosine (ms2i6A) is known to occur in bacterial tRNA43–44, as well as nuclear and mitochondrial RNA in some eukaryotes.45–46 Notably, N-glucosylated derivatives of N6-isopentenyladenosine and the side-chain hydroxylated zeatin (Figure 2c) are important plant hormones that play a unique role in the control of developmental processes in plants,47 and plant parasitic nematodes have recently been shown to produce cytokines to manipulate the host system and establish a long-term parasitic interaction.48 We further detected uric acid analogs(e.g. uglas#1) of puglas#1, whose structures were proposed based on MS/MS data and analogy to puglas#1.
In total, we detected about 30 novel ascarosylated glucoside derivatives with one or two additional substituents attached to the glucose (Figure 2b–e, Tables S2–S5). This includes ascarosides containing indole- and anthranilic acid glucosides as well as corresponding phosphoglucosides (Figure 2d). To confirm these structural assignments, we synthesized the indolyl derivative iglas#1 as a representative member of this compound family (Supporting Information). The non-ascaroside portions of these compounds, e.g. angl#1 and iglu#2, had been previously shown to emanate from C. elegans gut granules, a family of lysosome-like organelles, which contribute to a “fluorescent wave of death” propagating through the animal at death49 and recently have been shown to be required for the biosynthesis of previously identified modular ascarosides.50
Modular Octopamine Ascarosides in C. elegans L1 larvae
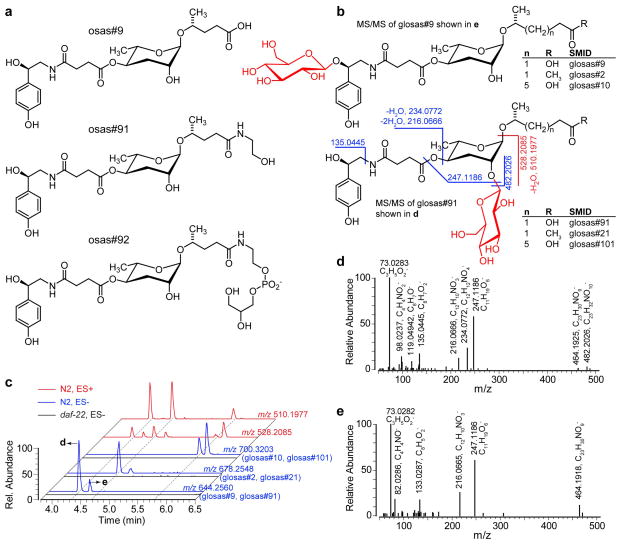
Among 74 metabolites that are downregulated or absent in Cel-daf-22 L1 samples, MS/MS analyses indicate that at least 20 represent ascaroside derivatives, of which 10 appear to incorporate an octopamine succinate moiety, as in the known osas#9, a dispersal signal that is specifically produced by starved L1 larvae.30 In addition to simple homologs of the known “osas” ascarosides, differing only in the length of the fatty acid sidechain, we detected ethanolamide, glycerophosphoethanolamide (GPE), and glucoside derivatives (Figure 3a, b, Tables S2, S4). As a representative member of this compound family, we confirmed the structure of osas#91 by coinjection with a synthetic sample (Figure S5). In case of the “osas” family glucosides, we detected two chromatographically well separated isomers for each of the osas#2, osas#9, and osas#10 derivatives (Figure 3c), which could result from the presence of the α- and β-anomers or from attachment of glucose to either 2′-position of ascarylose or the hydroxy group of octopamine. The glucose moiety does not appear to be attached to the carboxy end, as all “osas” glucosides produce a fragment with m/z 528.2085 in positive ionization MS/MS, which is derived from loss of the fatty acid side chain (Figure 3b), without concomitant loss of the glucose moiety (compare with ascarosyl glucosides, Figure 2b). Whereas all “osas” glucosides produce a fragment with m/z 528.2085, only the earlier eluting peak in each pair also yields a fragment m/z 510.1977 resulting from a loss of water (Figure 3c, Table S2, S4). This suggests that in the early eluting isomers the glucose is attached to ascarylose, leaving the elimination-prone octopamine hydroxy group unsubstituted, whereas in later eluting isomers, this hydroxy group is glucosylated (Figure 3b, d, e).
Figure 3.
a) Structures of osas#9 and examples of newly identified derivatives. b) Structures of two series of osas glucoside isomers with selected MS/MS fragmentations in ES− (blue) and ES+ (red) that distinguish isomers. c) EICs of osas glucosides. EICs in ES− (blue) correspond to glucosides of osas#9 (m/z 644.2560), osas#2 (m/z 678.2548) and osas#10 (m/z 700.3203). Neutral loss of fatty acid side chain in ES+ results in fragments common to all three osas glucosides (m/z 528.2085 for all glucosides and m/z 510.1977 after additional water loss, which is favored when the octopamine OH group is unsubstituted). d, e) MS/MS spectra of osas#9 glucosides in ES−. Absence of ions with m/z 135.0445, 234.0772, 482.2026 in e) suggests glycosylation of the octopamine hydroxy group in this isomer.
