Abstract
Background
Interactions between host immune cells and gut microbiota are crucial for the integrity and function of the intestine. How these interactions regulate immune cell responses in the intestine remains a major gap in the field.
Aim
We have identified the signalling lymphocyte activation molecule family member 4 (SLAMF4) as an immunomodulator of the intestinal immunity. The aim is to determine how SLAMF4 is acquired in the gut and what its contribution to intestinal immunity is.
Methods
Expression of SLAMF4 was assessed in mice and humans. The mechanism of induction was studied using GFPtg bone marrow chimaera mice, lymphotoxin α and TNLG8A-deficient mice, as well as gnotobiotic mice. Role in immune protection was revealed using oral infection with Listeria monocytogenes and Cytobacter rodentium.
Results
SLAMF4 is a selective marker of intestinal immune cells of mice and humans. SLAMF4 induction occurs directly in the intestinal mucosa without the involvement of the gut-associated lymphoid tissue. Gut bacterial products, particularly those of gut anaerobes, and gut-resident antigen-presenting cell (APC)TNLG8A are key contributors of SLAMF4 induction in the intestine. Importantly, lack of SLAMF4 expression leads the increased susceptibility of mice to infection by oral pathogens culminating in their premature death.
Conclusions
SLAMF4 is a marker of intestinal immune cells which contributes to the protection against enteric pathogens and whose expression is dependent on the presence of the gut microbiota. This discovery provides a possible mechanism for answering the long-standing question of how the intertwining of the host and gut microbial biology regulates immune cell responses in the gut.
Keywords: ANTIBIOTICS, ENTERIC INFECTIONS, GASTROINTESTINAL IMMUNE RESPONSE, INTESTINAL BACTERIA, MUCOSAL IMMUNITY
Significance of this study.
What is already known on this subject?
The constant changes imposed on the intestinal tract require different dynamics from the immune system at this site.
Signalling lymphocytic activation molecule (SLAM) receptors have an important role in the development of immune responses.
SLAMF4 is expressed by a small fraction (<8%) of haematopoietic-derived cells in the periphery, and most of its functions have been examined on natural killer (NK) cells.
What are the new findings?
SLAMF4 is expressed in the intestinal mucosa by conventional and natural T lymphocyte subsets, B cells and professional APCs (Mφ and DCs), in addition to NK cells and innate lymphoid cell types.
Gut bacterial products are efficient at increasing SLAMF4 expression on APCs but require the presence of gut-resident APCTNLG8A for SLAMF4 induction on lymphocytes.
SLAMF4 contributes to the regulation of gut immunity by promoting the production of proinflammatory cytokines during enteric infection.
How might it impact on clinical practice in the foreseeable future?
SLAMF4 is expressed by gut innate and adaptive immune cells involved in GI pathologies, and hence, this identification may expand the current list of targets that can facilitate the development of new intestinal mucosa-targeted therapeutics.
Our finding further supports the importance of a balanced gut microflora biodiversity in host immune homeostasis and suggests that prescribing oral antibiotics to patients, particularly those who are immunocompromised, has to be carefully weighed.
These findings suggest that phenotypical and functional analysis of SLAMF4 is warranted in human patients with immune-related intestinal diseases and may also lead to a better understanding of immune cell regulation mechanisms in human intestine.
Introduction
Gut microbes comprise more than 800 species that, as a whole, constitute the gut microbiota.1 In the intestinal tract, the microbiota contribute to the digestion of food, the provision of essential nutrients and to preventing the invasion of pathogens, as it represents the most frequent site of infection.1 2 To maintain this beneficial relationship, the mucosal immune system is likely to exert the means for tolerogenic regulation by inducing inhibitory molecules for immune signalling. On the other hand, because the gut is exposed to the environment, the risk of infection with exogenous pathogenic microorganisms is constant. Therefore, the mucosal immune system is likely to remain guarded and poised to turn on a quick attack on invasive pathogens by inducing activating molecules for immune signalling. However, the signalling molecules by which the gut immune system generates these simultaneously activating and inhibitory pathways, to switch between homeostatic, often immunosuppressive and barrier-protective, function and potent active immunity are not fully understood. In this regard, the most commonly accepted view is that such a dual function may occur as a result of the interactions between host immune cells and the gut microbiota.1 2
Natural killer cell receptors (NKR) are membrane proteins that provide specificity to NK cell responses in either an activating or inhibitory fashion.3 There are two major families of NKRs: NKRs that share homology with C-type lectins and killer cell Ig-like receptors, which include the signalling lymphocyte activation molecule family member 4, termed SLAMF4 (also known as CD244 and 2B4).3 4 The natural ligand for SLAMF4 is CD48, and in vitro engagement of SLAMF4 by CD48 induces cytotoxicity and cytokine secretion by human and mouse NK cells.5 6 The slamf4 gene can be alternately spliced into two protein products, differing in their intracellular domains, with affinities for adaptor molecules that initiate or inhibit signalling.7–10 One splice variant has a shorter intracellular domain and is activating, while the variant with the longer intracellular domain was shown to be inhibitory.7 8 Since there are two isoforms of SLAMF4 that differ in their signalling capacities, the relative amounts of these isoforms could dictate cell responsiveness to SLAMF4 ligation.7 8
Under normal physiological conditions, SLAMF4 is expressed by murine and human NK cells, but it is absent from most naïve CD4 and CD8T cells, B lymphocytes and neutrophils.7 11 12 However, other cell types such as mast cells, dendritic cells, skin γδ T cells, eosinophils and some activated CD8T cell subsets are SLAMF4+.11 13–16 In humans and mice, CD8+ T cells expressing SLAMF4 are absent from cord blood, and expression of SLAMF4 can be induced on only a small fraction of CD8+ T cells after in vitro activation or in vivo antigen challenge.10 17 Previously, we and others reported that in the steady-state condition, the vast majority (>95%) of conventional CD8+ T cells in the gut mucosa are SLAMF4+,18 19 while their peripheral counterparts are SLAMF4−.18 Because the intestine provides a unique environment for the development and function of immune cells,20–23 we sought to determine whether SLAMF4 expression in the gut mucosa extends to other immune cell populations, and if so, how it is acquired in the intestine and what its contribution to gut immunity is.
Results
SLAMF4 is a marker for intestinal immune cells
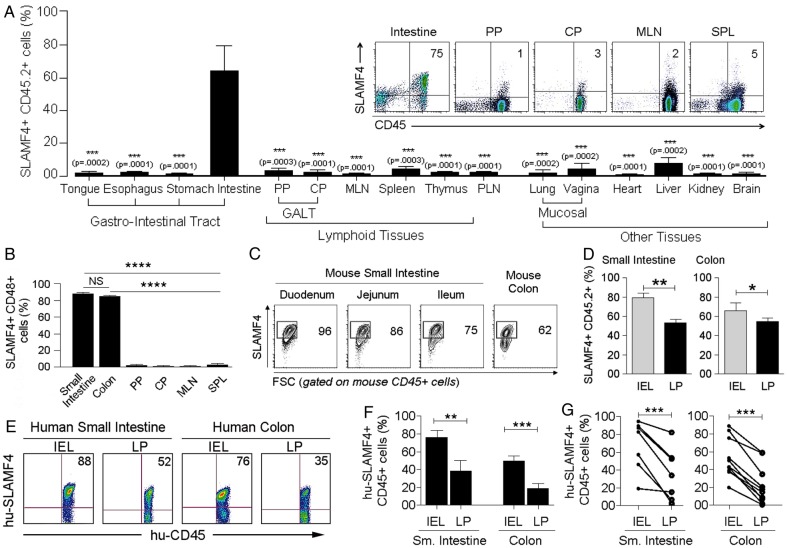
In the murine intestine, over 60% of haematopoietic-derived (CD45+) cells express SLAMF4, while this expression in other organs was <5% (figure 1A and see online supplementary figure S1a, b). In addition, over 80% of gut CD45+ cells were also positive for CD48, the only known ligand for SLAMF47 8 (figure 1B). SLAMF4 was highly expressed in both the intraepithelial (IEL) compartment and lamina propria (LP) of the small intestine and colon, but this expression was generally more enhanced among haematopoietic-derived cells of the IEL (∼70%) compartment than among those of the LP (∼50%) (figure 1C,D, and see online supplementary figures S2 and S3). In concordance with the murine data, SLAMF4 was found to be expressed at high levels in both human small intestine (n=10) and human colon (n=7) (figure 1E–G, and see online supplementary figure S4), suggesting that SLAMF4 can be used as a marker for intestinal immune cells in mice and humans.
Figure 1.
