Abstract
Objective
Cancer stem cells (CSCs) are responsible for tumour formation and spreading, and their targeting is required for tumour eradication. There are limited therapeutic options for advanced colorectal cancer (CRC), particularly for tumours carrying RAS-activating mutations. The aim of this study was to identify novel CSC-targeting strategies.
Design
To discover potential therapeutics to be clinically investigated as single agent, we performed a screening with a panel of FDA-approved or investigational drugs on primary CRC cells enriched for CSCs (CRC-SCs) isolated from 27 patients. Candidate predictive biomarkers of efficacy were identified by integrating genomic, reverse-phase protein microarray (RPPA) and cytogenetic analyses, and validated by immunostainings. DNA replication stress (RS) was increased by employing DNA replication-perturbing or polyploidising agents.
Results
The drug-library screening led to the identification of LY2606368 as a potent anti-CSC agent acting in vitro and in vivo in tumour cells from a considerable number of patients (∼36%). By inhibiting checkpoint kinase (CHK)1, LY2606368 affected DNA replication in most CRC-SCs, including RAS-mutated ones, forcing them into premature, lethal mitoses. Parallel genomic, RPPA and cytogenetic analyses indicated that CRC-SCs sensitive to LY2606368 displayed signs of ongoing RS response, including the phosphorylation of RPA32 and ataxia telangiectasia mutated serine/threonine kinase (ATM). This was associated with mutation(s) in TP53 and hyperdiploidy, and made these CRC-SCs exquisitely dependent on CHK1 function. Accordingly, experimental increase of RS sensitised resistant CRC-SCs to LY2606368.
Conclusions
LY2606368 selectively eliminates replication-stressed, p53-deficient and hyperdiploid CRC-SCs independently of RAS mutational status. These results provide a strong rationale for biomarker-driven clinical trials with LY2606368 in patients with CRC.
Keywords: CELL CYCLE CONTROL, CHEMOTHERAPY, COLORECTAL CANCER, DNA DAMAGE, DRUG DEVELOPMENT
Significance of this study.
What is already known on this subject?
Cancer stem cells (CSCs), the subpopulation of cells driving tumour initiation and spreading, are associated with cancer relapse, therapeutic resistance and poor patient prognosis thereby limiting the efficacy of antineoplastic regimens.
Patient-derived cancer models enriched for CSCs are successfully employed for discovering novel cancer drugs, predicting the clinical efficacy of novel antineoplastic regimens and rapidly translating these novelties into the clinical setting.
We and others previously showed that the therapeutic resistance of CSCs is due to their high efficiency in activating the DNA damage response (DDR) in the presence of DNA lesions, but further investigations are required to elucidate the status and molecular mechanisms of DDR in CSCs and how these insights can be exploited for cancer therapy.
Previous evidence indicated that the inhibition of the ataxia telangiectasia mutated serine/threonine kinase (ATM)-checkpoint kinase (CHK)2 or ataxia telangiectasia mutated and Rad3 related serine/threonine kinase (ATR)-CHK1 axis of DDR, alone or in combination with DNA-damaging chemotherapeutics, could kill cancer cells displaying genomic instability, but the effect on CSC survival needs further investigations.
What are the new findings?
We identified LY2606368 as a potent in vitro and in vivo anti-CSC agent able to kill a significant fraction (approximately a third) of our large panel of primary colorectal cancer (CRC) cells enriched for CSCs (CRC-SCs).
The mechanism of CRC-SC killing by LY2606368 involves the specific inhibition of CHK1, which alters the DNA replication and intra-S checkpoint of CRC-SCs, in turn resulting in the generation of an intolerable DNA damage burden and the consequent cell demise via replication catastrophe.
The fraction of CRC-SCs displaying signs of DNA replication stress, such as phosphorylation of RPA32 or ATM, deficiency in p53 and increased number of chromosomes (hyperdiploidy) was shown to be highly dependent on CHK1 activity and thus targetable with CHK1 inhibitors.
Boosting replication stress or increasing the number of chromosomes by employing DNA replication-perturbing or mitosis-perturbing agents was proven to be a strategy to sensitise formerly resistant CRC-SCs to LY2606368.
How might it impact on clinical practice in the foreseeable future?
Our results, together with the recent evidence on tolerable safety profiles and preliminary antineoplastic activity of LY2606368 as monotherapy in a phase I, non-randomised, open-label, dose-escalation trial in patients affected by advanced or metastatic solid tumours who underwent at least three previous lines of treatment, support the further clinical development of LY2606368.
The selective targeting of CHK1 by LY2606368 can be rapidly translated into the clinical settings for the eradication of tumours with replication-stressed, p53-deficient and hyperdiploid CRC-SCs, which can be easily identified by p53 sequencing combined with immunohistochemical analysis for replication stress markers (phosphorylation of ATM and RPA32) and ploidy status (measurement of nuclear area).
Our findings also guide for the further development of therapeutic regimens based on the induction of replication stress in patients with low DNA replication-stressed CRC (identified as reported above) so as to broaden the therapeutic use of LY2606368 to a large number of subjects.
Introduction
The management of patients with colorectal cancer (CRC) remains a major clinical challenge, owing to the high incidence of tumour relapse and development of treatment-refractory diseases.1 Despite the discovery of novel regimens combining biological therapies to the chemotherapy backbone over the past decade, intrinsic and acquired resistance remains a critical hurdle. The mechanisms of therapeutic resistance, often emerging after initial tumour responses, include (but are not limited to) mutation(s) in the gene encoding the target protein,2 induction of bypass or parallel cascade(s),3 restoration of the defective targeted pathway4 and outgrowth of pre-existing minor or dormant clones.5
The development of effective therapeutics against solid malignancies is further hampered by their considerable genetic, epigenetic, functional and phenotypical heterogeneity over space and time.6–8 This intratumour heterogeneity is reported to arise early in the development of CRC and feeds therapeutic resistance.9 Mounting evidence indicates that intratumour heterogeneity and therapeutic resistance is driven by transient/stable subsets of immature, undifferentiated cells known as cancer stem cells (CSCs).6 10 11 CSCs have been isolated from multiple malignancies, including CRC, by specific cell-surface markers or serum-free in vitro culture enrichment.12 13 This stem-like population is at the apex of the hierarchical organisation of the cancer system, where it constitutes a perpetual pool generating non-tumorigenic progeny with variable degree of differentiation and plasticity, and in dynamic crosstalk with the tumour microenvironment.10 CSCs are responsible for tumour initiation, and are involved in cancer progression, recurrence, metastasis and therapeutic failure.10 14 15 Underscoring their clinical importance, specific CSC-related gene-expression signatures have been associated to inferior clinical outcomes,6 16 17 and CRC patients with stem cell-like tumours have been documented to display lower 5-year disease-free survival rates than those having differentiated tumours.18 The therapeutic targeting of CSCs is thus advocated as essential for the development of effective drugs.
Multicellular spheroids (best known as tumourspheres) are a cancer model employed to efficiently purify, enrich and propagate patient-derived CSCs,13 hence representing a powerful tool for discovering anticancer drugs and companion predictive biomarkers. Considering the enormous potential of CSC-based assays as hypothesis-generating models for early clinical trials, we decided to screen freshly generated patient-derived CRC tumourspheres enriched for CSCs (hereafter referred to as CRC-SCs) with a panel of >300 drugs already approved or in clinical development.
Materials and methods
Cell lines, culture conditions and chemicals
Unless otherwise indicated, media, supplements and plasticware were purchased from Gibco-Thermo Fisher Scientific (Waltham, Massachusetts, USA) or Corning-Life Sciences (Corning, New York, USA). CSC isolation and culture from samples of human patients with CRC (obtained in accordance with the standards of the institutional Ethics Committee on human experimentation, authorisation CE5ISS-09/282) were performed as previously reported19 and described in online supplementary information. All CRC-SCs were validated for their capability to generate neoplasms faithfully phenocopying the original patient tumour when xenotransplanted into immunocompromised mice.20 Selected CRC-SCs belonging to the three categories (#1 and #4 for high; #3 and #6 for medium; and #8, #10 and #12 for low sensitive) were employed in most in vitro experiments. Of these CRC-SCs, #1, #3 and #8 were used in vivo. Additional CRC-SCs were employed depending on cell availability and status always being well balanced towards the point of LY2606368 sensitivity. Compounds were provided from Selleck Chemicals (Houston, Texas, USA) with the exception of aphidicolin, colchicine, Mps1-IN-3, nocodazole, reversine, thymidine, UCN-01 (all purchased from Sigma-Aldrich), LY2606368 (also known as prexasertib, provided from Eli Lilly, Indianapolis, Indiana, USA) and PV1019 (obtained from Calbiochem-Merck Millipore, Billerica, Massachusetts, USA).
gutjnl-2016-312623supp001.pdf (5.3MB, pdf)
Compound screening and measurement of cell proliferation/viability
To determine the IC50 or drug-combination efficiency, dissociated cells were seeded in 96-well plates (6×103 cells/100 µL medium/well), cultured for 24 hours and then treated as indicated in the figure legends. Cell viability/proliferation was determined by evaluating the ATP levels via CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, Wisconsin, USA) with a multimode reader (DTX-880; Beckman Coulter, Brea, California, USA). For drug screening, CRC-SCs were seeded onto 96-well plates. Triplicate plates for each CRC-SC/drug-library plate combination were generated. Upon treatment for 72 hours, CRC-SC proliferation/viability was determined by CellTiter-Glo assay. Normalised viability for each drug (VD) was obtained, for each plate, by referencing luminescence values (LD) to the averaged luminescence values of Dimethyl sulfoxide (DMSO)/controls (mLC) using the formula: VD=(LD/mLC)×100. The z scores (ZD) were calculated by referencing each normalised viability replicate (VD) to the grand mean (Gm) and grand SD (Gsd) of all compounds using the formula: ZD=(VD-Gm)/Gsd. Drug screening data were analysed using the ‘R’ software (R-Foundation for Statistical Computing, Vienna, Austria).
