Short abstract
Aims
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging tick-borne infectious disease. SFTS is epidemic in Asia, and its fatality rate is around 30% in Japan. The causative virus severe fever with thrombocytopenia syndrome virus (SFTSV) is a phlebovirus of the family Phenuiviridae (the order Bunyavirales). Although effective treatments are required, there are no antiviral agents currently approved for clinical use. Ribavirin and favipiravir were examined for their anti-SFTSV activity and found to be selective inhibitors of SFTSV replication in vitro. However, their activity was not sufficient. Therefore, it is mandatory to identify novel compounds active against SFTSV. To this end, we have established a safe and rapid assay system for screening selective inhibitors of SFTSV.
Methods
The virus was isolated from SFTS patients treated in Kagoshima University Hospital. Vero cells were infected with SFTSV and incubated in the presence of various concentrations of test compounds. After three days, the cells were examined for their intracellular viral RNA levels by real-time reverse transcription-PCR without extracting viral RNA. The cytotoxicity of test compounds was determined by a tetrazolium dye method.
Results
Among the test compounds, the antimalarial agent amodiaquine was identified as a selective inhibitor of SFTSV replication. Its 50% effective concentration (EC50) and cytotoxic concentration (CC50) were 19.1 ± 5.1 and >100 µM, respectively. The EC50 value of amodiaquine was comparable to those of ribavirin and favipiravir.
Conclusion
Amodiaquine is considered to be a promising lead of novel anti-SFTSV agents, and evaluating the anti-SFTSV activity of its derivatives is in progress.
Keywords: Bunyaviridae, inhibitors, replication
Introduction
Severe fever with thrombocytopenia syndrome (SFTS) is a recently identified tick-borne disease caused by SFTS virus (SFTSV). SFTSV is a novel phlebovirus of the family Phenuiviridae (the order Bunyavirales), which is epidemic in Asia including Japan.1-3 Main clinical features of SFTS are high fever, fatigue, gastrointestinal symptoms, such as vomiting and diarrhea, thrombocytopenia, and leukopenia. The elevation of serum aspartate aminotransferase, alanine aminotransferase, and lactate dehydrogenase is often observed.4 The case fatality rate of SFTS is around 10%, according to the previous report in China. However, it increases with increasing age of patients and can be as high as 30% in Japan. Therefore, it is desired to identify the novel compounds active against SFTSV.
The nucleoside analog ribavirin has a broad spectrum of antiviral activity against various emerging virus diseases.5 These include bunyavirus infections, such as Crimian-Congo hemorrhagic fever, Rift Valley fever, hemorrhagic fever with renal syndrome, and hantavirus pulmonary syndrome.6-8 Therefore, ribavirin was examined for its antiviral activity against SFTSV replication in vitro and found to be active.9,10 More recently, the nucleotide analog favipiravir was reported to selectively inhibit SFTSV replication in vitro and in vivo.11,12 Favipiravir has been licensed in Japan as a potent and selective inhibitor of influenza virus replication through the inhibition of viral RNA-dependent RNA polymerase activity.13 However, the antiviral activity of favipiravir appears to be optimized for influenza virus. Therefore, it is still mandatory to identify selective SFTSV inhibitors of which mechanism of action is different from that of favipiravir.
In the present study, we established a safe and rapid assay system for screening selective inhibitors of SFTSV in vitro. Using the assay system, we examined several compounds for their antiviral activity and found that the antimalarial drug amodiaquine and its derivatives were selective inhibitors of SFTSV replication.
Materials and methods
Compounds
Amodiaquine was synthesized according to the method described by Burckhalter et al.14 4-(7-Fluoroquinolin-4-ylamino)-2-diethylaminomethylphenol is a novel compound and was synthesized by the following procedure. Briefly, 4-amino-2-[(diethylamino)methyl]phenol dihydrochloride (133.6 mg, 0.500 mmol) was dissolved in dry ethanol (2.0 ml) and 4-chloro-7-fluoroquinoline (95.3 mg, 0.525 mmol) was added to the solution, and the reaction mixture was refluxed for a further 3 h. The solvent was removed under reduced pressure, and the residue was dissolved in distilled water. Careful addition of a dilute ammonium hydroxide (2%) liberated the free base as a solid. The solid was filtered off, washed with water, and dried for overnight under vacuum. Purification was achieved by recrystallization using methanol to give the compound. 4-(7-Bromoquinolin-4-ylamino)-2-diethylaminomethylphenol and 4-(7-iodoquinolin-4-ylamino)-2-diethylaminomethylphenol were synthesized according to the method described by Conroy et al.15 Ribavirin and favipiravir were purchased from Sigma-Aldrich (St. Louis, MO) and Selleckchem (Houston, TX), respectively. All compounds were dissolved in dimethyl sulfoxide at a concentration of 20 mM and stored –20°C until use.