Signalling lymphocyte activation molecule family member 4 (SLAMF4) is a marker of intestinal immune cells. (A) SLAMF4 expression was assessed on haematopoietic-derived (CD45+) cells in the Peyer's patches (PP) and caecal patch (CP) which belong to the gut-associated lymphoid tissue (GALT), mesenteric lymph nodes (MLN), spleen (SPL), peripheral lymph nodes (PLN), as well as other organs as indicated. Bar graph summarises SLAMF4 expression as means of % CD45 cells that are SLAMF4+. (Inset) Dot plots show SLAMF4 expression by CD45+ cells in different tissues. (B) Gut mucosal CD45+ cells express both SLAMF4 and its ligand CD48. Data are presented as means of % SLAMF4+CD45+ cells that are CD48+. Data shown in (A, B) are from ten experiments using two to three mice per experiment for intestinal tissues and four experiments using two to three mice per experiment for other organs. (C) Dot plots show SLAMF4 expression by CD45+ cells in the intraepithelial (IEL) compartment of different segments of the small intestine and colon. Numbers indicate % SLAMF4+CD45+ cells. (D) Murine IEL and lamina propria (LP) fractions were prepared from the small intestine and colon and stained for flow cytometry. Bar graphs show SLAMF4 expression as means of % CD45+ cells that are SLAMF4+ in the IEL compartment versus those in the LP. Data shown in (C, D) are from n=6. (E) SLAMF4 expression was assessed on human small intestine (n=7) and colon (n=10) samples. Dot plots show SLAMF4 expression on human CD45+ cells in the IEL and LP compartments. (F) Data are shown as means of % human CD45+ cells expressing SLAMF4 and (G) relationship between IEL and LP for each tissue sample. Error bars represent SEM. A two-tailed Student's t-test distribution with paired groups of samples was evaluated for statistical significance. A value of p>0.05 is considered not significant (NS); *p<0.05, **p<0.005, ***p<0.005, ****p<0.00005.
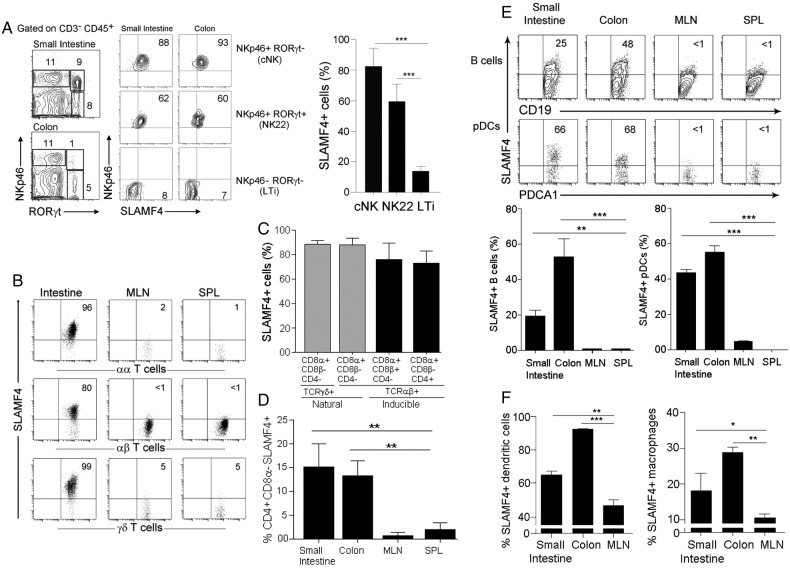
The finding that SLAMF4 is induced on most intestinal CD45+ cells raised a key question: which cell types express SLAMF4? To answer this question, we first assessed the expression of SLAMF4 on gut NK cell types to serve as positive control.7 As anticipated, we found that virtually all conventional (c) NK (NKp46+RORγt−) cells expressed SLAMF4 (figure 2A). Interestingly, while most NK-22 cells (NKp46+RORγt+) expressed SLAMF4, only a few LTi (NKp46−RORγt+) cells were positive for SLAMF4 expression (figure 2A). Thus, in conformity with the classification of SLAMF4 as an NK receptor,3 7 most cNK cells express SLAMF4. However, this expression was divergent among gut innate lymphoid cell subsets.
Figure 2.
Signalling lymphocyte activation molecule family member 4 (SLAMF4) is expressed by different immune cell types in the intestinal mucosa. (A) Lamina propria cells were first gated on CD45+CD3− cells. Dot plots show three NK cell subtypes identified based on the expression of NKp46 and the transcriptional factor RORγt. Numbers indicate the % of each cell subset (left) as well as % SLAMF4+ cells (right). Bar graph summarises % SLAMF4+ cells gated on each subtype. Data are from four experiments using three mice per experiment. (B) SLAMF4 expression was assessed on CD8αα+, CD8αβ+TCRαβ+ and TCRγδ+ T cells in the intestine, mesenteric lymph nodes (MLN) and spleen (SPL). Numbers indicate % of SLAMF4+ cells. (C) Summary data are shown as means of % gut CD8 T cell subsets that are SLAMF4+. Data are from four experiments using two mice per experiment. (D) Bar graph shows SLAMF4 expression on CD4+ cells in the intestine versus periphery. Data are presented as means of % CD4+ (CD3+CD4+CD8α−CD8β−) cells that are SLAMF4+. Data are from three experiments using three mice per experiment. (E) SLAMF4 expression on B cells (CD45+CD3-B220+CD19+) and plasmacytoid dendritic cells (pDCs, CD45+PDCA1+CD11c+). Below, bar graphs summarise FACS data. (F) Bar graphs show SLAMF4 expression on dendritic cells (DC, CD11c+CD19−CD3−F4/80−, left) and macrophages (Mφ, CD11c−CD11b+F4/80+, right). Data are shown as means of % DCs and Mφ that are SLAMF4+. Data shown in (E, F) are from three experiments using three to four mice per experiment. Data are shown as means±SEM. Error bars represent SEM. A two-tailed Student's t-test distribution with paired groups of samples was evaluated for statistical significance. *p<0.05, **p<0.005, ***p<0.0005.
Next, we examined the expression of SLAMF4 on T lymphocyte populations. Confirming our previous report,18 the vast majority (>80%) of γδTCR+, CD8αα+ and CD8αβ+ intestinal T cells were SLAMF4+ (figure 2B,C). In addition, SLAMF4 was also significantly expressed by gut mucosal CD4+ T cells, and this expression was eightfold to tenfold higher than the expression of SLAMF4 on peripheral CD4+ lymphocytes (figure 2D). Because T cells and NK cells alone cannot account for the high percentage of CD45+ cells expressing SLAMF4 in the intestinal mucosa, we examined whether other cell populations express SLAMF4. B lymphocytes and plasmacytoid dendritic cells (pDCs) are crucial for defence against bacterial and viral pathogens,24 25 and the gut is known to be a major reservoir in the body for these cell types.25 Previous studies showed that peripheral B cells lack SLAMF4 expression.11 12 Our phenotypic analysis of splenic B cells confirmed this observation (figure 2E). Strikingly, a significant portion of B cells in the gut mucosa expressed SLAMF4 (figure 2E). In addition, most intestinal pDCs were SLAMF4+ while their peripheral counterparts were SLAMF4− (figure 2E). Furthermore, over 60% of dendritic cells (DCs, CD11c+CD19−CD3−F4/80−) in the small intestine were SLAMF4+, and in the colon this expression was increased to >90% (figure 2F). Similarly, we found that some macrophages (Mφ, F4/80+CD11b+CD3−CD19−) expressed SLAMF4 in lymphoid organs (∼10%), and this expression was approximately threefold higher on intestinal macrophages (figure 2F and data not shown).
Together, these data show that SLAMF4 is expressed by a multitude of T lymphocyte subsets in the gut mucosa, in addition to NK and NK22 cells. In the periphery, NK cells express SLAMF4, but CD4 and CD8T cell expression of SLAMF4 is restricted to the intestine. We also show that professional APCs (Mφ and DCs) express higher levels of SLAMF4 in the gut mucosa when compared with those in the periphery, while SLAMF4 expression on B cells and pDCs is restricted to the intestinal mucosa. Therefore, while previous studies showed restricted SLAMF4 expression to a few cell types, the novelty of these findings is that SLAMF4 expression extends to most innate and adaptive immune cell populations of the intestinal mucosa.
Induction of SLAMF4 occurs directly in the intestinal mucosa
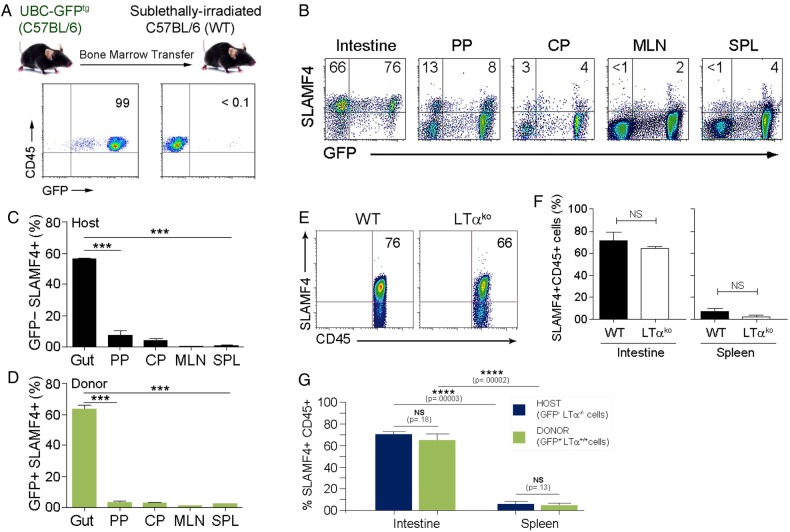
High expression of SLAMF4 among immune cells is a unique feature of the intestine (figures 1 and 2). Thus, we speculated that SLAMF4 might be induced on immune cells directly in the intestinal mucosa. To test this hypothesis, we generated bone marrow (BM) chimaeras using donor UBC-GFP transgenic (GFPtg) mice and sublethally irradiated major histocompatibility complex (MHC)-matched C57BL/6 recipients. Of note, GFPtg mice express GFP under the ubiquitin C promoter, and all haematopoietic-derived cells are GFP+26 (figure 3A). We found that GFP+ cells derived from donor cells expressed SLAMF4 at high frequencies in the intestinal mucosa (∼70%), but not in other tissues (<5%) (figure 3B, C). In addition, the GFP− host cells, expected to survive in sublethally irradiated recipient animals, also expressed SLAMF4 only at high frequencies in the intestinal mucosa (figure 3B, D), suggesting that the induction of SLAMF4 on intestinal CD45+ cells occurs directly in the gut mucosa.