Genetic and reverse-phase protein microarray analyses
For targeted deep DNA resequencing, the sequencing library was prepared with the Truseq Custom Amplicon Kit (Illumina, San Diego, California, USA), while sequencing was performed on a MiSeq instrument. FASTQ files were analysed with Miseq Reporter software using the somatic variant caller algorithm, and variant annotation was performed with the VariantStudio suite (all from Illumina). The OncoPrint was generated using custom ‘R’ scripting and complex heatmaps library (https://github.com/jokergoo/ComplexHeatmap). Microsatellite instability (MSI) analysis was performed with a reference panel of five fluorescent dye-labelled microsatellite primers (NR-21, BAT-25, MONO-27, NR-24, BAT-26) using the MSI Analysis System kit, Version_1.2 (Promega). Amplified fragments were detected by loading the PCR products for capillary electrophoresis using the ABI Prism 3500 Genetic Analyser and the POP-4 polymer (both from Applied Biosystems, Foster City, California, USA) according to manufacturer's instructions (Promega). MSI status was determined upon analysis with GeneMapper software, Version_4.1 (Applied-Biosystems). Reverse-phase protein microarray (RPPA) was performed as previously described.21
In vivo study
For xenograft studies, CRC-SCs were resuspended in 50% Matrigel Basement Membrane Matrix (BD Biosciences, Franklin Lakes, New Jersey, USA)/50% growth medium, and 5×105 cells were injected subcutaneously in the flank of mice as reported by De Angelis et al.20 When tumours were palpable (after 3/5 weeks), mice were randomised to control and treatment groups (10 mice/group) and treated subcutaneously with vehicle only (Captisol; CyDex Pharmaceuticals, La Jolla, California, USA), or 5 or 10 mg/kg of the mesylate salt formulation of LY2606368 (which has comparable in vitro activity and increased in vivo bioavailability). For in vivo rescue studies, CRC-SC-derived xenografts were harvested from vehicle-treated or LY2606368-treated mice (6 tumours/group), dissociated as single cells and injected into secondary mice as described above. All animals were left untreated and in vivo tumour growth and volumes were measured as reported by De Angelis et al.20
Immunohistochemistry (IHC)
Sections from formalin-fixed/paraffin-embedded CRC-SC-derived xenografts were stained as described in online supplementary information. Images were obtained using an Eclipse 55i microscope (Nikon, Melville, New York, USA) equipped with Eureka Interface System (Menarini, Florence, Italy). The levels of ataxia telangiectasia mutated serine/threonine kinase (ATM) and replication protein A (RPA)32 phosphorylation were evaluated in terms of proportion of expressing tumour cells (0–100%) and staining intensity (0:negative, 1+:weak, 2+:moderate, 3+:strong) and analysed as categorical variables. The two scores were multiplied (maximum=300) and the median score of all tumours was used to classify low-expressing and high-expressing CRC-SC-derived xenografts.
Statistical procedures
Unless otherwise specified, all experiments were performed in triplicate parallel instances and independently repeated at least three times, data were analysed with Microsoft Excel (Microsoft, Redmond, Washington, USA), GraphPad Prism (GraphPad Software, San Diego, California, USA) or ‘R’ software. Statistical significance of data from most in vitro and in vivo studies was evaluated by one-way or two-way ANOVA and Bonferroni multiple comparisons test (figures 1, 5, 7 and online supplementrary figures S2, S5–S9). Statistical analyses of RPPA and COMET data were performed by: Kruskal-Wallis non-parametrical test followed by Wilcoxon signed-rank test and p value adjustment using Benjamini-Hochberg's correction (false discovery rate, FDR); and/or factorial ANOVA (type-III sums of squares) followed by Tuckey's honestly significant difference (HSD) multiple comparison's test (figures 3, 4, 5 and online supplementary figures S4 and S10). The Pearson's χ2 test and Fisher’s exact test, when appropriate, were applied to evaluate the relationship between LY2606368 sensitivity and the phosphorylation of RPA32, ATM, the ploidy status, MSI status and tumour protein 53 (TP53/p53) mutational status (figures 4, 6 and online supplementary text) with SPSS software Version_21 (SPSS, Chicago, Illinois, USA).
Figure 1.
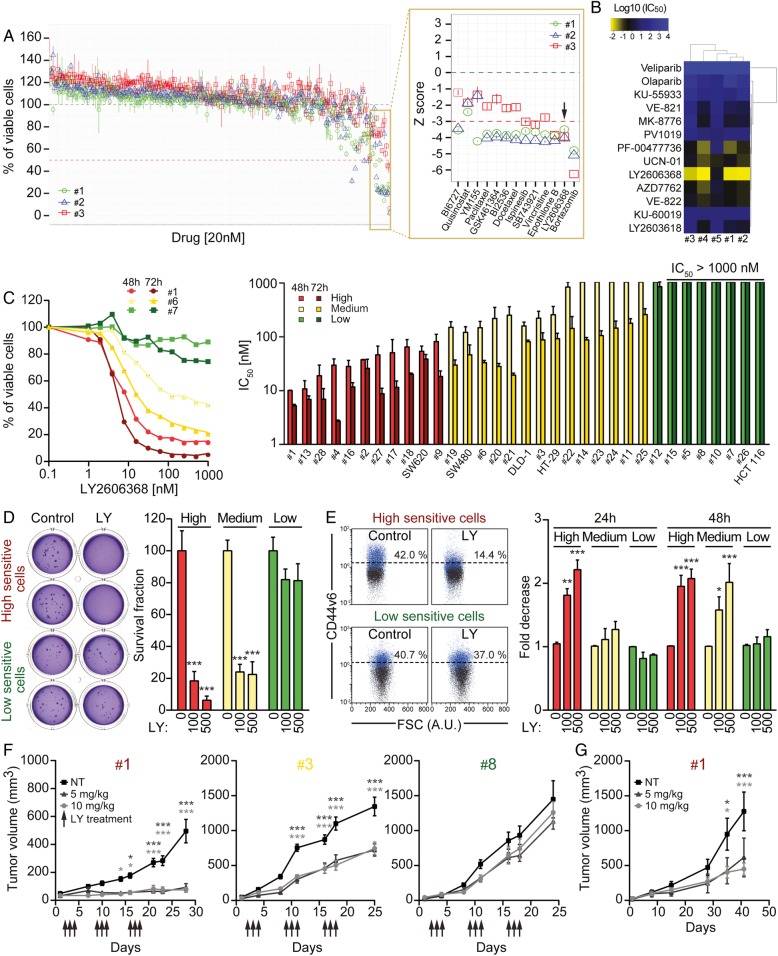
Identification of LY2606368 (LY) as a potent anti-colorectal cancer stem cells (CRC-SCs) agent, both in vitro and in vivo. (A) Three CRC-SCs (#1, #2 and #3) were left untreated or treated with a library of 305 compounds at 20 nM (see online supplementary table S2). After 72 hours, cell proliferation/viability was evaluated by means of CellTiter-Glo assay. Results are reported as means±SD of three technical replicates. In the magnification on the right, the corresponding normalised viability values for the most active compounds are shown as standardised z scores. Bortezomib, epothilone B and LY2606368 had the most potent anti-CSC activity as demonstrated by the reduction of cell viability below 50% or 3 SDs from the grand mean in all CRC-SCs analysed. (B and C) The depicted CRC-SCs (B,C) and American Type Culture Collection (ATCC) CRC cell lines (C) were left untreated or administered with a dose range of 13 specific pharmacological inhibitors of DNA damage response (DDR) players (B), or LY2606368 (C). Forty-eight hours (C) or 72 hours (B and C) later, CRC-SCs were assessed for their proliferation/viability by CellTiter-Glo assay. In (B), a hierarchical clustering of logarithmic scaled IC50 values obtained from dose-response curves in online supplementary figure S1 is illustrated. Panel (C) shows the dose-response curves of three representative CSCs (#1, #6 and #7) and a histogram reporting the IC50 values for the indicated CRC-SCs (means±SEM; n≥2). Bright and dark colour scales refer to end points at 48 hours and 72 hours, respectively. CRC-SCs were classified in three groups according to their sensitivity to LY2606368: high sensitive (IC50<100 nM at 48 hours and 72 hours of treatment), medium sensitive (IC50>100 nM at 48 hours of treatment and <500 nM at 72 hours of treatment), low sensitive/resistant (IC50>500 nM at 48 hours and 72 hours of treatment). (D) CRC-SCs from each class of LY2606368 sensitivity were left untreated or treated for 24 hours with LY2606368 at the depicted concentrations. Upon drug washout, CRC-SCs were seeded at low cell density and subjected to soft agar assay to evaluate their clonogenic potential. Cells were cultivated for up to 15 days in drug-free medium and then colonies were counted upon staining with crystal violet. Representative images of colonies formed from LY2606368-sensitive and LY2606368-resistant CRC-SCs as well as quantitative data (means±SEM; n≥3) obtained upon normalisation to plating efficiency of untreated cells are shown. At least three CRC-SCs from each group of LY2606368 sensitivity were employed. *p<0.05, **p<0.01, ***p<0.001 (two-way ANOVA and Bonferroni post hoc tests) as compared with the corresponding untreated CRC-SCs. (E) CRC-SCs differing for their LY2606368 sensitivity were left untreated or administered with LY2606368 as depicted, and then co-stained with the vital dye 4',6-diamidino-2-phenylindole (DAPI) and an antibody directed against the CRC-SC marker CD44v6 followed by cytofluorimetric analysis. Representative plots for one LY2606368-sensitive (#4) and one LY2606368-resistant (#7) CRC-SCs (numbers indicate the percentages of corresponding events) as well as a histogram reporting quantitative data (means±SEM; n=4) concerning the fold change decrease of CD44v6+ cells as compared with control conditions are reported. Only viable cells (ie, those excluding DAPI) were included in the analysis. At least two CRC-SCs from each group of LY2606368 sensitivity were employed. *p<0.05, **p<0.01, ***p<0.001 (two-way ANOVA and Bonferroni post hoc tests) as compared with the corresponding untreated CRC-SCs. AU, arbitrary units. (F) CRC-SC #1 (in vitro high sensitive), #3 (in vitro medium sensitive) and #8 (in vitro low sensitive) were injected subcutaneously into immunodeficient mice. When tumours were palpable, mice underwent subcutaneous injections of captisol (vehicle/control group, NT) or LY2606368 at 5 mg/kg or 10 mg/kg (three cycles of three consecutive days, bis in die, followed by 4 days of rest). Ten mice per group were employed. Arrows refer to LY2606368 injection. Tumour size was routinely monitored by means of a common calliper. Results are reported as means±SEM. *p<0.05, **p<0.01, ***p<0.001 (two-way ANOVA and Bonferroni post hoc tests) as compared with mice injected with the same CRC-SCs and treated with the vehicle. (G) CRC-SC #1-xenografted mice were treated as in (F). Upon the last treatment tumours were recovered and re-injected in immunodeficient mice (secondary xenografts). All secondary mice were left untreated and tumour growth (means±SEM) was routinely evaluated by means of a common calliper. *p<0.05, **p<0.01, ***p<0.001 (two-way ANOVA and Bonferroni post hoc tests) as compared with mice injected with #1-derived tumours recovered from vehicle-treated mice (NT). In (C–E) LY2606368-high, LY2606368-medium and LY2606368-low sensitive CRC-SCs are depicted in red, yellow and green, respectively.