Production of Modular Ascarosides is Highly Specific
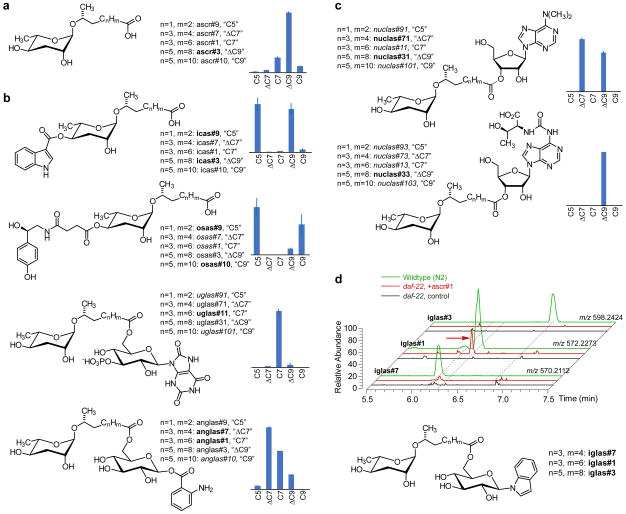
Whereas ascr#3, featuring a monounsaturated nine-carbon side chain (ΔC9), is the most abundant unmodified ascaroside detected in negative-ionization mode (Figure 4a), the sidechain lengths of ascarosides preferentially included in modular ascarosides are strongly structure-dependent (Figure 4b, c). For example, O-glucosyl anthranilic acid is preferentially combined with ΔC7 ascaroside (as in anglas#7), whereas O-glucosyl uric acid is almost exclusively combined with C7 ascaroside (as in uglas#11). Similarly, dimethyl adenosine is combined with ΔC7 and ΔC9 ascaroside (as in nuclas#71 and nuclas#31), whereas t6A is exclusively combined with ΔC9 ascaroside (as in nuclas#33). As suggested by previous work, 4′-modified ascarosides bearing an indolecarboxy moiety (icas,26) or octopamine succinate moiety (osas,30) also have specific side-chain profiles (Figure 4b). This high level of specificity is consistent with the recent finding that ascaroside-modifying enzymes may be sensitive to subtle structural features of their ascaroside substrates. For example, the acyl-CoA synthetase ACS-7 is involved in attachment of a indole carboxy moiety to the 4′ position of the C5 ascaroside ascr#9 (as in icas#9, Figure 4b) but does not contribute to the biosynthesis of corresponding derivatives of the ΔC9 and C9 ascarosides ascr#3 and ascr#10.50
Figure 4.
a) Relative abundances as determined by LC-MS of unsubstituted ascarosides. b) Relative abundances of selected families of 4′-modified ascarosides and ascaroside-glucoside combinations. c) Relative abundances of two families of ascarosylated nucleosides. d) LC-MS analysis of ascr#1 feeding experiment. EICs (ES−) of iglas#7 (m/z 570.2112), iglas#1 (m/z 572.2273) and iglas#3 (m/z 598.2424). These compounds are present in wildtype (N2) C. elegans (green) but absent in daf-22 medium (black). When synthetic ascr#1 is added to daf-22 cultures, the worms produce the ascr#1-derivative iglas#1 (red arrow) but not the corresponding ascr#7 or ascr#3 derivatives (red).
To independently confirm that compounds annotated as derivatives of ascr#1 do in fact incorporate ascr#1, and to test whether added ascaroside is incorporated specifically into the proposed modular structures, we performed a feeding experiment where we added synthetic ascr#1 to Cel-daf-22 cultures. Although these pβo-deficient worms are unable to produce short-chain ascarosides or change the length of their side chains, a previous study showed that ascaroside modifications that do not require pβo can still occur if the worms are fed with a synthetic ascaroside.15 Consistent with our predictions based on MS/MS, we detected peaks for modular ascarosides incorporating ascr#1, but not ascr#7 or ascr#3, in Cel-daf-22 samples fed with ascr#1 (Figure 4c). These ascr#1-derivatives include primarily compounds modified at the carboxy terminus, whereas only trace amounts of ascarosides featuring modifications at the 4′ (or 2′) position of the ascarylose were detected. Taken together, these observations further support the notion that the diverse modular structures we describe here are derived from specific enzymatic transformations.
Acylethanolamines and Glycerophosphoethanolamides
Approximately a third of all metabolites that are downregulated in Cel-daf-22 were annotated as fatty acid ethanolamides, glycerophosphoethanolamides (GPEs), and GPE glucosides (Figure 5a). We previously identified a subset of these GPEs and their glucosides in ethanol feeding experiments with starved C. elegans L1 larvae;51 however, it was unknown that these compounds are pβo dependent. N-acylethanolamines serve important signaling functions in C. elegans52–53 and other animals, including humans (e.g. as endocannabinoids,54–56). Izrayelit et al. reported that levels of eicosopentaenoic acid ethanolamide (EPEA) and a few other ethanolamides were reduced approximately 10-fold in C elegans daf-22 cultures compared to wildtype.21 This result was attributed to low amounts of available ethanolamine, which is instead consumed in the formation of ethanolamides of long-chain ascarosides in this mutant. We confirmed reduction of EPEA (Figure 5b) along with buildup of long-chain ethanolamide ascarosides in daf-22 growth medium, but our results paint a more complex picture. First, the effect of daf-22 loss-of-function on the levels of specific ethanolamides depends on developmental stage. For example, EPEA and several other ethanolamides are significantly downregulated in daf-22 growth medium, but are unaffected in daf-22 L1 samples (Figure 5b), yet ethanolamide derivatives of long-chain ascarosides still accumulate in L1 cultures (Table S6, S8). Second, even in samples from the same developmental stage, not all ethanolamides or GPEs are similarly affected. Some are almost completely absent in daf-22, others are diminished several fold but still present, whereas some are practically unaffected (Figure S6). In contrast to C. elegans, analysis of P. pacificus metabolome samples revealed few, if any, daf-22-dependent acylethanolamines.