Figure 3.
Signalling lymphocyte activation molecule family member 4 (SLAMF4) induction occurs directly in the gut mucosa and independently of the gut-associated lymphoid tissue (GALT). (A) B6.GFPtg bone marrow cells were adoptively transferred into sublethally irradiated B6 mice. Plots show GFP expression on donor (left) and host (right) CD45+ cells. Data are representative of n=10. (B) Eight weeks later, mice were euthanised to isolate cells from the gut mucosa, Peyer's patch (PP), caecal patch (CP), mesenteric lymph nodes (MLN) and spleen (SPL). Numbers indicate % of host (GFP−, top left) and donor (GFP+, top right) SLAMF4+ cells. Summary of data displayed in (B) are shown in (C) and (D), respectively. (E) Expression of SLAMF4 in the intestinal mucosa of B6.WT and B6.Ltαko mice. Data are representative of n=9. (F) Summary of data indicating SLAMF4 expression as means of % CD45+ cells in the intestine and spleen. (G) B6.GFPtg bone marrow cells were adoptively transferred into sublethally irradiated B6.Ltαko recipients as described in (A). Data are shown as means of % host (GFP−Ltα−/−) and donor (GFP+Ltα+/+) CD45.2+ cells that are SLAMF4+. Data are from three experiments using two (C,D,F) and three (g) mice per experiment. Data are shown as means±SEM. Error bars represent SEM. A two-tailed Student's t-test distribution with paired groups of samples was evaluated for statistical significance. p>0.05 is considered not significant (NS); *p<0.05, ***p<0.0005, ****p<0.00005. WT, wild type.
GFP+ cells migrating to the gut mucosa clearly become SLAMF4+ (figure 3A–D). However, it was unclear whether these cells were induced with SLAMF4 in the gut-associated lymphoid tissue (GALT) and mesenteric lymph nodes (MLN), and then migrated to the gut mucosa, or whether they were imprinted directly in the mucosa. To clarify this issue, we generated GFPtg BM chimaeras using sublethally irradiated MHC-matched C57BL/6 recipients lacking GALT and MLN, known as lymphotoxin-α (Ltα)-deficient mice.27 Our data showed no significant difference in the percentage of SLAMF4-expressing CD45+ cells of Ltαko mice when compared with their wild-type (WT) counterparts (figure 3E, F). Moreover, similar to GFP− host cells, GFP+ cells derived from GFPtg donor cells also expressed high levels of SLAMF4 in the intestinal mucosa of Ltαko recipients (figure 3G). Together, these data show that SLAMF4 expression on intestinal CD45+ cells occurs directly in the gut mucosa, and that GALT and MLN are largely dispensable for this process.
Intestinal microflora drive SLAMF4 expression
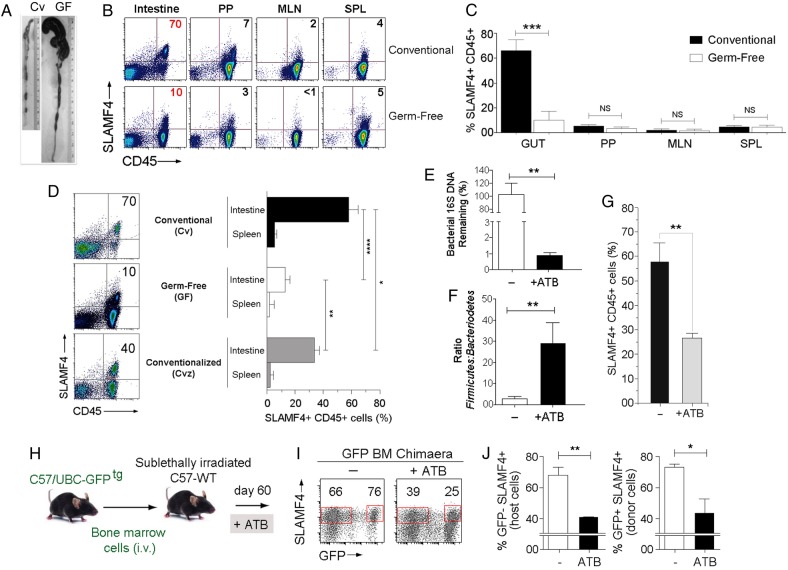
The intestine houses the largest population of commensal microorganisms in the body.1 2 We speculated that the difference noted in SLAMF4 expression between the gut mucosa and other organs might be related to the vast difference in numbers of commensal microbes available. Therefore, we compared the expression of SLAMF4 on CD45+ cells in the gut mucosa of conventionally (Cv) raised BALB/c mice to that of germ-free (GF) animals (figure 4A and28). SLAMF4 expression on haematopoietic-derived cells of the Peyer's patches (PP), MLN and spleen (SPL) was not significantly different between GF and Cv animals (figure 4B, C). Strikingly, SLAMF4 expression was almost completely abolished on intestinal CD45+ cells of GF mice (figure 4B, C), except on a small fraction (∼20%) constituted mainly by natural CD8αα T cells that remain SLAMF4+ (see online supplementary figure S5A).
Figure 4.
Signalling lymphocyte activation molecule family member 4 (SLAMF4) expression in the gut mucosa is dependent on the continued presence of the commensal microflora. (A) Representative photographs illustrating colon morphology in conventional (Cv) and germ-free (GF) BALB/c mice. (B) SLAMF4 expression on intestinal CD45+ cells is inhibited in GF animals, but there is no difference in SLAMF4 expression in Peyer's patches (PP), mesenteric lymph nodes (MLN) or spleen (SPL). (C) Summary of SLAMF4 expression on CD45+ cells shown in (B). (D) Dot plots show SLAMF4 expression on gut CD45+ cells in Cv, GF and conventionalised (Cvz) mice. Bar graph summarises data as means of % CD45+ cells that are SLAMF4+ in the intestine and spleen. Data shown in (B–D) are from three experiments using two mice per animal group per experiment. (E–G) B6 mice were left untreated (−) or treated with a combination of four antibiotics (ATB, ampicillin, metronidazole, neomycin, vancomycin) in the drinking water for 4 weeks. (E) Faecal DNA was made from intestinal contents and analysed for total bacterial contents. (F) Faecal DNA analysed for proportions (normalised to total bacteria) of Bacteroidetes and Firmicutes by qPCR. Data displayed are fold Bacteroidetes/Firmicutes. (G) Bar graph shows % gut CD45+ cells that remain SLAMF4+ after ATB treatment. Data shown in (E–G) are from five experiments using two mice per animal group per experiment. (H) B6.GFPtg bone marrow chimaera mice were left untreated or treated with ATB as described in (G). (I) Dot plots show SLAMF4 expression on intestinal cells of untreated and ATB-treated mice. Numbers indicate % SLAMF4+ cells. (J) Bar graphs summarise % CD45+ cells remaining SLAMF4+ among host (GFP−, right) and donor (GFP+, left) intestinal cells (n=5 per animal group). Overall data are shown as means±SEM. Error bars represent SEM. A two-tailed Student's t-test distribution with paired groups of samples was evaluated for statistical significance. *p<0.05, **p<0.005, ***p<0.0005, ****p<0.0001.
gutjnl-2016-313214supp001.pdf (3.3MB, pdf)
Next, we wanted to test whether SLAMF4 expression in the gut is plastic or developmental in nature. In the first set of experiments, we conventionalised GF mice to determine if SLAMF4 expression reappears in the gut mucosa. Our data revealed that recolonising the GF bowel with commensal microflora significantly increased SLAMF4 expression on intestinal CD45+ cells (figure 4D). However, the percentage of SLAMF4+ cells in conventionalised (Cvz) animals was still less than the percentage found in the gut of Cv animals. This is probably due to the fact that transferring faecal contents only partially replenishes GF mice with the total composition of the microflora, which is a known limitation of current procedures of gut microbiota transplantation.
In the second set of experiments, we tested whether altering the gut microflora of Cv mice through antibiotic (ATB) treatment could alter SLAMF4 expression in the gut mucosa (figure 4E–G, see online supplementary figures S5b and S6a). To this end, we treated Cv mice with a cocktail of four ATB (ampicillin, neomycin, metronidazole and vancomycin) for 4 weeks.29 While this treatment resulted in the ablation of most bacteria (figure 4E), the remaining bacteria were over-represented by the Firmicutes (a group consisting of Staphylococcus and Clostridium species among others)1 29 30 (figure 4F). Importantly, SLAMF4+ cells were significantly less frequent in the intestine of treated than untreated animals (figure 4G, see online supplementary figure S5b), suggesting that SLAMF4 expression can be altered by perturbation of normal microflora composition.
In the third set of experiments, we sought to investigate whether haematopoietic cells, recently developed in the gut mucosa and induced with SLAMF4, can also lose SLAMF4 expression when the gut microflora composition is disturbed. To this end, we treated GFPtg BM chimaera mice with ATB as indicated above (figure 4H). Our data revealed a significant reduction in SLAMF4+CD45+ cells among both the GFP+ donor cells and GFP− host cells in ATB-treated versus control chimaera mice (figure 4I, J). Together, these findings show that SLAMF4 expression on intestinal CD45+ cells depends on the continued presence of the normal gut microflora.
