Abstract
Genetic, neuroimaging, and gene expression studies suggest a role for oligodendrocyte (OLG) dysfunction in schizophrenia (SZ). Disrupted-in-schizophrenia 1 (DISC1) is a risk gene for major psychiatric disorders, including SZ. Overexpression of mutant truncated (hDISC1), but not full-length sequence of human DISC1 in forebrain influenced OLG differentiation and proliferation of glial progenitors in the developing cerebral cortex concurrently with reduction of OLG progenitor markers in the hindbrain. We examined gene and protein expression of the molecular determinants of hindbrain OLG development and their interactions with DISC1 in mutant hDISC1 mice. We found ectopic upregulation of hindbrain glial progenitor markers (early growth response 2 [Egr2] and NK2 homeobox 2 [Nkx2-2]) in the forebrain of hDISC1 (E15) embryos. DISC1 and Nkx2-2 were coexpressed and interacted in progenitor cells. Overexpression of truncated hDISC1 impaired interactions between DISC1 and Nkx2-2, which was associated with increased differentiation of OLG and upregulation of hindbrain mature OLG markers (laminin alpha-1 [LAMA1] and myelin protein zero [MPZ]) suggesting a suppressive function of endogenous DISC1 in OLG specialization of hindbrain glial progenitors during embryogenesis. Consistent with findings in hDISC1 mice, several hindbrain OLG markers (PRX, LAMA1, and MPZ) were significantly upregulated in the superior temporal cortex of persons with SZ. These findings show a significant effect of truncated hDISC1 on glial identity cells along the rostrocaudal axis and their OLG specification. Appearance of hindbrain OLG lineage cells and their premature differentiation may affect cerebrocortical organization and contribute to the pathophysiology of SZ.
Keywords: schizophrenia, oligodendrocytes, gene expression, DISC1, myelin
Introduction
Growing evidence supports the involvement of oligodendrocytes (OLG) and myelin in the pathophysiology of schizophrenia (SZ).1 The cause(s) of OLG abnormalities in SZ are unknown.
Disrupted-in schizophrenia 1 (DISC1) is a strong candidate gene for SZ and for a range of other mental disorders.2–5 Association between OLG and DISC1 was first reported in zebrafish6,7 and later confirmed in different mouse models.8,9 A genomic translocation that disrupts the expression of the DISC1 gene10 may lead to haploinsufficiency and/or the production of a truncated and functionally deficient DISC1 protein. Studies of DISC1 mutant or deficient mice have revealed behavioral and anatomical deficits.11–13 Reduced white matter integrity in frontal commissural and association fiber tracts was also associated with the t(1;11) structural variant of DISC1 in a Scottish pedigree.14
The current study used a transgenic mouse model with forebrain restricted inducible dominant negative expression of human truncated DISC1 (hDISC1).13 hDISC1 is associated with cell cycle abnormalities, upregulation of markers of OLGs and their precursors in the forebrain,8 suggesting expansion of glial progenitors during development. We examined gene and protein expression of the molecular determinants of OLG development and their possible interactions with DISC1 at different developmental points and in adult hDISC1 mice.
During early development (starting at E11.5 in mice15), OLG are derived from the ventral neuroepithelium along the entire rostrocaudal axis of the neural tube in successive waves in response to extracellular signals. OLG progenitors then migrate radially and tangentially into surrounding regions where they actively proliferate.16 The expression of transcription factors including Olig1/2, Nkx2-1, Nkx2-2, genetic-screened homeobox 2 (Gsh2), and empty spiracles 1 (Emx1) have been shown to regulate, at least in part, oligodendrogenesis of different populations of glial progenitors.17–20 These determinants of various glial populations appear to be restricted to specific territories along the dorsal-ventral axis of the entire telencephalon and diencephalon,21 except in the rostral hindbrain where some of them, such as Olig2 and Nkx2-2 overlap.22 In rostral hindbrain, Olig2/Nkx2-2 positive cells are colocalized with Egr2 which activates the expression of many myelin genes.23 This glial pool extends along the entire rostrocaudal length of the hindbrain24 from E10.5 to E13.5 days.25
In this study, we determined that the Nkx2-2 and Egr2 positive population of glial progenitors, which is normally restricted to the rostral brain, was increased in the forebrain of hDISC1 mice during embryogenesis. We determined direct interactions between Nkx2-2 and DISC1, and showed that the truncated hDISC1 is likely to impair these interactions influencing OLG differentiation. Positioning of hindbrain OLG lineage cells in the forebrain and their premature differentiation may affect cortical organization and myelination, providing new clues for the developmental mechanisms contributing to OLG dysfunction in SZ.
Material and Methods
Ethics Statement and Brain Specimens
Postmortem brains, donated by the next of kin of deceased subjects participating in studies of aging, early dementia and SZ, were received by the Mount Sinai School of Medicine Department of Psychiatry Brain Bank. All assessments were approved by the Icahn School of Medicine at Mount Sinai institutional review board and the next of kin of all tissue donors gave formal written consent for research use of the brain tissue. The specimen handling, neuropathology, and diagnostic systems used for classifying human postmortem brains have been described extensively.26–28 All animal procedures adhered to the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the Icahn School of Medicine at Mount Sinai and the Johns Hopkins University.