Cells and virus
Human hepatoma-derived Huh-7 and monkey kidney-derived Vero cells were used for experiments. The cells were cultured in Dulbecco’s modified Eagle medium (Nacalai Tesque, Kyoto, Japan) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin G, and 100 µg/ml streptomycin. A clinical strain of SFTSV was isolated from SFTS patients hospitalized and treated in Kagoshima University Hospital. The virus was propagated in Huh-7 cells, and its infectious titer was determined in a focus forming assay. Briefly, the cells were inoculated with serially diluted virus suspensions. After incubating for three days, viral foci were detected by immunostaining with an anti-SFTSV rabbit polyclonal antibody, as previously described.11 The virus stocks were stored at –80°C until use. All experiments with infectious SFTSV were performed in a biosafety level 3 (BSL3) facility in Kagoshima University, according to the institutional biosafety operating regulations and procedures.
Results
Detection and isolation of SFTSV from patients
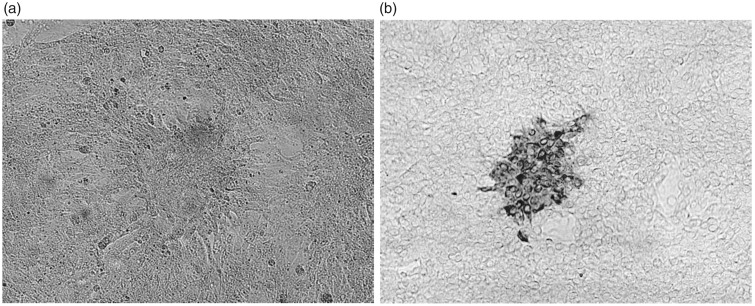
The serum samples were inoculated onto Vero cells. After incubating for three days, viral RNA was isolated from culture supernatants and amplified with reverse transcription (RT)-PCR using SFTSV-specific primer pairs.16 The cells were subjected to immunostaining with the anti-SFTSV antibody and observed microscopically (Figure 1). The culture supernatants were further inoculated onto Huh-7 cells to propagate the virus. After incubating for three days, the culture supernatants of infected Huh-7 cells were examined for their infectious titer by immunostaining and found to be 5 × 106 focus forming units (FFU)/ml (data not shown).
Figure 1.
SFTSV-infected Vero cells. (a) Microscopic observation (100×) and (b) immunostaining (100×).
Establishment of anti-SFTSV assay system
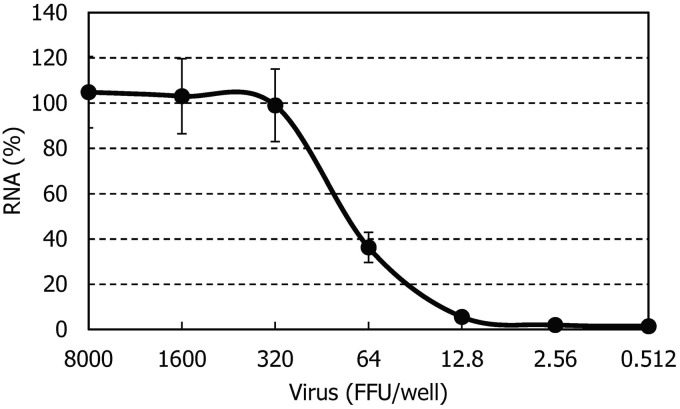
Vero cells (2 × 104 cells/well) were cultured in a microtiter plate. After incubating for 24 h, the cells were infected with various dilutions of SFTSV and further incubated in the presence of various concentrations of test compounds. After three days, the cells were washed once with PBS. The intracellular SFTSV RNA was quantified with real-time RT-PCR using TaqMan Gene Expression Cells-to-CT™ Kit (Thermo Fisher Scientific, Waltham, MA) and SFTSV-specific TaqMan primers/probe.17 The primers (5′-GGGTCC CTGAAGGAGTTGTAAA-3′ and 5′-TGCCTTCA CCAAGACTATCAATGT-3′) and probe (5′-FAM-TTCTGTCTTGCTGGCTCCGCGC-TAMRA-3′) were designed to target the conserved nucleic acid sequences in the S segment. Figure 2 shows a dose-dependent viral RNA amplification signal curve. The amplification signal was saturated, when the viral input was 320 FFU or higher. No signal was identified, when the input was less than 12.8 FFU. The amplification signal curve decreased with decreasing the input between 320 and 12.8 FFU. Therefore, 200 FFU (multiplicity of infection = 0.01) was used for the anti-SFTSV assay of compounds.