Role of TNLG8A costimulation in SLAMF4 induction
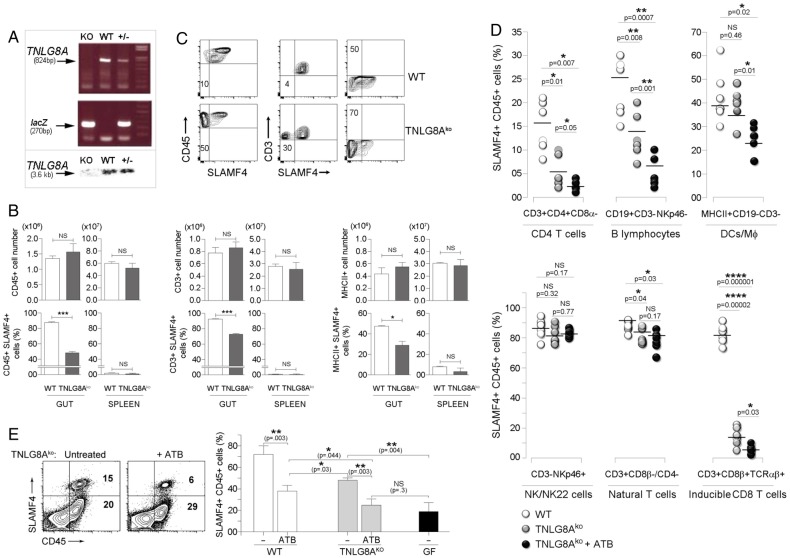
Previously, we showed that SLAMF4 expression on conventional CD8αβ+ T cells in the intestinal mucosa can be significantly diminished by injecting a blocking TNLG8A antibody.18 TNLG8A, the tumour necrosis factor ligand 8A, also known as CD70, is a costimulatory molecule that is constitutively expressed by gut-resident APCs.31 32 Thus, we hypothesised that SLAMF4 induction on conventional CD8T lymphocytes and potentially other cell types may require gut bacterial antigens to be presented to them in the context of TNLG8A costimulation. To test this possibility, we generated a TNLG8A-deficient mouse line using ES clones from the KOMP repository (figure 5A). Since differences in intestinal microbiota composition contribute to phenotypical and functional differences in mice,33 age-matched and sex-matched TNLG8Ako mice and WT littermates were cohoused to prevent potential intestinal microbiota changes. Overall analysis showed no significant difference in intestinal cell numbers between TNLG8Ako and WT littermates (figure 5B top). However, there was a significant decrease in SLAMF4 expression on intestinal cell populations of TNLG8Ako when compared with their WT counterparts (figure 5B bottom, C). Specifically, a lack of TNLG8A significantly reduced SLAMF4 expression on inducible CD8T cells (CD3+CD8β+TCRαβ+), CD4T cells (CD3+CD4+CD8α−TCRαβ+) and B cells (CD3-CD19+NKp46−), but had little or no effect on DC/Mφ (MHCII+CD19−CD3−) and NK/NK22 cells (CD3−CD19−NKp46+) (figure 5D). Still, the observed defect was only partial and did not fully recapitulate the phenotype observed in GF mice (figure 5E, left). Therefore, we tested whether disruption of the normal gut microbiota would have synergistic effects with TNLG8A gene deficiency. Following ATB treatment, the already decreased SLAMF4 expression in TNLG8Ako mice was further reduced to levels similar to those found in GF animals (figure 5E). With the exception of NK/NK22 cells and natural T lymphocytes (CD3+CD8β−CD4−), ATB treatment significantly reduced SLAMF4 expression on all other immune cell types (figure 5D). Collectively, data from figures 3–5 suggest that SLAMF4 expression is mostly dependent on gut microbial products that may require presentation in the context of TNLG8A costimulation in a way that does not require the GALT and MLN.
Figure 5.
Role of TNLG8A in signalling lymphocyte activation molecule family member 4 (SLAMF4) induction. (A) Lack of TNLG8A gene expression in knockout mice. Amplification of two PCR products, TNLG8A (824bp) and lacZ (270bp), revealed the deletion of the TNLG8A gene in knockout mice (top), and absence of the TNLG8A product (3.6 kb) by Southern blot analysis confirmed this gene deletion (bottom). (B) Pooled IEL and LP cells from wild-type (WT) and TNLG8Ako mice were analysed by flow cytometry. Bar graphs summarise SLAMF4 expression as means of numbers (top) and % (bottom) of total leucocytes (CD45+), T cells (CD3+MHCII−) and APCs (MHCII+CD3−) that are SLAMF4+. (C) Dot plots show SLAMF4 expression in the intestinal mucosa of WT and TNLG8Ako mice. Numbers indicate % SLAMF4− cells. Data shown in (A–C) are from three experiments using three mice per animal group per experiment. (D) WT and TNLG8Ako mice were left untreated or treated with ATB as described in figure 4E. Graphs summarise % SLAMF4+ cells in the intestinal mucosa of untreated WT (control) versus untreated and ATB-treated TNLG8Ako animals. Data show significant reduction of SLAMF4 expression on CD4T cells (CD3+CD4+CD8α−), inducible CD8T cells (CD3+CD8β+TCRαβ+), B cells (CD3−CD19+NKp46−) and DC/MΦ (CD3−CD19−MHCII+), but little or no decrease on natural CD8T cells (CD3+CD8α+CD8β−CD4−) and NK/NK22 cells (CD3−CD19−NKp46+). (E) Dot plots show SLAMF4 expression on gut leucocytes in untreated and ATB-treated TNLG8Ako animals. Bar graph summarises SLAMF4 expression as means of % intestinal CD45+ cells that are SLAMF4+ in untreated (−) and ATB-treated WT and TNLG8Ako mice in comparison with germ-free (GF) animals. Data are from three experiments using two (untreated) and three (ATB-treated) mice per animal group per experiment. Data are shown as means ±SD. Error bars represent SD. A two-tailed Student's t-test distribution with paired sample groups was evaluated for statistical significance. Value of p>0.05 was considered not significant (NS); *p<0.05, **p<0.005, ***p<0.0005, ****p<0.0001.
SLAMF4 imprinting on peripheral immune cells
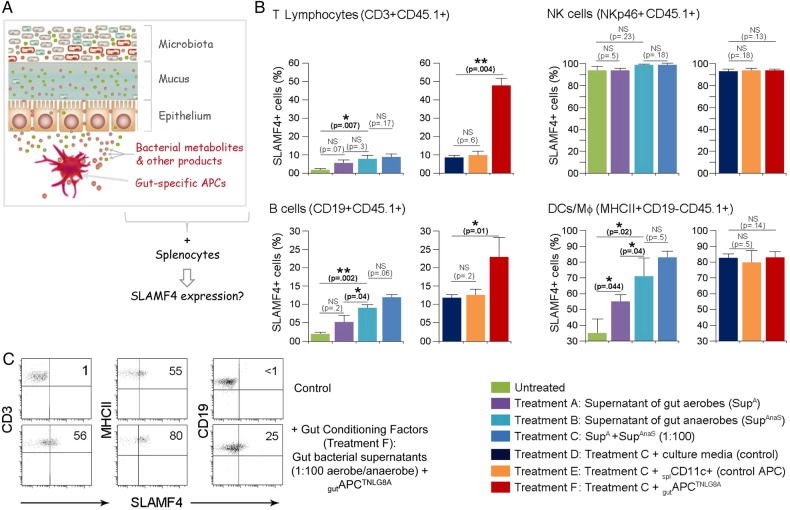
Secreted products, such as antigens34 and metabolites,22 35 36 by gut commensal bacteria are key contributors to the modulation of the host immune system. Therefore, we hypothesised that secreted gut microbial products and gut-resident APCTNLG8A could be key contributors to the induction of SLAMF4 expression on intestinal immune cells. We reasoned that if this hypothesis is true, then provision of these factors would enable the transfer of the gut phenotype (SLAMF4 expression) to systemic immune cells (figure 6A). To this end, we used in vitro culture systems of CD45.1 B6 splenocytes incubated with gut bacterial supernatants in the presence or absence of gutAPCTNLG8A or control APC (splenic CD11c+ cells). Briefly, supernatants of gut aerobes and anaerobes or control broths were used to stimulate magnetically purified CD45.1 splenocyte types (CD3+, CD19+, NKp46+ and MHCII+CD19−). Because the majority of the gut microbiota is composed of a mixture of anaerobes which dominate the aerobes by several orders of magnitude of at least up to 100-fold,37 we also tested the response to combined conditioned supernatants using a more physiological aerobe-to-anaerobe ratio of 1:100. Eighteen hours later, immune cell types were examined for SLAMF4 expression by flow cytometry. As anticipated, SLAMF4 expression on NK cells (NKp46+) was affected neither by stimulation with bacterial supernatants nor by the presence of APCs (figure 6B). In addition, stimulation with control broths exerted no detrimental effects on the cell viability and had no effect on SLAMF4 induction (see online supplementary figure S7). Importantly, we found that supernatants of anaerobes were more potent at inducing SLAMF4 than aerobic supernatants, and this phenotype was somewhat enhanced when a physiological aerobe-to-anaerobe ratio of 1:100 was used (figure 6B). Notably, SLAMF4 expression was found to be significantly increased on APCs (DCs and MΦ; MHCII+CD19−) when compared with T (CD3+) and B (CD19+) lymphocytes (figure 6B). However, provision of gutAPCTNLG8A drastically enhanced SLAMF4 expression on lymphocyte subsets, particularly T cells, but had little or no effect on APCs (figure 6B, C). Together, these data suggest that gut bacterial products, particularly those derived from gut anaerobes, are efficient at increasing SLAMF4 expression on APCs but require the presence of gutAPCTNLG8A for SLAMF4 induction on lymphocytes.