Demographics of SZ Cohort
The demographic characteristics of the SZ cohort are shown in supplementary table S1. For inclusion in the study, the 119 brains were selected from over 600 potential specimens.29 Samples were matched with controls subjects by age and brain pH. Donors with SZ had significantly longer postmortem intervals (PMI, P = .007), nevertheless RNA integrity was high (RIN ≥ 7) and equal between the comparison groups (P = .602).
Animal Models
Inducible hDISC1 (truncated—64 kD protein), or full-length (FL) hDISC1 mice were generated as described previously13 (for details see supplementary materials). Brains of embryonic E15 day mice were cut at the midbrain (coronal plane) resulting in 2 parts: frontal (forebrain) and posterior (hindbrain). For detailed description of dissections of postnatal day 0 (P0), P14, P21, and adult 1-year-old mice see supplementary material.
RNA Isolation and Reverse Transcription–Quantitative Polymerase Chain Reaction
The procedures for RNA isolation and cDNA synthesis from tissue aliquots have been described previously.29 The mRNA levels of selected genes were measured by quantitative polymerase chain reaction (qPCR) using TaqMan probes and primer sets (supplementary table S2) using an ABI Prism 7900HT Sequence Detection System. For relative quantification of mRNA expression, relative values of the examined genes were calculated using the standard curve method and were further normalized to the geometric means of endogenous control genes (RPLP0, GUSB, and PPIA for human or Gapdh and Ppia for mouse study) as described.29
Edu-Pulse Labeling
To study cell migration and proliferation, pregnant hDISC1 dams received i.p. injections of Edu (25 mg/kg body weight) in a solution of 10 mg/ml in PBS (pH 7.35). For 24 h pulse labeling, 6 pregnant dams were sacrificed and embryos at E16 were collected. Brain tissue disassociation, cell fixation, and permeabilization were carried out according to Click-It Edu assay manual (ThermoFisher). DNA was stained with 7-aminoactinomycin (AnaSpec, Inc.). Flow cytometry was performed on a BD Biosciences FACSCanto Flow Cytometer (see supplementary materials).
Immunohistochemistry
E15 embryos and P0 brains were removed, postfixed, and incubated overnight at 4°C. Tissues were cut in 12 µm serial sections. Primary antibodies used—rabbit a DISC1—AlexaFluor 488 (internal region; 1:200 v/v), mouse Nkx2-2-AF647 (1:100 v/v, both from Novus, CO); Ki-67-FITC (1:50 v/v, Miltenyi Biotec), and rabbit Egr2 (1:150 v/v, Aviva Systems Biology) (for details see supplementary materials). Stained sections were viewed and photographed using Carl Zeiss AxioImager Z1 microscope.
Proximity Ligation Assay
P0 mouse brains sections were processed according to the Duolink PLA manual (Sigma–Aldrich). Primary goat DISC1 (internal region, GeneTex, CA) is cross-reactive with mouse DISC1 (supplementary figure 6) and binds full length, but not truncated DISC1 allowing it to be used to study interaction between full length DISC1 and Nkx2.2. Mouse Nkx2-2 (clone 3E4, Thermo Scientific) antibodies (both 1:200 v/v) and rabbit Pcm1 (C-terminal, 1:100 v/v, Aviva System Biology) were used. Evaluation of the processed slides was performed using a Zeiss confocal Laser Scanning Microscope LSM700. PLA signals count was performed using ImageJ 1.50b software (Wayne Rasband, NIH) on 4 independent images for each experimental group.
In Situ Hybridization
2-plex QuantiGene ViewRNA Assay (Affymetrix) was used for simultaneous detection of human DISC1 (limited to the N-terminal region of DISC1 mRNA) and mouse Nkx2-2 (both from Affymetrix).
PrimeFlow RNA Assays
PrimeFlow RNA assay with oligo probes for mouse Nkx2-2 (488 nm) and human DISC1 (647 nm) (all from Affymetrix) was used for detection and measurement of cell type populations expressing Nkx2-2 and human DISC1. Flow cytometry was performed on a S3e cell flow cytometer (BioRad).
Western Blotting
Protein abundance was measured using western blotting as described before29 (for details see supplementary materials). Blots were incubated with antibodies: Egr2 (1:1000 v/v) and Nkx2-2 (1:250 v/v, both from Aviva Systems Biology); mouse anti-human GAPDH (Meridian Life Science) using SNAP i.d. (Millipore). Multiplex western blots were scanned, analyzed and quantitated on an Odyssey Imaging System (Li-Cor Biosciences).
Statistical Data Analysis
Comparison of the demographic variables was performed using separate 1-way analyses of variance (ANOVAs) or the nonparametric Mann–Whitney test as appropriate, based on deviation from normality. Pearson correlations were performed to examine the relationship of confounds: age, gender, tissue pH, and PMI with the outcome variables derived from qPCR experiments. A 2-tailed Student’s t-test was used to compare relative mRNA expression of analyzed genes in flow cytometry, qPCR experiments, in situ proximity ligation assays, TUNEL assay, and relative abundance of proteins in western blots. P-values less than .05 were deemed to be statistically significant. All procedures were performed using SPSS (IBM ver.22).