Figure 5.
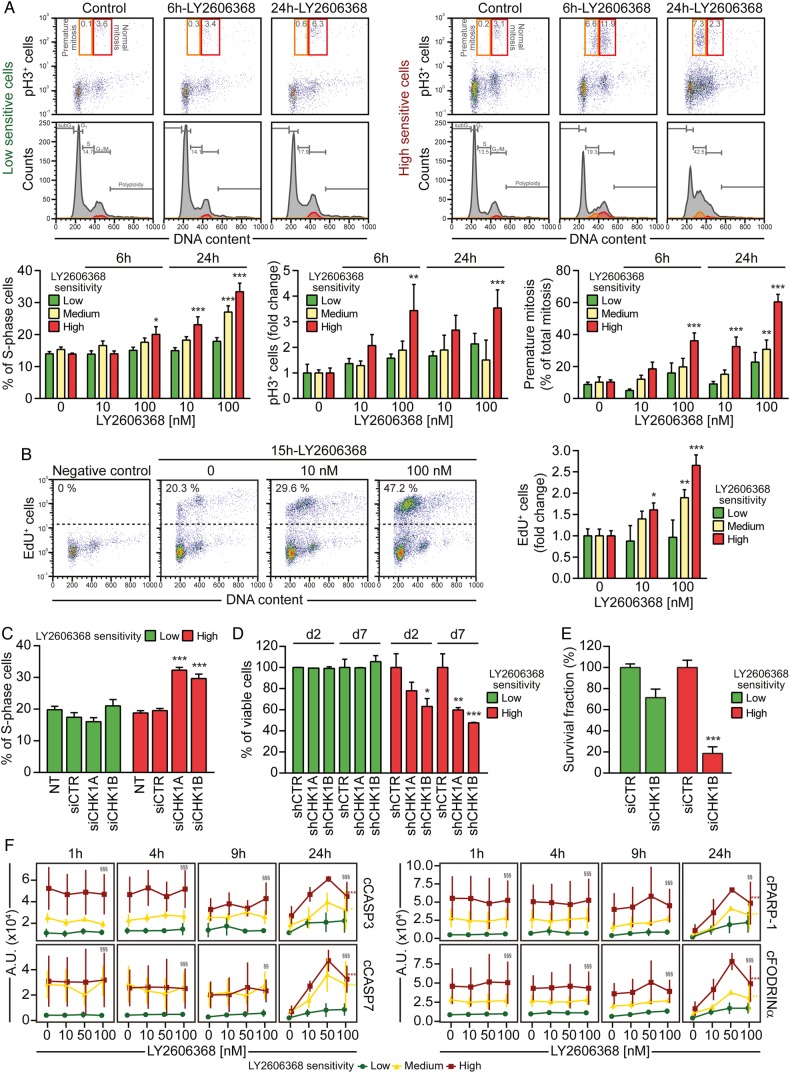
LY2606368 kills colorectal cancer stem cells (CRC-SCs) by inducing replication catastrophe and premature mitosis entry following checkpoint kinase (CHK)1 inhibition. (A) CRC-SCs from each class of LY2606368 sensitivity were left untreated or treated for 6 hours or 24 hours with LY2606368 at 10 nM or 100 nM, then fixed with ethanol and stained with the DNA dye 4',6-diamidino-2-phenylindole (DAPI) and an antibody directed against phospho-histone H3 (pH3_S10), for cytofluorimetric analysis of DNA content and mitotic cells. In the upper part, cell cycle profiles and dot plots of pH3+ cells with the indicated DNA content are shown for one representative high sensitive and low sensitive CRC-SC. Orange and red quadrants highlight premature mitosis (pH3+ cells with a <4n DNA content) and normal mitosis (pH3+ cells with a 4n DNA content) events, respectively. Numbers indicate the percentages of corresponding events. The histograms report quantitative data (means±SEM; n=4; at least three CRC-SCs from each group of LY2606368 sensitivity were employed) concerning the percentage or the fold change (as compared with control conditions) of S-phase, pH3+ cells and premature mitosis. Note that the sub-G1 fraction was excluded from the analysis. *p<0.05, **p<0.01, ***p<0.001 (two-way ANOVA and Bonferroni post hoc tests) as compared with the corresponding untreated CRC-SCs. (B) CRC-SCs differing for their LY2606368 sensitivity were left untreated or treated for 15 hours with LY2606368 as indicated, then fixed and labelled with the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU). Representative dot plots of EdU+ cells for LY2606368-high sensitive CRC-SCs and quantitative data (means±SEM; n=3; at least three CRC-SCs from each group of LY2606368 sensitivity were employed) for the three groups of sensitivity to LY2606368 are reported. Note that the sub-G1 fraction was excluded from the analysis. *p<0.05, **p<0.01, ***p<0.001 (two-way ANOVA test followed by Bonferroni post hoc test) as compared with the corresponding CRC-SCs left untreated. (C–E) CRC-SCs from each class of LY2606368 sensitivity were transfected with an unrelated small interfering (si) RNA (siCTR) or two specific siRNAs directed against CHK1 (siCHK1A and siCHK1B) (C and E) or, alternatively were transduced with lentiviral vectors expressing non-silencing short hairpin (sh) RNA (shCTR) or CHK1-targeting shRNAs (shCHK1A and shCHK1B) (D). Samples were collected, dissociated as single cells and then either analysed for their DNA content by flow cytometry upon staining with the DNA dye Hoechst 33342 (C), subjected to the cytofluorimetric assessment of mitochondrial membrane potential loss (a cell death-associated parameter) upon staining with Tetramethylrhodamine, methyl ester (TMRM) (D), or evaluated for their clonogenic potential by soft agar assay (E) as described in figure 1D. At least two CRC-SCs from a distinct group of LY2606368 sensitivity were employed in all the assays. Histograms reporting quantitative data (means±SEM; n≥3) concerning the percentage of S-phase cells or viable cells at the indicated time upon transfection/transduction are shown in (C) or (D), respectively. ‘d2’ and ‘d7’ refer to the day after the puromycin selection round. NT, non-transfected cells. In (C) the sub-G1 fraction was excluded from the analysis, while in (D) only the transduced cell population (green fluorescent protein+, GFP+ cells) was assessed. Quantitative data of clonogenicity (means±SEM; n≥2) obtained upon normalisation to plating efficiency of siCTR-transfected CRC-SCs are shown in (E). *p<0.05, **p<0.01, ***p<0.001 (two-way ANOVA and Bonferroni post hoc tests) as compared with the corresponding siCTR-transfected or shCTR-transduced CRC-SCs. (F) Reverse-phase protein microarray (RPPA) time-response and dose-response plots of cleaved Caspase-3 (cCASP3), cleaved Caspase-7 (cCASP7), cleaved poly(ADP-ribose) polymerase 1 (cPARP-1) and cleaved Fodrinα (cFODRINα) of CRC-SCs grouped by LY2606368 sensitivity. Results are shown as means±SD. For more insights on RPPA data statistical analysis refer to legend of figure 3B. LY2606368-high, LY2606368-medium and LY2606368-low sensitive CRC-SCs are depicted in red, yellow and green, respectively. A.U., arbitrary units.
Figure 7.