Figure 6.
Transfer of signalling lymphocyte activation molecule family member 4 (SLAMF4) expression on systemic immune cells. (A) The hypothesis that is being tested is that gut bacterial products and gut-specific APCs provide the environmental conditions for SLAMF4 induction in the intestine, and provision of these gut signals enables the transfer of this phenotype to peripheral immune cells. (B) T lymphocytes (CD3+), natural killer (NK) cells (NKp46+), B cells (CD19+) and other APCs (CD19-MHCII+) were magnetically purified from CD45.1 splenocytes. Subsequently, they were cultured with 8 U (normalised value of OD600×μL) of aerobe and anaerobe supernatants in the presence or absence of CD45.2+ APCs (splCD11c+ cells or gutAPCTNLG8A). Eighteen hours later, CD45.1+ cells were examined for the expression of SLAMF4 by flow cytometry. Data are shown as means of % CD45.1+ cells±SD. Error bars represent SD. Data are the summary of four experiments using two mice per treatment per experiment. (C) CD45.1 splenocyte types were treated with bacterial broths in the presence of (CD45.2+) splCD11c+ cells (control, upper) or with gut bacterial supernatants (1:100 anaerobe/aerobe) in the presence of gutAPCTNLG8 (bottom). Dot plots show expression of SLAMF4 on T (CD3+) cells, B (CD19+) cells and other APCs (CD19-MHCII+). Numbers indicate % SLAMF4+ cells. A two-tailed Student's t-test distribution with paired sample groups was evaluated for statistical significance. A value of p>0.05 was considered not significant (NS); *p<0.05, ***p<0.0005.
Lack of SLAMF4 renders mice more susceptible to enteric infections
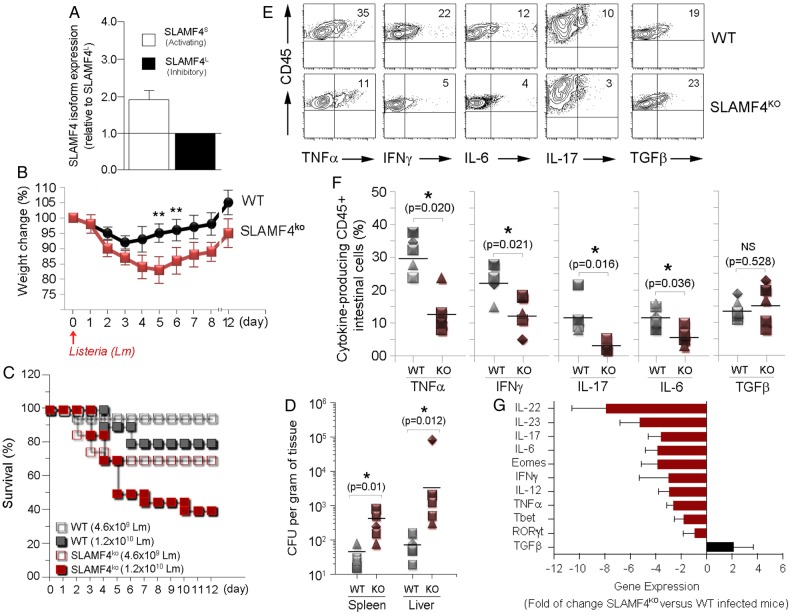
There are two distinct SLAMF4 proteins which are generated by the long (L) and short (S) isoforms.7–9 SLAMF4S is reported to be an activating receptor6 7 38 while SLAMF4L is known to be inhibitory.4 7 38 Due to the high levels of SLAMF4 expression in the gut, we were interested to know which isoform dominates in the intestine. To this end, we extracted RNA from magnetically purified intestinal CD45+ cells and performed QPCR for both splice variants. In the gut mucosa, approximately two-thirds of the expressed SLAMF4 transcripts are of the short isoform (figure 7A), suggesting that SLAMF4 expression in the intestine favours the activating splice variant of SLAMF4.
Figure 7.
Lack of signalling lymphocyte activation molecule family member 4 (SLAMF4) renders mice susceptible to Listeria monocytogenes (Lm) infection. (A) RNA was isolated from murine gut CD45+ cells. Subsequently, cDNA was generated, and qPCR was performed using primers for the activating (SLAMF4S) and inhibitory (SLAMF4L) isoform and normalised to β-actin expression. Each sample was performed in triplicate. (B–F) Wild-type (WT) and SLAMF4ko mice were infected orally with 0.46–1.2×1010 CFU of Lm and monitored for 12 days post infection. Results show (B) body weight change, (C) % survival and (D) bacterial load in the spleen and liver on day 6 post infection with 1.2×1010 CFU Lm. (E–G) Infected SLAMF4ko mice and WT littermates were euthanised on day 4.5 post infection. (E) Pooled intraepithelial and lamina propria suspensions were restimulated with Lm and incubated in the presence of protein transporter inhibitor GolgiStop overnight. Dot plots show cytokine-producing CD45+ cells. (F) Data are presented as means of % cytokine-producing CD45 cells±SD. Error bars represent SD. Data shown in (B–F) are from five experiments using 4–6 (SLAMF4ko mice) and 2–4 (WT littermates) per group per experiment. (g) Intestines were harvested and homogenised to obtain cDNA for the quantitative gene expression analysis of cytokines and transcriptional factors. Results are shown as the fold of change of each gene expression in Lm-infected SLAM4ko mice versus Lm-infected WT littermates. Data are from n=9 per animal group and expressed as means±SD. Error bars represent SD. Statistical significance was evaluated using a two-tailed Student's t-test (A) and Mann-Whitney test (B–D) distribution with paired groups. *p<0.05 was considered significant, and **p<0.005. IFN, interferon; IL, interleukin; TGF, transforming growth factor; TNF, tumour necrosis factor.
Since most gut immune cells express SLAMF4 and two-thirds of the expressed transcripts are of the activating isoform, we investigated whether SLAMF4 has protective functions against enteric pathogens. To address this issue, we orally infected age-matched and sex-matched groups of SLAMF4ko and WT mice with Listeria monocytogenes (Lm). Overall data showed significant weight loss (figure 7B) and increased mortality (figure 7C) with higher bacterial burden (figure 7D) in SLAMF4ko mice when compared with WT littermates. At the collective cellular level, immune susceptibility of SLAMF4ko mice to Lm was associated with decreased production of proinflammatory cytokines such as interferon (IFN)-γ, tumour necrosis factor (TNF)-α, interleukin (IL)-6, IL-17 and IL-12 (figure 7E–G, and see online supplementary figure S8b) nor from differences in intestinal microbiota composition since SLAMF4ko animals were cohoused with WT littermates.
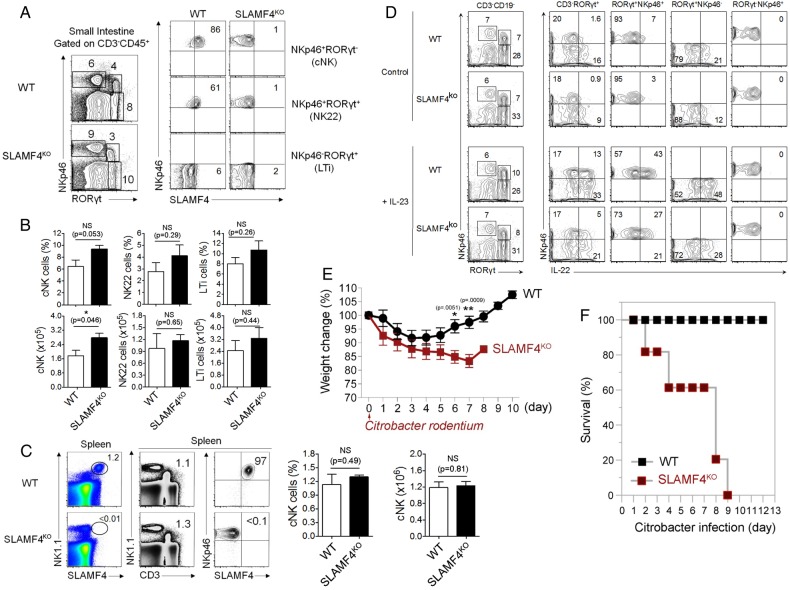
Another critical consequence resulting from the lack of SLAMF4 expression is the drastic reduction of IL-22 expression (figure 7G). This cytokine plays a critical role in mucosal immune defence that is mainly produced by group 3 innate lymphoid cells termed ILC3.39 In this regard, our analysis showed little or no difference in the number and percentage of cNK cells and ILC3 between SLAMF4ko and WT mice (figure 8A-C), suggesting that decreased levels of IL-22 expression in SLAMF4ko are not the result of reduced numbers of innate lymphoid cells in mice lacking SLAMF4. Accordingly, we also confirmed the reduction of IL-22 production by SLAMF4−/− ILC3 in vitro (figure 8C). Since IL-22 production by ILC3 cells depends on IL-23 production by gut CD103+ DCs,40 we also assessed whether IL-23 expression is altered in Lm-infected SLAMF4ko mice. Consistent with the alteration of IL-22 production, IL-23 expression was drastically reduced in SLAMF4ko mice compared with WT littermates (figure 7G), and this phenotype was not the result of reduced numbers of CD103+DCs in mice lacking SLAMF4 (data not shown). Based on these findings, we hypothesised that lack of SLAMF4 may lead to impaired IL22/IL23-mediated intestinal immunity. To test this hypothesis, we used the infection model of Citrobacter rodentium, which, unlike Lm,41 largely depends on IL22/IL23-mediated immunity.39 42 To this end, SLAMF4ko and WT mice were orally infected with C. rodentium and monitored for survival. While WT mice maintained a healthy body weight in response to infection, SLAMF4ko lost a considerable weight (figure 8E). Importantly, the failure of SLAMF4ko to resist C. rodentium resulted in premature death, as indicated by 100% in response to 2×1010 colony-forming unit (CFU) of the pathogen, while all WT animals efficiently resisted C. rodentium and survived this dose of infection (figure 8F).