Results
Maximum Effect of Truncated hDISC1 on the Expression of OLG Genes Occurred Prenatally
Expression of OLG marker genes was measured at different time points during development: embryonic (E15), birth (P0), peak of myelination (P14), weaning (P21), and adulthood (12 months) in mice. Data are summarized in figure 1A and supplementary table S3. The expression of all tested OLG markers (Mag, Cnp, Plp1) was significantly upregulated in the forebrain of E15, P0, and 12 mo. Sox10 levels were elevated at E15 and 12 mo. The upregulation of mRNA levels of OLG markers was supported by increased protein levels for Cnp and Pdgfra in the forebrain of hDISC1 mice.8 Sox10, Cnp, and Plp1 were also significantly increased in the hindbrain of E15 and in the cerebellum (Mag, Cnp, Plp1) of P0 mice. In contrast to early developmental changes, most of the OLG markers were unchanged during active myelination (P14) and at P21 day. Based on the developmental time points evaluated, the maximum effect of hDISC1 on the expression of OLG genes was observed at E15, coinciding with the peak of expression of endogenous30 and transgene hDISC1.13,31
Fig. 1.
Maximum effect of truncated hDISC1 on differentiation and proliferation of oligodendrocyte (OLG) occurred during the embryonic stage (E15). (A) Ratio plots of OLG markers during different developmental time points and in adult disrupted-in-schizophrenia 1 mice. Mean fold change values were plotted. N = 6–7/group. (B and C) Percentage of Edu-AF488 positive events in G0/G1 phase of cell cycle analyzed by flow cytometry (N = 3/group).
In contrast to truncated hDISC1 mice, mice overexpressing intact full-length human DISC1 (hDISC1-FL) under the control of CamK2A promoter showed no significant effect on tested cell-type specific markers, including OLG/OPC (Pdgfra, Sox10, CNP, ErbB3, Mag), astrocytic-Aldh1l1, and neuronal (Nrg1, Grin1) genes (supplementary table S4), but showed significant upregulation of endogenous mouse DISC1 mRNA levels. These findings support further the conclusion that the expression of truncated hDISC1, lacking a C-terminal domain and associated with the range of psychiatric disorders, may influence normal OLG development.
Proliferation Is Increased in hDISC1 Mice
To test whether elevated expression of OLG genes was associated with increased proliferation of progenitors in E15 brains we measured G0/G1-phase Edu labeled cells (24 h) by flow cytometry (figure 1B). The overall proliferation of neural progenitors was significantly increased ~30% (P = .03) in the brains of hDISC1 mice (figure 1C) relative to controls, suggesting further that hDISC1 strongly impacts the proliferation and differentiation of early neural progenitors. We next tested, whether the increased proliferation is associated with apoptosis, as it may trigger DNA damage responses. Coronal sections subjected to TUNEL assay did not show evident differences in TUNEL fluorescence intensity between controls and mutant mice (Supplementary figure 1). Therefore, the scaling up of progenitor cell proliferation in hDISC1 mice is unlikely to have been accompanied by excessive apoptosis.
Differential Effect of Truncated hDISC1 on Markers of Glial Progenitor Lineages
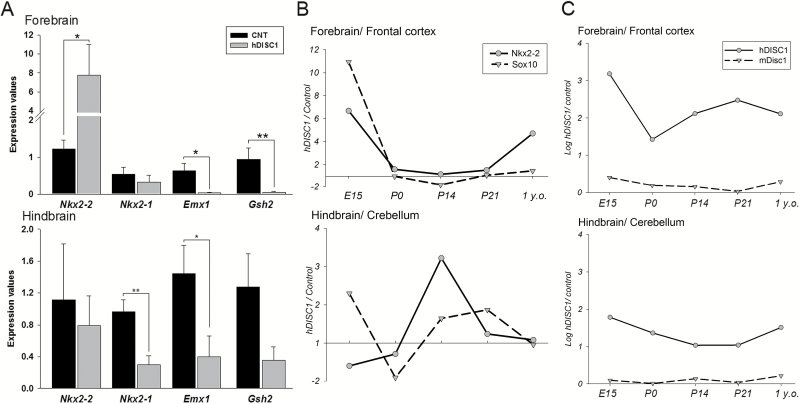
Four transcription factors: Nkx2-1, Nkx2-2, Emx1, and Gsh2 are known to be involved in specification of diverse subpopulations of glia in the telencephalon and diencephalon. Nkx2-2 mRNA levels were significantly upregulated (~7-fold; P = .043), while Emx1 and Gsh2 were significantly downregulated (P = .025 and P = .018, respectfully) in the forebrain of hDISC1 mice (figure 2A, upper). In contrast to the forebrain, hindbrain levels of Nkx2-1 and Emx1 were significantly decreased (P = .007 and P = .048, respectively), and Gsh2, Nkx2-2 levels were unchanged in the hDISC1 mice (figure 2A, lower). The developmental pattern of forebrain Nkx2-2 expression (figure 2B) was remarkably similar to the pattern of other OLG markers (figure 1A) and coincided with the maximum expression of hDISC1 (figure 2C). This dramatic increase of Nkx2-2 levels in the forebrain of hDISC1 mice was accompanied by concurrent reduction of Nkx2-2 levels in the hindbrain of E15 mice (figure 2B, lower), the region where Nkx2-2 normally specifies cell fate of OLG32 and interneurons33 during embryogenesis. Evaluation of Nkx2-2 protein levels showed biphasic increase peaked at P0 and in 1-year-old mice. Increased protein levels of Nkx2-2 (P = .002) in the forebrain of hDISC1 mice (supplementary figure 2) were paralleled by increased Nkx2-2 gene expression. Forebrain overexpression of complete hDISC1-FL had no effect on Nkx2-1, Nkx2-2, Emx1, and Gsh2 (supplementary table S4), further supporting specificity of the effect of truncated hDISC1 overexpression on glial progenitors.