Figure 5.
a) Generic structures of N-acylethanolamine, GPE, and GPE glucosides. b) Levels of EPEA (m/z 346.2741), arachidonic acid ethanolamides (AEA, m/z 348.2898) and other ethanolamides are significantly reduced in the metabolome of daf-22 growth medium, but largely unaffected in the L1 metabolome. The two peaks for AEA correspond to N- and O-acylethanolamines as determined by MS/MS (Tables S2–S9). c) Structures of npar#1 and ubas#6 previously identified from wildtype P pacificus. d) ES+ EICs for m/z 616.2368, showing two peaks present in Ppa-daf-22 but not in wildtype. Proposed structures for two isomers along with selected MS/MS fragmentations used in structure assignments are shown in e) and full ES+ MS/MS spectra for both isomers are shown in f) for phesad#1 and in g) for phesad#2.
pβo-Dependent Metabolites in P. pacificus
More than a dozen different pβo-dependent ascarosides have been described in P. pacificus.5 In addition, several paratosides, in which the configuration of the 2′-hydroxy group is inverted relative to ascarosides, have been shown to be pβo-dependent.29 Both ascarosides and paratosides serve important signaling functions in P. pacificus.5 Recently, untargeted metabolomics of Ppa-uar-1, a gene required for biosynthesis of ureidoisobutyrate containing ascarosides (ubas), revealed several additional ascarosides belonging to the ubas family, e.g. ubas#6 (Figure 6a) (Falcke et al., in review). Our Ppa-daf-22 comparative analysis revealed 17 more compounds whose MS/MS fragmentation patterns suggested that they are part of the ubas family (Table S11, S13). We verified that all of these peaks are absent not only in Ppa-daf-22 but also in Ppa-uar-1 cultures, which provides additional support for their structural assignments as ureidoisobutyrate-containing ascarosides.
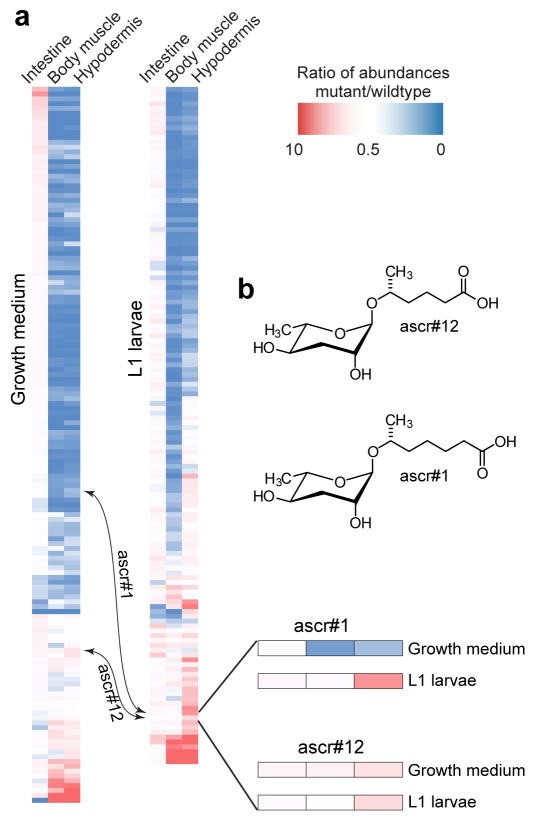
Figure 6.
Tissue specific daf-22 rescue in C. elegans (also see Figure S8). a) Heat maps showing expression of daf-22-dependent metabolites in tissue specific rescue lines relative to N2 levels in growth medium and L1 metabolomes (ES− MS). b) Enlargement showing ascr#12 and ascr#1 expression patterns. In contrast to most other metabolites, daf-22 expression in any tissue rescues ascr#12 to near wildtype levels. ascr#1 can be effectively synthesized in the hypodermis and body muscle in L1 larvae but not in adults.
Unlike C. elegans, the P. pacificus metabolome is rich in sulfated fatty acid derivatives, some of which elicit a fear-like response in C. elegans, a potential prey of the carnivorous P. pacificus (Pribadi et al., submitted). We found that biosynthesis of some, but not all, sulfated fatty acids is dependent on Ppa-daf-22 (Figure S7). Longer chain sulfates that accumulate as shunt metabolites in Ppa-daf-22 double mutants appear to be methyl ketones (Figure S7), which may be derived from non-enzymatic decarboxylation of β-keto acids, analogous to the production of long-chain methyl ketone ascarosides as shunt metabolites in Cel-daf-22 mutants.21
Production of the identified Ppa-daf-22-dependent compounds was largely abolished in Ppa-daf-22.1 single mutant, whereas the metabolome of the Ppa-daf-22.2 mutant was similar to wildtype, except for minor differences that did not reach significance. Therefore, it appears that pβo of ascarosides and other metabolites in P. pacificus relies primarily on Ppa-daf-22.1.
P. pacificus “Frustration Metabolites”
Substituted ascarosides are complex modular structures that, in addition to peroxisomal β-oxidation, integrate inputs from many other metabolic pathways.5, 50 Since the long chain ascarosides that accumulate as shunt metabolites in daf-22 mutants do not get decorated with additional building blocks in the same manner as short-chain ascarosides, one could wonder what happens to these building blocks when they have no place to attach. In Ppa-daf-22, we detected two shunt metabolites consisting of a phenylethylamine succinate moiety attached to an isomer of t6A, a modified nucleoside commonly found in tRNAs (Figure 5c–g). These metabolites are undetectable in wildtype P. pacificus cultures (Figure 5d), where the phenylethylamine module is incorporated in the ascaroside ubas#6 and a modified t6A is part of the npar#1 (Figure 5c). MS/MS spectra of the two shunt metabolite isomers indicates that they differ with regard to the point of attachment of the succinate, which is connected to either the hydroxy group of threonine or the pentose (Figure 5e–g). These “frustration metabolites” may result from coupling between the respective activated building blocks when their local concentrations exceed a certain threshold. Interestingly, the detected concentrations of the building blocks comprising these shunt metabolites - phenylethylamine succinate and t6A - are not significantly different between the wildtype and Ppa-daf-22 exo-metabolomes. We did not detect formation of similar “frustration metabolites” in C. elegans.