Figure 8.
Lack of signalling lymphocyte activation molecule family member 4 (SLAMF4) leads to impaired IL-22-mediated gut immunity. (A) Dot plots of lamina propria cells show three NK cell subtypes identified based on the expression of NKp46 and the transcriptional factor RORγt. Numbers indicate % of each cell type (left) and % SLAMF4+ cells (right) in wild-type (WT) and SLAMF4ko mice. (B) Bar graph summarises % and number of cells. (C) Dot plots of splenocytes harvested from WT and SLAMF4ko show conventional (C) NK cells (NKp46+CD3−) that are expressing SLAMF4. Bar graphs summarise % (left) and total number (right) of cells gated on cNK cells. Data are shown as means of % SLAMF4+ cNK cells±SD. Error bars represent SD. Data shown in (A–C) are from n=8 (WT) and n=6 (SLAMF4ko) mice. (D) IL-22 production by ILC3 was assessed by in vitro stimulation of lamina propria cells with or without 10 ng/mL of IL-23. Dot plots show IL-22 production by NKp46+ILC3 (NKP46+RORγt+) of WT and SLAMF4ko mice. Data are representative of three experiments using two mice per strain per experiment. (E,F) WT and SLAMF4ko mice were infected orally with 2×1010 CFU of Citrobacter rodentium and monitored for 12 days post infection. Results show (E) body weight change and (F) % survival of SLAMF4ko versus WT mice. Data were obtained from two independent experiments using five (WT) to eight (SLAMF4ko) mice per animal group per experiment. Data are shown as means±SD. Error bars represent SD. A two-tailed Student's t-test distribution with paired groups was evaluated for statistical significance. Statistical significance was evaluated using a two-tailed Student's t-test (A–C) and Mann-Whitney test (E, F) distribution with paired groups. A value of p>0.05 was considered not significant (NS); *p<0.05, **p<0.005.
Discussion
Gut microbes are critical for inducing SLAMF4 on intestinal immune cells, and their continued presence is required for the maintenance of this expression. We came to this conclusion when we compared the expression of SLAMF4 on CD45+ cells in the gut mucosa of conventionally raised and GF mice. Interestingly, GF animals exhibit a drastic but not a complete decline in SLAMF4 expression among gut immune cells. Notably, CD8αα+CD8β−CD4− T lymphocytes remain SLAMF4+ in the intestinal mucosa of GF animals. These cells, often referred to as natural or unconventional IELs, are induced in the thymus with gut-homing molecules to migrate directly from the thymus to the gut epithelium.43 Interestingly, thymic natural CD8αα, but not inducible CD8αβ, are SLAMF4+.44 Therefore, it appears that the acquisition of gut-specific markers in the thymus is not limited to the induction of gut-homing receptors but extends to the induction of SLAMF4. Whether SLAMF4 plays a role in the thymic development of natural IELs and/or their post-thymic maturation and transition to the gut epithelium is completely unknown and requires further investigation.
Ablation of the gut microflora, either through GF husbandry or extensive antibiotic treatment is efficient at reducing SLAMF4 expression in the gut mucosa. These examples are extreme as it is impossible for any individual to be GF, and most individuals will never be subjected to such a stringent ATB regimen, while most will take a course of a single ATB at some point in their lives. In this context, our findings showed that even subtle gut microflora changes, such as those caused by antibiotic monotherapies and weight gain, can alter SLAMF4 expression (see online supplementary figure S6). From our standpoint, these findings point to an intriguing observation that the overall balance in the composition of the gut microbial community, and probably the presence or absence of cues from key species capable of affecting SLAMF4 expression, is important for ensuring optimal induction of SLAMF4 in immune cells or alteration thereof at the intestine. Cues from gut microbes are known to modulate the type of immune response, with major consequences for immune cell phenotype and function in the intestine22 35 36 and at sites distant from the gut mucosa such as the brain34 and joints.45 These are striking examples of a transfer of information from the gut microbiota to the intestine and from the intestine to distant sites. It appears, however, that SLAMF4 induction in gut immune cells by the microbiota is a phenomenon that is confined to the intestine, and data suggested that this immune phenotype is not exported to distant organs. This is probably due to the fact that SLAMF4 expression requires cues from intestinal microbes (eg, more than one gut microbial component) as well as gut-resident APCs, and migrating cells from the intestinal mucosa to distant tissues may be unable to export this ensemble of signals to transfer the gut phenotype.
Several features distinguish intestinal immune cells from their peripheral counterparts. For instance, systemic administration of soluble proteins without adjuvant induces differentiated T cells in the mucosa, but anergy in peripheral tissues.20 In addition, while B cell maturation to immunoglobulin (Ig)-producing cells occurs in the germinal centres of lymphoid organs, gut mucosal B cells expressing AID (activation-induced cytidine deaminase required for IgA class-switch recombination) may mature to IgA-producing plasma cells in the LP outside the germinal centres of the GALT.21 Moreover, while macrophages provide a stimulatory environment in the periphery, these innate cells generally provide an inhibitory milieu in the gut mucosa.22 Furthermore, despite the apparent suppressive gut environment, T lymphocyte responses to infections are more robust and more prolonged in the gut mucosa than in the periphery.23 Given that SLAMF4 is expressed by gut lymphoid and myeloid cells, it will be important to understand the possible role of this gene in the maintenance of gut immune homeostasis. Many studies of SLAMF4 function in peripheral NK cells have characterised the receptor as being inhibitory.4 In humans, SLAMF4 seems to have a role supportive of activation.5 There is a paucity of reports detailing expression of SLAMF4 and its splice variants on other cell types.11 13 14 17 Such studies found that the activating SLAMF4 splice variant is to be preferred by activated CD8+ T cells.10 17 These results are similar to our findings showing that expression of the activating splice variant is favoured in the intestine. In this regard, the high-level expression of the activating splice variant in the gut mucosa could be a mechanism by which intestinal immune cells counterbalance the suppressive gut environment to fight oral pathogens. Indeed, our studies suggest that SLAMF4 licenses gut mucosal immune cells to mount inflammatory responses against enteric pathogens by promoting proinflammatory cytokines production. Since a bi-directional signalling can occur through SLAMF4 and CD4817 46 47 and over 60% of gut immune cells are positive for both CD48 and SLAMF4, different scenarios of directional signalling may occur in the gut mucosa. For instance, SLAMF4 may (1) deliver signals to modulate the immune functions of neighbouring CD48+ intestinal cells, which as a whole constitute 86% of haematopoietic-derived cells of the gut mucosa, (2) receive signals from neighbouring CD48+ intestinal cells to stimulate SLAMF4+ immune cells or (3) simultaneously receive and deliver signals to modulate immune responses at an individual cell level and cell-type level, and at a cell community level to promote overall gut homoeostasis. Nevertheless, our understanding of the function of SLAMF4 is far more complex as other factors, in addition to the expression of splice variants, can determine whether the activating or inhibitory form can take a minor role. Such factors include the density and glycosylation changes of SLAMF4,48 the phosphorylation of the intracellular domain17 and the availability of signalling adaptors at the cell-type level.6 8 9 17 Currently, the status of these factors in the intestine is completely unknown.
In summary, the discovery that gut symbiotic bacteria induce intestinal immune cells with the SLAMF4 receptor exemplifies host–microbe communication within the intestine and further supports the importance of a balanced gut microflora biodiversity in host immune functions. There are, however, gaps in our knowledge that require greater understanding of the mechanisms upstream SLAMF4 through which gut microbial products act. We also do not know the extent to which SLAMF4 is significant in the competence of individual gut immune lineages (eg, whether SLAMF4 expression in only certain gut immune cell types is important for intestinal immune defence) as well as the extent to which the observed splice variant bias is functionally significant in intestinal immune functions. Still, the identification of SLAMF4 on most gut innate and adaptive immune cells involved in GI pathologies is an important discovery as it may expand the current list of targets that can facilitate the development of new intestinal mucosa-targeted therapeutics. In addition, the identification of SLAMF4 on human gut immune cells suggests that phenotypical and functional analysis of SLAMF4 is warranted in human patients with immune-related intestinal diseases and may lead to a better understanding of immune cell regulation mechanisms in human gut.