Fig. 2.
Expression of NK2 homeobox 2 (Nkx2-2) was dramatically upregulated in forebrain regions during development. (A) Regional gene expression levels of oligodendrocyte lineage markers in hDISC1 mice and controls (E15). Data are expressed as geometric means ± SEM (N = 5–6/group; *P ≤ .05; **P ≤ .01). (B) Ratio plot of normalized Nkx2-2 and Sox10 gene expression. (C) Log ratio plot of normalized hDISC1 and mouse disrupted-in-schizophrenia 1 gene expression. Mean fold change values of hDISC1 vs control were plotted. N = 6–7/group.
Double Positive hDISC1/Nkx2-2 Cell Population Is Increased in Forebrain of hDISC1 Mice
To test co-expression of Nkx2-2 and hDISC1 in embryonic progenitor cells we performed in situ hybridization. Expression of both hDISC1 and Nkx2-2 transcripts was detected in the same cell nuclei in the subventricular zone (VZ/SVZ) of hDISC1 mice (figure 3A). Immunohistochemistry experiments on the same sections determined that Nkx2-2 protein is also co-localized with DISC1 in the VZ/SVZ (figure 3B) of hDISC1 mice. DISC1 staining showed both cytoplasmic and nuclear localization, and perinuclear localization for Nkx2-2 protein. Interestingly, in developing spinal cord of E15 embryos (supplementary figure 3) Nkx2-2 immunostaining did not overlap with mouse endogenous DISC1 immunolabeled cells, suggesting that spinal cord progenitors may derive from a different cell lineage(s), than those detected in forebrain regions.
Fig. 3.
Double positive NK2 homeobox 2 (Nkx2-2)/hDISC1 cell population was increased in forebrain of hDISC1 embryo (E15). (A) 2-plex ISH for Nkx2-2 and hDISC1 in coronal sections from mutant hDISC1 embryo shows hDISC1 is co-expressed with Nkx2-2. (B) Double immunohistochemistry for NKX2-2 and disrupted-in-schizophrenia 1, cell nuclei were counterstained with DAPI. Scale bar: 10 µm. (C) Nkx2-2 (488 nm) and hDISC1 (647 nm) cell populations in the E15 brains were measured by PrimeFlow RNA assay. (D) The percentage of double labeled Nkx2-2 and hDISC1 cells (Panel C, gate R4). N = 4/group; ***P < .001; **P < .01.
Double positive cell population expressing hDISC1 and Nkx2-2 in the forebrain and hindbrain (E15 embryos) were additionally measured by PrimeFlow RNA assay using mRNA specific probes for mouse Nkx2-2 (488 nm) and human DISC1 (647 nm) fluorescence (figure 3C). The cell population positive for both Nkx2-2 and hDISC1 was increased ~16.8-fold in forebrain (10.5% of total cells, P = 5.8E-06; figure 3D) of mutant hDISC1 embryos relative to controls. The hindbrain double positive population was increased only 2.5-fold in the hDISC1 mice (2.4% of total cells, P = .002; figure 3D).
DISC1 and Nkx2-2 Form Complexes Disrupted by Truncated hDISC1
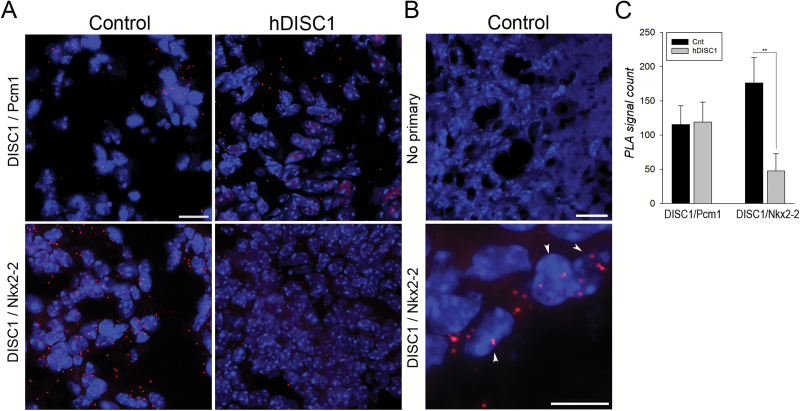
To test protein–protein interactions between DISC1 and Nkx2-2, we employed a proximity ligation assay (PLA). Brain sections from P0 mice, ie, at the peak of Nkx2-2 protein expression (figure 4A and 4B) confirmed the previously reported interactions between DISC1 and pericentriolar material 1 (Pcm1).34 Unchanged positive PLA signals for DISC1–Pcm1 complexes in frontal cortex from both control and mutant mice (figure 4A and 4C) indicated lack of hDISC1 influence on DISC1/Pcm1 interactions. Robust PLA signals for DISC1/Nkx2-2 complexes were detected in the frontal cortex of control mice, but were dramatically diminished (~3.7-fold, P = .002) in hDISC1 mice (figure 4A and 4C). In control mice, DISC1/Nkx2-2 complexes were detected not only in the cytoplasm, but also within the nuclei (figure 4B, lower). We additionally validated binding of DISC1 and Nkx2-2 in protein lysates from brains of control mice by co-IP/western blotting (supplementary figure 5C).
Fig. 4.