Figure 2.
Dose-dependent viral RNA amplification signal curve. Vero cells (2 × 104/well) were cultured in a 96-well plate for 24 h in the absence of compounds. After incubation, the cells were infected with the indicated amount of STFSV and further incubated. After three days, the amount of intracellular viral RNA was determined by the real-time RT-PCR method using TaqMan Gene Expression Cells-to-CT™ Kit. The experiment was carried out in triplicate and means ± SD are shown.
The cytotoxicity of compounds was examined in parallel with their antiviral activity, which was based on the viable cell number of mock-infected cells determined by a tetrazolium dye method. Briefly, Vero cells (2 × 104 cells/well) were cultured in a microtiter plate. After incubating for 24 h, the cells were exposed to various concentrations of test compounds and further incubated. After three days, 100 µl of culture medium was removed from each well, and 10 µl of water-soluble tetrazolium dye solution (Dojindo, Kumamoto, Japan) was added. After incubating for 2 h, the absorbance was read at two wavelengths (450 and 690 nm) with a microplate reader.
Anti-SFTSV activity of amodiaquine derivatives
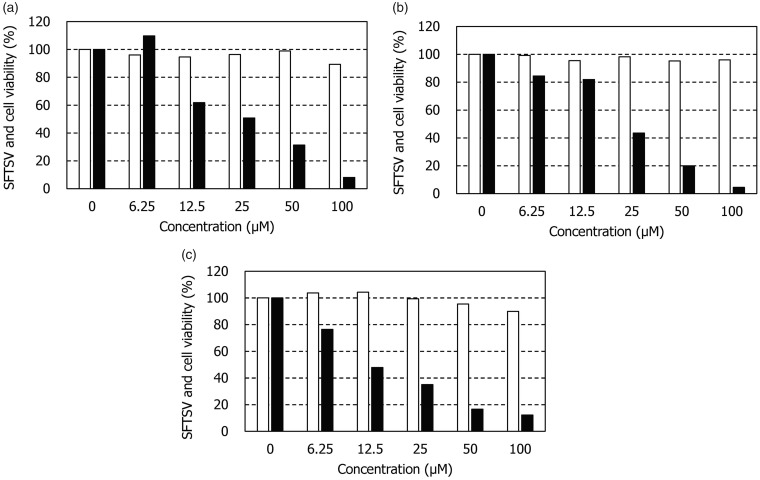
Several nucleoside and nonnucleoside compounds were examined for their anti-SFTSV activity in Vero cells. Ribavirin and favipiravir were used as reference compounds. As previously reported, both compounds reduced the intracellular SFTSV RNA level in a dose-dependent manner (Figure 3(a) and (b)). Ribavirin and favipiravir did not reduce the viability of mock-infected Vero cells at concentrations up to 100 μM. These results indicate that the present assay method functions for evaluating the anti-SFTSV activity of compounds. Among the test compounds, the antimalarial agent amodiaquine was identified as a selective inhibitor of SFTSV replication (Figure 3(c)). The anti-SFTSV activity of amodiaquine was almost equivalent to that of favipiravir. The 50% effective concentrations (EC50) of ribavirin, favipiravir, and amodiaquine were 40.1 ± 16.3, 25.0 ± 9.3, and 19.1 ± 5.1 μM, respectively (Table 1). Their 50% cytotoxic concentrations (CC50) were > 100 μM, indicating amodiaquine is a selective inhibitor of SFTSV replication.
Figure 3.
Anti-SFTSV activity and cytotoxicity of ribavirin, favipiravir, and amodiaquine. Vero cells (2 × 104/well) were cultured in a 96-well plate for 24 h in the absence of compounds. After incubation, the cells were mock-infected or infected with STFSV at a multiplicity of infection of 0.01 and further incubated in the presence of various concentrations of (a) ribavirin, (b) favipiravir, and (c) amodiaquine. After three days, the number of viable mock-infected cells (open columns) and the intracellular viral RNA levels of infected cells (closed columns) were determined by the tetrazolium dye and real-time RT-PCR methods, respectively. The experiment was carried out in triplicate, and representative results are shown.SFTSV: severe fever with thrombocytopenia syndrome virus.