Methods
Mice
C57BL/6 (B6), B6.CD45.1, BALB/c, UBC-GFPtg, Ltαko and diet-induced obesity (DIO) mice were purchased from Jackson Laboratory. SLAMF4ko mice were kindly provided to us by Dr Dorothy Yuan, UT Southwestern Medical Center at Dallas. The Rutgers Transgenic Core provided the service of the rederivation of this strain. Age-matched and sex-matched SLAMF4ko and WT mice were cohoused to prevent potential intestinal microbiota changes. All mice were maintained in microisolator cages under specific pathogen-free conditions and used when they were 7–12 weeks of age. Germ-free BALB/c mice were purchased from the Medical University of South Carolina Gnotobiotic Animal Research Core Facility. All animals were housed in temperature-controlled, water-controlled and humidity-controlled cages that alternated between 12-hour light and dark cycles. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Rutgers University and Michigan University.
Generation of TNLG8A-deficient mice
The experimental set-up is described in detail in the online supplementary methods.
gutjnl-2016-313214supp002.pdf (107.3KB, pdf)
Generation of GFP BM chimaera mice
The experimental set-up is described in detail in the online supplementary methods.
Isolation of haematopoietic cells from mucosal tissues
The experimental set-up is described in detail in the online supplementary methods.
Isolation of haematopoietic cells from human intestines
The experimental set-up is described in detail in the online supplementary methods.
Isolation of haematopoietic cells from other tissues
We employed purification techniques described in Refs.49–51 The experimental set-up is described in the online supplementary methods.
Flow cytometry
Clones of SLAMF4 antibodies, B6 Alloantigen (BD Biosciences) and eBio244F4 (eBioscience) for C57BL-6 mice, and C9.1 for BALB-c mice were used in this study. The experimental set-up is described in detail in the online supplementary methods.
Gating strategy
Leucocyte types were identified based on the methods of Faucher et al by using similar orientating and specific gates as previously described.52 Details are described in the online supplementary methods.
Quantitative gene expression
QPCR reactions for the long and short splice variants of SLAMF4 were performed as described in Ref. 38. QPCR reactions for cytokines and transcriptional factors were performed using primers described in online supplementary table S1. The experimental set-up is described in detail in the online supplementary methods.
Sequences of oligonucleotide primers for analysis of cytokines and transcriptional factors. Primer sequence sets used in this study were obtained from Invitrogen
gutjnl-2016-313214supp003.pdf (232.3KB, pdf)
Oral infection with Listeria monocytogenes and Cytobacter rodentium
The experimental set-up is described in detail in the online supplementary methods.
Survival assays
The experimental set-up is described in detail in the online supplementary methods.
Direct intracellular staining
We employed the in vivo approach described in Ref. 53. The experimental set-up is described in detail in the online supplementary methods.
Antibiotic treatment
We employed the in vivo approach described in Ref. 29. The experimental set-up is described in detail in the online supplementary methods.
Conventionalisation of germ-free mice with normal gut microflora
The experimental set-up is described in detail in the online supplementary methods.
DNA isolation from mouse faecal material and QPCR
QPCR primers and conditions for Firmicutes, Bacteroidetes and all bacteria were described previously.30 The experimental set-up is described in detail in the online supplementary methods.
Culture of gut bacteria
Microbial supernatants were prepared as previously described54 using the oxygen scavenger OxyRase AnaSelect (OxyRase, Ohio) for anaerobic cultures.55–57 The experimental set-up is described in detail in the online supplementary methods.
Isolation of APCs and stimulation of splenocytes
To purify gut-resident APCTNLG8A, we employed the approach described in Ref. 31. Stimulation with gut bacterial supernatants was carried out as described in Ref. 54. The experimental set-up is described in detail in the online supplementary methods.
In vitro stimulation of LP cells
The experimental set-up is described in detail in the online supplementary methods.
Statistics
Software programs Microsoft Excel for PC and Prism for Mac (GraphPad, La Jolla, California, USA) were used to calculate the arithmetic mean, SEM and/or SD for each group of mice. Statistical significance was evaluated using a two-tailed Student's t-test or Mann-Whitney test distribution with paired groups of samples. A value of p<0.05 was considered significant.
Study approval
Animal handling and procedures were conducted according to animal protocols approved by the review committee at the Rutgers University and the facility used by the Child Health Institute to house mice as well as the review committee at the Michigan University. Testing of human intestinal samples was conducted according to IRB protocol approved by the review committee at Rutgers University and the Robert Wood Johnson Medical School. Written informed consent was excluded since the human samples were discarded after bowel surgery.
Acknowledgments
The authors thank Dr Yufang Shi (Child Health Institute of NJ, Rutgers University, New Brunswick, New Jersey, USA) for his critical reading of the manuscript; Dr Tae Joon Won (University of Michigan, Ann Arbor, Michigan, USA) for help with intestinal phenotype; Amira Esseghir, Mariel Watkins and Gaurav Singh for technical assistance with faecal QPCR and FCAP analysis. The authors thank Dr Dorothy Yuan (UT Southwestern Medical Center at Dallas, Texas, USA) for kindly providing SLAMF4ko mice which were originally given to her by Dr Michael Bennett (The University of Texas Southwestern Medical Center at Dallas, Dallas, Texas). The authors also thank Dr Bhaumik for rederiving this mouse strain and help with generating TNLG8Ako mice. They also acknowledge the support of the NIH-R01AI083642 to YL, the Robert Wood Johnson Foundation (grant # 67038) for the Child Health Institute of NJ and the Robert Wood Johnson Foundation (grant # 581534) to AL.
Footnotes
Contributors: AC, DS, MT, YJ, BC and AL performed the experiments. AL, DS, AC, YS, YL and BC analysed the data. YL provided reagents. AL and YL designed the experiments. AL, DS and AC wrote the manuscript.
Funding: NIH (R01AI083642 to YL, The Robert Wood Johnson Foundation_SG (grant # 581534 to AL), The Robert Wood Johnson Foundation (grant # 67038 for the Child Health Institute of NJ).
Competing interests: None declared.
Ethics approval: The review committee at Rutgers University and the Robert Wood Johnson Medical School.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: We do not have unpublished data from the study. However, as soon as the paper is accepted, we would wish to make our results available to the scientific community. In addition, we would welcome collaboration with others. Our plan includes also presentations at national and international scientific meetings. In addition, important data might be posted on our institutional website and published through our institutional newsletter.
References
- 1.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 2010;10:159–69. 10.1038/nri2710 [DOI] [PubMed] [Google Scholar]
- 2.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009;9:313–23. 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 2008;9:495–502. 10.1038/ni1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee KM, McNerney ME, Stepp SE, et al. 2B4 acts as a non-major histocompatibility complex binding inhibitory receptor on mouse natural killer cells. J Exp Med 2004;199:1245–54. 10.1084/jem.20031989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakajima H, Colonna M. 2B4: an NK cell activating receptor with unique specificity and signal transduction mechanism. Hum Immunol 2000;61:39–43. 10.1016/S0198-8859(99)00170-6 [DOI] [PubMed] [Google Scholar]
- 6.Bida AT, Upshaw Neff JL, Dick CJ, et al. 2B4 utilizes ITAM-containing receptor complexes to initiate intracellular signaling and cytolysis. Mol Immunol 2011;48:1149–59. 10.1016/j.molimm.2011.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schatzle JD, Sheu S, Stepp SE, et al. Characterization of inhibitory and stimulatory forms of the murine natural killer cell receptor 2B4. Proc Natl Acad Sci USA 1999;96:3870–5. 10.1073/pnas.96.7.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chlewicki LK, Velikovsky CA, Balakrishnan V, et al. Molecular basis of the dual functions of 2B4 (CD244). J Immunol 2008;180:8159–67. 10.4049/jimmunol.180.12.8159 [DOI] [PubMed] [Google Scholar]
- 9.Wang N, Calpe S, Westcott J, et al. Cutting edge: the adapters EAT-2A and -2B are positive regulators of CD244- and CD84-dependent NK cell functions in the C57BL/6 mouse. J Immunol 2010;185:5683–7. 10.4049/jimmunol.1001974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNerney ME, Lee KM, Kumar V. 2B4 (CD244) is a non-MHC binding receptor with multiple functions on natural killer cells and CD8+ T cells. Mol Immunol 2005;42:489–94. 10.1016/j.molimm.2004.07.032 [DOI] [PubMed] [Google Scholar]
- 11.Romero X, Benitez D, March S, et al. Differential expression of SAP and EAT-2-binding leukocyte cell-surface molecules CD84, CD150 (SLAM), CD229 (Ly9) and CD244 (2B4). Tissue Antigens 2004;64:132–44. 10.1111/j.1399-0039.2004.00247.x [DOI] [PubMed] [Google Scholar]