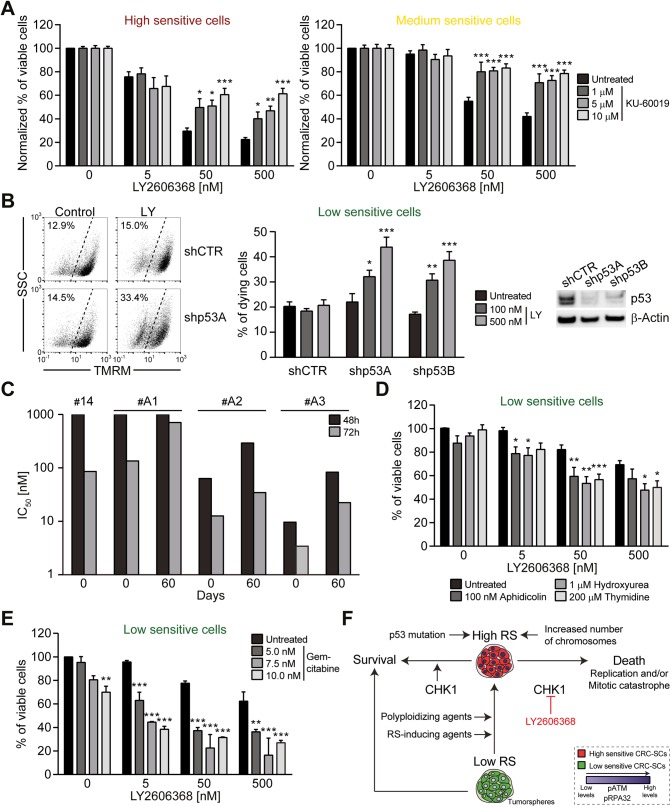
Strategies to modulate the sensitivity of colorectal cancer stem cells (CRC-SCs) to LY2606368. (A) Three representative LY2606368-high or LY2606368-medium sensitive CRC-SCs were left untreated or were treated with LY2606368 and/or the ataxia telangiectasia mutated serine/threonine kinase (ATM) inhibitor KU-60019 at the indicated dose. Upon 48 hours, CRC-SC proliferation/viability were assessed by CellTiter-Glo assay (means±SEM; n=5). The percentage of viable cells shown for co-treatments with KU-60019 and LY2606368 is normalised over the correspondent treatment with KU-60019 as single agent. Note that KU-60019 alone decreases CRC-SC viability of 0%, ∼9% and ∼27% at doses 1 µM, 5 µM and 10 µM, respectively. *p<0.05, **p<0.01, ***p<0.001 (two-way ANOVA and Bonferroni post hoc test) as compared with the corresponding CRC-SCs left untreated or treated only with LY2606368. (B) Representative p53-proficient CRC-SCs displaying resistance to LY2606368 (LY) were transduced with lentiviral vectors expressing non-silencing short hairpin (sh) RNA (shCTR) or p53-targeting shRNAs (shp53A and shp53B). Upon 5 days of selection with 1.5 µg/mL puromycin, cells were amplified, seeded and then left untreated or treated with the indicated concentration of LY2606368. Finally, cells were subjected to the assessment of mitochondrial membrane potential loss (a cell death-associated parameter) by flow cytometry upon staining with Tetramethylrhodamine, methyl ester (TMRM). Only the transduced cell population (GFP+ cells) was evaluated. Representative plots (numbers refer to the percentage of TMRMlow cells), quantitative data (means±SEM; n≥3), and western blot analyses performed with antibodies recognising the p53 or β-actin (whose levels were monitored to ensure equal loading of lanes) are reported. *p<0.05, **p<0.01, ***p<0.001 (two-way ANOVA and Bonferroni post hoc tests) as compared with CRC-SCs transduced with the same shRNA but left untreated. (C) CRC-SCs #14 were left untreated or treated with nocodazole for 48 hours, washed and then cultured in standard medium. Upon 2 weeks, cells were subcloned by limiting dilution to isolate diploid (2n) and tetraploid (4n) clones. The ploidy of proliferating clones was assessed as in figure 6C. The IC50 values of the parental cells (#14), one diploid (#A1) and two tetraploid clones (#A2 and #A3) were calculated by CellTiter-Glo assay after LY2606368 administration as indicated. Results from a single polyploidising series are reported. Note that following ∼30 days of culture #A2 and #A3 clones spontaneously reverted to near-to-diploidy decreasing their sensitivity to LY2606368. (D and E) Three representative LY2606368-low sensitive CRC-SCs were left untreated or treated with the indicated concentration of LY2606368 alone or together with sublethal doses of aphidicolin, hydroxyurea or thymidine (C), or three doses of gemcitabine (D). Cell viability, as assessed by CellTiter-Glo assay after 72 hours of treatment, is reported as mean±SEM (n≥3). *p<0.05, **p<0.01, ***p<0.001 (two-way ANOVA and Bonferroni post hoc tests) as compared with the corresponding CRC-SCs left untreated or treated only with LY2606368. (F) Proposed model accounting for LY2606368 sensitivity of CRC-SCs. RS, replication stress; pATM, phosphorylated ATM; pRPA32, phosphorylated RPA32.
Figure 3.
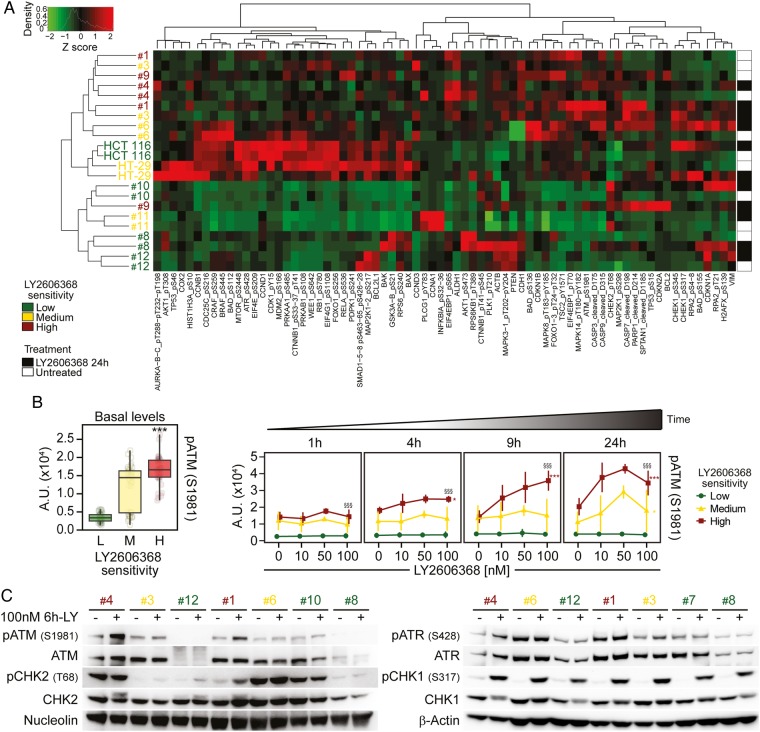
Phosphorylation of ataxia telangiectasia mutated serine/threonine kinase (ATM) as a marker of colorectal cancer stem cells (CRC-SCs) sensitivity to LY26063668. (A and B) Nine representative CRC-SCs, three from each group of LY2606368 sensitivity, and the ATCC cell lines HCT 116 and HT-29 were left untreated or treated with LY2606368 at low doses (10 nM, 50 nM and 100 nM) for 1 hour, 4 hours, 9 hours or 24 hours and then subjected to reverse-phase protein microarray (RPPA) analysis. Panel (A) represents the hierarchical clustering of RPPA results obtained on untreated CRC-SCs (white) and CRC-SCs exposed for 24 hours to 100 nM LY2606368 (black). In the left part of (B), basal levels of phosphorylated (p)ATM (pATM_S1981) in untreated CRC-SCs at all time points were pooled for each sensitivity group and box-plotted. Statistical analysis: Wilcoxon signed-rank test followed by p value adjustment by false discovery rate (FDR) for the comparison of high versus low sensitive CRC-SCs (*p<0.05, **p<0.01, ***p<0.001). In (B), on the right, the full time-dependent and dose-dependent RPPA kinetics of pATM are shown as means±SD of CRC-SCs grouped by LY2606368 sensitivity. Statistical analysis: factorial ANOVA design followed by p value adjustment (Tuckey's honestly significant difference (HSD)) for the comparison of (1) 100 nM LY2606368-treated versus untreated CRC-SCs of the same sensitivity group (*p<0.05, **p<0.01, ***p<0.001) or (2) high versus low sensitive CRC-SC treated with 100 nM LY2606368 (§p<0.05, §§p<0.01, §§§p<0.001). A.U., arbitrary units. (C) The illustrated CRC-SCs were left untreated (–) or administrated with 100 nM LY2606368 (LY) for 6 hours (+) and then subjected to western blot analyses with antibodies recognising the phosphorylated or total forms of ATM, checkpoint kinase (CHK)2, ataxia telangiectasia mutated and Rad3 related serine/threonine kinase (ATR) or CHK1. Nucleolin or β-actin levels were monitored to ensure equal loading of lanes. Note that six CRC-SCs, two from each sensitivity class, were the same used in RPPA studies. One representative western blot is shown. For the densitometric and statistical analysis of the western blots, refer to online supplementary figure S5 and online supplementary information. LY2606368-high (H), LY2606368-medium (M) and LY2606368-low (L) sensitive CRC-SCs are depicted in red, yellow and green, respectively.
Figure 4.