Table 1.
Anti-SFTSV activity and cytotoxicity of ribavirin, favipiravir, and amodiaquine in Vero cells.
| Compound | EC50 (μM) | CC50 (μM) |
|---|---|---|
| Ribavirin | 40.1 ± 16.3 | >100 |
| Favipiravir | 25.0 ± 9.3 | >100 |
| Amodiaquine | 19.1 ± 5.1 | >100 |
Note: All data represent mean ± SD for three separate experiments.
EC50: 50% effective concentration; CC50: 50% cytotoxic concentration; SFTSV: severe fever with thrombocytopenia syndrome virus.
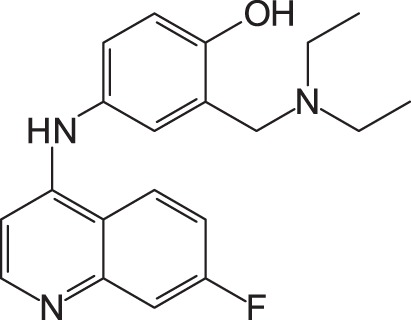
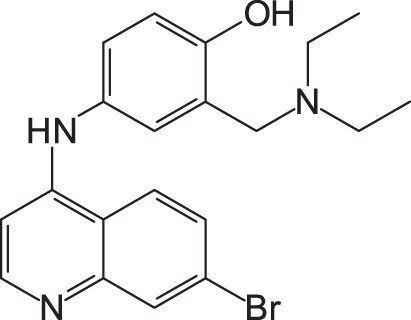
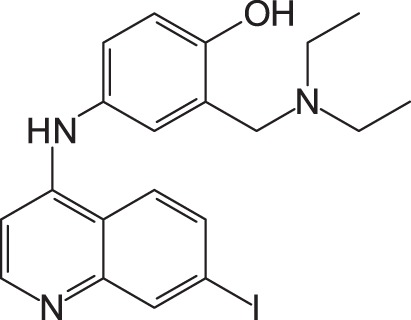
To gain more active inhibitors, several amodiaquine derivatives were synthesized and examined for their activity. When the chloride of amodiaquine was substituted for another halogen molecule, such as fluorine, bromine, and iodine, all derivatives also displayed the anti-SFTSV activity (Table 2). Among the derivatives, the iodine derivative appeared to be the most active. Its EC50 and CC50 were 15.6 ± 4.9 and >100 μM, respectively.
Table 2.
Anti-SFTSV activity and cytotoxicity of amodiaquine derivatives in Vero cells.
| Compound | Structure | EC50 (µM) | CC50 (µM) |
|---|---|---|---|
| 1 (Amodiaquine) |
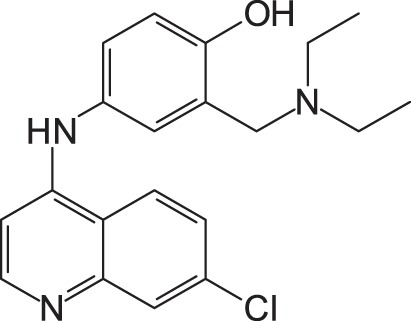
|
19.1 ± 5.1 | >100 |
| 2 |
|
36.6 ± 9.3 | >100 |
| 3 |
|
31.1 ± 16.8 | >100 |
| 4 |
|
15.6 ± 4.9 | >100 |
Note: All data represent mean ± SD for three separate experiments.
1: 4-(7-Chloroquinolin-4-ylamino)-2-diethylaminomethylphenol; 2: 4-(7-Fluoroquinolin-4-ylamino)-2-diethylaminomethylphenol; 3: 4-(7-Bromoquinolin-4-ylamino)-2-diethylaminomethylphenol; 4: 4-(7-Iodoquinolin-4-ylamino)-2-diethylaminomethylphenol; EC50: 50% effective concentration; CC50: 50% cytotoxic concentration; SFTSV: severe fever with thrombocytopenia syndrome virus.