Mutant hDISC1 disrupts endogenous disrupted-in-schizophrenia 1(DISC1)–NK2 homeobox 2 (Nkx2-2) protein interactions. (A) Upper: positive DISC1/Pcm1 interactions in control and hDISC1 mice (P0). Lower: positive DISC1/Nkx2-2 interactions in control; DISC1/Nkx2-2 complexes were drastically reduced in hDISC1 mouse. (B) Upper: PLA probes—negative control. Lower: intranuclear DISC1/Nkx2-2 complexes in controls are marked by the arrowheads. Scale bar: 10 µm. (C) PLA signals counts for complexes of DISC1 with Pcm1 and Nkx2-2. Data are expressed as mean ± SD. **P < .01.
Fig. 5.
Hindbrain oligodendrocyte (OLG) markers are significantly upregulated in forebrain of hDISC1 mice and in superior temporal cortex from persons with schizophrenia (SZ) compare to controls. (A) E15 mRNA levels of early growth response 2 (Egr2), laminin alpha-1 (LAMA1), and myelin protein zero (MPZ), and in frontal cortex of 1-year-old mice (C). (B) Brain regional Egr2 protein levels at E15 (top), and at P0 (bottom). N = 5–6/group. (D) mRNA levels of hindbrain OLG markers: PRX, LAMA1, and MPZ in BA22 in SZ (gray bars) and in controls (NL, black bars). N = 59/group. *P ≤ .05; * P ≤ .05; **P ≤ .01. Data are expressed as mean ± SEM.
Hindbrain Mature OLG Markers Are Upregulated in the Forebrain of hDISC1 Mice
To validate specialization of Nkx2-2 positive progenitors into differentiated hindbrain OLGs,35,36 we measured the expression of known markers of hindbrain OLGs, including early growth response 2 (Egr2), myelin protein zero (Mpz), and laminin alpha 1 (LAMA1). Gene expression level of Egr2 and Mpz were strongly upregulated (~4- and 55-fold, p = 0.009 and p = 0.005, respectively) in the forebrain, but not in the hindbrain of E15 mutant mice (figure 5A), while only MPZ remained significantly increased (P = .019) in the frontal cortex of 12 months. hDISC1 mice (figure 5C). Out of 3 hindbrain OLG markers, only LAMA1 showed significant downregulation (~3-fold, P = .036) in the hindbrain of hDISC1 mice. Increased protein levels of Egr2 paralleled gene expression levels in the forebrain at E15 and P0 (P = .042 and P = .004, respectively; figure 5B). Double immunofluorescence staining detected Egr2 in the telencephalon VZ/SVZ (supplementary figure 4) along with Nkx2-2 (supplementary figure 4), confirming their colocalization. Double immunofluorescence staining also showed that Nkx2-2 is co-localized with the proliferation marker, Ki-67 in the VZ/SVZ (supplementary figure 5), confirming association of Nkx2-2 with proliferating progenitor cells. The proportion of double positive Nkx2-2 and Ki-67 cells was significantly increased (P = .024) in hDISC1 mice (supplementary figure 5D).
Hindbrain OLG Markers Are Upregulated in the Superior Temporal Cortex in SZ
We next examined expression of hindbrain specific genes, LAMA1, MPZ, and PRX-periaxin (figure 5D) in BA22 of persons with SZ. All 3 genes showed significantly upregulated levels of mRNA in BA22 from persons with SZ relative to controls (PRX, P = .049; LAMA1, P = .003, and MPZ, P = 0.03). Two of these markers (LAMA1 and MPZ) were also increased in the forebrain regions of E15 hDISC1 mice (figure 5A). These data suggest increased ectopic expression of markers of hindbrain OLGs in the temporal cortex of persons with SZ, similarly to the changes documented in the forebrain of hDISC1 mice. Sample pH, PMI, RNA integrity number, and age of the donors did not correlated significantly with gene expression levels.
Antipsychotic medication exposure is a confounding factor in postmortem studies of SZ. The subjects with SZ included in the study had been treated with antipsychotic medication for most of their lives. However, 6 of the subjects with SZ had been free of neuroleptics from 10 weeks to 5 years prior to death. Removal of neuroleptics “free” subjects with SZ from the comparison between controls and SZ cohort did not affect the observed differences in gene expression (data not shown). Nevertheless, given the smaller group size of neuroleptic “free” subjects (accounting only for ~10% of the entire SZ cohort), the possibility of antipsychotic medication exposure confounds cannot be entirely ruled out.
Discussion
SZ is a complex disorder with multiple genetic associations including a specific DISC1 haplotype2,3,10 and independent associations with several DISC1 interacting proteins.37 Regional cortical myelin/OLG related abnormalities in SZ have been replicated in a number of independently conducted studies (reviewed in Haroutunian et al1). Katsel et al8 and others6,7,9,38 have shown previously that DISC1, in addition to its role in neuronal function, can affect OLG and peripheral glia differentiation. In this study, we evaluated the effect(s) of forebrain restricted expression of truncated hDISC1, a mouse model of the translocation DISC1 haplotype in SZ on oligodendrogenesis and specification of diverse glial lineage progenitor pools. We have presented convergent lines of evidence that support the role of DISC1 in differentiation of OLG, the proliferation of OLG progenitors and DISC1’s potential involvement in the neurobiology of SZ.