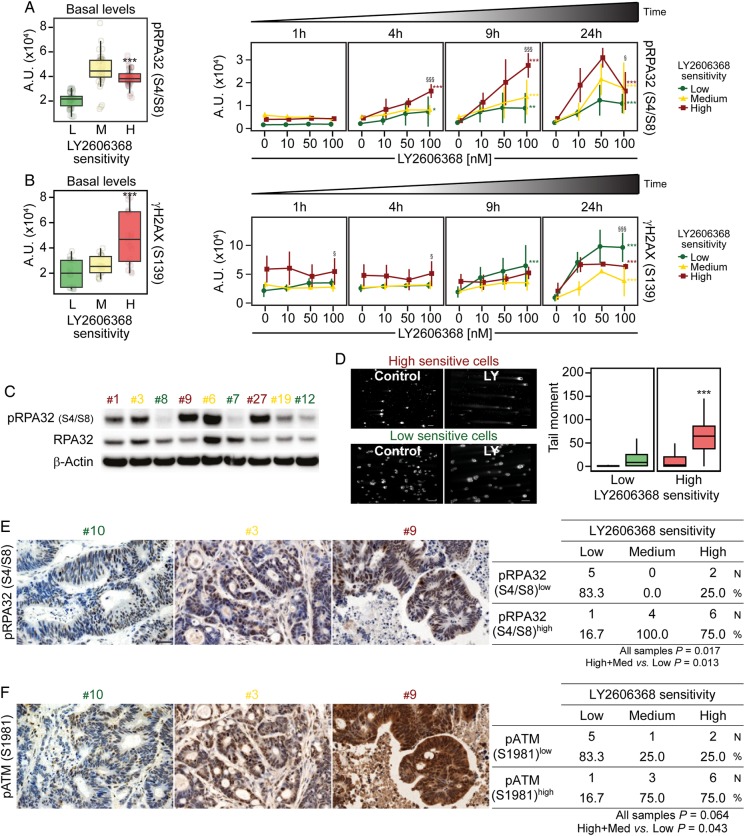
Endogenous DNA damage and replication stress predict the response of colorectal cancer stem cells (CRC-SCs) to LY2606368, both in vitro and in vivo. (A–C) reverse-phase protein microarray (RPPA) box plots or time-response and dose-response plots (A and B) and western blot analyses (untreated conditions) performed with antibodies recognising the phosphorylated (p) form of RPA32 (pRPA32_S4/S8) and total RPA32 (C) on a representative panel of CRC-SCs. In (A) and (B), the box plots on the left represent basal levels of pRPA32_S4/S8 or γH2AX of untreated CRC-SCs pooled for each sensitivity group, while line plots on the right report the RPPA kinetics of pRPA32_S4/S8, and γH2AX shown as means±SD of CRC-SCs grouped by LY2606368 sensitivity. For more insights on RPPA data statistical analysis refer to legend of figure 3B. A.U., arbitrary units. In (C) β-actin levels were monitored to ensure equal loading of lanes. One representative western blot is shown. For the densitometric and statistical analyses of the western blots, refer to online supplementary figure S5 and online supplementary information. (D) CRC-SCs from each class of LY2606368 (LY) sensitivity were left untreated or treated for 24 hours with 100 nM LY2606368, and then subjected to single cell gel electrophoresis (also known as COMET assay) to measure DNA lesions. The panel shows representative microphotographs of one sensitive CRC-SC (#4) and one resistant CRC-SC (#7) as well as box plots of the tail moments of at least 100 nuclei per experimental point measured by image analysis. Scale bar=100 µm. Statistical analysis: Wilcoxon signed-rank test followed by p value adjustment by false discovery rate (FDR) for the comparison of high versus low sensitive CRC-SCs (*p<0.05, **p<0.01, ***p<0.001). (E and F) Representative IHC images obtained from CRC-SC-derived xenograft sections stained with antibodies recognising pRPA32_S4/S8 (E) and phosphorylated ATM (pATM_S1981) (F) (scale bar=30 μm). Upon IHC analyses specimens were divided into two groups, one with low staining (pRPA32_S4/S8low or pATM_S1981low) and one with high staining (pRPA32_S4/S8high or pATM_S1981high) according to the median intensity values and by scoring on a continuous scale of 0–300. Numbers in the tables refer to the number (N) and the percentage (%) of samples in each group. The association between phosphorylation levels and LY2606368 sensitivity in all samples was evaluated by Pearson's χ2 test, whereas the comparison of high+medium (med) versus low sensitive CRC-SCs by Fisher's exact test. p values <0.05 were considered statistically significant. LY2606368-high (H), LY2606368-medium (M) and LY2606368-low (L) sensitive CRC-SCs are depicted in red, yellow and green, respectively.
Figure 6.
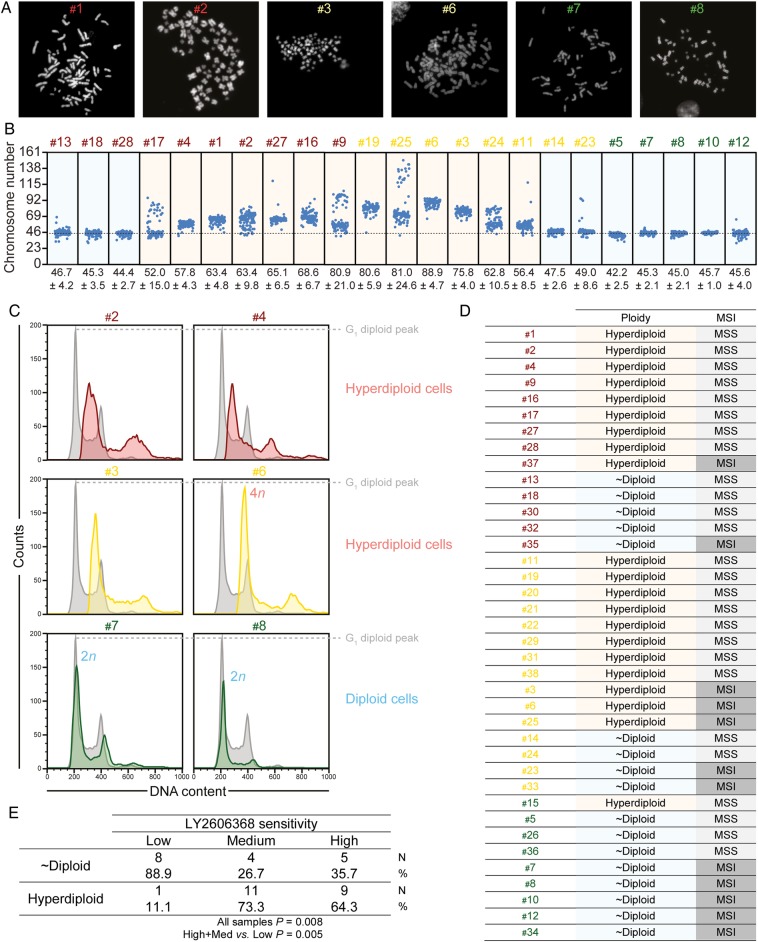
Hyperdiploidy is a cytogenetic marker of colorectal cancer stem cells (CRC-SCs) sensitivity to LY2606368. (A and B) A panel of CRC-SCs displaying high, medium or low sensitivity to LY2606368 (total=23) was treated for 4 hours with 5 μM colchicine and then subjected to metaphase spreading upon fixation and staining with Hoechst 33342. Representative metaphase images of six CRC-SCs and dot plots reporting the modal chromosome number (means±SEM; n=100) of all the CRC-SCs analysed are shown in (A) and (B), respectively. (C–E) A panel of CRC-SCs (total=38) was dissociated as single cells and then either stained with the DNA dye Hoechst 33342 or fixed with ethanol and stained with the DNA dye propidium iodide (C–E). Alternatively, their DNA was extracted for determining microsatellite instability (MSI) status (D). Representative cell cycle profiles of six CRC-SCs are shown in (C), while a table summarising the ploidy status, LY2606368 sensitivity and MSI status of all CRC-SCs analysed is reported in (D). The statistical analysis of the association between ploidy and LY2606368 sensitivity is illustrated in (E). HCT 116 cells (chromosome number=45; grey profiles) was used as internal diploid control. In (E), numbers in the table refer to the number (N) and the percentage (%) of samples in each group. The association between ploidy status and LY2606368 sensitivity in all samples was evaluated by Pearson's χ2 test, whereas the comparison of high+medium (med) versus low sensitive CRC-SCs by Fisher's exact test. p values <0.05 were considered statistically significant. The classes of sensitivity to LY2606368 are colour coded as follows: high/red, medium/yellow and low/green.
gutjnl-2016-312623supp003.pdf (170.8KB, pdf)
The detailed description of all materials and methods are reported in online supplementary information.
Results
Identification of the checkpoint kinase (CHK)1/CHK2 inhibitor LY2606368 as a potent in vitro and in vivo anti-CSC agent
To discover novel compounds targeting CSCs, we took advantage of a large collection of primary CRC-SCs12 characterised for the mutational status of recurrently CRC-altered genes, whose mutational frequency is consistent with that reported in The Cancer Genome Atlas (TCGA) (see online supplementary table S1) (http://cancer.sanger.ac.uk/cosmic/).22 Throughout this study only early passage CRC-SCs were used.
gutjnl-2016-312623supp002.pdf (91.3KB, pdf)
For the initial screening, we selected three CRC-SCs differing for Kirsten rat sarcoma viral oncogene homolog (KRAS) status, one wild type (#1) and two mutated (#2 and #3). Moreover, we manually curated a library of 305 clinically relevant (as mostly US Food and Drug Administration (FDA)-approved or investigational) compounds targeting most cancer-related pathways (see online supplementary table S2). CRC-SCs were treated with the full drug library at 20 nM and assessed 72 hours later for their proliferation/viability by measuring ATP levels. This high-throughput drug-sensitivity assay allowed us to identify three compounds exerting the most potent anti-CSC activity (figure 1A). The novel CHK1-CHK2 inhibitor LY260636823 was selected for further investigation, considering that: (1) the ATM-CHK2 and ATM and Rad3 related serine/threonine kinase (ATR)-CHK1 axes of the DNA damage response (DDR) were shown to be involved in CSC therapeutic resistance;24 25 and (2) LY2606368 demonstrated acceptable safety profile and preliminary evidence of antineoplastic activity in a recent phase I, dose-expansion study,26 and is being employed in ongoing phase I and phase II clinical trials (https://clinicaltrials.gov/).