- 12.De Salort J, Sintes J, Llinàs L, et al. Expression of SLAM (CD150) cell-surface receptors on human B-cell subsets: from pro-B to plasma cells. Immunol Lett 2011;134:129–36. 10.1016/j.imlet.2010.09.021 [DOI] [PubMed] [Google Scholar]
- 13.Munitz A, Bachelet I, Fraenkel S, et al. 2B4 (CD244) is expressed and functional on human eosinophils. J Immunol 2005;174:110–18. 10.4049/jimmunol.174.1.110 [DOI] [PubMed] [Google Scholar]
- 14.Schuhmachers G, Ariizumi K, Mathew PA, et al. 2B4, a new member of the immunoglobulin gene superfamily, is expressed on murine dendritic epidermal T cells and plays a functional role in their killing of skin tumors. J Invest Dermatol 1995;105:592–6. 10.1111/1523-1747.ep12323533 [DOI] [PubMed] [Google Scholar]
- 15.Georgoudaki AM, Khodabandeh S, Puiac S, et al. CD244 is expressed on dendritic cells and regulates their functions. Immunol Cell Biol 2015;93:581–90. 10.1038/icb.2014.124 [DOI] [PubMed] [Google Scholar]
- 16.Elishmereni M, Fyhrquist N, Singh Gangwar R, et al. Complex 2B4 regulation of mast cells and eosinophils in murine allergic inflammation. J Invest Dermatol 2014;134:2928–37. 10.1038/jid.2014.280 [DOI] [PubMed] [Google Scholar]
- 17.Waggoner SN, Kumar V. Evolving role of 2B4/CD244 in T and NK cell responses during virus infection. Front Immunol 2012;3:377 10.3389/fimmu.2012.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laouar A, Manocha M, Wan M, et al. Cutting edge: distinct NK receptor profiles are imprinted on CD8T cells in the mucosa and periphery during the same antigen challenge: role of tissue-specific factors. J Immunol 2007;178:652–6. 10.4049/jimmunol.178.2.652 [DOI] [PubMed] [Google Scholar]
- 19.O'Keeffe MS, Song JH, Liao G, et al. SLAMF4 is a negative regulator of expansion of cytotoxic intraepithelial CD8+ T cells that maintains homeostasis in the small intestine. Gastroenterology 2015;148:991–1001.e1004. 10.1053/j.gastro.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SK, Reed DS, Olson S, et al. Generation of mucosal cytotoxic T cells against soluble protein by tissue-specific environmental and costimulatory signals. Proc Natl Acad Sci USA 1998;95:10814–19. 10.1073/pnas.95.18.10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fagarasan S, Kinoshita K, Muramatsu M, et al. In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature 2001;413:639–43. 10.1038/35098100 [DOI] [PubMed] [Google Scholar]
- 22.Chang PV, Hao L, Offermanns S, et al. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA 2014;111:2247–52. 10.1073/pnas.1322269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pope C, Kim SK, Marzo A, et al. Organ-specific regulation of the CD8T cell response to Listeria monocytogenes infection. J Immunol 2001;166:3402–9. 10.4049/jimmunol.166.5.3402 [DOI] [PubMed] [Google Scholar]
- 24.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol 2012;12:821–32. 10.1038/nri3322 [DOI] [PubMed] [Google Scholar]
- 25.Tezuka H, Abe Y, Asano J, et al. Prominent role for plasmacytoid dendritic cells in mucosal T cell-independent IgA induction. Immunity 2011;34:247–57. 10.1016/j.immuni.2011.02.002 [DOI] [PubMed] [Google Scholar]
- 26.Schaefer BC, Schaefer ML, Kappler JW, et al. Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cell Immunol 2001;214:110–22. 10.1006/cimm.2001.1895 [DOI] [PubMed] [Google Scholar]
- 27.De Togni P, Goellner J, Ruddle NH, et al. Abnormal development of peripheral lymphoid organs in mice deficient in lymphotoxin. Science 1994, 264:703–7. 10.1126/science.8171322 [DOI] [PubMed] [Google Scholar]
- 28.Fox J, Barthold S, Davisson M, et al. The mouse in biomedical research In: The Mouse of Biomedical Research. Fox J, ed. Vol. 3, 2nd edn San Diego, CA: Elsevier Academic Press, 2006. [Google Scholar]
- 29.Ismail AS, Severson KM, Vaishnava S, et al. Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci USA 2011;108:8743–8. 10.1073/pnas.1019574108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo X, Xia X, Tang R, et al. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett Appl Microbiol 2008;47:367–73. 10.1111/j.1472-765X.2008.02408.x [DOI] [PubMed] [Google Scholar]
- 31.Laouar A, Haridas V, Vargas D, et al. CD70+ antigen-presenting cells control the proliferation and differentiation of T cells in the intestinal mucosa. Nat Immunol 2005;6:698–706. 10.1038/ni1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atarashi K, Nishimura J, Shima T, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature 2008;455:808–12. 10.1038/nature07240 [DOI] [PubMed] [Google Scholar]
- 33.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009;139:485–98. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochoa-Repáraz J, Mielcarz DW, Wang Y, et al. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol 2010;3:487–95. 10.1038/mi.2010.29 [DOI] [PubMed] [Google Scholar]
- 35.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013;504:446–50. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 36.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016;16:341–52. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris MA, Reddy CA, Carter GR. Anaerobic bacteria from the large intestine of mice. Appl Environ Microbiol 1976;31:907–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kambayashi T, Assarsson E, Chambers BJ, et al. Cutting edge: regulation of CD8(+) T cell proliferation by 2B4/CD48 interactions. J Immunol 2001;167:6706–10. 10.4049/jimmunol.167.12.6706 [DOI] [PubMed] [Google Scholar]
- 39.Zheng Y, Valdez PA, Danilenko DM, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 2008;14:282–9. 10.1038/nm1720 [DOI] [PubMed] [Google Scholar]
- 40.Kinnebrew MA, Buffie CG, Diehl GE, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity 2012;36:276–87. 10.1016/j.immuni.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham AC, Carr KD, Sieve AN, et al. IL-22 production is regulated by IL-23 during Listeria monocytogenes infection but is not required for bacterial clearance or tissue protection. PLoS ONE 2011;6:e17171 10.1371/journal.pone.0017171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Killig M, Glatzer T, Romagnani C. Recognition strategies of group 3 innate lymphoid cells. Front Immunol 2014:5:142 10.3389/fimmu.2014.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith P, MacDonald T, Blumberg R, eds. Principles of mucosal immunology. 1st edn New York, NY: Garland Science, 2012. [Google Scholar]
- 44.Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat Immunol 2004;5:597–605. 10.1038/ni1070 [DOI] [PubMed] [Google Scholar]
- 45.Wu HJ, Ivanov II, Darce J, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010;32:815–27. 10.1016/j.immuni.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Assarsson E, Kambayashi T, Schatzle JD, et al. NK cells stimulate proliferation of T and NK cells through 2B4/CD48 interactions. J Immunol 2004;173:174–80. 10.4049/jimmunol.173.1.174 [DOI] [PubMed] [Google Scholar]
- 47.Lee KM, Bhawan S, Majima T, et al. Cutting edge: the NK cell receptor 2B4 augments antigen-specific T cell cytotoxicity through CD48 ligation on neighboring T cells. J Immunol 2003;170:4881–5. 10.4049/jimmunol.170.10.4881 [DOI] [PubMed] [Google Scholar]
- 48.Margraf-Schönfeld S, Böhm C, Watzl C. Glycosylation affects ligand binding and function of the activating natural killer cell receptor 2B4 (CD244) protein. J Biol Chem 2011;286:24142–9. 10.1074/jbc.M111.225334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fischer HG, Reichmann G. Brain dendritic cells and macrophages/microglia in central nervous system inflammation. J Immunol 2001;166:2717–26. 10.4049/jimmunol.166.4.2717 [DOI] [PubMed] [Google Scholar]
- 50.Austyn JM, Hankins DF, Larsen CP, et al. Isolation and characterization of dendritic cells from mouse heart and kidney. J Immunol 1994;152:2401–10. [PubMed] [Google Scholar]
- 51.Pillarisetty VG, Miller G, Shah AB, et al. GM-CSF expands dendritic cells and their progenitors in mouse liver. Hepatology 2003;37:641–52. 10.1053/jhep.2003.50074 [DOI] [PubMed] [Google Scholar]
- 52.Faucher JL, Lacronique-Gazaille C, Fré, et al. ‘6 markers/5 colors’ extended white blood cell differential by flow cytometry. Cytometry A 2007;71:934–44. 10.1002/cyto.a.20457 [DOI] [PubMed] [Google Scholar]
- 53.Liu F, Whitton JL. Cutting edge: re-evaluating the in vivo cytokine responses of CD8+ T cells during primary and secondary viral infections. J Immunol 2005;174:5936–40. 10.4049/jimmunol.174.10.5936 [DOI] [PubMed] [Google Scholar]
- 54.Sinsimer D, Esseghir A, Tang M, et al. The common prophylactic therapy for bowel surgery is ineffective for clearing Bacteroidetes, the primary inducers of systemic inflammation, and causes faster death in response to intestinal barrier damage in mice. BMJ Open Gastroenterol 2014;1:e000009 10.1136/bmjgast-2014-000009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joseph JK, Bunnachak D, Burke TJ, et al. A novel method of inducing and assuring total anoxia during in vitro studies of O2 deprivation injury. J Am Soc Nephrol 1990;1:837–40. [DOI] [PubMed] [Google Scholar]
- 56.Wiggs LS, Cavallaro JJ, Miller JM. Evaluation of the oxyrase OxyPlate anaerobe incubation system. J Clin Microbiol 2000;38:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gannon C, Copeland JC. An anaerobic microbiology system for the clinical laboratory. Am Clin Lab 1999;18:18–19. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2016-313214supp001.pdf (3.3MB, pdf)
gutjnl-2016-313214supp002.pdf (107.3KB, pdf)
Sequences of oligonucleotide primers for analysis of cytokines and transcriptional factors. Primer sequence sets used in this study were obtained from Invitrogen
gutjnl-2016-313214supp003.pdf (232.3KB, pdf)