Discussion
Since the identification of the first case of SFTS in 2012, more than 250 cases have been reported so far in Japan.18,19 In particular, Kagoshima prefecture, where our university is located, is one of the most epidemic areas of SFTS in Japan. Therefore, we attempted to isolate SFTSV from patients and establish a simple and safe assay system for screening of compounds against the virus. SFTSV can grow in a variety of cell lines, including Vero and Huh-7 cells. However, it was reported that SFTSV hardly induced cytopathic effect in infected cells, except for the macrophage cell line DH82.1 In fact, we also observed that our clinical isolates could propagate well in Vero and Huh-7 cells without apparent morphological change (Figure 1(a) and data not shown). Therefore, immunostaining with an anti-SFTSV antibody was necessary for detecting the infected cells (Figure 1(b)).
To evaluate the antiviral activity of compounds, several assay methods have been established and used for their screening. For anti-SFTSV assays, the viral yield reduction assay is most commonly used. It is based on the measurement of infectious virus titer of culture supernatants in the absence or presence of compounds by a focus forming assay.9-11 This method is reliable and reproducible but not suitable for screening of compounds, since serial dilution of culture supernatants, overlay with agar or methylcellulose, and immunostaining with a virus-specific antibody are required to complete the assay. It is also possible to measure SFTSV RNA in culture supernatants by real-time RT-PCR.10 However, the extraction of viral RNA from each culture supernatant is necessary, suggesting that the method is not realistic to deal with a large number of assay samples. Furthermore, considering the high virulence of SFTSV, which has to be handled in the BSL3 facility, the infectious virus particles in assay samples should be inactivated at an early step of the assay procedures.
Based on the above, we have decided to directly measure the intracellular viral RNA level without extracting RNA. The infected cells exposed to various concentrations of test compounds can be treated with lysis buffer after washing with PBS and subjected to real-time RT-PCR without extracting RNA, when TaqMan Gene Expression Cells-to-CT™ Kit is used for the assay. The treatment of the cells with lysis buffer inactivates infectious virus particles, so that the subsequent experiments can be performed without the BSL3 facility. In addition, all manipulations from cell lysis to real-time RT-PCR can be carried out in a microplate, which has already been applied to our anti-hepatitis C virus assay in replicon cells.20,21 Only four days are required to complete the assay, even when the cells are infected with the virus at a low multiplicity of infection (0.01). Furthermore, culture medium change or replenishment is not necessary during the assay period. Taken together, it is clear that the established anti-SFTSV assay is rapid, sensitive, and safe.
Using the assay system, we have examined a number of compounds for their anti-SFTSV activity in vitro. Among the compounds, amodiaquine was identified as a selective inhibitor of SFTSV (Figure 3(c)). Amodiaquine is a drug approved for treatment of malaria in clinic, while it is also known to inhibit dengue virus and Ebola virus replication in vitro and in vivo, respectively.22,23 Sakurai et al.24 recently showed that 14 derivatives including amodiaquine itself were potent and selective inhibitors of Ebola virus replication in vitro. We also tested several amodiaquine derivatives and found to be active against SFTSV replication (Table 2). Although the mechanism (target molecule) of amodiaquine for inhibition of SFTSV replication remains unknown, it is possible that amodiaquine inhibits SFTSV replication through a mechanism similar to that against dengue virus.22 Further studies, such as time-of-addition experiments and combination with other antivirals, are in progress to elucidate the mechanism of action of amodiaquine.
In conclusion, we have established a safe and rapid assay system for screening selective inhibitors of SFTSV. Using this system, we have found that amodiaquine and its derivatives are selective inhibitors of SFTSV replication in vitro. Thus, the compounds should be further pursued for their clinical potential as anti-SFTS agents.
Acknowledgements
The authors are thankful to the patients and staff at the Hematology and Immunology, Kagoshima University Hospital.
Authors’ contribution
MB and MT carried out the experimental work. MB and MO analyzed the data. NS synthesized amodiaquine derivatives. NA collected sera from SFTS patients, while MS provided the anti-SFTSV rabbit polyclonal antibody as well as useful information of the virus. MB wrote the manuscript.
Declaration of conflicting interests
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: MB, MT, and NS are the inventors of a patent application on amodiaquine derivatives as anti-SFTSV agents.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by JSPS KAKENHI Grant number JP26460559 and Center for Clinical and Translational Research of Kyushu University (2016, A-77).