Further confirming its potent anti-CRC-SC activity, in a dose-response study LY2606368 resulted the most effective DDR inhibitor in killing CSCs (IC50<13 nM in four out of five CRC-SCs) (figure 1B and online supplementary figure S1). When we extended our analyses to multiple CRC-SCs, we confirmed the dose-dependent and time-dependent anti-CRC-SC activity of LY2606368, and classified CRC-SCs in three categories according to their LY2606368-sensitivity: high sensitive, medium sensitive, low sensitive/resistant, with approximately a third of samples for each group (figure 1C and online supplementary table S3). Administration of LY2606368 significantly decreased the clonogenic potential of medium sensitive and high sensitive but not that of resistant CRC-SCs (figure 1D), and dwindled the fraction of cells expressing specific colorectal CSC markers—CD44v6 and ephrin B2—or displaying elevated WNT activity exclusively in LY2606368-sensitive CRC-SCs (figure 1E and online supplementary figure S2). Consistent with our in vitro approach, the in vivo growth of #1-derived xenografts and, to a lesser extent, that of xenografted #3 were arrested by the administration of LY2606368, whereas that of #8-derived xenografts was unaffected by LY2606368 (figure 1F). To confirm the decrease in the CSC fraction by LY2606368 administration, we recovered high sensitive CRC-SCs from LY2606368-treated mice and reimplanted cells to form secondary heterotopic xenografts. Such tumours displayed less clonogenicity and growth delay in comparison to those generated by CRC-SCs recovered from vehicle-treated mice (figure 1G).
gutjnl-2016-312623supp004.pdf (34.7KB, pdf)
Taken together, these results demonstrate that LY2606368 efficiently targets a large subset of CRC-SCs in vitro and in vivo by preferentially depleting CSCs.
Impact of DDR on LY2606368 sensitivity
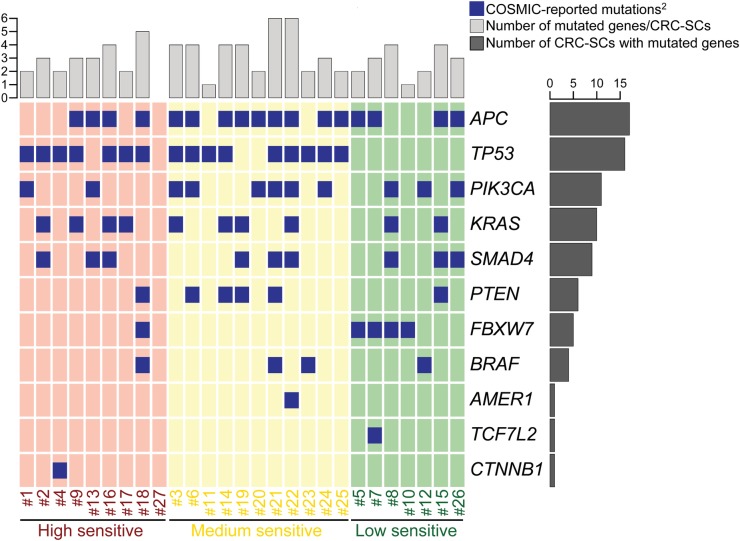
When we correlated drug sensitivity with genome sequencing data, we observed that mutations of TP53 were the most significant biomarker predicting CRC-SCs sensitivity to LY2606368 (p=0.001, Pearson's χ2 test, all samples analysed) (figure 2). However, a small percentage (20%, 4/20) of high sensitive and medium sensitive CRC-SCs did not show TP53 mutation indicating the existence of additional contributing factors.
Figure 2.
Oncoprint of mutations for the 16 most commonly mutated genes in colorectal cancer (CRC) found in CRC stem cells (CRC-SCs) by deep sequencing1. 1http://cancer.sanger.ac.uk/cosmic, online supplementary table S1. 2No Catalogue Of Somatic Mutations In Cancer (COSMIC) mutations were found for: ACVR1B, KIAA1804, MAP2K4, NRAS, SMAD2.
To identify other clinically relevant pharmacodynamic biomarkers of LY2606368 sensitivity, we thus performed a time-course RPPA with a panel of antibodies covering tumour-relevant pathways (see online supplementary table S4). We found that the constitutional activation of the DDR was crucial in conferring sensitivity to LY2606368 (figure 3A and online supplementary figure S3). Indeed, basal levels of ATM and ATR activating phosphorylation were significantly higher in LY2606368-high sensitive CRC-SCs than in LY2606368-resistant ones (box plots on the left in figure 3B and online supplementary figure S4). Moreover, LY2606368 administration induced a specific time-dependent phosphorylation of ATM (but not ATR) in LY2606368-responding (ie, high+medium sensitive) cells but not in unresponsive CRC-SCs (kinetics on the right in figure 3B and online supplementary figure S4) and an early increase of CHK1 phosphorylation in all CRC-SCs analysed (see online supplementary figure S4). A further investigation of the status of the ATR-CHK1 and ATM-CHK2 axis by western blot confirmed the significant high basal level of ATM phosphorylation in LY2606368-responding CRC-SCs as well as activation of ATM on LY2606368 administration in high sensitive CRC-SCs (figure 3C and online supplementary figures S5). Corroborating the relevance of ATM, whole-exome sequencing performed in a restricted number of CRC-SCs revealed that ATM was more frequently mutated in resistant (4/5) than in medium (2/5) or high sensitive (0/6) CRC-SCs (see online supplementary table S5).
gutjnl-2016-312623supp005.pdf (61.7KB, pdf)
gutjnl-2016-312623supp006.pdf (36.6KB, pdf)
Altogether, these findings indicate that the phoshorylation of ATM is a potential marker of sensitivity to LY2606368 in CRC-SCs.
Basal levels of endogenous DNA damage dictate the response of CRC-SCs to LY2606368
We then performed extensive analyses of DNA damage levels by means of RPPA, western blot and immunofluorescence studies. These analyses revealed that basal phosphorylation levels of RPA2/RPA32, a signalling intermediate of ongoing DNA replication stress (RS) response,27 and H2A histone family, member X (γH2AX), a double strand break (DSB)-sensitive marker, were significantly higher in LY2606368-sensitive than in LY2606368-resistant CRC-SCs (box plots on the left in figure 4A–C and see online supplementary figures S6–S8). These results indicate an elevated amount of basal endogenous DNA damage and RS in these cells. In addition, LY2606368 significantly induced DNA damage (figure 4D) and increased the levels of RPA32 phosphorylation and γH2AX in CRC-SCs (box plots on the right in figure 4A, B and see online supplementary figure S6). IHC analyses performed on paraffin-embedded sections of >15 CRC-SCs-derived xenografts confirmed a significant association between LY2606368 sensitivity and phosphorylation of ATM or RPA32 (p=0.043 or 0.013, respectively; high+medium vs low) (figure 4E, F).
Altogether, these results indicate that high basal levels of RS coupled to ATM phosphorylation represent reliable in vitro and in vivo markers for predicting the response of CRC-SCs to LY2606368.
Mechanisms of CSCs killing by, and of resistance to, LY2606368
We then explored the impact of LY2606368 on CRC-SC cell cycle progression. We observed that LY2606368 affected cell cycle distribution selectively in LY2606368-sensitive (high+medium) CRC-SCs by enriching the percentage of cells with a DNA content between 2n and 4n (figure 5A). S-phase accumulation was accompanied by a significant augmentation of the mitotic cell fraction (pH3+) in high but not medium sensitive or low sensitive CRC-SCs (figure 5A and online supplementary figure S4E). Moreover, upon LY2606368 exposure a considerable percentage of pH3+ cells in high+medium sensitive CRC-SCs displayed <4n DNA content, while the fraction of premature mitoses did not significantly vary among LY2606368-unresponsive CRC-SCs (figure 5A). LY2606368 induced a significant increase in the percentage of DNA-replicating (5-ethynyl-2′-deoxyuridine (EdU)+) cells only in responsive CRC-SCs (figure 5B), suggesting the deregulation of the replication process. These results, which are reminiscent of those found in the absence of CHK1,23 28 indicate that CHK1 is the main target of LY2606368. Accordingly, the depletion of CHK1 induced an accumulation of cells with a DNA content between 2n and 4n (figure 5C), triggered cell death (figure 5D and see online supplementary figure S9), and impaired the clonogenic potential (figure 5E) exclusively in LY2606368-sensitive CRC-SCs. The absence of the p53-dependant G1 checkpoint forces S-phase entry in the presence of DNA damage. In line with this evidence, the expression levels of the p53 target cyclin-dependent kinase inhibitor 1A (CDKN1A/p21) were higher in resistant than sensitive cells (see online supplementary figure S10). This confirms that p53 deficiency is a marker of LY2606368 sensitivity and that the p53 pathway protects from the lethal effect of LY2606368. CRC-SCs responding to LY2606368 could not endure cell cycle deregulation and eventually die for the activation of the caspase-dependent pathway of apoptosis (figure 5F).