Tissue Specificity of daf-22 Function in C. elegans
In C. elegans, daf-22 is primarily expressed in the intestine, hypodermis, and body muscle. Previous studies showed that intestinal daf-22 expression rescues expression of three dauer-inducing ascarosides,17 but whether other tissues are involved in the biosynthesis of ascarosides and other daf-22 dependent molecules remained unclear. To address this question, we analyzed integrated transgenic lines in C. elegans, in which daf-22 expression was rescued using tissue-specific promoters. We analyzed L1 and growth medium exo-metabolomes from these strains focusing on the identified daf-22 dependent metabolites, including previously described known ascarosides. Overall, intestinal daf-22 expression almost completely rescued wildtype metabolic profile, whereas expression in the hypodermis and body wall muscle did not rescue biosynthesis of most daf-22-depndent compounds (Figures 6a, S8). Nevertheless, for some metabolites daf-22 expression in any of the three tissues resulted in similarly high levels, e.g. for ascr#12, an ascaroside whose biological function has not been studied (Figure 6b). There are also compounds for which tissue specificity differed significantly between starved L1 and growth medium samples, this is the case e.g. for ascr#1 (Figure 6a, b). These results demonstrate that, although most daf-22 dependent metabolites are produced primarily in the intestine, other tissues contribute significantly for a subset of compounds, including specific ascarosides. It is possible that the observed differences in tissue specificity in part reflect differences in the expression profiles of the three acox genes known to participate in ascaroside biosynthesis, which have different substrate preferences.16 In addition, differences in the pools of long-chain fatty acids and the availability of other building blocks in different tissues may underlie some of the observed tissue-specific differences as well as the developmental plasticity in contributions of specific tissues.
DISCUSSION
In humans, peroxisomal β-oxidation is essential for metabolism of branched and very long chain fatty acids and biosynthesis of bile acids. Several mutations in mammalian pβo enzymes lead to severe abnormalities and disease.57 In nematodes, pβo has been primarily known for its role in shortening fatty acid side chains in the biosynthesis of the ascarosides, a large family of signaling molecules that regulate many aspects of nematode life history.5, 29 Here we show that pβo disruption has profound direct and indirect effects on numerous metabolic pathways and leads to rewiring of a large portion of C. elegans and P. pacificus metabolism. We detected several hundred compounds that are partially or fully abolished in daf-22 and a similar number of metabolites that accumulate in Cel-daf-22 or Ppa-daf-22 mutants. Given that we used fairly stringent criteria – metabolites had to be detected in all replicates and at least five-fold upregulated compared to wildtype – and given that the auto-mated peak detection algorithms we relied upon are imperfect,31 these numbers likely represent only a fraction of the total pβo-dependent metabolome.
In wild type (N2) C. elegans, we discovered 86 new ascarosides, uncovering unexpected structural diversity that may be part of the vocabulary nematodes use in chemical communication. Some structures provided additional evidence for the involvement of metabolic pathways that previously had been implicated in the biosynthesis of modular ascarosides. For example, glutamate derivatives of ascr#8 and hydroxylated pasa#9 (Figure 2a) lend additional support to the hypothesis that these ascarosides originate from folate and kynurenine catabolic pathways, respectively.9, 25 Other structures demonstrate that pβo-dependent ascaroside biosynthesis intersects with additional primary metabolic pathways. For example, our results show that modified tRNA and rRNA-derived nucleobases as well as oxidized purines are incorporated into modular ascarosides in C. elegans (Figure 2b) That such ascaroside-nucleoside combinations may serve as signaling molecules is suggested by the role of the likely tRNA-derived npar#1, which is part of the P. pacificus dauer pheromone. Identification of the ascaroside gluconucleoside puglas#1 led to the finding that C. elegans produces large quantities of the N-glucosyl 2-methylthio-N6-isopentenyladenine, which is likely derived from 2-methylthio-N6-isopentenyladenosine in RNA.46 N-glucosylated derivatives of N6-isopentenyladenosine and the related zeatin (Figure 2) are important plant hormones.47 Given that plant parasitic nematodes use zeatin derivatives to manipulate their plant hosts,48 it seems possible that N-glucosyl 2-methylthio-N6-isopentenyladenenine or the corresponding ascaroside derivative puglas#1 plays a role in the complex ecology of C. elegans, which involves interactions with diverse microbial communities.58
In addition to novel ascaroside derivatives, we detected a large number of other daf-22 dependent fatty acid derivatives, including free fatty acids, diacids (Table S2–S17), as well as N-acylethanolamides and GPEs (Figure 5, S6). Many of these are completely absent in daf-22 worms, indicating that pβo is strictly required for their biosynthesis or of corresponding fatty acid precursors. The finding that daf-22 deletion has dramatically different impacts on amounts of structurally very similar metabolites (see Figure 5, S6) suggests that subtle structural differences may route corresponding precursors to distinct metabolic pathways. For example, some substrates may be processed via both mitochondrial and peroxisomal β-oxidation, whereas others are only processed by pβo.
Most of the identified ascarosides are life stage-specific, for example starved C. elegans L1 larvae produce a wide range of ascarosides incorporating octopamine succinate, most of which are absent in growth medium samples. Conversely, most glucosylated ascarosides were found only in growth medium, but not in L1 samples. This specificity demonstrates the necessity to explore metabolism-related phenotypes at multiple developmental stages and under varying environmental conditions. In addition, tissue-specific production of metabolites must be considered. Even though most ascarosides are produced in the intestine, it is interesting that a specific subset of ascarosides is also produced in muscle and the hypodermal cells, including ascr#12, an unusual ascaroside of unknown function that has an even number of carbons in its sidechain.
Despite some common features, the pβo-dependent metabolomes of C. elegans and P. pacificus differ substantially. For example, both P. pacificus and C. elegans incorporate modified RNA nucleosides into ascarosides, but specific modifications and the ways of attachment differ. Moreover, daf-22-dependent acylethanolamines and GPEs are abundant in the C. elegans metabolome, both in terms of the number of distinct structures and their overall amounts, whereas in P. pacificus most of these compounds could not be detected. These results highlight the extent to which output of intersecting primary metabolic pathways may vary between different nematode species.