The main findings of this study are: (1) evaluation of 2 different transgene models of human DISC1 (truncated and full-length) determined that only the overexpression of the truncated hDISC1 influences glial progenitors and OLGs prenatally (at E15). (2) Expression of Nkx2-2 and Egr2, which specify hindbrain OLG patterning during development, was strongly and aberrantly increased in forebrain regions of transgenic embryos. (3) Gene expression of transcription factors typifying forebrain glial progenitor lineages were either downregulated (Gsh2 and Emx1), or unchanged (Nkx2-1). (4) Mouse DISC1 and truncated hDISC1 were colocalized and interacted with Nkx2-2 in hindbrain glial progenitor cells. (5) Truncated hDISC1 impaired the formation of endogenous DISC1–Nkx2-2 complexes and that may influence early OLG differentiation. (6) Consistent with hDISC1 mice, upregulated ectopic expression of hindbrain/peripheral myelin markers were detected postmortem in the superior temporal cortex of persons with SZ.
The essential purpose of OLG is myelination of axons. This occurs predominantly in the first weeks of life in rodents. OLG development, however, starts during late embryogenesis well before the onset of myelination. The expression of transcription factors, including Olig1/2, Nkx2-1, Gsh2, and Emx1 have been shown to regulate oligodendrogenesis of different populations of progenitors,17–20 which appear to be restricted to specific territories along the dorsal-ventral axis of the telencephalon and diencephalon.21 In the rostral hindbrain, OLG progenitors are characterized by 2 transcription factors, Olig2 and Nkx2-2, which are colocalized in 2 rhombomeres: r3 and r524 with the zinc finger protein Egr2 that activates expression of many myelin genes and function as the master regulator of PNS myelination.23 Nkx2-2 positive migratory cells are known to co-express multiple OLG markers32 and Nkx2-2 itself is a key regulator of OLG specification and is involved in the temporal control of OLG differentiation.39 These progenitor pools spread along the entire rostrocaudal length of the hindbrain from E10.5 to E13.5 days.25 The ectopic expression of Nkx2-2, and hindbrain OLGs markers: Egr2 and Mpz in the forebrain of E15 hDISC1 mice suggests abnormal rostral migration of hindbrain progenitors. It is also possible, that the changes reported here are results of abnormal expression of hindbrain specification markers in the forebrain. Interestingly, dorsally derived Emx1 and ventral Gsh2 forebrain progenitor pools may be reduced in the hDISC1 mice as evidenced by downregulation of Gsh2 and Emx1 in hDISC1 mice, and in human neural precursor cells with genomic disruption of DISC1 near the site of the translocation identified in a Scottish pedigree.40
DISC1 has previously been directly implicated in the regulation of neural migration in the developing brain7,41,42 by modulating microtubular dynamics in centrosome assembly.34,43 Here, we showed that DISC1 interacts with Nkx2-2 in a manner similar to its interaction with PCM1 at the centrosome.34 Two flanking domains of DISC1, N-terminal (1–348 aa), and C-terminal (601-C) are involved in binding to PCM1.34 There were no significant changes in PLA signals that reveal DISC1/PCM1 interactions (figure 4A and 4C) in mice expressing hDISC1 lacking the C-terminal domain. While, DISC1/Nkx2-2 interactions were disrupted by the expression of hDISC1 lacking the C-terminal region. It is not clear how truncated hDISC1 interferes with DISC1/Nkx2-2 complexes. The recruitment of another binding partner(s), which facilitates DISC1/Nkx2-2 complex formation and is possibly targeted by truncated hDISC1, cannot be ruled out. Additional evaluation of the binding domains of DISC1 will resolve this ambiguity.
A recent study in mice showed that expression of DISC1 in postnatal OLG progenitor cells negatively regulated OLG differentiation, while overexpression of human DISC1 lacking the C-terminal domain (1–598 aa) promoted OLG differentiation presumably by competing with and inhibiting expression of endogenous DISC1.9 DISC1’s role in transcriptional regulation is supported by DISC1 localization to the nucleus44 and evidence that it can bind to transcription factors.43 In contrast, Nkx2-2 upregulation has been previously linked to precocious OLG differentiation attributed to a suppressive effect of Nkx2-2 on PDGFRA receptor signaling, which promotes proliferation of glial progenitors.35 Therefore, we hypothesize that normally DISC1 interacts with Nkx2-2 to suppress OLG differentiation by sequestrating Nkx2-2 protein and preventing Nkx2-2 from initiating OLG fate specification in embryonic glial progenitor cells. Truncated hDISC1 disrupts these complexes and liberates Nkx2-2, which initiates transcription signatures sufficiently to induce early OLG differentiation.45 Thus, DISC1 can control cell-fate determination in OLG progenitors during embryonic brain development.
Augmented cell-fate specialization during embryogenesis can dramatically affect cortical cytoarchitecture in the developing brain. For example, 2 main exiting populations of neurons (slowly and rapidly exiting) that undergo cell division are distinguished in the SVZ/intermediate zone.46 These progenitors are destined to migrate into deep and superficial cortical layers, respectfully. Thus, premature cell-fate specialization accompanied by a rapid exit from cell division can disrupt migration of differentiating progenitors into the predesignated cortical layer and may contribute to the pathology of developmental disorders.
Findings from postmortem human studies showed increased ectopic expression of hindbrain OLG related genes in temporal cortex of subjects with SZ. Although, subjects included in the human postmortem study did not have cytogenetic abnormalities similar, or related, to the t(1;11) structural variant of DISC1, significant upregulation of genes characteristic to hindbrain/peripheral glial pools in the temporal cortex could be a consequence of faulty signaling mechanisms participating in glial progenitors patterning during early development.