Altogether these results indicate that LY2606368 kills CRC-SCs by inhibiting CHK1 resulting in the deregulation of DNA replication, impairment of cell cycle checkpoints and lethal replication catastrophe.
Impact of the ploidy status on the sensitivity of CRC-SCs to LY2606368
We then evaluated the impact of chromosomal content on LY2606368 activity. Through metaphase spreading, we found a heterogeneous modal chromosome number with ∼57% (13/23) CRC-SCs exceeding the near-to-diploid set (>50, ie, hyperdiploid) (figure 6A, B). Cell cycle profiling by flow cytometry confirmed the significant association between LY2606368 sensitivity and increased chromosome number (p=0.005; Pearson's χ2 test, high+medium vs low) (figure 6C–E). Thus, almost all hyperdiploid CRC-SCs (∼95.2%, 20/21) were high+medium sensitive to LY2606368, whereas 88.9% (8/9) of resistant CRC-SCs were near-to-diploid (figure 6D, E). As opposed to ploidy, MSI status was not significantly associated to the response of CRC-SCs to LY2606368 (p=0.108; Pearson's χ2 test, high+medium vs low) (figure 6D). Consistent with the role of p53 in controlling ploidy,29 ∼81% (13/16) of TP53-mutated CRC-SCs were hyperdiploid, while ∼73% (8/11) TP53 wild type CRC-SCs displayed near-to-diploid karyotypes (association TP53 mutation and ploidy status: p=0.043; Pearson's χ2 test) (figure 2). Taken together these results indicate that hyperdiploidy is a cytogenetic marker of LY2606368 sensitivity.
Strategies to modulate CRC-SC sensitivity to LY2606368
We finally assessed the relevance of the predictive markers identified above. We first evaluated the impact of ATM activity demonstrating that the pharmacological inhibition of ATM partially protected medium/high sensitive CRC-SCs from LY2606368 (figure 7A). Thereafter, we assessed the contribution of p53 showing that the constitutive knockdown of p53 via transduction of shRNA-expressing lentiviral vectors sensitised previously resistant CRC-SCs to LY2606368 (figure 7B). To analyse whether an artificial increase in cellular ploidy might confer sensitivity to LY2606368, we generated clones bearing a double DNA content (hereafter called tetraploid) from one p53-mutated (#14) and three p53-proficient (#7, #8, #10) near-to-diploid CRC-SCs with an optimised protocol.30 We obtained two tetraploid clones only from CRC-SC #14, both displaying an augmented sensitivity to LY2606368 (figure 7C). Corroborating the positive impact of RS on LY2606368 sensitivity, by perturbing DNA replication with low/sublethal doses of aphidicolin, hydroxyurea or thymidine, we were able to significantly sensitise previously non-responding CRC-SCs to LY2606368 (figure 7D). Along similar lines, LY2606368 cytotoxicity was highly increased in LY2606368-resistant CRC-SCs by coadministering gemcitabine (figure 7E).
These results confirm the reliability of the phosphorylation of ATM, p53 deficiency, hyperdiploidy and high RS as markers of LY2606368 sensitivity, thus validating our experimental strategy.
Discussion
In this study, after a pharmacological screening with therapeutic compounds on CRC-SCs, LY2606368 was identified as a potent anti-CRC-SC single agent acting independently of RAS mutational status. We also demonstrated that CHK1 targeting by LY2606368 potently killed CRC-SCs from approximately a third of tumours by preferentially depleting the CSC fraction. Cell death occurred via a mechanism involving the alteration and deregulation of DNA replication, which resulted in the generation of an intolerable DNA damage burden and induction of replication catastrophe.31 By correlating drug sensitivity with data coming from genome sequencing, RPPA and cytogenetic analyses, we identified four predictive and interlinked markers of CRC-SC sensitivity to LY2606368: (1) high basal levels of RS markers; (2) overactivation of the DDR player ATM; (3) mutations of TP53; and (4) hyperdiploidy (figure 7F).
Compelling evidence indicates that the DDR acts as a barrier during oncogenesis by limiting genomic instability induced by RS.32 Deregulated DDR pathways are frequently found in human neoplasms, where they are linked to increased genomic instability and therapeutic failure and/or sensitivity.22 33 34 Moreover, it is becoming evident that, once established, tumours overactivate the DDR to endure high levels of RS.35 CSCs efficiently activate the DDR24 25 and are responsible for tumour resilience. From a therapeutic perspective, here, we demonstrated that CRC-SCs displaying RS are highly dependent on the activity of CHK1, a kinase frequently overexpressed in neoplasms and regulating DNA replication, RS response and cell cycle progression.36 37 We surmise that the essential role of CHK1 in CRC-SCs is to maintain a high but tolerable level of RS (figure 7F). This ‘threshold level’ hypothesis is supported by two observations. First, LY2606368-mediated inhibition of CHK1 increased the level of RS and induced a lethal replication catastrophe31 exclusively in CRC-SCs responding to LY2606368. This occurred through a mechanism involving: (1) unscheduled DNA replication accompanied by the slowing down of the replication process and RS, ultimately leading to an excess of stretches of single-stranded DNA recognised by phosphorylated RPA32;31 (2) the deregulation of S-phase progression due to the impairment of the CHK1-dependent intra-S checkpoint; and (3) replication fork collapse eventually resulting in the accumulation of DSBs during replication. This evidence confirms recent observations demonstrating that CHK1, which is a key player for the correct execution and regulation of DNA replication, rather than CHK2, which operates mainly during the G2-M transition, is the major target of LY2606368.23 It also proves that LY2606368 induces DNA damage and, at the same time, inhibits the DDR. Second, strategies aimed at perturbing replication (eg, gemcitabine coadministration) sensitised non-responding CRC-SCs to LY2606368. These results, which are in line with previous observations,38 39 suggest that the activity of CHK1 becomes essential in CRC-SCs as a means to cope with enhanced RS.
In this preclinical study, CRC-SC sensitivity to LY2606368 is associated with the phosphorylation of ATM, suggesting a crucial role of ATM in coordinating the RS response, which is in line with previous findings.40 Accordingly, we found a significant association between the phosphorylation of ATM and RPA32, and responsiveness of CRC-SCs to LY2606368. These results confirm the crosstalk between the ATM-CHK2 and ATR-CHK1 cascade.27 Reportedly, ATR-CHK1 inhibitors displayed synthetic lethality with deficiency in other DDR players.41 42 Here, we provide evidence that the inhibition of ATM protects CRC-SCs to LY2606368, suggesting a functional role of ATM in the mechanism of CRC-SC killing by LY2606368. Further experiments are needed to elucidate the precise link between ATM and CHK1.
We also provided evidence of a cytogenetic cause of RS in tumours. CRC-SCs having a hyperdiploid chromosome set showed higher levels of RS than near-to-diploid CRC-SCs, which explains their exquisite sensitivity to LY2606368. We surmise that the imbalance/augmentation in copy number of multiple chromosomes is behind the increased RS displayed by hyperdiploid cells.43 44 Moreover, a hyperdiploid karyotype may lead to the deregulation of cancer-related proteins involved in RS response. In this context, hyperdiploid CRC-SCs always harbour mutation(s) in TP53, which may promote RS by favouring the generation/tolerance of hyperdiploidy or diminishing DNA repair efficiency. Accordingly, we demonstrated that the absence of p53 significantly increased the cytotoxicity caused by the inhibition of CHK1. Moreover, the few diploid CRC-SCs sensitive to LY2606368 mostly presented TP53 mutations and high RS levels, while p53-proficient near-to-diploid CRC-SCs constitutionally activated the p53 pathway to limit RS and LY2606368 toxicity. These findings are coherent with previous observations reporting enhanced sensitivity to CHK1 inhibition of tetraploid and/or p53-deficient cancer cells.45 46
Patient-derived cancer models are promising tools for drug discovery and clinical efficacy prediction.47–50 The major strength of this preclinical study is the vast collection of molecularly and functionally characterised CRC-SCs. This enabled us to (1) identify a clinically relevant agent with broad anti-CSC activity at low nanomolar doses, and effective against KRAS-mutated, non-hypermutated and/or p53-deficient CRC-SCs, (2) uncover and validate in vitro and in vivo candidate predictive markers of CRC-SC responsiveness, and (3) design strategies to overcome the intrinsic resistance to this therapeutic regimen. Importantly, LY2606368 demonstrated tolerable safety profiles and preliminary evidence of antineoplastic activity in a recent phase I, non-randomised, open-label, dose-escalation trial in patients affected by advanced or metastatic solid tumours who underwent at least three previous lines of treatment.26 In this study, partial response and disease stabilisation was obtained in 4.4% and 33.3% of patients, respectively. LY2606368 is currently employed alone or combined with other therapeutic agents in ongoing phase I or phase II studies also in patients with CRC (eg, NCT02203513, NCT02124148; https://clinicaltrials.gov/). Our preclinical study supports the further clinical development of LY2606368 as we demonstrated its potent anti-CSC activity in a subset of CRC-SCs and uncovered for the first time biomarkers associated with its efficacy in CRC. The prospective validation of the predictive value of these biomarkers may thus provide valuable background to the definition of prediction nomograms. In this context, the identification of reliable markers of CSCs and enhanced/ongoing RS response will allow to prospectively distinguish CSCs and replication-stressed CSCs within the tumour mass and thus confirm the true potential of this anti-CSC strategy.