CONCLUSION
Structural annotation of the very large number of metabolites that can be detected with LC-MS systems poses one of the most significant challenges toward uncovering the network of chemical processes underlying biology, and approaches are needed that supersede traditional one-by-one compound identification.38 Our work shows that high-resolution MS/MS-based comparative metabolomic analysis of mutants in lynchpin metabolic pathways, here pβo, can enable large-scale structural annotation of unknown metabolites. In our workflow, we first relied on similarity analysis of high-resolution MS/MS data to propose structures for families of metabolites (Figure S2, S3, Table S2–S17). For compound families with particularly interesting structures, we then confirmed structures via total synthesis or isolation followed by 2D NMR spectroscopic analysis. Within the constraints of current LC-MS systems and software, a systematic annotation of large parts of the metabolomes of C. elegans and other model organisms, each likely representing over 10,000 unknown metabolites, seems feasible using this approach.
MATERIALS AND METHODS
Nematode Strains
Wild type C. elegans (N2, Bristol), FCS1 daf-22(ok693), wild type P. pacificus RS2333, RS2770 Ppa-daf-22.1(tu489);Ppa-daf-22.2(tu504),29. Strains PS7211 [daf-22(ok693); syIs456 [Pvha-6::gfp::daf-22 (10 ng/uL); Pmyo-2::dsRed (5ng/uL)] ] (intestinal expression), PS7212 [daf-22(ok693); syIs457 [Pmyo-3::gfp::daf-22 (10 ng/uL); Pmyo-2::dsRed (5ng/uL)] (body wall muscle expression), and PS7213 (daf-22(ok693); syIs458 [Pdpy-7::gfp::daf-22 (10 ng/uL); Pmyo-2::dsRed (5ng/uL)]] (hypodermal expression) are described in detail in A. E. A., A. B.A., F.C.S., and P.W.S., in preparation. daf-22(ok693) was a gift from H. Y. Mak. Worms were maintained on Nematode Growth Medium (NGM) plates seeded with E. coli HB101.
Metabolite Naming
All newly identified compounds were assigned four-to-six letter “SMID”s (a search-compatible, Small Molecule IDentifier)—e.g., “icas#3” or “ascr#10.” The SMID database (www.smid-db.org) is an electronic resource maintained in collaboration with WormBase (www.wormbase.org). A complete list of SMIDs can be found at www.smid-db.org/browse and example structures for different SMIDs at www.smid-db.org/smidclasses.
Nematode Culture and Extraction
We started by seeding synchronized L1 larvae obtained by bleaching of 2–3 6cm maintenance plates onto six 10cm plates at 1200–1500 L1s/plate. 2.5 days later (at 20°C) we washed worms and eggs off the plates and bleached them (8 ml H2O + 2 ml bleach + 0.3 ml 10M NaOH for 6 min). After two washes with M9 buffer eggs were resuspended in 3 ml M9 and allowed to hatch and synchronize (24–34h at 20°C). Liquid cultures were started with synchronized L1 larvae obtained as described above and grown at 22°C, 220 rpm in S-complete medium supplemented with 2% (w/w) E. coli HB101. For a small-scale liquid culture, we inoculated 25 ml S-complete in a 250ml flask with 7·104 synchronized L1s and added 1 ml 50% E. coli stock suspension. We monitored the worm culture during the next 2 days and added E. coli as it became depleted. After ca. 60 h growth we spun down the worms, collected the growth medium, and bleached the worm pellet to yield ca. 106 eggs (100 μl egg pellet). Eggs were allowed to hatch in 10 ml of M9 buffer (22 °C, 220 rpm shaking) and L1 medium was collected 24 h later. Growth and L1 medium were frozen, lyophilized, and extracted with methanol at room temperature overnight. The extracts were dried in vacuo, resuspended in 100–150 μL methanol and analyzed by LC/MS. All cultures were grown in three replicates on different days.
Mass Spectrometric Analysis
High resolution LC-MS analysis was performed on a Dionex 3000 UPLC coupled with a Thermo Q Exactive high-resolution mass spectrometer equipped with a HESI ion source as described previously.29 Metabolites were separated using water–acetonitrile gradient on Agilent Zorbax Eclipse XDB-C18 column (150 mm × 2.1 mm, particle size 1.8 μm) maintained at 40 °C. Solvent A: 0.1% formic acid in water; Solvent B: 0.1% formic acid in acetonitrile. A/B gradient started at 5% B for 5 min after injection and increased linearly to 100% B at 12.5 min, using a flow rate 0.5 ml/min. Mass spectrometer parameters: spray voltage 2.9 kV, capillary temperature 320 °C, prober heater temperature 300 °C; sheath, auxiliary, and spare gas 70, 2, and 0, respectively; S-lens RF level 55, resolution 140,000 at m/z 200, AGC target 1·106. The instrument was calibrated daily with positive and negative ion calibration solutions (Thermo-Fisher). Each sample was analyzed in negative and positive modes with m/z range 200 to 1000. We used peaks of known abundant ascarosides (such as ascr#1 and ascr#3) to monitor mass accuracy, chromatographic peak shape, and instrument sensitivity for each batch of samples.