References
- 1.Yu X, Liang M, Zhang S, et al. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med 2011; 364: 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim K, Yi J, Kim G, et al. Severe fever with thrombocytopenia syndrome, South Korea, 2012. Emerg Infect Dis 2013; 19: 1892–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi T, Maeda K, Suzuki T, et al. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J Infect Dis 2014; 209: 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu B, Liu L, Huang X, et al. Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan province, China: discovery of a new bunyavirus. PLoS Pathog 2011; 7: e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Clercq E andLi G.. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev 2016; 29: 695–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidwell RW andSmee DF.. Viruses of the Bunya- and Togaviridae families: potential as bioterrorism agents and means of control. Antiviral Res 2003; 57: 101–111. [DOI] [PubMed] [Google Scholar]
- 7.Jonsson CB Hooper J andMertz G.. Treatment of hantavirus pulmonary syndrome. Antiviral Res 2008; 78: 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ascioglu S, Leblebicioglu H, Vahaboglu H, et al. Ribavirin for patients with Crimean-Congo haemorrhagic fever: a systematic review and meta-analysis. J Antimicrob Chemother 2011; 66: 1215–1222. [DOI] [PubMed] [Google Scholar]
- 9.Shimojima M, Fukushi S, Tani H, et al. Effect of ribavirin on severe fever with thrombocytopenia syndrome virus in vitro. Jpn J Infect Dis 2014; 67: 423–427. [DOI] [PubMed] [Google Scholar]
- 10.Lee MJ, Kim K-H, Yi J, et al. In vitro antiviral activity of ribavirin against severe fever with thrombocytopenia syndrome virus. Korean J Intern Med 2017; 32: 731–737 [DOI] [PMC free article] [PubMed]
- 11.Tani H, Fukuma A, Fukushi S, et al. Efficacy of T-705 (favipiravir) in the treatment of infections with lethal severe fever with thrombocytopenia syndrome virus. mSphere 2016; 1: e00061–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gowen BB, Westover JB, Miao J, et al. Modeling severe fever with thrombocytepenia syndrome virus infection in golden Syrian hamsters: importance of STAT2 in preventing disease and effective treatment with favipiravir. J Virol 2017; 18: e01942–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sangawa H, Komeno T, Nishikawa H, et al. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob Agents Chemother 2013; 57: 5202–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burckhalter JH DeWald HA andTendick FH.. An alternate synthesis of camoquin. J Am Chem Soc 1950; 72: 1024–1025. [Google Scholar]
- 15.Conroy AE Mosher HS andWhitmore FC.. Heterocyclic basic compounds. XII. 7-Bromo- and 7-iodoquinolines. J Am Chem Soc 1949; 71: 3236–3237. [Google Scholar]
- 16.Yoshikawa T, Fukushi S, Tani H, et al. Sensitive and specific PCR systems for detection pf both Chinese and Japanese severe fever with thrombocytopenia syndrome virus strains and prediction of patient survival based on viral load. J Clin Microbiol 2014; 52: 3325–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui L, Ge Y, Xu G, et al. Detection of severe fever with thrombocytopenia syndrome virus by reverse transcription – cross-priming amplification coupled with vertical flow visualization. J Clin Microbiol 2012; 50: 3881–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito T, Fukushima K, Umeki K, et al. Severe fever with thrombocytopenia syndrome in Japan and public health communication. Emerg Infect Dis 2015; 21: 487–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Institute of Infectious Diseases in Japan. SFTS cases in Epidemiological Surveillance of Infectious Diseases (Japanese), www.niid.go.jp/niid/ja/sfts/3143-sfts.html (2017, accessed 10 July 2017).
- 20.Salim MTA, Aoyama H, Sugita K, et al. Potent and selective inhibition of hepatitis C virus replication by novel phenanthridinone derivatives. Biochem Biophys Res Commun 2011; 415: 714–719. [DOI] [PubMed] [Google Scholar]
- 21.Ito W, Toyama M, Okamoto M, et al. Isolation and characterization of hepatitis C virus resistant to a novel phenanthridinone derivatives. Antiviral Chem Chemother 2016; 24: 148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boonyasuppayakorn S, Reichert ED, Manzano M, et al. Amodiaquine, an antimalarial drug, inhibits dengue virus type 2 replication and infectivity. Antiviral Res 2014; 106: 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gignoux E, Azman AS, de Smet M, et al. Effect of artesunate-amodiaquine on mortality related to Ebola virus disease. N Engl J Med 2016; 374: 23–32. [DOI] [PubMed] [Google Scholar]
- 24.Sakurai Y, Sakakibara N, Toyama M, et al. Identification of novel amodiaquine derivatives as anti-Ebola virus compounds. In: The 30th international conference on antiviral research, Atlanta, USA, 21–25 May 2017, abstract no. 134.