Adult glial progenitors that undergo differentiation into mature OLGs in vivo can also exhibit peripheral signature of myelin-forming cells in the CNS of rodents.47,48 Stimulation of adult glial progenitor differentiation by antipsychotic medication treatment has been documented previously in hippocampus and in the prefrontal cortex.49 However, given the similarity of findings in hDISC1 mice and persons with SZ, attribution of changes observed in SZ to exposure to antipsychotic medications seems unlikely.
Overall, this study has uncovered a new function for DISC1 in early oligodendrogenesis, ie, modulation of the migration and proliferation of glial progenitors and their differentiation into OLGs. Overexpression of truncated hDISC1 has been associated with ectopic expression of hindbrain glial progenitors and their premature OLG specification, which can ultimately lead to the abnormalities in myelin structural integrity. The similarity in the ectopic expression of hindbrain OLG-associated transcription factors in the forebrain of persons with SZ suggests that similar mechanisms may influence OLG fate and specification in SZ. Given the critical role of myelin in maintaining the rapid conduction and synchronous timing of neural networks, abnormalities in axonal myelination can contribute to modulation of synaptic efficacy and disconnectivity of the neural circuits involved in SZ.50,51
Supplementary Material
Supplementary data are available at Schizophrenia Bulletin online.
Funding
These studies were supported by National Institute of Health grant (MH097997 to P.K. and M.P.; sub-award) and by the JJ Peters VA Mental Illness Research, Education and Clinical Center (V.H.).
Supplementary Material
Acknowledgments
Conflict of interest: The authors declare no competing financial interest. Author contributions: P.K., M.P., and V.H. drafted and revised the manuscript, conceptualized the study, and performed molecular and statistical analyses; P.F., S.K., Y.J., and W.T. performed molecular studies/assays; M.P., Y.J., and C.Y. maintained animal sample collection and experiments; S.R and P.F. performed flow cytometry experiments and analysis. V.H. contributed postmortem brain samples.
References
- 1. Haroutunian V, Katsel P, Roussos P, Davis KL, Altshuler LL, Bartzokis G. Myelination, oligodendrocytes, and serious mental illness. Glia. 2014;62:1856–1877. [DOI] [PubMed] [Google Scholar]
- 2. St Clair D, Blackwood D, Muir W et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. [DOI] [PubMed] [Google Scholar]
- 3. Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders–cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Macgregor S, Visscher PM, Knott SA et al. A genome scan and follow-up study identify a bipolar disorder susceptibility locus on chromosome 1q42. Mol Psychiatry. 2004;9:1083–1090. [DOI] [PubMed] [Google Scholar]
- 5. Hennah W, Thomson P, McQuillin A et al. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14:865–873. [DOI] [PubMed] [Google Scholar]
- 6. Wood JD, Bonath F, Kumar S, Ross CA, Cunliffe VT. Disrupted-in-schizophrenia 1 and neuregulin 1 are required for the specification of oligodendrocytes and neurones in the zebrafish brain. Hum Mol Genet. 2009;18:391–404. [DOI] [PubMed] [Google Scholar]
- 7. Drerup CM, Wiora HM, Topczewski J, Morris JA. Disc1 regulates foxd3 and sox10 expression, affecting neural crest migration and differentiation. Development. 2009;136:2623–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Katsel P, Tan W, Abazyan B et al. Expression of mutant human DISC1 in mice supports abnormalities in differentiation of oligodendrocytes. Schizophr Res. 2011;130:238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hattori T, Shimizu S, Koyama Y et al. DISC1 (disrupted-in-schizophrenia-1) regulates differentiation of oligodendrocytes. PLoS One. 2014;9:e88506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Millar JK, Christie S, Anderson S et al. Genomic structure and localisation within a linkage hotspot of Disrupted In Schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol Psychiatry. 2001;6:173–178. [DOI] [PubMed] [Google Scholar]
- 11. Clapcote SJ, Lipina TV, Millar JK et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. [DOI] [PubMed] [Google Scholar]
- 12. Hikida T, Jaaro-Peled H, Seshadri S et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104:14501–14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pletnikov MV, Ayhan Y, Nikolskaia O et al. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry 2008;13:173–186. [DOI] [PubMed] [Google Scholar]
- 14. Whalley HC, Dimitrova R, Sprooten E et al. Effects of a balanced translocation between chromosomes 1 and 11 disrupting the DISC1 locus on white matter integrity. PLoS One. 2015;10:e0130900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pringle NP, Richardson WD. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development. 1993;117:525–533. [DOI] [PubMed] [Google Scholar]
- 16. Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–419. [DOI] [PubMed] [Google Scholar]
- 18. Kessaris N, Pringle N, Richardson WD. Specification of CNS glia from neural stem cells in the embryonic neuroepithelium. Philos Trans R Soc Lond B Biol Sci. 2008;363:71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chapman H, Waclaw RR, Pei Z, Nakafuku M, Campbell K. The homeobox gene Gsx2 controls the timing of oligodendroglial fate specification in mouse lateral ganglionic eminence progenitors. Development. 2013;140:2289–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vallstedt A, Klos JM, Ericson J. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron. 2005;45:55–67. [DOI] [PubMed] [Google Scholar]