Feature Detection and Characterization
LC/MS RAW files from triplicate wildtype and daf-22 samples were converted to mzXML (profile mode) using MSConvert (ProteoWizard), followed by analysis using a customized XCMS R-script based on matchFilter algorithm to extract all features.59 Some data sets were also analyzed with the centWave XCMS algorithm60 and MS-DIAL.34 Resulting tables of all detected features were used to compute daf-22/wildtype peak area ratios. To select differential features, we applied a filter retaining entries with peak area ratios larger than 5 (up in daf-22) or smaller than 0.2 (down in daf-22). We manually curated the resulting list to remove false positive entries, i.e. features that upon manual inspection of raw data were not differential. For the features that were verified to be differential, we examined elution profiles, isotope patterns, and MS1 spectra to find molecular ions and remove adducts, fragments, and isotope peaks. Remaining masses were put on the inclusion list for MS/MS (ddMS2) characterization. Positive and negative mode data were processed separately. In both cases we checked if a feature had a corresponding peak in the opposite ionization mode, since fragmentation spectra in different modes often provide complementary structural information. Parameters for MS/MS were MS1 resolution 70,000, AGC target 1·106, maximum IT (injection time) 250 ms, MS2 resolution 35,000, AGC target 2·105, maximum IT 125 ms, isolation window 0.8 m/z, stepped NCE (normalized collision energy) 20, 40, 60 or 20, 40, 80, underfill ratio 2.0%, dynamic exclusion 1 s. Top 5 masses were chosen for MS2 for each scan.
Molecular Networking
A molecular network was created using the online workflow at GNPS37 and visualized in Cytoscape. The data was filtered by removing all MS/MS peaks within +/− 17 Da of the precursor ion. MS/MS spectra were window filtered by choosing only the top 6 peaks in the +/− 50 Da window throughout the spectrum. The data was then clustered with MS-Cluster with a parent mass tolerance of 0.02 Da and a MS/MS fragment ion tolerance of 0.02 Da to create consensus spectra. Consensus spectra that contained less than two spectra were discarded. A network was then created where edges were filtered to have a cosine score above 0.5 and more than 3 matched peaks. Further edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top 10 most similar nodes.
Public Deposition of MS Data
MS1 data for nematode samples analyzed in this study have been uploaded to the GNPS web site (massive.ucsd.edu). MassIVE ID numbers for the P. pacificus and C. elegans data sets are MSV000081803 and MSV000081802, respectively.
Preparative Chromatography
Medium from N2 mixed-stage culture (total volume 7 liters) was lyophilized and extracted with methanol. Dried methanol extract was loaded on Celite and fractionated on medium pressure reverse phase chromatography (86g C18 Combiflash RediSep column). A 0.1% aqueous acetic acid-acetonitrile solvent gradient was used at a flow rate of 60 ml/min, starting with acetonitrile content of 0% for 10 min which was increased to 100% over a period of 82 min. After assaying fractions by UPLC-MS, the relevant fractions were combined, solvent removed, and the residue was further fractionated on preparative reverse-phase HPLC using an Agilent Zorbax Eclipse XDB-C8 column (9.4 × 250 mm, 5 μm particle diameter) and 0.1% aqueous formic acid-acetonitrile solvent gradient at a flow rate of 3.6 ml/min. We assayed all fractions by LC/MS to confirm correct fractionation. Solvent from the fraction containing compound of interest was removed in vacuo and the residue was subjected to the second round of preparative HPLC on ACE 5 C18-AR column (10 × 250 mm) using the same solvent gradient, which yielded a sample for NMR analysis (CD3OD, 800 MHz).
Ascr#1 Feeding Experiment
Mixed stage daf-22(ok693) worms from a populated 10 cm NGM agar plate seeded with E. coli OP50 were washed into two flasks containing 10 ml of S-complete medium and 2% HB101. One flask additionally contained 10 μM of synthetic ascr#1. HB101 was added on days 1, 3 and 5 for a 7-day culture period, while shaking at 22 °C, 220 rpm. The medium was then collected, processed, and analyzed by high-resolution UPLC-MS as described above.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported in part by the NIH (GM113692, GM088290 to FCS, and T32GM007616 to A.E.A.) and HHMI.
We are grateful to David Kiemle for help with acquiring NMR spectra.
ABBREVIATIONS
- LC
liquid chromatography
- MS
mass spectrometry
- GPE
glycerophosphoethanolamide
- EIC
extracted ion chromatogram
Footnotes