Acknowledgments
The authors thank Emanuela Pilozzi for providing the paraffin-embedded xenograft sections, and Paola Di Matteo, Roberto Ricci and Stefano Guida for technical assistance. In vivo experiments were performed at Plaisant Castel Romano and Istituto Superiore di Sanità (Rome, Italy).
Footnotes
Contributors: GM, MS, AS, GR designed and performed experiments, analysed and interpreted data; FC, SS, MM, SV, MLDA, CAA, SDF, SB, GDL performed experiments; MP, EP, FDN, MF analysed data and performed bioinformatic studies; FS, MM-S analysed data and performed statistical analysis; MM, AZ, GS, MB, MT designed experiments; IV obtained funding, supervised the project, designed and performed experiments, analysed and interpreted data and wrote the manuscript; RDM obtained funding, supervised the project, designed experiments and wrote the manuscript.
Funding: This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC, MFAG 2013 grant number 14641 to IV, 5 per Mille grant number 9979 to GS, MT and RDM), Ministero Italiano della Salute (grant number RF_GR-2011-02351355 to IV), Ministero Italiano dell'Istruzione, dell'Universita e della Ricerca (MIUR, Programma per i Giovani Ricercatori ‘Rita Levi Montalcini’ 2010 to IV and Fondo per gli Investimenti della Ricerca di Base, FIRB, grant number RBAP11WCRZ-005 U54 2010 to RDM). AS is supported by AIRC (Start-Up 2016 #18418). GM is supported by AIRC (Triennial Fellowship ‘Antonietta Latronico’ 2014). AZ is supported by AIRC (grant number 15749). SDF is an AIRC fellowship recipient for 2016.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014;64:104–17. 10.3322/caac.21220 [DOI] [PubMed] [Google Scholar]
- 2.Montagut C, Dalmases A, Bellosillo B, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nat Med 2012;18:221–3. 10.1038/nm.2609 [DOI] [PubMed] [Google Scholar]
- 3.Bardelli A, Corso S, Bertotti A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov 2013;3:658–73. 10.1158/2159-8290.CD-12-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouwman P, Aly A, Escandell JM, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 2010;17:688–95. 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreso A, O'Brien CA, van Galen P, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 2013;339:543–8. 10.1126/science.1227670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalerba P, Kalisky T, Sahoo D, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 2011;29:1120–7. 10.1038/nbt.2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014;46:225–33. 10.1038/ng.2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010;141:69–80. 10.1016/j.cell.2010.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sottoriva A, Kang H, Ma Z, et al. A Big Bang model of human colorectal tumor growth. Nat Genet 2015;47:209–16. 10.1038/ng.3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell 2014;14:275–91. 10.1016/j.stem.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 11.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature 2013;501:328–37. 10.1038/nature12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature 2007;445:111–15. 10.1038/nature05384 [DOI] [PubMed] [Google Scholar]
- 13.Zeuner A, Todaro M, Stassi G, et al. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell 2014;15:692–705. 10.1016/j.stem.2014.11.012 [DOI] [PubMed] [Google Scholar]
- 14.Todaro M, Gaggianesi M, Catalano V, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014;14:342–56. 10.1016/j.stem.2014.01.009 [DOI] [PubMed] [Google Scholar]
- 15.Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer 2013;13:727–38. 10.1038/nrc3597 [DOI] [PubMed] [Google Scholar]
- 16.de Sousa E Melo F, Colak S, Buikhuisen J, et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 2011;9:476–85. 10.1016/j.stem.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 17.Merlos-Suárez A, Barriga FM, Jung P, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011;8:511–24. 10.1016/j.stem.2011.02.020 [DOI] [PubMed] [Google Scholar]
- 18.Dalerba P, Sahoo D, Paik S, et al. CDX2 as a Prognostic Biomarker in Stage II and Stage III Colon Cancer. N Engl J Med 2016;374:211–22. 10.1056/NEJMoa1506597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricci-Vitiani L, Pallini R, Biffoni M, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature 2010;468: 824–8. 10.1038/nature09557 [DOI] [PubMed] [Google Scholar]
- 20.De Angelis ML, Zeuner A, Policicchio E, et al. Cancer stem cell-based models of colorectal cancer reveal molecular determinants of therapy resistance. Stem Cells Transl Med 2016;5:511–23. 10.5966/sctm.2015-0214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marziali G, Signore M, Buccarelli M, et al. Metabolic/proteomic signature defines two glioblastoma subtypes with different clinical outcome. Sci Rep 2016;6:21557 10.1038/srep21557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King C, Diaz HB, McNeely S, et al. LY2606368 causes replication catastrophe and antitumor effects through CHK1-dependent mechanisms. Mol Cancer Ther 2015;14:2004–13. 10.1158/1535-7163.MCT-14-1037 [DOI] [PubMed] [Google Scholar]
- 24.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006;444: 756–60. 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- 25.Bartucci M, Svensson S, Romania P, et al. Therapeutic targeting of Chk1 in NSCLC stem cells during chemotherapy. Cell Death Differ 2012;19:768–78. 10.1038/cdd.2011.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong D, Infante J, Janku F, et al. Phase I study of LY2606368, a checkpoint kinase 1 inhibitor, in patients with advanced cancer. J Clin Oncol 2016;34:1764–71. 10.1200/JCO.2015.64.5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maréchal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res 2015;25:9–23. 10.1038/cr.2014.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam MH, Liu Q, Elledge SJ, et al. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell 2004;6:45–59. 10.1016/j.ccr.2004.06.015 [DOI] [PubMed] [Google Scholar]
- 29.Vitale I, Galluzzi L, Senovilla L, et al. Illicit survival of cancer cells during polyploidization and depolyploidization. Cell Death Differ 2011;18:1403–13. 10.1038/cdd.2010.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castedo M, Coquelle A, Vivet S, et al. Apoptosis regulation in tetraploid cancer cells. EMBO J 2006;25:2584–95. 10.1038/sj.emboj.7601127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toledo LI, Altmeyer M, Rask MB, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 2013;155:1088–103. 10.1016/j.cell.2013.10.043 [DOI] [PubMed] [Google Scholar]
- 32.Gorgoulis VG, Vassiliou LV, Karakaidos P, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 2005;434:907–13. 10.1038/nature03485 [DOI] [PubMed] [Google Scholar]
- 33.Aguilera A, García-Muse T. Causes of genome instability. Annu Rev Genet 2013;47:1–32. 10.1146/annurev-genet-111212-133232 [DOI] [PubMed] [Google Scholar]
- 34.Pearl LH, Schierz AC, Ward SE, et al. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer 2015;15:166–80. 10.1038/nrc3891 [DOI] [PubMed] [Google Scholar]
- 35.Norquist B, Wurz KA, Pennil CC, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol 2011;29:3008–15. 10.1200/JCO.2010.34.2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol 2014;16:2–9. 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer 2014;134:1013–23. 10.1002/ijc.28226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks K, Oakes V, Edwards B, et al. A potent Chk1 inhibitor is selectively cytotoxic in melanomas with high levels of replicative stress. Oncogene 2013;32:788–96. 10.1038/onc.2012.72 [DOI] [PubMed] [Google Scholar]
- 39.Murga M, Campaner S, Lopez-Contreras AJ, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol 2011;18:1331–5. 10.1038/nsmb.2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNeely S, Conti C, Sheikh T, et al. Chk1 inhibition after replicative stress activates a double strand break response mediated by ATM and DNA-dependent protein kinase. Cell Cycle 2010;9:995–1004. 10.4161/cc.9.5.10935 [DOI] [PubMed] [Google Scholar]
- 41.Al-Ahmadie H, Iyer G, Hohl M, et al. Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy. Cancer Discov 2014;4:1014–21. 10.1158/2159-8290.CD-14-0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwok M, Davies N, Agathanggelou A, et al. Synthetic lethality in chronic lymphocytic leukaemia with DNA damage response defects by targeting the ATR pathway. Lancet 2015;385(Suppl 1):S58 10.1016/S0140-6736(15)60373-7 [DOI] [PubMed] [Google Scholar]
- 43.Burrell RA, McClelland SE, Endesfelder D, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013;494:492–6. 10.1038/nature11935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vitale I, Manic G, Senovilla L, et al. Karyotypic aberrations in oncogenesis and cancer therapy. Trends in Cancer 2015;1:124–35. 10.1016/j.trecan.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 45.Ma CX, Cai S, Li S, et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Invest 2012;122:1541–52. 10.1172/JCI58765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toledo LI, Murga M, Zur R, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol 2011;18:721–7. 10.1038/nsmb.2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertotti A, Migliardi G, Galimi F, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov 2011;1:508–23. 10.1158/2159-8290.CD-11-0109 [DOI] [PubMed] [Google Scholar]
- 48.van de Wetering M, Francies HE, Francis JM, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015;161:933–45. 10.1016/j.cell.2015.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Francescangeli F, Patrizii M, Signore M, et al. Proliferation state and polo-like kinase1 dependence of tumorigenic colon cancer cells. Stem Cells 2012;30:1819–30. 10.1002/stem.1163 [DOI] [PubMed] [Google Scholar]
- 50.Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009;138:645–59. 10.1016/j.cell.2009.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2016-312623supp001.pdf (5.3MB, pdf)
gutjnl-2016-312623supp003.pdf (170.8KB, pdf)
gutjnl-2016-312623supp002.pdf (91.3KB, pdf)
gutjnl-2016-312623supp004.pdf (34.7KB, pdf)
gutjnl-2016-312623supp005.pdf (61.7KB, pdf)
gutjnl-2016-312623supp006.pdf (36.6KB, pdf)