- 23. Schneider-Maunoury S, Topilko P, Seitandou T et al. Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell. 1993;75:1199–1214. [DOI] [PubMed] [Google Scholar]
- 24. Miguez A, Ducret S, Di Meglio T et al. Opposing roles for Hoxa2 and Hoxb2 in hindbrain oligodendrocyte patterning. J Neurosci. 2012;32:17172–17185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Balderes DA, Magnuson MA, Sussel L. Nkx2.2:Cre knock-in mouse line: a novel tool for pancreas- and CNS-specific gene deletion. Genesis. 2013;51:844–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haroutunian V, Perl DP, Purohit DP et al. Regional distribution of neuritic plaques in the nondemented elderly and subjects with very mild Alzheimer disease. Arch Neurol. 1998;55:1185–1191. [DOI] [PubMed] [Google Scholar]
- 27. Davis KL, Mohs RC, Marin D et al. Cholinergic markers in elderly patients with early signs of Alzheimer disease. JAMA. 1999;281:1401–1406. [DOI] [PubMed] [Google Scholar]
- 28. Naslund J, Haroutunian V, Mohs R et al. Correlation between elevated levels of amyloid beta-peptide in the brain and cognitive decline. JAMA. 2000;283:1571–1577. [DOI] [PubMed] [Google Scholar]
- 29. Katsel P, Tan W, Fam P, Purohit DP, Haroutunian V. Cell cycle checkpoint abnormalities during dementia: a plausible association with the loss of protection against oxidative stress in Alzheimer’s disease [corrected]. PLoS One. 2013;8:e68361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ozeki Y, Tomoda T, Kleiderlein J et al. Disrupted-in-schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A. 2003;100:289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ayhan Y, Abazyan B, Nomura J et al. Differential effects of prenatal and postnatal expressions of mutant human DISC1 on neurobehavioral phenotypes in transgenic mice: evidence for neurodevelopmental origin of major psychiatric disorders. Mol Psychiatry. 2011;16:293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qi Y, Cai J, Wu Y et al. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. [DOI] [PubMed] [Google Scholar]
- 33. Briscoe J, Sussel L, Serup P et al. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398:622–627. [DOI] [PubMed] [Google Scholar]
- 34. Kamiya A, Tan PL, Kubo K et al. Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Arch Gen Psychiatry. 2008;65:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu Q, Zhao X, Zheng K et al. Genetic evidence that Nkx2.2 and Pdgfra are major determinants of the timing of oligodendrocyte differentiation in the developing CNS. Development. 2014;141:548–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Clark JK, O’keefe A, Mastracci TL, Sussel L, Matise MP, Kucenas S. Mammalian Nkx2.2+ perineurial glia are essential for motor nerve development. Dev Dyn. 2014;243:1116–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Porteous DJ, Thomson PA, Millar JK et al. DISC1 as a genetic risk factor for schizophrenia and related major mental illness: response to Sullivan. Mol Psychiatry. 2014;19:141–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boyd PJ, Cunliffe VT, Roy S, Wood JD. Sonic hedgehog functions upstream of disrupted-in-schizophrenia 1 (disc1): implications for mental illness. Biol Open. 2015;4:1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zou Y, Jiang W, Wang J et al. Oligodendrocyte precursor cell-intrinsic effect of Rheb1 controls differentiation and mediates mTORC1-dependent myelination in brain. J Neurosci. 2014;34:15764–15778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Srikanth P, Han K, Callahan DG et al. Genomic DISC1 disruption in hiPSCs alters Wnt signaling and neural cell fate. Cell Rep. 2015;12:1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kamiya A, Kubo K, Tomoda T et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. [DOI] [PubMed] [Google Scholar]
- 42. Mao Y, Ge X, Frank CL et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (disrupted-in-schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–1608. [DOI] [PubMed] [Google Scholar]
- 44. Sawamura N, Ando T, Maruyama Y et al. Nuclear DISC1 regulates CRE-mediated gene transcription and sleep homeostasis in the fruit fly. Mol Psychiatry. 2008;13:1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang J, Pol SU, Haberman AK, Wang C, O’Bara MA, Sim FJ. Transcription factor induction of human oligodendrocyte progenitor fate and differentiation. Proc Natl Acad Sci U S A. 2014;111:E2885–E2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tabata H, Yoshinaga S, Nakajima K. Cytoarchitecture of mouse and human subventricular zone in developing cerebral neocortex. Exp Brain Res. 2012;216:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keirstead HS, Blakemore WF. The role of oligodendrocytes and oligodendrocyte progenitors in CNS remyelination. Adv Exp Med Biol. 1999;468:183–197. [DOI] [PubMed] [Google Scholar]
- 48. Akiyama Y, Honmou O, Kato T, Uede T, Hashi K, Kocsis JD. Transplantation of clonal neural precursor cells derived from adult human brain establishes functional peripheral myelin in the rat spinal cord. Exp Neurol. 2001;167:27–39. [DOI] [PubMed] [Google Scholar]
- 49. Newton SS, Duman RS. Neurogenic actions of atypical antipsychotic drugs and therapeutic implications. CNS Drugs. 2007;21:715–725. [DOI] [PubMed] [Google Scholar]
- 50. Friston KJ. The disconnection hypothesis. Schizophr Res. 1998;30:115–125. [DOI] [PubMed] [Google Scholar]
- 51. Davis KL, Stewart DG, Friedman JI et al. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Arch Gen Psychiatry. 2003;60:443–456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.