Supporting Information. Synthetic procedures and NMR spectroscopic data of synthetic and isolated compounds, Supporting Figures S1–S8 and Tables S1–S18. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lodhi IJ, Semenkovich CF. Cell Metab. 2014;19(3):380–92. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wanders RJA, Waterham HR. Ann Rev Biochem. 2006;75(1):295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 3.Nyathi Y, Baker A. Biochim Biophys Acta. 2006;1763(12):1478–95. doi: 10.1016/j.bbamcr.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 4.Baker A, Graham IA, Holdsworth M, Smith SM, Theodoulou FL. Trends Plant Sci. 2006;11(3):124–32. doi: 10.1016/j.tplants.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 5.von Reuss SH, Schroeder FC. Nat Prod Rep. 2015;32(7):994–1006. doi: 10.1039/c5np00042d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsueh YP, Mahanti P, Schroeder FC, Sternberg PW. Curr Biol. 2013;23(1):83–6. doi: 10.1016/j.cub.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manosalva P, Manohar M, von Reuss SH, Chen S, Koch A, Kaplan F, Choe A, Micikas RJ, Wang X, Kogel KH, Sternberg PW, Williamson VM, Schroeder FC, Klessig DF. Nat Commun. 2015;6:7795. doi: 10.1038/ncomms8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao L, Zhang X, Wei Y, Zhou J, Zhang W, Qin P, Chinta S, Kong X, Liu Y, Yu H, Hu S, Zou Z, Butcher RA, Sun J. Nat Commun. 2016;7:12341. doi: 10.1038/ncomms12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yim JJ, Bose N, Meyer JM, Sommer RJ, Schroeder FC. Org Lett. 2015;17(7):1648–51. doi: 10.1021/acs.orglett.5b00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bose N, Ogawa A, von Reuss SH, Yim JJ, Ragsdale EJ, Sommer RJ, Schroeder FC. Angew Chem Int Ed Engl. 2012;51(50):12438–43. doi: 10.1002/anie.201206797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choe A, Chuman T, von Reuss SH, Dossey AT, Yim JJ, Ajredini R, Kolawa AA, Kaplan F, Alborn HT, Teal PE, Schroeder FC, Sternberg PW, Edison AS. Proc Natl Acad Sci USA. 2012;109(51):20949–54. doi: 10.1073/pnas.1218302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noguez JH, Conner ES, Zhou Y, Ciche TA, Ragains JR, Butcher RA. ACS Chem Biol. 2012;7(6):961–6. doi: 10.1021/cb300056q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Reuss SH, Dolke F, Dong C. Anal Chem. 2017;89(19):10570–7. doi: 10.1021/acs.analchem.7b02803. [DOI] [PubMed] [Google Scholar]
- 14.Izrayelit Y, Srinivasan J, Campbell SL, Jo Y, von Reuss SH, Genoff MC, Sternberg PW, Schroeder FC. ACS Chem Biol. 2012;7(8):1321–5. doi: 10.1021/cb300169c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Reuss SH, Bose N, Srinivasan J, Yim JJ, Judkins JC, Sternberg PW, Schroeder FC. J Am Chem Soc. 2012;134(3):1817–24. doi: 10.1021/ja210202y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Feng L, Chinta S, Singh P, Wang Y, Nunnery JK, Butcher RA. Proc Natl Acad Sci USA. 2015;112(13):3955–60. doi: 10.1073/pnas.1423951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butcher RA, Ragains JR, Li W, Ruvkun G, Clardy J, Mak HY. Proc Natl Acad Sci USA. 2009;106(6):1875–9. doi: 10.1073/pnas.0810338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludewig AH, Izrayelit Y, Park D, Malik RU, Zimmermann A, Mahanti P, Fox BW, Bethke A, Doering F, Riddle DL, Schroeder FC. Proc Natl Acad Sci USA. 2013;110(14):5522–7. doi: 10.1073/pnas.1214467110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maures TJ, Booth LN, Benayoun BA, Izrayelit Y, Schroeder FC, Brunet A. Science. 2014;343(6170):541–4. doi: 10.1126/science.1244160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aprison EZ, Ruvinsky I. Curr Biol. 2016;26(20):2827–33. doi: 10.1016/j.cub.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 21.Izrayelit Y, Robinette SL, Bose N, von Reuss SH, Schroeder FC. ACS Chem Biol. 2013;8(2):314–9. doi: 10.1021/cb3004644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong PY, Jung M, Yim YH, Kim H, Park M, Hong E, Lee W, Kim YH, Kim K, Paik YK. Nature. 2005;433(7025):541–5. doi: 10.1038/nature03201. [DOI] [PubMed] [Google Scholar]
- 23.Butcher RA, Fujita M, Schroeder FC, Clardy J. Nat Chem Biol. 2007;3(7):420–2. doi: 10.1038/nchembio.2007.3. [DOI] [PubMed] [Google Scholar]
- 24.Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PEA, Malik RU, Edison AS, Sternberg PW, Schroeder FC. Nature. 2008;454(7208):1115–8. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg PW, Schroeder FC. Proc Natl Acad Sci USA. 2009;106(19):7708–13. doi: 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srinivasan J, von Reuss SH, Bose N, Zaslaver A, Mahanti P, Ho MC, O’Doherty OG, Edison AS, Sternberg PW, Schroeder FC. PLoS Biol. 2012;10(1):e1001237. doi: 10.1371/journal.pbio.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong C, Dolke F, von Reuss SH. Org Biomol Chem. 2016;14(30):7217–25. doi: 10.1039/c6ob01230b. [DOI] [PubMed] [Google Scholar]
- 28.Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Biochemistry. 2004;43(45):14332–9. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 29.Markov GV, Meyer JM, Panda O, Artyukhin AB, Claassen M, Witte H, Schroeder FC, Sommer RJ. Mol Biol Evol. 2016;33(10):2506–14. doi: 10.1093/molbev/msw090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Artyukhin AB, Yim JJ, Srinivasan J, Izrayelit Y, Bose N, von Reuss SH, Jo Y, Jordan JM, Baugh LR, Cheong M, Sternberg PW, Avery L, Schroeder FC. J Biol Chem. 2013;288(26):18778–83. doi: 10.1074/jbc.C113.477000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers OD, Sumner SJ, Li S, Barnes S, Du X. Anal Chem. 2017;89(17):8689–95. doi: 10.1021/acs.analchem.7b01069. [DOI] [PubMed] [Google Scholar]
- 32.Myers OD, Sumner SJ, Li S, Barnes S, Du X. Anal Chem. 2017;89(17):8696–703. doi: 10.1021/acs.analchem.7b00947. [DOI] [PubMed] [Google Scholar]
- 33.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. Anal Chem. 2012;84(11):5035–9. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, Arita M. Nat Methods. 2015;12(6):523–6. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garg N, Kapono C, Lim YW, Koyama N, Vermeij MJ, Conrad D, Rohwer F, Dorrestein PC. Int J Mass Spectrom. 2015;377:719–7. doi: 10.1016/j.ijms.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC. Proc Natl Acad Sci USA. 2012;109(26):E1743–E52. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu WT, Crusemann M, Boudreau PD, Esquenazi E, Sandoval-Calderon M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu CC, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel J, Murphy BT, Gerwick L, Liaw CC, Yang YL, Humpf HU, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, Klitgaard A, Larson CB, PCAB, Torres-Mendoza D, Gonzalez DJ, Silva DB, Marques LM, Demarque DP, Pociute E, O’Neill EC, Briand E, Helfrich EJN, Granatosky EA, Glukhov E, Ryffel F, Houson H, Mohimani H, Kharbush JJ, Zeng Y, Vorholt JA, Kurita KL, Charusanti P, McPhail KL, Nielsen KF, Vuong L, Elfeki M, Traxler MF, Engene N, Koyama N, Vining OB, Baric R, Silva RR, Mascuch SJ, Tomasi S, Jenkins S, Macherla V, Hoffman T, Agarwal V, Williams PG, Dai J, Neupane R, Gurr J, Rodriguez AMC, Lamsa A, Zhang C, Dorrestein K, Duggan BM, Almaliti J, Allard PM, Phapale P, Nothias LF, Alexandrov T, Litaudon M, Wolfender JL, Kyle JE, Metz TO, Peryea T, Nguyen DT, VanLeer D, Shinn P, Jadhav A, Muller R, Waters KM, Shi W, Liu X, Zhang L, Knight R, Jensen PR, Palsson BO, Pogliano K, Linington RG, Gutierrez M, Lopes NP, Gerwick WH, Moore BS, Dorrestein PC, Bandeira N. Nat Biotechnol. 2016;34(8):828–37. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dias DA, Jones OA, Beale DJ, Boughton BA, Benheim D, Kouremenos KA, Wolfender JL, Wishart DS. Metabolites. 2016;6(4) doi: 10.3390/metabo6040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffin BE, Jarman M, Reese CB, Sulston JE, Trentham DR. Biochemistry. 1966;5(11):3638–49. [Google Scholar]
- 40.Sharma S, Lafontaine DL. Trends Biochem Sci. 2015;40(10):560–75. doi: 10.1016/j.tibs.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 41.Deutsch C, El Yacoubi B, de Crecy-Lagard V, Iwata-Reuyl D. J Biol Chem. 2012;287(17):13666–73. doi: 10.1074/jbc.M112.344028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jurecka A, Zikanova M, Kmoch S, Tylki-Szymańska A. J Inher Metab Dis. 2015;38(2):231–42. doi: 10.1007/s10545-014-9755-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrullo LA, Gallagher PJ, Elseviers D. Mol Gen Genet. 1983;190(2):289–94. doi: 10.1007/BF00330653. [DOI] [PubMed] [Google Scholar]
- 44.Wilson RK, Roe BA. Proc Natl Acad Sci USA. 1989;86(2):409–13. doi: 10.1073/pnas.86.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Globisch D, Pearson D, Hienzsch A, Bruckl T, Wagner M, Thoma I, Thumbs P, Reiter V, Kneuttinger AC, Muller M, Sieber SA, Carell T. Angew Chem Int Ed Engl. 2011;50(41):9739–42. doi: 10.1002/anie.201103229. [DOI] [PubMed] [Google Scholar]
- 46.Reiter V, Matschkal DMS, Wagner M, Globisch D, Kneuttinger AC, Müller M, Carell T. Nucl Acids Res. 2012;40(13):6235–40. doi: 10.1093/nar/gks240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaller GE, Bishopp A, Kieber JJ. Plant Cell. 2015;27(1):44–63. doi: 10.1105/tpc.114.133595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siddique S, Radakovic ZS, De La Torre CM, Chronis D, Novak O, Ramireddy E, Holbein J, Matera C, Hutten M, Gutbrod P, Anjam MS, Rozanska E, Habash S, Elashry A, Sobczak M, Kakimoto T, Strnad M, Schmulling T, Mitchum MG, Grundler FM. Proc Natl Acad Sci USA. 2015;112(41):12669–74. doi: 10.1073/pnas.1503657112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coburn C, Allman E, Mahanti P, Benedetto A, Cabreiro F, Pincus Z, Matthijssens F, Araiz C, Mandel A, Vlachos M, Edwards SA, Fischer G, Davidson A, Pryor RE, Stevens A, Slack FJ, Tavernarakis N, Braeckman BP, Schroeder FC, Nehrke K, Gems D. PLoS Biol. 2013;11(7):e1001613. doi: 10.1371/journal.pbio.1001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panda O, Akagi AE, Artyukhin AB, Judkins JC, Le HH, Mahanti P, Cohen SM, Sternberg PW, Schroeder FC. Angew Chem Int Ed Engl. 2017;56(17):4729–33. doi: 10.1002/anie.201700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Artyukhin AB, Yim JJ, Cheong Cheong M, Avery L. Sci Rep. 2015;5:10647. doi: 10.1038/srep10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Folick A, Oakley HD, Yu Y, Armstrong EH, Kumari M, Sanor L, Moore DD, Ortlund EA, Zechner R, Wang MC. Science. 2015;347(6217):83–6. doi: 10.1126/science.1258857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lucanic M, Held JM, Vantipalli MC, Klang IM, Graham JB, Gibson BW, Lithgow GJ, Gill MS. Nature. 2011;473(7346):226–9. doi: 10.1038/nature10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Marzo V, Matias I. Nat Neurosci. 2005;8(5):585–9. doi: 10.1038/nn1457. [DOI] [PubMed] [Google Scholar]
- 55.Schwartz GJ, Fu J, Astarita G, Li X, Gaetani S, Campolongo P, Cuomo V, Piomelli D. Cell Metab. 2008;8(4):281–8. doi: 10.1016/j.cmet.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu J, Gaetani S, Oveisi F, Lo Verme J, Serrano A, Rodriguez De Fonseca F, Rosengarth A, Luecke H, Di Giacomo B, Tarzia G, Piomelli D. Nature. 2003;425(6953):90–3. doi: 10.1038/nature01921. [DOI] [PubMed] [Google Scholar]
- 57.Wanders RJ. Mol Genet Metab. 2004;83(1–2):16–27. doi: 10.1016/j.ymgme.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 58.Schulenburg H, Felix MA. Genetics. 2017;206(1):55–86. doi: 10.1534/genetics.116.195511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. Anal Chem. 2006;78(3):779–87. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 60.Tautenhahn R, Bottcher C, Neumann S. BMC Bioinform. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.